Abstract
The hypothesis that dichromatic behavior on a clinical anomaloscope can be explained by the complement and arrangement of the long- (L) and middle-wavelength (M) pigment genes was tested. It was predicted that dichromacy is associated with an X-chromosome pigment gene array capable of producing only a single functional pigment type. The simplest case of this is when deletion has left only a single X-chromosome pigment gene. The production of a single L or M pigment type can also result from rearrangements in which multiple genes remain. Often, only the two genes at the 5′ end of the array are expressed; thus, dichromacy is also predicted to occur if one of these is defective or encodes a defective pigment, or if both of them encode pigments with identical spectral sensitivities. Subjects were 128 males who accepted the full range of admixtures of the two primary lights as matching the comparison light on a Neitz or Nagel anomaloscope. Strikingly, examination of the L and M pigment genes revealed a potential cause for a color-vision defect in all 128 dichromats. This indicates that the major component of color-vision deficiency could be attributed to alterations of the pigment genes or their regulatory regions in all cases, and the variety of gene arrangements associated with dichromacy is cataloged here. However, a fraction of the dichromats (17 out of 128; 13%) had genes predicted to encode pigments that would result in two populations of cones with different spectral sensitivities. Nine of the 17 were predicted to have two pigments with slightly different spectral peaks (usually≤2.5 nm) and eight had genes which specified pigments identical in peak absorption, but different in amino acid positions previously associated with optical density differences. In other subjects, reported previously, the same small spectral differences were associated with anomalous trichromacy rather than dichromacy. It appears that when the spectral difference specified by the genes is very small, the amount of residual red-green color vision measured varies; some individuals test as dichromats, others test as anomalous trichromats. The discrepancy is probably partly attributable to testing method differences and partly to a difference in performance not perception, but it seems there must also be cases in which other factors, for example, cone ratio, contribute to a person's ability to extract a color signal from a small spectral difference.
Keywords: Color vision deficiency, Cone photopigments, Pigment optical density, Genetics
Introduction
The common, dichromatic forms of inherited color-vision deficiency fall into two categories, protanopia and deuteranopia. The former is associated with the absence of functional long-wavelength-sensitive (L) cones, and the latter with the absence of functional middle-wavelength-sensitive (M) cones. The genes that encode the L and M cone photopigments lie in a head-to-tail tandem array on the X-chromosome. Humans with normal color vision have an array that contains an L gene followed by one or more M genes (Nathans et al., 1986b). The human L and M photopigment genes are greater than 98% identical in nucleotide sequence, and the arrays undergo both equal and unequal homologous recombination, producing new gene arrangements that underlie the common inherited forms of red-green color-vision deficiency (Nathans et al., 1986a). Since the pioneering work of Nathans et al., there have been several key advances in our understanding of the mechanisms that control the absorption spectra of the L and M cone photopigments and in the regulation of L and M gene expression that have aided in understanding the genetic basis for color-vision deficiency.
Together, amino acid positions 277 and 285, encoded by exon 5 of the L and M genes, are responsible for producing the majority of the spectral difference between L and M pigments. Thus, the sequence of exon 5 can be used to determine whether a gene encodes a pigment of the L-class with a wavelength of peak absorbance (λmax) near 560 nm, or of the M-class with peak near 530 nm. Substitutions at five other polymorphic positions encoded by exons 2, 3, and 4 of the genes shift the λmaxof L-class pigments and a subset of them also shift the λmaxof M-class pigments. The λmaxof pigments of known sequence have been estimated by a variety of in vitro (Merbs & Nathans, 1992, 1993; Asenjo et al., 1994) and in vivo methods (Neitz et al., 1995; Sharpe et al., 1998; Carroll et al., 2002), and this information can be used to predict the difference in λmaxbetween pigments encoded by different genes.
Recent evidence has demonstrated that, with some exceptions (Sjoberg et al., 1998), only the first two genes in the X-chromosome visual pigment gene array are expressed (Hayashi et al., 1999; Hayashi et al., 2001; Neitz et al., 2003). Long-distance polymerase chain reaction (PCR) allows amplification of the polymorphic exons from the first gene in the array and from all of the downstream genes together, and of exons 3 through 6 of the last gene in the array. These PCR products can be directly sequenced, and by a deductive process the sequences of the polymorphic exons from the first two genes in the array can often be determined.
The purpose of this study was to test the hypothesis that the majority of cases of dichromacy, diagnosed using a conventional clinical anomaloscope, can be explained simply by the complement and arrangement of L and M photopigment genes on the X-chromosome. The hypothesis specifically predicts that a male will be dichromatic if he has an array in which (1) there is only one visual pigment gene, (2) the first two genes encode pigments with identical absorption spectra, or (3) one of the genes in the first two positions has a mutation that prevents expression of the gene or function of the encoded pigment. We tested this hypothesis by examining the photopigment gene arrays in 128 dichromats, and found that in every case the complement and arrangement of the photopigment genes predicted a color-vision deficient phenotype. For 111 subjects, the gene arrays predicted strictly the production of a single X-chromosome photopigment and thus a dichromatic phenotype. For 17 subjects, the genes predicted the presence of two pigments with slightly different absorption spectra; thus, even though the genes predicted a color-vision defect, color-matching performance was slightly worse than might have been predicted by the genetic results.
Materials and methods
Phenotyping
Color-vision phenotype was assessed using standard color-vision tests including the Ishihara pseudoisochromatic plates, the AOHRR, FM100, and the panel D15. Color matching was performed using either a Neitz anomaloscope (Type I) or a Nagel anomaloscope (model OT1). Only males who accepted the entire range of mixtures of the primary test lights as matching the standard comparison light were accepted as dichromats. 73 deuteranopes and 55 protanopes were identified.
Genotyping
Genomic DNA was isolated from whole blood or from a cheek swab. The relative number of L and M genes per X-chromosome array was determined using two real-time quantitative PCR assays that have been described in detail elsewhere (Neitz & Neitz, 2001). One assay was used to estimate the ratio of L to M genes, and another was used to estimate the ratio of first to downstream genes. For each assay, quadruplicate reactions were done for each subject, and the gene ratio estimates reported in Tables 2 and 3 are the averages of the quadruplicate reactions. In addition, control samples of known gene ratios were run in parallel with the experimental samples in order to monitor the reliability of the assays. The 95% confidence interval for the gene ratio estimates is ± 4%.
Table 2.
Multigene deuteranopes
| L gene sequencesd | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Subject ID |
% L genesc |
% Downstream genes |
No. L genes |
No. M genes |
Exon 2 65111 116 |
Exon 3 153 171 174 178 180 |
Exon 4 203 230 233 236 |
M gene mutationd |
Downstream gene promoter |
Last gene |
Predicted Δλmax (nm) |
| 110a | 48.87 | 52.09 | 1 | 1 | ND | ND | ND | IVYLIAVATSV | not done | not done | 0 |
| 152a | 39.92 | 43.85 | 1 | 1 | ND | ND | ND | C203R | not done | not done | |
| 61a | 49.38 | 49.67 | 1 | 1 | T I S | M I A I S | C I A M | C203R | not done | not done | |
| JD051380b | 50.98 | 53.04 | 1 | 1 | T I S | L V A I A | C I A M | C203R | not done | not done | |
| JG092977b | 45.76 | 53.26 | 1 | 1 | T I S | L V A I A | C I A M | C203R | not done | not done | |
| BS030727b | 38.13 | 56.3 | 1 | 1 | T I S | ND | C I A M | C203R | not done | not done | |
| 165a | 32.07 | 65.62 | 1 | 2 | ND | ND | ND | C203R | not done | not done | |
| TL020777b | 33.74 | 71.91 | 1 | 2 | T I S | L V A I A | C I A M | C203R | not done | not done | |
| TT041171b | 30.28 | 73.54 | 1 | 2-3 | ND | L V A I A | C I A M | C203R | not done | not done | |
| TF050275b | 34.25 | 68.73 | 1 | 2 | T I S | L V A I A | C I A M | P187S | not done | not done | |
| MF041877b | 105.94 | 49.55 | 2 | 0 | T I S | M V V V A | C I A V | not done | not done | 0 | |
| DF081184b | 105.62 | 50.32 | 2 | 0 | T I S | M V V V A | C I A V | not done | not done | 0 | |
| 176a | 101.78 | 40.34 | 2 | 0 | T I S | L V A I A | C I A M | not done | not done | 0 | |
| RE080644b | 65.4 | 62.93 | 2 | 1 | T I S | L V A I S | C I A M | none found | not done | M | 0 |
| FT21730b | 60.44 | 74.69 | 2-3 | 1 | I V Y | L V A I S | C I A M | none found | not done | M | 0 |
| 94a | 47.9 | 76.15 | 2 | 2 | T I S | M V A I A | C I A M | none found | not done | not done | 0 |
| RG100741b | 102.91 | 70.97 | 3 | 0 | T I S | M V A I A | C I A M | T -483 C, G -21 A | not done | 0 | |
| 145a | ND | 59.75 | 2 | 1 | N D I/V S/Y | M V A I A | C I A M | none found | G & A @ -21 | not done | 2.5 |
| 35a | 53.12 | 79.63 | 2-3 | 2-3 | T/I I/V S/Y | L/M V A I A | C I A M | not done | NORMAL | not done | 2.5 |
| 130a | 61.1 | 55.69 | 2 | 1 | T/I I/V S/Y | M V A I A | C T S V | not done | NORMAL | M | 2.5 |
| 171a | 62.42 | 71.06 | 2 | 1 | T I/V S/Y | L V A I S | C I A M | not done | NORMAL | M | 2.5 |
| BC072327b | 48.92 | 71.27 | 2 | 1-2 | T/I I/V S/Y | L/M V A I A | C I A M | none found | NORMAL | M | 2.5 |
| JB011640b | 52.27 | 76.61 | 2 | 2 | T I/V S/Y | L/M V V V A | C I A V/M | none found | NORMAL | M | 2.5 |
| MJ102679b | ND | 75.24 | ≥2 | ≥1 | T/I I/V S/Y | L/M V A I A | C I A M | none found | NORMAL | not done | 2.5 |
| 138a | ND | 63.15 | 2 | 1 | T/I I/V S/Y | L V A I A/S | C T/I S/A V/M | not done | NORMAL | not done | >0 |
| 34a | ND | 70.1 | 2 | 1 | T/I I/V S/Y | L/M V A I A/S | C T/I S/A V/M | not done | NORMAL | not done | >0 |
Rayleigh match obtained with Nagel Type 1 anomaloscope.
Rayleigh match obtained with Neitz OT1 anomaloscope.
No Data, unreliable real-time PCR.
Single letter amino acid code: A = alanine, C = cysteine, I = isoleucine, L= leucine, M = methionine, R = arginine, S = serine, T = threonine, V = valine, Y = tyrosine, ND = not determined.
Table 3.
Multigene protanopes
| M gene sequencesc | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Subject ID | % L genes |
% Downstream genes |
No. L genes |
No. M genes |
Exon 2 65111116 |
Exon 3 153171174176180 |
Exon 4 204230233 236 |
Mutation | Predicted Δλmax (nm) |
| JB122273b | 31.01 | 73.61 | 1 | 4 | I V Y | M V A I A | C T S V | L gene last | 0 |
| RS072630b | 43.3 | 53.79 | 1 | 1 | not done | not done | not done | TISLIAVAIAMd | 0 |
| DT090543b | 1.87 | 65.79 | 0 | 3 | T/I I/V S/Y | L V A I A | C/R T S V | C203R | 0 |
| 54a | 1.57 | 47.22 | 0 | 2 | T/I I/V S/Y | L V A I A | C/R T S V | C203R | 0 |
| 48a | 0.56 | 62.65 | 0 | 3 | I V Y | L/M V A I A | C/R T S V | C203R | 0 |
| GP041934b | 0 | 51.92 | 0 | 2 | I V Y | M V A I A | C T S V | none found | 0 |
| BS070784b | 0 | 55.3 | 0 | 2 | I V Y | M V A I A | C T S V | none found | 0 |
| 167a | 7.73 | 55.81 | 0 | 2 | I V Y | M V A I A | C T S V | none found | 0 |
| BW112726b | 1.06 | 56.57 | 0 | 2 | I V Y | M V A I A | CT S V | none found | 0 |
| SS091021b | 0 | 82.78 | 0 | 5 | I V Y | M V A I A | C T S V | none found | 0 |
| VJ022026b | 0 | 70.37 | 0 | 3 | I V Y | M V A I A | C T S V | none found | 0 |
| 27a | 0 | 71.87 | 0 | 3 | I V Y | M V A I A | C T S V | none found | 0 |
| JH082574b | 0.83 | 62.82 | 0 | 3 | I V Y | M V A I A | C T S V | none found | 0 |
| 166a | 8.09 | 57.84 | 0 | 2 | I V Y | M V A I A | C T S V | none found | 0 |
| 20a | 0 | 48.59 | 0 | 2 | I V Y | M V A I S | C T S V | none found | 0 |
| DM081241b | 0.17 | 71.62 | 0 | 3 | I V Y | M V A V A | C T S V | G -425 Ae | 0 |
| 141a | 0 | 51.15 | 0 | 2 | I V Y | M V V V A | C T S V | none found | 0 |
| AT052178b | 0 | 48.16 | 0 | 2 | I V Y | M V A/V I/V A | C T S V | none found | 0 |
| 174a | 6.4 | 55.36 | 0 | 2 | I V Y | M V A/V I/V A | C T S V | G -425 Ae | 0 |
| CW121741b | 0.62 | 48.74 | 0 | 2 | T I/V S/Y | L/M V A I A | C T S V | none found | 0 |
| RW061526b | 0.14 | 50.72 | 0 | 2 | T/I I/V S/Y | L V A I/V A | C T S V | none found | 0 |
| 10a | 3.05 | 47.17 | 0 | 2 | T/I I/V S/Y | L/M V A I A | C T S V | none found | 0 |
| 105a | 4.22 | 50.73 | 0 | 2 | T/I I/V S/Y | L/M V A I A | C T S V | none found | 0 |
| RL052633b | 2.14 | 54.51 | 0 | 2 | T/I I/V S/Y | L/M V A I A | C T S V | none found | 0 |
| 178a | 3.25 | 57.15 | 0 | 2 | T/I I/V S/Y | L/M V A I S | C T S V | none found | 0 |
| 139a | 4.58 | 48.22 | 0 | 2 | T/I I/V S/Y | L/M V A/V I/V A | C T S V | none found | 0 |
| TT010182b | 0 | 48.13 | 0 | 2 | T/I I/V S/Y | M V A I A | C T S V | none found | 0 |
| JH101146b | 1.28 | 47.84 | 0 | 2 | T/I I S/Y | M V V V A | C T S V | G -425 Ae | 0 |
| BM072774b | 1.3 | 54.98 | 0 | 2 | T/I I/V S/Y | M V A I S | C T S V | A -71 Ce | 0 |
| CB050289b | 1.03 | 67.23 | 0 | 3 | T/I I/V S/Y | L/M V A I A | C T S V | none found | 0 |
| JW092022b | 0.57 | 67.28 | 0 | 3 | T/I I/V S/Y | L/M V A I A | C T S V | none found | 0 |
| JB111483b | 0.16 | 64 | 0 | 3 | T/I I/V S/Y | M V A I A | C T S V | none found | 0 |
| HH122516b | 0 | 64.67 | 0 | 3 | T/I I/V S/Y | M V A I A | C T S V | none found | 0 |
| 49a | 0.39 | 66.38 | 0 | 3 | T/I I/V S/Y | M V A I A | C T S V | none found | 0 |
| 33a | 0.36 | 63.54 | 0 | 3 | T/I I/V S/Y | M V A I/V A | C T S V | none found | 0 |
| 126a | 2.92 | 66.67 | 0 | 3 | T/I I/V S/Y | M V V V A | C T S V | none found | 0 |
| 135a | 0.28 | 42.29 | 0 | 2 | T/I I/V S/Y | L/M V A I A | C T/I S/A V/M | none found | 1.5 |
Rayleigh matches were obtained with a Nagel Type I anomaloscope.
Rayleigh matches were obtained with a Neitz OT1 anomaloscope.
Single letter amino acid code: A = alanine, C = cysteine, I = isoleucine, L= leucine, M = methionine, R = arginine, S = serine, T = threonine, V = valine, Y = tyrosine.
Single letter amino acid code indicating amino acid found at positions 65, 111, 116, 153, 171, 174, 178, 180, 230, 233, and 236 of L pigment gene.
Mutation in promoter region of downstream gene, numbering convention counts backward from mRNA start site which is +1.
For subjects who had both L and M pigment genes on the X-chromosome, as indicated by the results of the real-time quantitative PCR assays, the M and L photopigment genes were separately and specifically amplified using primer pairs 1 and 2 and the PCR conditions indicated in Table 1. Specificity for L genes was conferred by primer 3′ redx5 (Table 1) and for M genes by primer 3′ greenx5 (Table 1), which hybridized to sequences within exon 5 of L or M genes, respectively. The PCR products obtained with primer pairs 1 and 2 were used in another round of PCR to amplify exons 2, 3, and 4 using primer pairs 3, 4, and 5, respectively (Table 1). For a subset of subjects, exons 1, 6, and 5 were amplified nonspecifically from all genes in the array using primer pairs 6, 7, and 8 (Table 1). Primer pairs 3 through 8 amplify individual exons including the intron/exon junctions. Also, for a subset of subjects, an 823-bp fragment containing the promoter region of all downstream genes in the array was amplified using primer pair 9 (Table 1). For those subjects for whom the results of the real-time quantitative PCR assay indicated the presence of only L or only M pigment genes, the exons of the genes were amplified using primer pairs 3 through 8 as indicated in Table 1.
Table 1.
PCR primers and conditions
| Primer | ||||||
|---|---|---|---|---|---|---|
| Pair | Name | Sequence | Position | Concentration | Region amplified | PCR conditions |
| new5′x2 | 5′GTCTCTGGCTTGAGGGACAG | intron 1, 180 bp upstream of exon 2 | 300 nm | intron 1-exon 5, M genes only | 94°C 5 min; 94°C 1 min, | |
| 1 | 62°C 30 sec, 72°C 5 min, | |||||
| 3′greenx5 | 5′CAGCAGAATGCCAGGACCATC | M gene exon 5 | 300 nm | 37 cycles; 72°C 20 min, 4°C hold | ||
| new5′x2 | 5′GTCTCTGGCTTGAGGGACAG | intron 1, 180 bp upstream of exon 2 | 300 nm | intron 1-exon 5, L genes only | 94°C 5 min; 94°C 1 min, | |
| 2 | 62°C 30 sec, 72°C 5 min, | |||||
| 3′redx5 | 5′GCAGTACGCAAAGATCATCACC | L gene exon 5 | 300 nm | 37 cycles; 72°C 20 min, 4°C hold | ||
| 5′exon2 | 5′CTCGAATTCGGTGCTGCAGCCGAGCTCC | 55bp upstream of exon 2 | 600 nm | exon 2 | 95°C 9 min 1 cycle; | |
| 3 | 94°C 45 sec, 68°C 1 min 40 cycles; | |||||
| 3′exon2 | 5′CTCGAATTCGAGCCTGGGCCCCGACTGGC | 30 bp downstream of exon 2 | 600 nm | 72°C 10 min, 4°C hold | ||
| In2ul36x3F | 5′CAGAGTCTGACCCTGCCCACT | intron 2, 136 bp upstream of exon 3 | 300 nm | exon 3 | 94°C 5 min; 94°C 45 sec, | |
| 4 | 61°C 45 sec, 72°C 45 sec 30 cycles; | |||||
| In3dn46x3R | 5′TGTCGTTTTTTCCACCTCAGTCC | intron 3, 46 bp downstream of exon 3 | 300 nm | 72°C 10 min, 4°C hold | ||
| In3up23x4F | 5′TTGAGGGCAGAGCAGCTTAGG | intron 3, 23bp upstream of exon 4 | 300 nm | exon 4 | 94°C 5 min; 94°C 45 sec, | |
| 5 | 62°C 45 sec, 72°C 45 sec 30 cycles; | |||||
| In4dn62x4R | 5′TGGCTGCCGGCCCTTC | intron 4, 62bp dwnstrm of exon 4 | 300 nm | 72°C 10 min, 4°C hold | ||
| 340-362 | 5′CAGCCACCCAGCCTCCAC | 160 bp upstream of ATG start codon, all genes | 300 nm | exon 1 | 95°C 9 min 1 cycle; 94°C 30 sec, | |
| 6 | 61°C 15 sec, 72°C 30 sec, 30 cycles; | |||||
| In1dn35x1R | 5′AGTCCCAGGCCCAATTAAGAGAT | intron 1, 35 bp dwnstrm of exon 1 | 300 nm | 72°C 7 min, 4°C hold | ||
| In5up42x6F | 5′GGAGAGGTGGCCAAAGCCC | intron 5, 42bp upstrm of exon 6 | 300 nm | exon 6 | 94°C 5 min; 94°C 30 sec, | |
| 7 | 63°C 15 sec, 72°C 30 sec 30 cycles; | |||||
| In6dn51x6R | 5′ACCCTTCCCTGCTCTGCTCAA | intron 6, 51bp dwnstrm of exon 6 | 300 nm | 72°C 10 min, 4°C hold | ||
| 5′exon5 | 5′ACGGTATTTTGAGTGGGATCTGCT | intron 4, 35bp upstream of exon 5 | 300 nm | exon 5 | 94°C 9 min; 94°C 45 sec, | |
| 8 | 59°C 45 sec. 72°C 45, 37 cycles; | |||||
| 3′exon5 | 5′TCCACCCCCCGACTCACTATCC | intron 5, 40 bp downstream of exon 5 | 300 nm | 72°C 10 min, 4°C hold | ||
| upstrmgrnFd | 5′CCTGCAAGTGGGAATCTAAACAGA | 793 bp upstream of start codon of downstream genes | 100 nm | downstream gene promoter | 95°C 5 min 1 cycle; 94°C 1 min, | |
| 9 | 60°C 1 min sec, 72°C 1.5 min, | |||||
| 565-545exl | 5′TGGGTGCTGTCCTCATAGCTG | exon 1, 70 bp downstream of start codon | 300 nm | 40 cycles; 72°C 10 min, 4°C hold | ||
For primer pairs 3, 6, and 8, the AmpliTaq Gold PCR kit was used; for all other primer pairs the XL-PCR kit was used in conjunction with AmpliWax Gems. Each PCR reaction was done in a final volume of 100 µl, and the concentration of each primer in the reaction is given in Table 1. The concentrations of all other reaction components were those recommended by the manufacturer for the individual kits.
The PCR products obtained with primer pairs 3 through 9 were amplified for direct sequencing with the same primers except that each forward primer was tagged with the M13-21 primer sequence, and all reverse primers were tagged with the M13 reverse primer sequence. DNA sequencing was done using dye-primer AmpiTaq FS sequencing kit with the M13-21 primer or the M13 reverse primer. Reactions were analyzed on an ABI 310 Prism Genetic Analyzer. The 823-bp PCR product containing the downstream gene promoter was also sequenced using dye-terminator chemistry with primer LMprm-190F (5′ CCAGCAAATCCCTCTGAGC). A 531-bp DNA fragment encompassing the promoter of the first gene in the array and extending 70 bp into exon 1 of the first gene was amplified and sequenced as described elsewhere (McMahon et al., 2003).
Identification of the first gene and the last gene in the array as encoding an L or M pigment was performed for selected subjects using previously described methods (Kainz et al., 1998; Hayashi et al., 1999). Whether a gene encodes an L- or an M-class pigment was determined by identifying the amino acids specified at positions 277 and 285 of the opsin; both positions are encoded by exon 5. L-class pigments specify tyrosine at position 277 and threonine at position 285, whereas M-class pigments specify phenylalanine at position 277 and alanine at position 285.
Experiments involving human subjects were conducted in accordance with the principles embodied in the declaration of Helsinki, and were approved by Institutional Review Boards at the Medical College of Wisconsin, and the University of California—Davis.
Results
The interpretation of the results of the real-time quantitative PCR assays to estimate the relative ratio of L:M genes and the ratio of first:downstream genes has been described in detail elsewhere (Neitz & Neitz, 2001). Briefly, the results are expressed as the percentage of genes in the array that are downstream of the first gene, and the percentage of L plus M genes that are L. Each estimate is the average of the results from four separate real-time quantitative PCR reactions, with a 95% confidence interval of ±4%. An estimate of 0% downstream genes indicates an array that contains a single visual pigment gene, an estimate of 50% downstream genes indicates an array with two genes, and a value of 66% downstream genes indicates an array with three genes. An estimate of 100% L genes indicates an array that contains only L genes, and an estimate of 0% L genes indicates an array with only M genes. Intermediate values, such as 50% L genes, indicate an array with equal numbers of L and M genes. Thus, an estimate of 50% L genes and 50% downstream genes is interpreted as an array with one L and one M gene, an estimate of 33% L genes and 66% downstream genes is interpreted as an array with one L and two M genes.
Results from real-time quantitative PCR indicated 0% downstream genes and 100% L genes for 47 deuteranopes, and 0% downstream genes and 0% L genes for 18 protanopes, identifying these 65 subjects as single-gene dichromats. The remaining 26 deuteranopes and 37 protanopes were estimated to have arrays with two or more visual pigment genes, as indicated by a value of more than 40% downstream genes. A variety of genetic analyses were performed to characterize the complement and arrangement of genes in the arrays for the multigene dichromats.
The results of the genetic analyses for the deuteranopes are summarized in Table 2. For four deuteranopes (subjects 145, 138, MJ102679, & 34), the values from real-time PCR for the % L genes were not reliable; however, PCR products were obtained with both primer pairs 1 and 2 (Table 1) indicating that each of these individuals had arrays containing both L and M genes (Table 2). Direct sequencing of the PCR products revealed that all four subjects had at least two different genes encoding L-class pigments (Table 2). For example, subject 138 was estimated to have 63% downstream genes, a value expected for an array containing three genes, and he had at least two L gene sequences (Table 2) and one M gene sequence. Thus, we concluded that he had an array containing two L and one M genes. A similar logic was used to estimate the number of L and M genes in the arrays of the other three subjects for whom a reliable estimate of the ratio of L:M genes was not obtained. The reason for the occasional failure in the reliability of the assay is probably due to a slightly lower quality of DNA obtained from some subjects.
The results of long-distance PCR to characterize the first gene in the array and real-time quantitative PCR to estimate the relative number and ratio of L:M genes indicated that nine of the deuteranopes (Table 2, subjects 110, 152, 61, JD051380, JG092977, BS030727, 165, TL020777, TT041171, & TF050275) had arrays that contained one L pigment gene, which was in the first position, followed by one or more M pigment genes. This structure is typical of arrays found in men with normal color vision. Since deuteranopes lack an M cone contribution to vision, we hypothesized that the M genes in these arrays contained mutations that either prevented expression or function of M pigment, and we tested this hypothesis by sequencing the M genes. Eight of the subjects had M genes containing a mutation that substituted the amino acid arginine for cysteine at position 203 (C203R) in the photopigment molecule (Table 2 & Fig. 1). This mutation was previously identified as being involved in deuteranopia (Bollinger et al., 2001), deuteranomaly (Winderickx et al., 1992), and blue-cone monochromacy (Nathans et al., 1989, 1993). It has been shown to disrupt proper folding of the photopigment molecule by preventing the formation of a conserved and essential disulfide bond (Karnik et al., 1988; Karnik & Khorana, 1990; Kazmi et al., 1997). Of the eight deuteranopes who had C203R-containing M genes, five of them had only one M gene; however, three of them had multiple M genes, all of which had the C203R mutation.
Fig. 1.
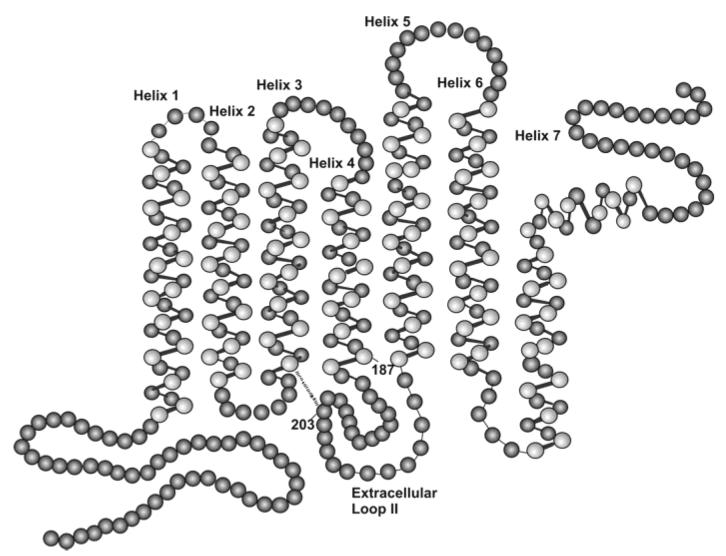
Location of inactivating amino acid substitutions in M photopigments in dichromats. Circles represent amino acids with the amino terminus in the lower left and carboxyl terminus in the upper right. The seven transmembrane alpha helical segments are labeled (helix 1-6). Amino acid positions 203 and 187 are indicated. The disulfide bond that is normally formed by cysteine residues at positions 126 and 203 is indicated by a dashed line.
Subject TF050275 (Table 2) had one normal M gene and one that contained a mutation that substituted the amino acid serine for proline at position 187 (P187S) of the photopigment molecule (Fig. 1). Long-distance PCR and DNA sequencing revealed that neither the first nor the last gene in the array had the mutation, so by deduction the second gene in the array must have it. Proline 187 is 100% conserved across the 212 vertebrate visual pigments in the G-protein coupled receptor database (www.gpcr.org), and it produces a significant bend at the extracellular end of the fourth transmembrane alpha helical segment (Palczewski et al., 2000). Replacing this conserved proline residue with serine very likely disrupts the proper folding and function of the photopigment molecule.
Subject 110 had an M gene that encoded a pigment with an unusual combination of amino acids at the polymorphic positions specified by exon 3, as shown in Table 2. This combination of normal polymorphisms (leucine 153, isoleucine 171, alanine 174, valine 178, alanine 180) has not been observed in the more than 300 L and M pigment genes that we have sequenced from individuals with normal color vision (Neitz & Neitz, unpublished results) and has only been observed in association with blue-cone monochromacy (pedigree H in Nathans et al., 1989). This unusual combination was observed in the affected members of pedigree H who had a single X-chromosome visual pigment gene, and it cosegregated with absence of function of the corresponding cones in 11 of 11 meioses. Thus, it seems reasonable to conclude that this unusual combination of polymorphisms is itself an inactivating mutation or it is tightly linked to one.
Sixteen deuteranopes had arrays containing more than one gene encoding an L-class pigment, and all but four of these also had M genes (Table 2). The L genes were sequenced to determine whether individuals had genes encoding pigments predicted to differ in λmax. There are 11 polymorphic amino acid positions among L-class pigments (Table 2); substitutions at four of them produce spectral variants (Merbs & Nathans, 1992; Asenjo et al., 1994; Carroll et al., 2002). Substitutions at positions 116 and 180 produce shifts of about 1.5 nm and 3.5 nm, respectively. Substitutions at 230 and 233 together produce a shift of about 4 nm. Thus, if two L pigments differ at one or more of these positions, they are expected to differ in λmax by a magnitude that depends on the specific sites that differ between the pigments. Seven of the 16 deuteranopes with multiple L genes (MF041877, DF081184, 176, RE080644, FT21730, 94, & RG100741) had arrays in which all of the L genes encoded pigments that were the same at the spectral tuning positions (Table 2). Three of these subjects also had M genes (Table 2). Two of the subjects (RE080644 & FT21730) had only one M gene, which long-distance PCR and sequencing showed to be at the 3′ end of the arrays. One subject, RG100741, had novel nucleotide changes upstream of the downstream genes (Fig. 2). Whether these interfered with proper expression of the second gene in the array is not known.
Fig. 2.
Nucleotide polymorphisms upstream of downstream genes in deuteranopes and protanopes. The sequence of the downstream gene promoter region extending from −493 to +1 found in men with normal color vision, where the +1 position is the transcriptional start site. Subject identification numbers are given at the top left. Subjects RG100741 and 145 were deuteranopes, and subjects JH101146, DM081241, 174, and BM072774 were protanopes. A period indicates positions of identity between the subject and the color normal sequence. Nucleotide differences are shown. One subject's downstream genes were polymorphic at position −21, indicated by an R (R is the IUB code for A or G).
The other nine deuteranopes (145, 138, 130, 171, BC072327, JB011640, MJ102679, 34, & 35) had multiple genes encoding L-class pigments that differed at one or more of the spectrally active positions (Table 2). No inactivating mutations were found in the coding regions or the intron/exon junctions of the L genes for these nine subjects. However, one subject (145) had a novel base change in the promoter region of at least one of his downstream genes, as shown in Fig. 2. Whether this change disrupts expression of the affected gene is not known. Of the deuteranopes with L genes encoding pigments expected to differ in λmax, most of them were predicted to differ by only about 1.5 nm (Table 2). For two subjects, the L pigments differ by at least 1 nm (138 and 34); however, we cannot make a more accurate prediction because we do not know how the substitutions were distributed across the two genes. Previous work indicates that the presence of two L pigments that differ in λmax by a small amount can support anomalous trichromacy (Neitz et al., 1996; Shevell et al., 1998).
All of the deuteranopes who had two L genes that were predicted to differ in λmax also had M pigment genes. Usually only the first two genes in the array are expressed. Thus, to explain why these subjects have a color-vision deficiency requires either that the M genes not be expressed due to their position in the array or due to the presence of an inactivating mutation. Sequencing of the M genes in these subjects did not reveal an inactivating mutation in the coding sequences, the intron/exon junctions or the promoter regions. Long-distance PCR and DNA sequencing showed that four of the deuteranopes had an M pigment gene in the last (3′ -most) position in the array (Table 2, subjects 171, BC072327, JB011640, & MJ102679). Subjects 171 and 130 had arrays which contained only one M pigment gene, and these results indicated that it was in a nonexpressed position. The two other deuteranopes (BC072327 & JB011640) had arrays with more than one M gene, and although one was in a nonexpressed position, it is not possible to determine whether the other one was also. Subjects 138 and 34 both had arrays estimated to have two L and one M genes. Long-distance PCR and sequencing indicated that the first gene in the arrays from these subjects encoded an L pigment, but attempts to amplify the last gene in the arrays were unsuccessful.
Table 3 summarizes the results for the 37 protanopes estimated to have X-chromosome arrays containing more than one visual pigment gene. All but two of the subjects had arrays that lacked genes encoding an L pigment. One exception was subject JB122273, who was estimated to have one L and four M genes. Sequence analysis revealed that the first gene in his array encoded an M-class pigment, while the last gene encoded an L-class pigment. Thus, subject JB122273 had an unusual gene order, in which the gene encoding the L-class pigment was not in the usual position at the 5′ end of the array, but instead was at the 3′ end of the array and was preceded by at least two M genes meaning the L gene was in a non-expressed position. Sequence analysis of his M genes indicated that they all encoded the identical M pigment. The other protanope with an array containing an L gene was RS072630, who was estimated to have one L and one M gene, with the L gene being first in the array. Since protanopia is characterized by the absence of functional L cones, we hypothesized that the L pigment gene was either not expressed or encoded a defective pigment. The sequence of RS072630's L gene revealed that the combination of amino acids at the polymorphic positions specified by exon 3 was the same unusual combination observed in deuteranopic subject 110 and previously observed in blue-cone monochromacy, and which we argue to either represent an inactivating mutation or be tightly linked to one.
For all other protanopes listed in Table 3, direct sequence analysis of exons 2, 3, and 4 of the X-chromosome visual pigment genes was performed in order to determine whether individuals had genes that encoded M pigments predicted to differ in λmax. The amino acid positions at which substitutions shift the spectrum of M-class pigments are position 180 encoded by exon 3, and positions 230 and 233 encoded by exon 4 (Merbs & Nathans, 1993; Asenjo et al., 1994). Three protanopes (subjects DT090543, 54, & 48) were discovered to have the C203R mutation in at least one of their M genes (Table 3). Twelve protanopes (GP041934, BS070784, 167, BW112726, SS091021, VJ022026, 27, JH082574, 166, 20, DM081241, & 141) had genes that specified M pigments that were identical in the regions encoded by exons 2, 3, and 4, and so encoded pigments expected to be identical in spectral absorbance. Two of the protanopes (AT052178 & 174) had genes encoding M pigments expected to be identical in spectral absorbance because they differed only at amino acid positions 174 and 178 where substitutions have been shown not to shift the λmax(Asenjo et al., 1994). One protanope (135) had two genes encoding M-class pigments predicted to differ in λmaxby about 2 nm. Seventeen protanopes (Table 3, CW121741, RW061526, 10, 105, RL052633, 178, 139, TT010182, JH101146, BM072774, CB050289, JW092022, JB111483, HH122156, 49, 33, & 126) had genes specifying pigments that differed only at exon 2 encoded positions that do not shift the λmaxof M-class pigments but have been proposed as candidates for altering the pigment optical density, thereby producing enough of a difference in spectral sensitivity to support anomalous trichromacy (Shevell & He, 1992; Neitz et al., 1999). Seven of these subjects (Table 3) were estimated to have three genes encoding M-class pigments. We cannot determine the relative order of these genes, but it is possible that the genes in the first two positions encode pigments that are identical in amino acid sequence; alternatively the first two genes may specify pigments that differ only by exon 2-encoded polymorphisms.
If the exon 2 polymorphisms produce a difference in spectral sensitivity that can be used for color discrimination, then one hypothesis to explain the dichromatic phenotype in these 16 protanopes is that there is an inactivating mutation in one of their photopigment genes. However, sequence analysis of the coding region and the intron/exon junctions did not reveal a mutation in any of the subjects. Sequence analysis of the promoters of the first gene and of the downstream genes in each array did reveal two subjects with nucleotide changes compared to the downstream gene promoter sequence found in arrays underlying normal color vision (McMahon et al., 2004). One subject, BM072774, had an adenine (A) to cytosine (C) transversion at position —71 (Fig. 2) of his downstream gene. This mutation was recently reported to be associated with color-vision deficiency (Ueyama et al., 2003). One subject (JH101146) had a novel mutation in a downstream gene promoter that was a guanine (G) to adenine (A) transition at position —425 (Fig. 2), but whether this sequence change alters the expression of the downstream genes is not known. This same nucleotide change was found in the downstream genes of two other protanopes (DM081241 & 174, Table 3) whose genes encoded pigments predicted to have identical absorption spectra.
If exon 2 polymorphisms produce a difference in spectral absorbance, we expect to find protanomalous subjects who have genes encoding M-class pigments that differ only at exon 2-encoded polymorphic positions. Thus, for comparison with the dichromats that are the focus of this study, we have included a sample of individuals who tested as protanomalous trichromats under the same testing conditions in which the dichromats were identified. Table 4 summarizes the genetic data and color-matching results for eight males identified as protanomalous on the basis of their Rayleigh match results. Five of the subjects (31, 40, 46, 146, & 159) had genes encoding M-class pigments predicted to differ in λmax, thereby explaining why they behave as anomalous trichromats. Three subjects (240, 226, & 288) had genes encoding pigments that differ only in exon 2-encoded positions, but not at positions that shift the λmax, which is consistent with the hypothesis that exon 2-encoded amino acid differences produce a difference in the pigment optical density that can be used for color discrimination.
Table 4.
Protanomalous subjects with M genes differing only in exon 2
| M gene sequencesa | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Subject ID | % L genes | % Downstream genes | No. L genes | No. M genes | Exon 2 65111 116 |
Exon 3 153171 174178180 |
Exon 4 203 230233 236 |
Predicted Δλmax (nm) | Rayleigh match rangeb |
| 240 | 0 | 48.92 | 0 | 2 | T/I I/V S/Y | M V A I A | C T S V | 0 | 62-63 |
| 226 | 0 | 56.81 | 0 | 2 | T/I I/V S/Y | M V A I A | C T S V | 0 | 54-65 |
| 288 | 0 | 83.78 | 0 | 5 | T/I I/V S/Y | M V A I A | C T S V | 0 | 55-69 |
| 31 | 0 | 71.73 | 0 | 3-4 | I V Y | M V A I S | C T/I S/A V/M | 2-3 | 45-55 |
| 40 | 0 | 62.38 | 0 | 3 | T/I I/V S/Y | L/M V A I A/S | C T/I S/A V/M | 2-7 | 50-63 |
| 46 | 0 | 47.19 | 0 | 2 | T/I I/V S/Y | L/M V A I A/S | C T S V | 4 | 51-67 |
| 146 | 0 | 41.68 | 0 | 2 | T/I I/V S/Y | M V V V A | C T S/A V/M | 2-3 | 56-60 |
| 159 | 0 | 56.05 | 0 | 2 | T/I I/V S/Y | L/M V A I A/S | C T/I S/A V/M | 2-7 | 53-65 |
Single letter amino acid code: A = alanine, C = cysteine, I = isoleucine, L = leucine, M = methionine, S = serine, T = threonine, Y = Tyrosine, V = valine.
Nagel Units, Rayleigh match done on Nagel Type 1 anomaloscope.
Discussion
In this study, we tested the hypothesis that dichromatic behavior on a clinical anomaloscope could be explained by the specific complement and arrangement of L and M photopigment genes on the X-chromosome. It is expected that a male will behave as a dichromat if he has an X-chromosome visual pigment gene array that falls into one of the following three categories: (1) all but one gene has been deleted, (2) one of the two genes at the 5′ end of the array contains an inactivating mutation that interferes with expression of the gene or function of the encoded pigment, or (3) the first two genes encode pigments that do not differ in spectral absorbance. For all 128 dichromats in this study, the examination of the complement and arrangement of the photopigment genes on the X-chromosome revealed a potential cause for color-vision deficiency. However, a fraction of the dichromats (17 out of 128; 13%) had genes that were predicted to encode two pigments with small differences in spectral absorbance. In these individuals, the red-green color vision that would be expected to be afforded by the small pigment spectral absorbance differences was not sufficient for them to reject either of the monochromatic primaries as matching the comparison light when tested using a clinical anomaloscope.
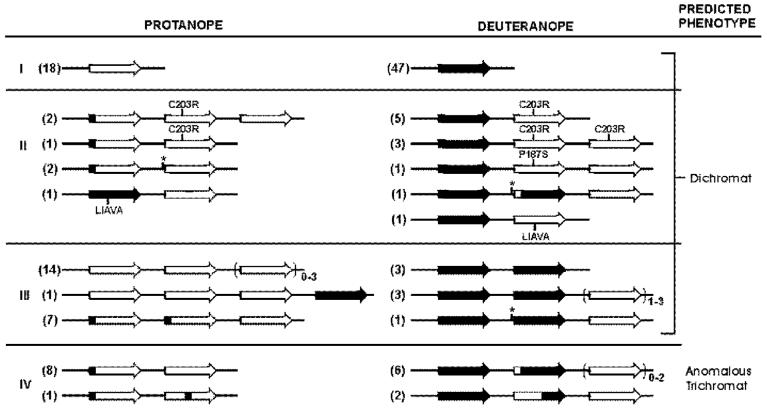
The array structures deduced from the genetic data for each subject are summarized in Fig. 3 and the arrays are grouped according to the three categories predicting dichromacy listed above and an additional category where the genes predicted pigments with a small spectral difference. Single-gene arrays were found in 64% of the deuteranopes and 33% of the protanopes, whereas multigene arrays were found in 36% of the deuteranopes and 67% of the protanopes.
Fig. 3.
Summary of genotypes in dichromats and the phenotypes predicted from genetics. See the text in the Discussion for description of category I, II, and III designations. Category IV contains arrays that did not fall into one of the other three categories. Black arrows represent genes encoding L pigment, white arrows represent genes encoding M pigments, arrows that are both black and white with a black arrowhead indicate genes encoding L-class pigments that differ in peak sensitivity compared to the pigment encoded by the black arrow, arrows that are black and white with a white arrowhead indicate genes encoding M-class pigments that differ in peak sensitivity compared to the pigment encoded by the white arrow. The numbers in parenthesis to the left of the arrays indicates the number of subjects with the given array structure. Arrows enclosed within parentheses indicates that subjects varied with regard to the number of genes of the type indicated by the arrow over the range of numbers given in the subscript to the right outside the parentheses.
The arrays in 85% of multigene deuteranopes had both L and M genes, 15% (4 individuals) lacked M genes. All four multigene deuteranopes who lacked M pigment genes had L genes encoding spectrally identical pigments (category III, Fig. 3), thus a dichromatic phenotype was expected.
Of the deuteranopes who had M genes, 50% had one L pigment gene, and M genes that encoded defective pigments (category II, Fig. 3). The most common defect was the C203R mutation, which was found in 31% of the multigene deuteranopes and 11% of all deuteranopes in this study. Two novel inactivating mutations were also identified; P187S and a specific combination of polymorphic amino acids encoded by exon 3.
The other 50% of the deuteranopes who had M genes had multiple L genes (categories III & IV, Fig. 3). The identity of the first gene in each array was determined directly for all subjects, and the identity of the last gene in the array was determined for a subset of subjects. The identity of the second gene was assigned by a combination of deductive reasoning in interpreting the genetic results and parsimony. For example, if a subject had a three-gene array, with two L and one M genes, and the first and last genes encoded an L and an M pigment, respectively, then by deduction the second gene in the array must encode an L-class pigment. If a subject had two M and two L genes, and the first and last genes encoded L and M pigments, respectively, and an inactivating mutation was not found in the M genes, then the most parsimonious explanation for the color-vision defect is that the second gene in the array encodes an L pigment. There were 12 deuteranopic subjects with multiple L genes and one or more M genes; the position of the M genes in a nonexpressed position was directly shown for six subjects (Table 3), and presumed for the rest. For four of these subjects (categories II & III, Fig. 3), the L genes encoded spectrally identical pigments, thus accounting for their dichromatic behavior. One of these subjects (category II, Fig. 3) also had a novel nucleotide substitution upstream of one of his downstream genes, which potentially could interfere with expression of the second gene in the array, thereby producing a dichromatic phenotype. For the other eight subjects (category IV, Fig. 3), the L genes encoded pigments predicted to differ in spectral peak. For most, the predicted spectral separation between the L-class pigments was ≤2.5 nm, and for two we could not determine a precise value, except that the spectral separation was predicted to be ≥1 nm). Two of these subjects in category IV (Fig. 3) may have more than two L genes, and if the first two in the array encode spectrally identical pigments, then a dichromatic phenotype would be predicted.
Only 1% of all protanopes (2/55) and ∼5% of multigene protanopes had arrays with L pigment genes. In one case the array contained one L and one M gene, and the L gene contained an inactivating mutation (category II, Fig. 3); in the other case the array contained four M genes that encoded identical M pigments, and the L gene was in a nonexpressed position (category III, Fig. 3).
Six percent of all protanopes in this study and 8% of the multigene protanopes had the C203R mutation in an M gene (category II, Fig. 3). This is the first report of the C203R mutation in protan gene arrays. One protanope had only two genes in his array, one with the mutation and one without it, thus dichromacy is the expected phenotype. Two protanopes had one gene with the mutation and two without it. For one of them (48), the encoded pigments did not differ at amino acid positions expected to alter spectral absorbance, and dichromacy is the expected phenotype. For the other, the first gene in the array does not have the C203R mutation, and there are three possible arrangements of the genes, two of which would result in dichromacy. That is, if the first gene encodes a functional pigment and the second one encodes the C203R pigment or the first two genes encode functional pigments with the same amino acid sequence, a dichromatic phenotype is expected.
Two protanopes (category II, Fig. 3) had novel nucleotide substitutions upstream of the downstream genes. Both had arrays containing only two genes, so dichromacy is the predicted phenotype if the promoter mutations interfere with expression of the downstream genes. Alternatively, if the promoter mutations have no effect on expression, the encoded pigments will differ only at exon 2-encoded sites, which is also the case for 15 other multigene protanopes. Seven of the 15 had three-gene arrays, and thus it is possible that the first two genes in the array encode pigments with identical amino acid sequences, and therefore identical spectral properties (category III, Fig. 3). The remaining eight subjects had two-gene arrays that differed in exon 2 (category IV, Fig. 3). Fourteen of the protanopes had multiple M genes that encoded the identical M pigments (category III, Fig. 3), thus explaining their dichromatic phenotype.
Polymorphic amino acid positions encoded by exon 2 were previously identified as candidates for altering the photopigment optical density. In a previous study, photopigment bleaching experiments and extended Rayleigh matching demonstrated that some protanopes have red-green color vision based on pigments that differ in optical density but not λmax. In that study, subjects who had exon 2-encoded dimorphisms behaved as protanomalous and subjects who behaved as dichromats did not have exon 2 differences or differences in exon 3 or 4 known to shift λmax. The probability that such an association could have occurred by chance is very low (<0.005) suggesting that the exon 2 differences were responsible for the optical density difference. Those earlier experiments were done using custom-built optical systems (Neitz et al., 1999), not clinical anomaloscopes, and part of the testing was done using large bright stimulus fields which maximize a subject's ability to demonstrate color-discrimination capacity. We propose that the protanopes with pigments which differ only at exon 2-specified positions would behave as protanomalous trichromats if tested on a comparable custom-built optical system which optimizes the detection of weak red-green color vision. In addition, we did identify three males who behaved as protanomalous when tested with the clinical anomaloscope and had M pigments which differed only at polymorphic amino acid positions encoded by exon 2. What we have come to appreciate from the experiments reported here is that when the spectral differences between pigments become small there is heterogeneity in performance on the clinical anomaloscope. Some individuals with pigments that have small differences in λmax test as dichromats and others test as anomalous trichromats. We propose that the red-green color vision of the protanomalous subjects in Table 4 arises from an optical density difference; however, just like the small differences in λmax predicted for other dichromats the signal produced by the optical density difference is not so strong that everyone with the difference will behave as an anomalous trichromat when tested on a clinical anomaloscope.
One protanope (category IV, Fig. 3) and nine deuteranopes had genes encoding pigments predicted to differ in λmax and at least eight other protanopes had exon 2 differences that have been associated with pigment optical density differences, thus, these subjects performed somewhat worse on the anomaloscope than their genes might predict. For subjects who have two cone types that differ only by a very small amount in spectral sensitivity, there are individual factors that might contribute to performance being worse than anticipated from the cone pigment complement. Untrained psychophysical subjects exhibit large differences in performance. Certainly, some people will agree that two lights match even though they perceive a small color difference while others will accept nothing but a perfect match. There must also be effects of anxiety, motivation, and attention that contribute to performance on the anomaloscope. However, in addition to the performance issues, physiological factors must also underlie true differences in discrimination ability among people who have pigments with small spectral differences. For example, a huge skew in cone ratio does not seem to produce a large loss in color-discrimination performance in a person with normal L and M pigments (Neitz et al., 2002); however, the same skewed cone ratio in a person with a very small pigment spectral sensitivity difference may diminish the color signal enough to cause the person to behave as a dichromat rather than an anomalous trichromat on a clinical anomaloscope.
Acknowledgments
This work was supported by NIH grants EY09303, AG04058, National Eye Institute Core Grants EY 12576 and EY01931, Research to Prevent Blindness, and the David and Ruth S. Coleman Charitable Foundation. J.S. Werner is a Jules and Doris Stein RPB Professor, and M. Neitz is the recipient of an RPB Senior Scientific Investigator Award. J. Neitz and M. Neitz are the recipients of an Alcon Research Institute Award.
References
- Asenjo AB, Rim J, Oprian DD. Molecular determinants of human red/green color discrimination. Neuron. 1994;12:1131–1138. doi: 10.1016/0896-6273(94)90320-4. [DOI] [PubMed] [Google Scholar]
- Bollinger K, Bialozynski C, Neitz J, Neitz M. The importance of deleterious mutations of M pigment genes as a cause of color vision defects. Color Research and Application. 2001;26:S100–S105. [Google Scholar]
- Carroll J, Neitz M, Neitz J. Estimates of L:M cone ratio from ERG flicker photometry and genetics. Journal of Vision. 2002;2:531–542. doi: 10.1167/2.8.1. [DOI] [PubMed] [Google Scholar]
- Hayashi T, Motulsky AG, Deeb SS. Position of a ‘green-red’ hybrid gene in the visual pigment array determines colour-vision phenotype. Nature Genetics. 1999;22:90–93. doi: 10.1038/8798. [DOI] [PubMed] [Google Scholar]
- Hayashi T, Yamaguchi T, Kitahara K, Sharpe LT, Jägle H, Yamade S, Ueyama H, Motulsky AG, Deeb SS. The importance of gene order in expression of the red and green visual pigment genes and in color vision. Color Research and Application. 2001;26 [Google Scholar]
- Kainz PM, Neitz M, Neitz J. Molecular genetic detection of female carriers of protan defects. Vision Research. 1998;38:3365–3369. doi: 10.1016/s0042-6989(97)00366-0. [DOI] [PubMed] [Google Scholar]
- Karnik SS, Khorana HG. Assembly of functional rhodopsin requires a disulfide bond between cysteine residues 110 and 187. Journal of Biological Chemistry. 1990;265:17520–17524. [PubMed] [Google Scholar]
- Karnik SS, Sakmar TP, Chen H-B, Khorana HG. Cysteine residues 110 and 187 are required for the formation of correct structure in bovine rhodopsin. Proceedings of the National Academy of Sciences of the U.S.A. 1988;85:8459–8463. doi: 10.1073/pnas.85.22.8459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazmi MA, Sakmar TP, Ostrer H. Mutation of a conserved cysteine in the X-linked cone opsins causes color vision deficiencies by disrupting protein folding and stablilty. Investigative Ophthalmology and Visual Science. 1997;38:1074–1081. [PubMed] [Google Scholar]
- McMahon C, Neitz J, Neitz M. Comparison of human and monkey pigment gene promoters to evaluate DNA sequences proposed to govern L:M cone ratio. In: Mollon JD, Knoblauch K, Pokorny J, editors. Normal and Defective Colour Vision. Oxford University Press; Oxford, UK: 2003. pp. 51–59. [Google Scholar]
- McMahon C, Neitz J, Neitz M. Evaluating the human X-chromosome pigment gene promoter sequences as predictors of L:M cone ratio variation. Journal of Vision. 2004;2:12–24. doi: 10.1167/4.3.7. [DOI] [PubMed] [Google Scholar]
- Merbs SL, Nathans J. Absorption spectra of the hybrid pigments responsible for anomalous color vision. Science. 1992;258:464–466. doi: 10.1126/science.1411542. [DOI] [PubMed] [Google Scholar]
- Merbs SL, Nathans J. Role of hydroxyl-bearing amino acids in differentially tuning the absorption spectra of the human red and green cone pigments. Photochemistry and Photobiology. 1993;58:706–710. doi: 10.1111/j.1751-1097.1993.tb04956.x. [DOI] [PubMed] [Google Scholar]
- Nathans J, Davenport CM, Maumenee IH, Lewis RA, Hejtmancik JF, Litt M, Lovrien E, Weleber R, Bachynski B, Zwas F, Klingaman R, Fishman G. Molecular genetics of blue cone monochromacy. Science. 1989;245:831–838. doi: 10.1126/science.2788922. [DOI] [PubMed] [Google Scholar]
- Nathans J, Maumenee IA, Zrenner E, Sadowski B, Sharpe LT, Lewis RA, Hansen E, Rosenberg P, Schwartz M, Hecken-lively JR, Trabousli E, Klingaman R, Bech-hansen NT, LaRouche GR, Pagon RA, Murphy WH, Weleber RG. Genetic heterogeneity among blue-cone monochromats. American Journal of Human Genetics. 1993;53:987–1000. [PMC free article] [PubMed] [Google Scholar]
- Nathans J, Piantanida TP, Eddy RL, Shows TB, Hogness DS. Molecular genetics of inherited variation in human color vision. Science. 1986a;232:203–210. doi: 10.1126/science.3485310. [DOI] [PubMed] [Google Scholar]
- Nathans J, Thomas D, Hogness DS. Molecular genetics of human color vision: The genes encoding blue, green, and red pigments. Science. 1986b;232:193–202. doi: 10.1126/science.2937147. [DOI] [PubMed] [Google Scholar]
- Neitz J, Neitz M, Kainz PM. Visual pigment gene structure and the severity of human color vision defects. Science. 1996;274:801–804. doi: 10.1126/science.274.5288.801. [DOI] [PubMed] [Google Scholar]
- Neitz J, Neitz M, He JC, Shevell SK. Trichromatic color vision with only two spectrally distinct photopigments. Nature Neuro-science. 1999;2:884–888. doi: 10.1038/13185. [DOI] [PubMed] [Google Scholar]
- Neitz J, Carroll J, Yamauchi Y, Neitz M, Williams DR. Color perception in mediated by a plastic neural mechanism that is adjustable in adults. Neuron. 2002;35:783–792. doi: 10.1016/s0896-6273(02)00818-8. [DOI] [PubMed] [Google Scholar]
- Neitz M, Neitz J. A new test for mass screening of school age children for red-green color vision defects. Color Research and Application. 2001;26:S239–S249. [Google Scholar]
- Neitz M, Neitz J, Jacobs GH. Genetic basis of photopigment variations in human dichromats. Vision Research. 1995;35:2095–2103. doi: 10.1016/0042-6989(94)00306-8. [DOI] [PubMed] [Google Scholar]
- Neitz M, Bollinger K, Neitz J. Middle wavelength sensitive photopigment gene expression is absent in deuteranomalous colour vision. In: Mollon JD, Knoblauch K, Pokorny J, editors. Normal and Defective Colour Vision. Oxford University Press; Oxford, UK: 2003. pp. 318–327. [Google Scholar]
- Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, LeTrong I, Teller DC, Okada T, Stenkamp RE, Yamamoto M, Miyano M. Crystal structure of rhodopsin: A G-protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- Sharpe LT, Stockman A, Jägle H, Knau H, Klausen G, Reitner A, Nathans J. Red, green, and red-green hybrid pigments in the human retina: Correlations between deduced protein sequences and psychophysically measured spectral sensitivities. Journal of Neuroscience. 1998;18:10053–10069. doi: 10.1523/JNEUROSCI.18-23-10053.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevell SK, He JC. Dual Rayleigh-like color matches sensitive to individual differences in cone photopigments. Advances in Color Vision: Journal of the Optical Society of America, Technical Digest Series. 1992;4:20–22. [Google Scholar]
- Shevell SK, He JC, Kainz PM, Neitz J, Neitz M. Relating color discrimination to photopigment genes in deutan observers. Vision Research. 1998;38:3371–3376. doi: 10.1016/s0042-6989(97)00434-3. [DOI] [PubMed] [Google Scholar]
- Sjoberg SA, Neitz M, Balding SD, Neitz J. L-cone pigment genes expressed in normal colour vision. Vision Research. 1998;38:3213–3219. doi: 10.1016/s0042-6989(97)00367-2. [DOI] [PubMed] [Google Scholar]
- Ueyama H, Li Y-H, Fu G-L, Lertrit P, Atchaneeyasakul L, Oda S, Tanabe S, Nishida Y, Yamade S, Ohkubo I. An A-71C substitution in a green gene at the second position in the red/green visual-pigment gene array is associated with deutan color-vision deficiency. Proceedings of the National Academy of Sciences of the U.S.A. 2003;100:3357–3362. doi: 10.1073/pnas.0637437100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winderickx J, Sanocki E, Lindsey DT, Teller DY, Motulsky AG, Deeb SS. Defective colour vision associated with a missense mutation in the human green visual pigment gene. Nature Genetics. 1992;1:251–256. doi: 10.1038/ng0792-251. [DOI] [PubMed] [Google Scholar]