Abstract
We have previously shown that the retinoblastoma protein (pRb) can activate expression of Runx2-dependent, bone-specific genes in cultured cells. We now show that pRb also plays a role early in osteogenesis, and that in primary RB1−/− calvarial cells there is an increased osteoprogenitor pool. To understand pRb's function in vivo, we generated a conditional RB1-KO mouse in which pRb expression is efficiently extinguished in osteoblasts. These animals display an apparent developmental defect in bones, most strikingly in the calvaria. Cultured RB1−/− calvarial osteoblasts fail to cease proliferation upon reaching confluence or following differentiation. Re-plating assays of primary RB1−/− calvarial cells after differentiation showed a clear adipogenic ability with increased multipotency. RB1−/− osteoblasts display a severe reduction in levels of mRNAs expressed late in differentiation. In this study, we present strong evidence that pRb has multiple regulatory roles in osteogenesis. Furthermore, in the absence of RB1−/− there is a larger pool of multipotent cells compared with the WT counterpart. This increased pool of osteoprogenitor cells may be susceptible to additional transforming events leading to osteosarcoma, and is therefore key to understanding RB1 as a target in malignancy.
Keywords: differentiation, retinoblastoma protein, osteoprogenitors
The retinoblastoma protein (pRb) is a tumor suppressor whose regulatory pathway is targeted in most human cancers (1, 2). pRb and its related pocket proteins, family members p107 and p130, mediate a number of cell processes including cell cycle progression, differentiation, and apoptosis. pRb functions in every cell type to control the exit from G1 by regulating the E2F family of transcription factors. This process of repressing E2F family members prevents S-phase progression and promotes cell cycle exit during differentiation and senescence programs (3–7). Although the pRb pathway is affected in many human cancers, it is targeted directly in only a subset of tumors. Individuals with inherited heterozygous loss of the RB1 gene develop retinoblastoma and have as much as a 1,000 fold increase in incidence of osteosarcoma (7–9). More rarely, RB1 is targeted in small-cell lung carcinoma, bladder carcinomas, and other tumors. This suggests that tumors exhibiting the highest frequency of pRb loss may require pRb function in multiple cell cycle exit programs discrete from specific mediation of S-phase gene expression.
Numerous studies have addressed the role of pRb in cellular differentiation. These include myogenesis (10, 11,) chondrogenesis (12), adipogenesis (4, 13), and osteogenesis (14). To better understand the role of pRb in tumor formation in a tissue in which it is commonly deleted, we decided to focus on pRb's role in bone development and osteosarcoma formation. Osteosarcomas are often highly aggressive neoplasms that rapidly progress and eventually recur and give rise to distant metastases, primarily to the lung. Osteosarcoma is the most common primary malignant tumor of bone apart from myeloma (15), and 81% of osteosarcomas are either poorly differentiated or undifferentiated (16). Osteosarcoma is characterized by the direct formation of bone or osteoid by the tumor cells (15). This implies that osteosarcoma is dependent on events related to commitment to the osteoblast lineage. Not surprisingly, mutation of the RB1 locus occurs in as many as 60% of all osteosarcomas (>80% in pediatric cases) and is considered an essential step in tumor formation (17).
Several lines of evidence indicate pRb is required for normal osteogenesis. Osteoblast differentiation can be inhibited in conditionally immortalized cells by the viral oncoproteins SV40 large T antigen and E1A 12S, which specifically target the pocket proteins (18). Further, work with the human osteosarcoma cell line SAOS2 has shown that re-expression of pRb alone is sufficient to force senescence and drive the expression of markers reminiscent of osteoblast differentiation (6, 7). The ability of pRb to promote osteoblast differentiation may be separable from its role as a regulator of the E2F family of transcription factors, as re-introduction of a pRb mutant (R661W), lacking the canonical E2F binding capacity, is capable of arresting and differentiating SAOS2 cells (6).
Osteogenesis is a multistep process regulated by several different transcription factors. Runx2 is the “master regulator” of this molecular pathway, required for mineralization of the skeleton and transcriptional regulation of many genes key to osteogenesis (19, 20). We have previously shown that hypo-phosphorylated pRb physically interacts with RUNX2 in vitro (14). This interaction augments RUNX2-dependent transcription from osteoblast-specific promoters. In the absence of pRb, BMP2-treated mouse embryonic fibroblasts (MEFs) do not terminally differentiate as measured by mineralization and expression of bone-specific genes. Interestingly, the early marker of differentiation, alkaline phosphatase (ALP), showed similar relative induction between RB1−/− and RB1+/+ MEFs. From this work we concluded that pRb acts as a transcriptional co-activator of Runx2 functioning in late osteogenic differentiation (14).
In this study, we present strong evidence that, in addition to pRb's role late in osteoblast differentiation, it also plays a critical role early in commitment to the osteoblast lineage. Furthermore, in the absence of RB1−/− there is a larger pool of multipotent cells compared with the WT counterpart. This increased pool of osteoprogenitor cells may be susceptible to additional transforming events leading to osteosarcoma, and therefore key to understanding RB1 as a target in malignancy.
Results
Bone-Targeted Knockout of the Retinoblastoma Gene.
Homozygous null RB1−/− mice die at E11.5 to E13.5, before mineralization of the skeleton is achieved. Therefore, to overcome the early placental defects that contribute to embryonic lethality in germline RB1-null mice (21), and to ablate RB1 efficiently in the bone, we used the Cre/lox site directed recombination system. Mice in which exon 19 of RB1 is flanked by loxP sites (22) were crossed with a transgenic mouse strain that expresses the Cre recombinase under the collagen 1a1 promoter, 3.6Col1a1, which expresses highly in the bone (23). 3.6Col1a1Cre;RBf19/f19 mice are born at the expected Mendelian ratio but the animals die at birth as a result of collateral RB1 deletion in vital tissues. Skeletons of the RB1−/− mice were examined by differential staining of cartilage (Alcian blue) and bone and other mineralized tissues (alizarin red) [supporting information (SI) Fig. S1 and SI Materials and Methods]. RB1-KO mice display morphological and mineralization defects in the xiphoid process of the sternum, defects in the appearance of ossification centers of the digits of the fore- and hind-limbs, and a mineralization deficiency in the apical portion of the cranial vault. To determine if the mineralization defect seen in the calvaria represented an absence of osteoblasts in the apical area, we checked for expression of the early marker of differentiation, ALP activity. We found staining along the entire circumference of the calvaria in both WT and RB1−/− animals (Fig. S2 and SI Materials and Methods). However, when staining for mineral by the Von Kossa method, we failed to detect it at the apex of the calvaria in RB1−/− mice (Fig. S2 and SI Materials and Methods) as anticipated from the results in Fig. S1. These phenotypes of RB1−/− animals are consistently observed, but it is not clear whether they result from cell autonomous alteration in osteoblast function, or if there is an influence of additional RB1−/− tissues, as bone is subject to various autocrine and paracrine signaling events. Therefore, we elected to isolate and interrogate the effects of pRb loss on osteoblasts in vitro by isolating cells from the calvaria.
Cultured Primary Osteoblasts from RB1−/− Mice Show Altered Proliferation and Mineral Formation Abilities at High Cell Density.
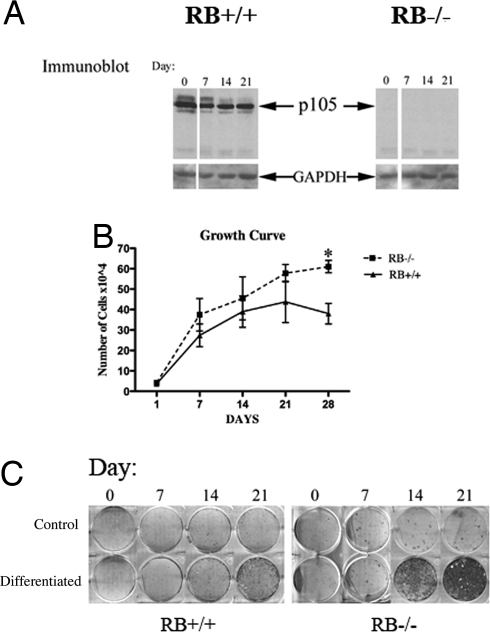
There are two methods of bone formation: endochondral or intramembranous ossification. Both forms of bone arise from mesenchymal cells, but the endochondral method, involved in most skeletal long bones, is complex and involves the formation of a cartilage template that is later replaced by bone (24). Intramembranous bone formation, mainly found in the flat bones of the skull, is less understood but is arguably simpler in that the cells of these condensations differentiate directly into bone-forming osteoblasts (24). The murine calvaria are a rich source of intramembranous osteoblasts that are readily cultured and analyzed in vitro. To interrogate the mineralization defect of the RB1-KO animals, calvarial osteoblasts from E18.5 animals were cultured. These cells were first examined for pRb expression by immunoblotting, which revealed a complete absence of the protein (Fig. 1A). We then measured proliferation rates of RB1−/− osteoblasts and litter-mate control WT cells. As anticipated from work in RB1−/− MEFs (25), these proliferation analyses showed that sub-confluent RB1−/− osteoblasts proliferate at a rate similar to that observed for their WT counterparts (Fig. 1B). However, the RB1−/− osteoblasts continue to proliferate after reaching high cell densities, whereas the WT osteoblasts cease proliferation by day 14. These data are further supported by BrdU incorporation studies, which demonstrated persistent S phase in confluent RB1−/− cultures, but not in WT control cells (Fig. S3 and SI Materials and Methods). In contrast, Ki67 staining of calvaria obtained from embryonic day 18 mice showed no discernible differences in proliferative fraction (Fig. S2 and SI Materials and Methods), consistent with a minimal role for RB loss in altering proliferation in cell populations already able to undergo rapid proliferation.
Fig. 1.
Proliferative properties and increased mineral deposition in cultured primary calvarial osteoblasts (A). Immunoblot analysis of extracts of cultured osteoblasts shows the absence of pRb in col3.6;RB1lox/lox mice. (B) Osteoblasts were isolated from the calvaria of E18.5 animals and differentiated in α-MEM media supplemented with ascorbic acid and β-glycerophosphate. Total cell numbers were determined at 1, 7, 14, 21, and 28 days after plating at an initial density of 500 cells/cm2. *, P < 0.05. (C) Mineral deposition was detected using alizarin red at 0, 7, 14, and 21 days following addition of differentiation medium.
Primary osteoblasts supplied with differentiation media in vitro activate an osteogenic gene transcription program and form mineralized nodules, which is thought to correspond to mature osteoblast functions in vivo. To determine the differentiation capacity and timing of mineral formation in RB1−/− cultures, mutant and control WT litter-mate cells were seeded at the same number, grown to confluence, and treated with media supplemented with ascorbic acid and β-glycerophosphate. Using this standard protocol of osteoblast differentiation, it is generally observed that WT cells take approximately 21 days to form mineralized nodules as detected by alizarin red staining (26). RB1−/− osteoblasts consistently showed earlier and more intense mineralization than those from their WT litter-mates (Fig. 1C). This increase in mineralization may result in part from the higher cell density in the RB1−/− osteoblast cultures. In differentiation media, osteoblasts have been shown to thrive on autocrine signaling for growth and differentiation in cases of high cell density (27). Therefore, the increased density of RB1−/− cell cultures may generate a more potent differentiation environment. Alternatively or in addition, the RB1−/− calvaria osteoblast preparations may contain an increased number of osteoprogenitor cells. Therefore, we set out to distinguish between the proliferative and differentiation defects by measuring and comparing osteoprogenitors present in RB1−/− and WT litter-mate primary osteoblast cultures.
Osteoblasts Lacking RB1 Display Enhanced Osteogenic Potential.
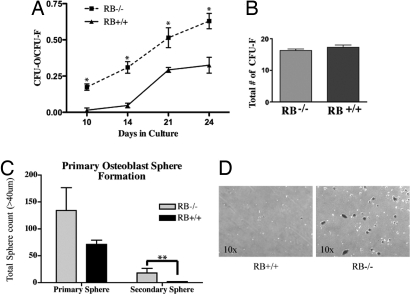
As is the case in many developing tissues, osteoblasts arise through commitment of a multipotent stem cell, in this case a mesenchymal stem cell, to the osteoblast lineage to produce an osteoprogenitor cell (OPC). Considerable progress in identifying cell surface markers on analogous stem and progenitor cells has been made in some tissues, such as blood and breast, but such makers are not well defined in the osteoblast system. Thus, a functional, in vitro assay is at present the best way to measure OPC cell numbers and function. One such method to measure OPCs is by a colony formation assay (28). It has been shown that the number of nodules that form can be used as an approximate index of the number of OPCs present in the original cell population. The idea is that a single initiating cell, referred to as a colony forming unit (CFU-O), gives rise to all of the cells necessary for nodule formation (28). To measure CFU-Os present in RB1−/− cultures, osteoblasts were plated at a low density to allow individual cells to form individual colonies. These plates were then stained for ALP activity, an early marker of the osteoblast lineage. The ratio of ALP-positive colonies (i.e., CFU-Os) to the total number of colonies formed (i.e., CFU-Fs) is determined by subsequent crystal violet staining, allowing a determination of the fraction of colonies with osteogenic potential. Colonies of sufficient size for analysis formed 10 days after plating. The ratio of CFU-O to CFU-F was approximately twofold greater in the cultures from RB1−/− mice relative to those from WT (Fig. 2A). This increase in the ratio of CFU-O to CFU-F is not accompanied by an overall increase in the total number of colonies (Fig. 2B), which is consistent with the similar proliferation rates of sub-confluent RB1−/− and WT osteoblasts (Fig. 1B). There was an earlier appearance of ALP-positive colonies in the RB1−/− cultures, which suggests an acceleration of osteoblast differentiation or that an increased fraction of osteoblasts in RB1−/− calvaria had advanced to the point of ALP expression at the time of cell harvest.
Fig. 2.
Quantitation of osteoprogenitors in primary calvarial osteoblast cultures. (A) Colony assay to determine osteoprogenitor cell numbers. Primary osteoblasts were seeded at low density and cultured for 24 days. Colonies of CFU-O from single cells were stained for ALP activity, and the CFU-F was determined by crystal violet staining. A colony was counted if it was a minimum of 10–20 mm in diameter. A minimum of 10 cells per colony had to be ALP-positive to be counted as one CFU-O. The ratio of CFU-O to CFU-F was increased in the cultures from RB−/− mice (dashed line) relative to those of their control litter-mates (solid line). *, P < 0.05. (B) Total number of colonies formed by osteoblasts derived from RB1−/− and WT animals. (C) Sphere formation assay for progenitor/multipotent cells in the calvarial cell population. Primary osteoblasts were seeded at 1.5 × 104 cells/cm2 on ultra-low adherent plates. Spheres were counted after 3 and 6 days. Primary spheres were collected, counted, trypsinized, and re-plated onto ultra-low adherent plates. Secondary spheres were counted after 6 days **, P < 0.05. (D) Image of primary spheres from RB−/− and RB-WT calvaria cells.
The CFU-O assay described is consistent with an increase in OPCs in RB1−/− osteoblast cultures. To obtain independent evidence of this and to extend our analysis of progenitor numbers and function in these cultures, we next performed a sphere formation assay. Sphere formation assays have long been used to show progenitor/multipotent cell populations in the human and murine mammary epithelial system (29–31). Several studies suggest that bone marrow derived cells can also produce spheres (32, 33), but to our knowledge there is no work that shows cells from murine calvaria can or should be able to form spheres. To determine if calvarial osteoblast preparations contained sphere-forming ability, cells were seeded on ultra-low adherent tissue culture plates in AlphaMEM medium supplemented with 10% FBS. Three to four days later, sphere formation was evident either in the presence or absence of RB1 (Fig. 2D). Interestingly, there is a noticeable difference in sphere number in RB1−/− primary calvarial preparations compared with those from WT litter-mates (Fig. 2C). These spheres were isolated, disassociated, and re-plated on non-adherent plates. Twice as many secondary spheres were generated from RB1−/− primary cultures compared with those from WT litter-mates. The overall reduced number of secondary spheres may result from suboptimal culture conditions, as we have yet to determine the nature of these “osteospheres,” and/or these osteoprogenitors may have a limited passage capability. For example, additional growth supplements must be added to the media to maintain optimal levels of mammosphere formation (34). These data strongly suggest that primary calvarial cells in the absence of RB1 have an increased osteoprogenitor cell population that may also consist of a population of cells that are able to self-renew and form spheres, and may be multipotent in nature. To further assess whether the cells are multipotent, we next determined if the cells could be differentiated into other cell types.
RB1−/− Osteoblasts Can Differentiate into Osteoblast and Adipose Cells.
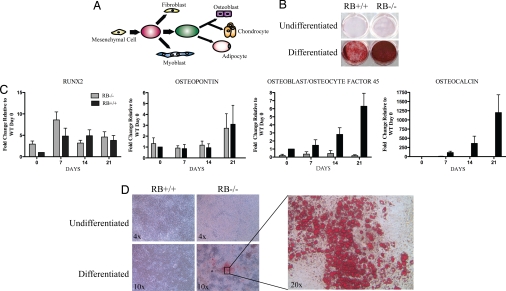
Mesenchymal stem cells are able to differentiate into several cell types including adipocytes, chondrocytes, and osteoblasts. Some evidence suggests that mesenchymal stem cells from bone marrow can also give rise to myoblasts and fibroblasts, and possibly to non-mesenchymal lineages (35) (Fig. 3A). To determine if RB1−/− calvarial osteoblast preparations contain multipotent cells, we treated the primary cells with standard differentiation protocols for osteoblasts, adipocytes, or myoblasts. As shown earlier, RB1−/− cells treated with osteoblast differentiation medium display increased mineral formation as determined by alizarin red S stain (Fig. 3B). To extend characterization of this osteoblastic differentiation, we analyzed the expression of several mRNAs encoding osteoblast-specific genes. We found that expression of Runx2 mRNA as well as that of osteopontin, a gene expressed at medium to late times after initiation of osteoblast differentiation, is unaffected in the presence or absence of RB1 (Fig. 3C). In contrast, osteoblast/osteocyte factor 45 (OF45), a late marker tightly linked to mineralization (36), was found to be poorly expressed in RB1−/− osteoblasts in comparison to WT osteoblasts. Similarly, osteocalcin, another late marker of osteoblast differentiation, is detectable in differentiated RB1−/− osteoblasts, but at levels vastly reduced compared with those in WT cells (Fig. 3C). Thus, expression of genes acting late in osteoblast differentiation is severely impaired in RB1−/− osteoblasts, just as was previously observed in BMP2-treated RB1−/− MEFs (14). However, increased CFU-O number and persistent expression of genes expressed at early to middle times after initiation of differentiation supports the notion that functional osteoprogenitors may be present at increased numbers in RB1−/− calvarial preparations.
Fig. 3.
Primary calvaria cells from RB1−/− mice can be differentiated into osteoblasts and adipose cells. (A) Schematic drawing of mesenchymal cell differentiation. (B) Primary RB−/− and RB+/+osteoblasts from the calvarial were grown to confluence and differentiated for 28 days with osteoblast differentiation media. Cells were stained with alizarin red. (C) RT-PCR for Runx2, osteopontin, OF45, and osteocalcin. mRNA was prepared from cells treated as in B. (D) Primary RB−/− and RB+/+ osteoblasts from calvaria were grown to confluence and differentiated for 19 days with adipose differentiation media. The cells were then stained with oil red O. The images are a representative of three cell preparations for each genotype.
Next we asked if the progenitor cells present in RB1−/− calvarial cell cultures were able to differentiate into cells other than osteoblasts. We focused on the adipocyte lineage, as this cell type is thought to arise from a common progenitor proximal to osteoblast commitment, as is depicted in Fig. 3A. To stimulate adipogenesis, cells from WT or RB1−/− calvaria were cultured in adipogenic medium (insulin, isobutylmethylxanthine, and dexamethasone). No adipogenesis was observed in WT osteoblast cultures, indicating that progenitor cells in these cultures may be committed to the osteoblast lineage. A similar resistance to adipogenic media has been reported in MEFs (4). In contrast, RB1−/− calvarial osteoblast cultures demonstrated robust differentiation to adipocytes as revealed by oil red O staining (Fig. 3D). These results clearly show that primary calvaria cells in the absence of RB1 are able to differentiate into cell lineages other than osteoblasts, supporting the persistence or accumulation of multipotent progenitor cells in the skulls of mutant animals.
pRb Is Needed for Terminal Cell Cycle Exit.
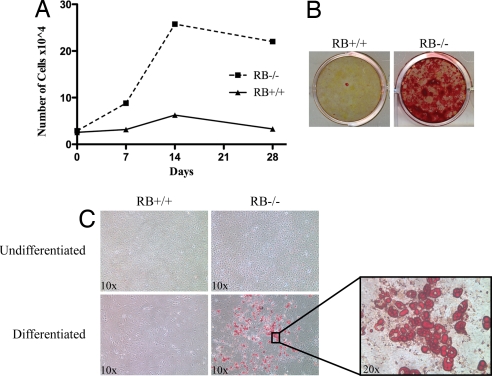
The data described suggest that loss of RB1−/− in calvarial osteoblasts impairs late gene expression and increases the number and potential of progenitor cells. To determine how RB1 loss might alter the proliferative capacity of differentiating progenitor cells, we next asked if pRb is involved in mediating permanent cell cycle arrest, a feature often associated with “terminal” differentiation. To this end, we trypsinized osteoblast cultures that had been in a confluent, differentiating condition for 28 days, re-plated these cells at the starting sub-confluent dilution, and performed a proliferation assay. Cell cycle analysis indicated that neither RB1−/− nor WT cultures contained proliferating cells at this time (data not shown). WT osteoblasts numbers failed to expand after re-plating. However, RB1−/− osteoblasts proliferated and repopulated the dish (Fig. 4). Further, when we assayed for mineral formation after addition of ascorbic acid and β-glycerophosphate as a measure of differentiation, RB1−/− cells stained robustly but WT cells failed to differentiate. In addition, to determine if cells maintained multi-potency, the re-plated cells were treated with adipogenic medium for 14 days. Similar to the initial primary cell preparations, RB1−/− cells demonstrated substantial differentiation to adipocytes, as revealed by oil red O staining (Fig. 4C). Therefore, pRb appears to be necessary for permanent cell cycle arrest of osteoblasts subjected to differentiation-inducing agents in culture.
Fig. 4.
Re-plated RB1−/− osteoblasts retain the ability to proliferate and differentiate into osteoblasts and adipose cells. Osteoblasts were allowed to differentiate as described in Fig. 2. After 28 days in differentiation media, cells were trypsinized, re-seeded, and differentiated anew. (A) RB1−/− osteoblasts proliferated in culture and were capable of differentiating as measured by mineral formation visualized with alizarin red (B). These results were observed twice. (A) WT cells failed to grow after re-plating, and showed no response to differentiating agents (B). (C) The trypsinized cells were re-plated and differentiated with adipogenic medium for 14 days. RB1−/− cells were capable of forming adipocytes, as determined by oil red O staining, whereas RB1+/+ cells showed no adipocyte-like cells.
Discussion
We have previously reported that direct interaction between pRb and Runx2 is required for osteocalcin expression late in osteoblast differentiation stimulated by BMP2 treatment of MEFs (14). We now show that pRb may also play a role early in osteogenesis, and that in primary RB1−/− calvarial cells there is an increased osteoprogenitor pool. Loss of RB1 in the osteoprogenitors results in mineralization defects in both endochondral and intramembranous bones. In vitro, RB1−/− cells treated with differentiation medium mineralize significantly earlier than their WT counterparts, in seeming contradiction to the in vivo observation of reduced mineralization. However, in keeping with our previously published work (14), we found that induction of the expression of terminal markers of osteoblast differentiation, osteocalcin, and OF45 was severely impaired in RB1−/− cells (Fig. 3C). It is interesting to note that the osteocalcin-KO mouse model showed an osteopetrotic phenotype, suggesting osteocalcin is a negative regulator of bone mineralization (37). Further, OF45 has also been shown to limit bone formation in vivo (38). Therefore, the absence of the expression of these genes may in part explain the increased mineral formation seen in RB1−/− calvarial osteoblasts in vitro.
Both increased number of CFU-O and increased mineralization are consistent with persistence of calvarial osteoblasts in a proliferative, progenitor-like state. Continuous, strong differentiation signals in vitro appear to stimulate mineral deposition, and we predict that similar effects may be observed in vivo in older animals lacking RB1−/− in bone. However, the early death of the animals used in our analyses prevents us from asking if excessively proliferative osteoblasts persist in RB1−/− animals throughout life or if these cells eventually succeed in mineralizing the affected areas. Future experiments using partially penetrant cre alleles expressed specifically in the bone may help to identify an excess of proliferative osteoprogenitors and altered mineralization in older animals. In addition to generating a higher number of cells with characteristics of osteoprogenitors (i.e., ALP-positive CFU-O number and sphere forming activity in addition to increased mineralization), RB1−/− calvarial cells showed additional progenitor-like changes in comparison to WT cells. First, whereas WT cells subjected to 3 to 4 weeks of differentiation lost all proliferative capacity upon re-plating, RB1−/− cells were readily able to expand and produce a high number of mineralization competent cells, suggesting that at least some RB1−/− calvarial cells resisted the cell cycle exit displayed by differentiated WT cells. Second, RB1−/− calvarial cells showed a clear adipogenic ability (although no change in myogenic ability) consistent with increased multi-potency.
Whether RB1−/− osteoblasts “back up” to display properties of multipotent progenitors or if loss of RB1 delays progression of a pool of multipotent progenitors normally found in calvarial condensates requires further study. Similarly, the ability of re-plated, differentiated RB1−/− osteoblasts to proliferate and mineralize may represent a “de-differentiation” of cells that had responded to differentiation media or alternatively may result from persistence of progenitor-like cells in these cultures. Indeed, our preliminary studies suggest that sphere formation and adipogenic abilities are present in re-plated cells following differentiation of RB1−/− cells. Finally, we note that, although several different primary cell preparations for each genotype were examined for each differentiation assay with equivalent results, there is the possibility that these cultures contain a mixed population of progenitors individually able to generate osteoblasts or adipocytes. The possibility of obtaining multiple RB1−/− cell types from these animals is supported by our observation that RB1 is lost in all tissues of animals expressing 3.6-Col1a1-Cre, and this expression begins at approximately embryonic day 10. To begin to examine this issue, we have established immortalized cell lines that are clonally derived from the primary cultures and have initially found that RB1−/−, but not WT cell lines, also possesses both adipogenic and osteogenic properties, consistent with the immortalization of a single multipotent progenitor cell (data not shown).
How pRb acts to attenuate early osteoblast progression and/or proliferation is not known. However, it is noteworthy that the multiple functions of pRb as an inhibitor of progenitor proliferation, mediator of progression, and effector of late differentiation and cell cycle exit would limit the size of the osteoprogenitor pool. Loss of pRb would then be expected to increase the progenitor pool while reducing terminal differentiation and cell cycle exit. Overall, the data presented here suggest that pRb function acts to regulate several aspects of osteoprogenitor and osteoblast proliferation and differentiation in vivo. We propose that pRb is a major coordinator of proliferation and differentiation events throughout bone development. Furthermore, in the absence of RB1, there is a larger pool of multipotent cells compared with the WT counterpart. This increased pool of osteoprogenitor cells may be susceptible to additional transforming events leading to osteosarcoma, and indeed loss of RB1 in the mouse has recently been shown to cooperate with p53 loss to generate bone tumors (39, 40). Studies of pRb-null progenitors such as those performed here are thus key to understanding RB1 as a target in malignancy.
Materials and Methods
Animals.
The flox19-RB1 mice were obtained from the laboratory of Doug Hanahan (San Francisco, CA) and maintained in a C57BL/6 background. 2.3- and 3.6-Col1a1-Cre transgenic mice were obtained from the laboratory of Barbara Kream (Farmington, CT). The F1 RBf19/WT;Col1a1-Cre mice were self-crossed and the resulting F2 mice were used in our studies.
Calvarial Cell Preparation and Culture.
Calvariae (topmost skull bones) from 18.5-day embryos were removed, subjected to a series of collagenase digestions at 37 °C, pooled, and then plated on one 10-cm dish per embryo in α-MEM medium (Invitrogen). Cells were maintained in α-MEM plus 10% FBS and 1% penicillin-streptomycin (Gibco; basic media). Primary cells were differentiated using 0.01 M β-glycerophosphate, 100 mM l-ascorbic acid, and 0.1 μM dexamethasone (Sigma) in basic media for osteoblast differentiation. Cells were stained with 2% alizarin red. For adipose differentiation, 0.1 μM dexamethasone, 0.5 mM isobutylmethylxanthine, and 5 μg/ml h-insulin (Sigma) were added to basic media. Cells were stained with oil red O as described later.
Immunoblotting.
Immunoblot analysis was performed as described previously (7). The following antibodies were used: human monoclonal antiRB: 245 (PharMingen) and anti-GAPDH (Chemicon). Horseradish peroxidase-conjugated secondary antibody was used (Jackson ImmunoResearch) and signal was detected by ECL (NEN).
Oil Red O Staining.
Oil red O was prepared as described (41). The final staining solution was 0.2% oil red O in 60% isopropanol (working solution). Cells were washed twice with PBS solution and fixed with 10% buffered formalin phosphate (Fisher) for at least 1 h at room temperature. The cells were washed twice with water, then stained for 2 h with oil red O working solution. The cells were washed with water, and the excess water was evaporated at room temperature. The stained cells were imaged under a microscope.
Quantitative Real-Time PCR.
RNA was extracted from primary osteoblasts using TRIzol (Invitrogen) according to the manufacturer's protocol. Ten nanograms of total RNA was used for first-strand cDNA synthesis using the iScript cDNA synthesis kit (Bio-Rad) according to the manufacturer's instructions. Quantitative PCR was carried out by employing the QuantiTect SYBR green PCR kit (Qiagen) and using 1 μl of the cDNA reaction. Primer sequences and PCR conditions are available on request. Relative quantification of gene expression was carried out by the comparative CT method (42).
Colony Formation Assay.
Colony formation assay was performed as described previously (43). Primary calvarial osteoblasts were re-seeded the day after isolation at a concentration of 500 cells/cm2. After 10 days in differentiation media, cells were fixed and stained for ALP activity with 10 mg naphthanol AS-MX (Sigma), 20 mg Fast Blue BB salt (Sigma), 1 ml N,N-dimethylformaide (Sigma), in 19 ml of 0.1 M Tris (pH 9.2) to detect CFU-O. Colonies greater than 1 mm in diameter were counted with a dissecting microscope. CFU-fibroblasts were detected by then staining the same plates with 0.2% crystal violet in 2% ethanol for 1 h. Plates were washed with water and then air-dried.
Sphere Formation Assay.
Primary cells from mouse calvaria were seeded at 1 × 105 cells/6 cm2 on an ultra-low attachment dish (Corning) in AlphaMEM (Invitrogen) plus 10% FBS and 1% penicillin-streptomycin (Gibco). Primary cells were counted after 3 days in culture, trypsinized, strained through a 40-μm BD Falcon strainer, and re-plated. Secondary spheres were counted on day 6.
Supplementary Material
Acknowledgments.
The authors thank C. Kuperwasser and P. Yelick for helpful comments. This work was supported by National Institutes of Health Grant AG020208 (to P.W.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0805925105/DCSupplemental.
References
- 1.Weinberg RA. Oncogenes and tumor suppressor genes. CA Cancer J Clin. 1994;44:160–170. doi: 10.3322/canjclin.44.3.160. [DOI] [PubMed] [Google Scholar]
- 2.Mulligan G, Jacks T. The retinoblastoma gene family: cousins with overlapping interests. Trends Genet. 1998;14:223–229. doi: 10.1016/s0168-9525(98)01470-x. [DOI] [PubMed] [Google Scholar]
- 3.Alexander K, Hinds PW. Requirement for p27(KIP1) in retinoblastoma protein-mediated senescence. Mol Cell Biol. 2001;21:3616–3631. doi: 10.1128/MCB.21.11.3616-3631.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen PL, Riley DJ, Chen Y, Lee WH. Retinoblastoma protein positively regulates terminal adipocyte differentiation through direct interaction with C/EBPs. Genes Dev. 1996;10:2794–2804. doi: 10.1101/gad.10.21.2794. [DOI] [PubMed] [Google Scholar]
- 5.Schneider JW, Gu W, Zhu L, Mahdavi V, Nadal-Ginard B. Reversal of terminal differentiation mediated by p107 in Rb-/- muscle cells. Science. 1994;264:1467–1471. doi: 10.1126/science.8197461. [DOI] [PubMed] [Google Scholar]
- 6.Sellers WR, et al. Stable binding to E2F is not required for the retinoblastoma protein to activate transcription, promote differentiation, and suppress tumor cell growth. Genes Dev. 1998;12:95–106. doi: 10.1101/gad.12.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tiemann F, Hinds PW. Induction of DNA synthesis and apoptosis by regulated inactivation of a temperature-sensitive retinoblastoma protein. EMBO J. 1998;17:1040–1052. doi: 10.1093/emboj/17.4.1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Horowitz JM, et al. Frequent inactivation of the retinoblastoma anti-oncogene is restricted to a subset of human tumor cells. Proc Natl Acad Sci USA. 1990;87:2775–2779. doi: 10.1073/pnas.87.7.2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sandberg AA, Bridge JA. Updates on the cytogenetics and molecular genetics of bone and soft tissue tumors: osteosarcoma and related tumors. Cancer Genet Cytogenet. 2003;145:1–30. [PubMed] [Google Scholar]
- 10.Novitch BG, Mulligan GJ, Jacks T, Lassar AB. Skeletal muscle cells lacking the retinoblastoma protein display defects in muscle gene expression and accumulate in S and G2 phases of the cell cycle. J Cell Biol. 1996;135:441–456. doi: 10.1083/jcb.135.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gu W, et al. Interaction of myogenic factors and the retinoblastoma protein mediates muscle cell commitment and differentiation. Cell. 1993;72:309–324. doi: 10.1016/0092-8674(93)90110-c. [DOI] [PubMed] [Google Scholar]
- 12.Cobrinik D, et al. Shared role of the pRB-related p130 and p107 proteins in limb development. Genes Dev. 1996;10:1633–1644. doi: 10.1101/gad.10.13.1633. [DOI] [PubMed] [Google Scholar]
- 13.Hansen JB, et al. Retinoblastoma protein functions as a molecular switch determining white versus brown adipocyte differentiation. Proc Natl Acad Sci USA. 2004;101:4112–4117. doi: 10.1073/pnas.0301964101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thomas DM, et al. The retinoblastoma protein acts as a transcriptional coactivator required for osteogenic differentiation. Mol Cell. 2001;8:303–316. doi: 10.1016/s1097-2765(01)00327-6. [DOI] [PubMed] [Google Scholar]
- 15.Schajowicz F, Sissons HA, Sobin LH. The World Health Organization's histologic classification of bone tumors. A commentary on the second edition. Cancer. 1995;75:1208–1214. doi: 10.1002/1097-0142(19950301)75:5<1208::aid-cncr2820750522>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 16.Unni KK, Dahlin DC. Osteosarcoma: pathology and classification. Semin Roentgenol. 1989;24:143–152. doi: 10.1016/0037-198x(89)90010-2. [DOI] [PubMed] [Google Scholar]
- 17.Toguchida J, et al. Preferential mutation of paternally derived RB gene as the initial event in sporadic osteosarcoma. Nature. 1989;338:156–158. doi: 10.1038/338156a0. [DOI] [PubMed] [Google Scholar]
- 18.Feuerbach D, Loetscher E, Buerki K, Sampath TK, Feyen JH. Establishment and characterization of conditionally immortalized stromal cell lines from a temperature-sensitive T-Ag transgenic mouse. J Bone Miner Res. 1997;12:179–190. doi: 10.1359/jbmr.1997.12.2.179. [DOI] [PubMed] [Google Scholar]
- 19.Komori T, et al. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755–764. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- 20.Otto F, et al. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997;89:765–771. doi: 10.1016/s0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- 21.Wu L, et al. Extra-embryonic function of Rb is essential for embryonic development and viability. Nature. 2003;421:942–947. doi: 10.1038/nature01417. [DOI] [PubMed] [Google Scholar]
- 22.Marino S, Vooijs M, van Der Gulden H, Jonkers J, Berns A. Induction of medulloblastomas in p53-null mutant mice by somatic inactivation of Rb in the external granular layer cells of the cerebellum. Genes Dev. 2000;14:994–1004. [PMC free article] [PubMed] [Google Scholar]
- 23.Liu F, et al. Expression and activity of osteoblast-targeted Cre recombinase transgenes in murine skeletal tissues. Int J Dev Biol. 2004;48:645–653. doi: 10.1387/ijdb.041816fl. [DOI] [PubMed] [Google Scholar]
- 24.Kronenberg HM. Developmental regulation of the growth plate. Nature. 2003;423:332–336. doi: 10.1038/nature01657. [DOI] [PubMed] [Google Scholar]
- 25.Sage J, et al. Targeted disruption of the three Rb-related genes leads to loss of G(1) control and immortalization. Genes Dev. 2000;14:3037–3050. doi: 10.1101/gad.843200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aubin JE. Advances in the osteoblast lineage. Biochem Cell Biol. 1998;76:899–910. [PubMed] [Google Scholar]
- 27.Tumber A, Meikle MC, Hill PA. Autocrine signals promote osteoblast survival in culture. J Endocrinol. 2000;167:383–390. doi: 10.1677/joe.0.1670383. [DOI] [PubMed] [Google Scholar]
- 28.Bellows CG, Aubin JE. Determination of numbers of osteoprogenitors present in isolated fetal rat calvaria cells in vitro. Dev Biol. 1989;133:8–13. doi: 10.1016/0012-1606(89)90291-1. [DOI] [PubMed] [Google Scholar]
- 29.Dontu G, et al. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17:1253–1270. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liao MJ, et al. Enrichment of a population of mammary gland cells that form mammospheres and have in vivo repopulating activity. Cancer Res. 2007;67:8131–8138. doi: 10.1158/0008-5472.CAN-06-4493. [DOI] [PubMed] [Google Scholar]
- 31.Shackleton M, et al. Generation of a functional mammary gland from a single stem cell. Nature. 2006;439:84–88. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- 32.Sun S, et al. Isolation of mouse marrow mesenchymal progenitors by a novel and reliable method. Stem Cells. 2003;21:527–535. doi: 10.1634/stemcells.21-5-527. [DOI] [PubMed] [Google Scholar]
- 33.Shiota M, et al. Isolation and characterization of bone marrow-derived mesenchymal progenitor cells with myogenic and neuronal properties. Exp Cell Res. 2007;313:1008–1023. doi: 10.1016/j.yexcr.2006.12.017. [DOI] [PubMed] [Google Scholar]
- 34.Woodward WA, Chen MS, Behbod F, Rosen JM. On mammary stem cells. J Cell Sci. 2005;118:3585–3594. doi: 10.1242/jcs.02532. [DOI] [PubMed] [Google Scholar]
- 35.Pereira RF, et al. Cultured adherent cells from marrow can serve as long-lasting precursor cells for bone, cartilage, and lung in irradiated mice. Proc Natl Acad Sci USA. 1995;92:4857–4861. doi: 10.1073/pnas.92.11.4857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gowen LC, et al. Targeted disruption of the osteoblast/osteocyte factor 45 gene (OF45) results in increased bone formation and bone mass. J Biol Chem. 2003;278:1998–2007. doi: 10.1074/jbc.M203250200. [DOI] [PubMed] [Google Scholar]
- 37.Ducy P, et al. Increased bone formation in osteocalcin-deficient mice. Nature. 1996;382:448–452. doi: 10.1038/382448a0. [DOI] [PubMed] [Google Scholar]
- 38.Petersen DN, Tkalcevic GT, Mansolf AL, Rivera-Gonzalez R, Brown TA. Identification of osteoblast/osteocyte factor 45 (OF45), a bone-specific cDNA encoding an RGD-containing protein that is highly expressed in osteoblasts and osteocytes. J Biol Chem. 2000;275:36172–36180. doi: 10.1074/jbc.M003622200. [DOI] [PubMed] [Google Scholar]
- 39.Walkley CR, et al. Conditional mouse osteosarcoma, dependent on p53 loss and potentiated by loss of Rb, mimics the human disease. Genes Dev. 2008;22:1662–1676. doi: 10.1101/gad.1656808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Berman SD, et al. Metastatic osteosarcoma induced by inactivation of Rb and p53 in the osteoblast lineage. Proc Natl Acad Sci USA. 2008;105:11851–11856. doi: 10.1073/pnas.0805462105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morito S, et al. Insulin signaling in adipocytes differentiated from mouse stromal MC3T3–G2/PA6 cells. Biol Pharm Bull. 2005;28:2040–2045. doi: 10.1248/bpb.28.2040. [DOI] [PubMed] [Google Scholar]
- 42.Fink L, et al. Real-time quantitative RT-PCR after laser-assisted cell picking. Nat Med. 1998;4:1329–1333. doi: 10.1038/3327. [DOI] [PubMed] [Google Scholar]
- 43.Sooy K, Sabbagh Y, Demay MB. Osteoblasts lacking the vitamin D receptor display enhanced osteogenic potential in vitro. J Cell Biochem. 2005;94:81–87. doi: 10.1002/jcb.20313. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.