Abstract
Background
DNA-bound transcription factors recruit an array of coregulatory proteins that influence gene expression. We previously demonstrated that DNA functions as an allosteric modulator of estrogen receptor α (ERα) conformation, alters the recruitment of regulatory proteins, and influences estrogen-responsive gene expression and reasoned that it would be useful to develop a method of isolating proteins associated with the DNA-bound ERα using full-length receptor and endogenously-expressed nuclear proteins.
Results
We have developed a novel approach to isolate large complexes of proteins associated with the DNA-bound ERα. Purified ERα and HeLa nuclear extracts were combined with oligos containing ERα binding sites and fractionated on agarose gels. The protein-DNA complexes were isolated and mass spectrometry analysis was used to identify proteins associated with the DNA-bound receptor. Rather than simply identifying individual proteins that interact with ERα, we identified interconnected networks of proteins with a variety of enzymatic and catalytic activities that interact not only with ERα, but also with each other. Characterization of a number of these proteins has demonstrated that, in addition to their previously identified functions, they also influence ERα activity and expression of estrogen-responsive genes.
Conclusion
The agarose gel fractionation method we have developed would be useful in identifying proteins that interact with DNA-bound transcription factors and should be easily adapted for use with a variety of cultured cell lines, DNA sequences, and transcription factors.
Background
Estrogen receptor α (ERα) is a ligand-inducible transcription factor involved in regulating expression of estrogen-responsive genes [1]. Upon binding hormone, ERα undergoes a conformational change, binds to estrogen response elements (EREs) residing in target genes, and initiates changes in gene expression. We and others have demonstrated that, in addition to the hormone-induced change in ERα conformation, the receptor undergoes another conformational change, which is induced by binding of the receptor to individual ERE sequences [2-7]. Thus, both hormone and DNA induce conformational changes in ERα structure.
ERα does not function in isolation, but serves as a nucleating factor to recruit numerous coregulatory proteins required to effectively modulate transcription. In fact, much of what we know about regulation of estrogen-responsive genes has come through the identification of ERα-associated coregulatory proteins and elucidation of mechanisms by which they influence ERα-mediated transactivation. The majority of ERα-associated coregulatory proteins have been identified through their interaction with a discrete functional domain of the receptor, most commonly the ligand binding domain (Reviewed in [8,9]. The p160 proteins steroid receptor coactivator 1 (SRC-1), transcription intermediary factor 2 (TIF-2), and amplified in breast cancer 1 (AIB1) interact with ERα in a hormone-dependent manner and enhance ERα-mediated transcription [10-17]. Both SRC-1 and AIB1 as well as CREB binding protein (CBP) and p300/CBP-associated factor (pCAF) possess intrinsic histone acetyltransferase activity that has been implicated in enhancing gene expression by modifying chromatin structure [18-24]. A large complex of proteins identified on the basis of its interaction with the thyroid hormone and vitamin D receptors has been designated as the thyroid hormone receptor associated protein (TRAP) or vitamin D receptor interacting protein (DRIP) complex [25-27]. DRIP205/TRAP 220, which anchors the DRIP/TRAP complex to nuclear receptors, interacts with ERα in a ligand-dependent manner and enhances transcription [28,29]. In addition to the numerous coactivators that enhance ERα-mediated transcription, the corepressors nuclear receptor corepressor (NCoR) and silencing mediator for RXR and TR (SMRT) bind to the antiestrogen-occupied receptor and inhibit ERα-mediated transcription by recruiting protein complexes containing Sin3 and histone deacetylases [30-34]. Thus, ERα-associated coregulatory proteins have positive and negative effects on the ability of the receptor to activate transcription.
To better understand how ERα regulates transcription of estrogen-responsive genes, we developed a novel method to isolate proteins associated with the DNA-bound receptor, which utilizes full-length ERα and endogenously-expressed nuclear proteins and takes into account DNA- and ligand-induced changes in receptor conformation. This method should be useful in isolating regulatory proteins associated with other DNA-bound transcription factors and could yield important new information about mechanisms regulating gene expression.
Results
Characterization of protein-ERα-ERE complexes
To isolate novel proteins that associate with ERα and might influence estrogen-responsive gene expression, we developed a method that relied on the segregation of proteins on agarose gels and was based on the capacity of these proteins to associate with the ERE-bound receptor. Using this method, we were able to take into consideration DNA-induced modulation of ERα conformation, which we have demonstrated alters recruitment of coregulatory proteins to the DNA-bound receptor [2-5]. E2 was also included to ensure that ligand-induced changes in receptor conformation were considered.
As seen in Fig. 1, when radiolabeled, ERE-containing oligos were fractionated on an agarose gel, neither ERα (lane 2) nor HeLa nuclear extracts (lane 3) alone produced a discrete protein-DNA complex, but when both ERα and HeLa nuclear extracts were included, a distinct, higher order protein-DNA complex was present (lane 4). The ability of an ERα-specific antibody (lane 6), but not a nonspecific antibody (lane 5), to supershift the protein-DNA complex indicated that the receptor was present in the complex and that interaction of the ERα antibody with the complex was specific. Furthermore, the ability of unlabeled ERE-containing oligos (lane 8), but not oligos containing a nonspecific DNA sequence (lane 7) to compete with the radiolabeled ERE-containing oligos confirmed the specificity of the receptor-DNA interaction. As an additional control, we utilized radiolabeled oligos that contained a nonspecific DNA sequence. While a protein-DNA complex was formed with the ERE-containing oligos, no complex was observed with the oligos containing a nonspecific DNA sequence (data not shown). Thus, this agarose-based gel fractionation method allowed us to isolate proteins that were ERα and ERE specific.
Figure 1.
Small-scale agarose gel electrophoresis. 32P-labeled, ERE-containing oligos were incubated without (lane 1) or with ERα (lanes 2, 4–8) and/or HeLa nuclear extracts (NX, lanes 3–8). Nonspecific (NS) antibody (lane 5), ERα-specific antibody (lane 6), unlabeled oligos containing a nonspecific (NS) DNA sequence (lane 7), or unlabeled ERE-containing oligos (lane 8) were added to the binding reactions to confirm that the complexes formed were specific. 17β-estradiol (E2) was included in all binding reactions. Complexes were resolved on an agarose gel and visualized by autoradiography.
Large scale isolation of protein-ERα-ERE complexes
Once we defined the gel conditions required for formation of specific protein-ERα-ERE complexes, the next step was to increase the sample size so that sufficient amounts of protein would be available for isolation and identification. For these large-scale reactions, ERα-specific antibody was utilized to stabilize the protein-ERα-ERE complex and unlabeled oligos were used to avoid unintentional exposure of equipment to radioactive probe during the subsequent isolation and identification steps. However, small-scale samples containing radiolabeled oligos, purified ERα, HeLa nuclear extracts, and an ERα-specific antibody were run in adjacent lanes so that the position of the protein-ERα-ERE complexes in the gel could be determined (Fig. 2A, lanes 1 and 4).
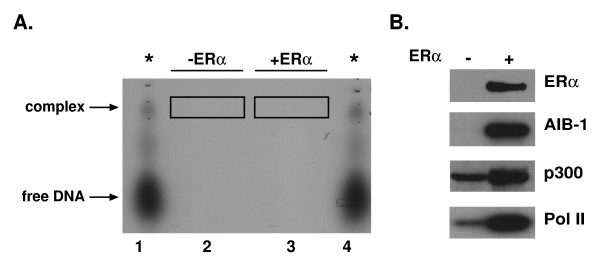
Figure 2.
Large-scale agarose gel electrophoresis and complex analysis. A. Large-scale reactions containing unlabeled ERE-containing oligos were incubated with HeLa nuclear extracts and an ERα-specific antibody in the absence (lane 2) or presence (lane 3) of purified ERα. Small-scale reactions containing 32P-labeled ERE-containing oligos, HeLa nuclear extracts, an ERα-specific antibody, and purified ERα were also prepared and run in parallel to indicate the location of the protein-DNA complexes (*, lanes 1 and 4). E2 was included in all binding reactions. Complexes were resolved on an agarose gel and were visualized in the wet gel by autoradiography. Gel regions comigrating with the 32P-labeled protein-ERα-ERE complexes were excised (boxed areas) and contained unlabeled DNA and associated proteins without (lane 2) or with (lane 3) ERα and ERα-specific antibody. B. Proteins were isolated from the excised agarose gel pieces and subjected to Western blot analysis with antibodies directed against ERα, AIB-1, p300, or Pol II.
Large-scale binding reactions containing unlabeled ERE-containing oligos, HeLa nuclear extracts, ERα, and an ERα-specific antibody were fractionated on one preparative-sized lane of an agarose gel and the gel region comigrating with the radiolabeled complexes was excised (lane 3, boxed area). Although distinct complexes were detected in our agarose gels when ERα, HeLa nuclear extracts, and ERα-specific antibody were included in the binding reactions (lanes 1 and 4), it seemed possible that some proteins might comigrate with the protein-ERα-ERE complex, but not actually be associated with it. Thus, a large-scale binding reaction containing the unlabeled ERE-containing oligos, HeLa nuclear extracts, and ERα-specific antibody, but no ERα, was processed in parallel and served as a negative control. The gel region comigrating with the radiolabeled protein-ERα-ERE complexes was also excised (lane 2, boxed area).
Initially, acetone or isopropanol precipitation was utilized to concentrate the proteins eluted from the agarose gel slices (data not shown). However, we found this method was unacceptable since it did not efficiently precipitate some proteins including ERα. By using a nebulizer column, which pulverizes the gel matrix and extracts the liquid and proteins, the protein recovery was far more efficient.
Identification of known coregulatory proteins in the protein-DNA complexes
To determine whether previously identified coregulatory proteins were associated with the ERα-ERE complex or merely comigrated with it, Western analysis was carried out. As expected, ERα was detected when the purified receptor was included in the binding reaction with HeLa nuclear extracts, but not when it was omitted (Fig. 2B). AIB-1, a known p160 coactivator of ERα-mediated transcription [12], was present in the complex when ERα had been added to the binding reaction, but not when it was omitted. Although p300 and RNA polymerase II (Pol II, Refs. [35-37] were detected in the absence of the receptor, significantly more p300 and Pol II were detected when ERα had been included in the reaction. Thus, the complexes we isolated were comprised of ERα and transcription factors that are known to be involved in regulating estrogen-responsive gene expression. Furthermore, the effective association of the coregulatory proteins with the complex was dependent upon the presence of ERα.
Identification of coregulatory proteins associated with the ERE-bound ERα
Although we had shown that previously identified coregulatory proteins were present in our protein-ERα-ERE complexes, the objective in these experiments was to identify novel proteins associated with the ERE-bound ERα. Mass spectrometry analysis was used to identify proteins present in gel regions that comigrate with the radiolabeled protein-ERα-ERE complexes (Fig. 2A, boxed areas). Numerous proteins involved in DNA replication and repair, chromatin remodeling, protein folding/stabilization, protein degradation, translation initiation and elongation, apoptosis, oxidative stress response, and signal transduction were identified (Table 1 and Additional file 1). While some proteins were identified in the absence and in the presence of ERα, significantly more peptides were recovered in the presence of ERα, as was observed in Fig. 2B with p300 and Pol II, reflecting a higher abundance of these proteins. The fact that we identified the same proteins in two or more experiments (see Additional file 1) suggests that the methods we used were reproducible. However, the most important validation of this method has come through functional characterization of these proteins. At this point, we have characterized the activity of 15 proteins associated with the DNA-bound ERα (Table 1) and found that each of these proteins influences estrogen-responsive gene expression [38-47] and unpublished data).
Table 1.
Proteins associated with the DNA-bound ERα
| Protein | # of discrete peptides | % a.a. sequence identified | Effect on ERα-mediated transcription | References Cited |
| 3-methyladenine DNA glycosylase (MPG) | 3 | 14 | Decrease | [40] |
| apurinic endonuclease-1 (APE1) | 3 | 18 | Gene specific | Curtis and Nardulli, Submitted |
| flap endonuclease-1 (FEN1) | 3 | 13 | Gene specific | [42] |
| high mobility group protein-2 (HMG-2) | 2 | 13 | Increase | [49,50,67] |
| nonmetastatic protein 23 homolog 1 (NM23-H1) | 5 | 35 | Decrease | [45] |
| proliferating cell nuclear antigen (PCNA) | 9 | 57 | Increases basal | [43] |
| protein disulfide isomerase (PDI) | 7 | 19 | Gene specific | [41] |
| pp32 | 5 | 19 | Decrease | [39] |
| retinoblastoma associated protein 46 (RbAp46) | 6 | 18 | Gene specific | [47] |
| retinoblastoma associated protein 48 (RbAp48) | 6 | 18 | Decrease | [47] |
| rho-GDP dissociation inhibitor α (RhoGDIα) | 7 | 57 | Gene specific | [44] |
| superoxide dismutase 1 (SOD1) | 9 | 86 | Increase | [46] |
| template activating factor 1β (TAF-Iβ) | 16 | 38 | Decrease | [38] |
| thioredoxin (Trx) | 4 | 46 | Gene specific | Rao and Nardulli, In Preparation |
| thioredoxin reductase (TrxR) | 14 | 54 | Gene specific | Rao and Nardulli, In Preparation |
Selected ERα-associated proteins were characterized. The effect of each protein on ERα-mediated transcription was analyzed by transient transfection and/or RNA interference assays.
Isolation of protein-ERα-ERE complexes using other cell lines
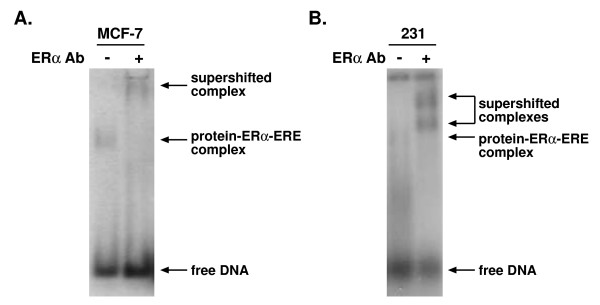
The agarose gel fractionation method we developed is not restricted in the type of cells utilized. We have used this method to form large protein-DNA complexes with nuclear extracts from MCF-7 breast cancer cells, which express endogenous ERα (Fig. 3A). Inclusion of an ERα-specific antibody supershifted the complex formed with these extracts. Interestingly, although we were unable to form a stable protein-ERα-ERE complex with purified ERα and nuclear extracts from MDA-MB-231 human breast cancer cells, which do not express ERα (Fig. 3B), inclusion of an ERα-specific antibody helped to stabilize protein-DNA complex formation. In fact, we routinely include ERα-specific antibodies to help stabilize our protein-ERα-ERE complexes.
Figure 3.
Small-scale agarose gel electrophoresis using MCF-7 and MDA-MB-231 nuclear extracts. 32P-labeled ERE-containing oligos were incubated with nuclear extracts from MCF-7 breast cancer cells, which endogenously express ERα (A), or nuclear extracts from MDA-MB-231 breast cancer cells and purified ERα (B). An ERα-specific antibody was included in the binding reaction as indicated. Complexes were resolved on agarose gels and visualized by autoradiography.
Discussion
We have developed a method of isolating stable protein-DNA complexes, the formation of which requires ERα, the ERE, and nuclear proteins. A number of factors were considered in establishing this methodology. First, full-length human ERα and endogenously-expressed nuclear proteins were utilized. Second, allosteric modulation of receptor conformation by DNA and hormone was taken into account by isolating proteins associated with the DNA-bound, E2-occupied ERα. It is, after all, the estrogen-occupied, DNA-bound receptor that recruits coregulatory proteins and initiates changes in transcription. By considering both E2- and DNA-induced changes in receptor conformation, we were able to identify proteins that are involved in transcriptional control and gain new insight to help define how changes in gene expression occur. Third, because traditional polyacrylamide gel shift assays do not allow large protein-DNA complexes to enter the gel [38,39,41-43,48-50], agarose gels were employed to isolate large molecular weight complexes containing ERα, ERE-containing oligos, and nuclear coregulatory proteins. In addition, low ionic strength buffer and ERα-specific antibody were used to stabilize protein-ERα-ERE complexes during the extended period of electrophoresis required. Finally, a nebulizer spin column utilized for isolating proteins from the agarose gel significantly raised the signal to noise ratio and was critical in recovering ERα and its associated proteins.
The electrophoretic agarose gel fractionation method has distinct advantages over other methods we previously used to isolate ERα-associated proteins. ERα pull-down assays were useful in identifying HeLa nuclear proteins associated with the flag-tagged ERα, but the number of proteins identified using this method was limited [38,39]. DNA affinity assays, which we used to identify a DNA glycosylase that associates with the ERE-bound ERα [40], were limited by the fact that numerous nuclear proteins bound to the ERE-containing oligos and/or agarose beads in the absence of ERα and produced a background that made it difficult to distinguish specific from nonspecific proteins. The agarose gel fractionation method allowed us to isolate a suite of ERα-associated proteins and significantly decreased the proportion of nonspecifically-bound proteins.
Isolation of interconnected protein networks
At first glance, it might appear that much of what we have done has been to identify a number of individual proteins that interact with ERα and influence estrogen-responsive gene expression. However, one of the most fascinating findings from our agarose gel-based approach was that rather than simply identifying individual proteins that interact with ERα, we identified interconnected networks of proteins with a variety of enzymatic and catalytic activities that interact not only with ERα, but also with each other (Table 2).
Table 2.
ERα-associated protein interactions
| Protein | Interacting Protein | Reference(s) |
| APE1 | ERα | Curtis and Nardulli, Submitted |
| FEN1 | [68] | |
| HMG-2 | [61,69] | |
| NM23-H1 | [61] | |
| PCNA | [68,70] | |
| pp32 | [61] | |
| TAF-Iβ | [61] | |
| FEN1 | APE1 | [68] |
| ERα | [42] | |
| PCNA | [67,71-74] | |
| HMG-2 | APE1 | [61,69] |
| ERα | [49,75] | |
| NM23-H1 | [61] | |
| pp32 | [61] | |
| TAF-Iβ | [61,69] | |
| MPG | ERα | [40] |
| PCNA | [70] | |
| NM23-H1 | APE1 | [61] |
| ERα | [45] | |
| HMG-2 | [61] | |
| pp32 | [61] | |
| TAF-Iβ | [60] | |
| pp32 | APE1 | [61] |
| ERα | [39] | |
| HMG-2 | [61] | |
| NM23-H1 | [61] | |
| PCNA | [76] | |
| TAF-Iβ | [51-53,59,60] | |
| PCNA | APE1 | [68,70] |
| ERα | [43] | |
| FEN1 | [67,71-74] | |
| MPG | [70] | |
| pp32 | [76] | |
| TAF-Iβ | [76] | |
| TAF-Iβ | APE1 | [61] |
| ERα | [38] | |
| HMG-2 | [61,69] | |
| NM23-H1 | [60] | |
| pp32 | [51-53,59,61] | |
| PCNA | [76] | |
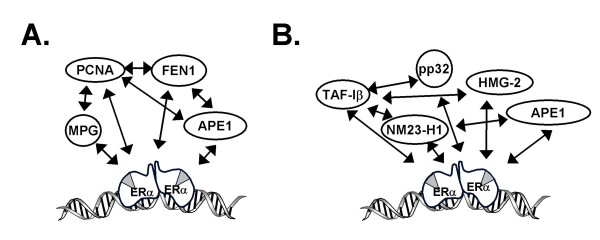
Two examples of the ERα-associated protein networks we isolated are illustrated in Fig. 4. We identified four proteins involved in DNA repair, 3-methyladenine DNA glycosylase (MPG), apurinic endonuclease 1 (APE1), proliferating cell nuclear antigen (PCNA), and flap endonuclease, (FEN1), each of which was associated with the DNA-bound ERα and influences estrogen-responsive gene expression (Refs. [40,42,43] and C. Curtis and A. Nardulli, unpublished data) These proteins form an interactive complex of proteins (Fig. 4A and Table 2) that together are involved in base excision repair (BER). Another complex of proteins we isolated were previously identified as the SET or INHAT complex [51-53], which is comprised of template activating factor Iβ (TAF-Iβ), pp32, high mobility group protein 2 (HMG-2), APE1, and nonmetastatic protein homolog 1 (NM23-H1). These proteins form an interactive group involved in determining cell fate by initiating DNA repair or caspase-independent apoptosis (Fig. 4B and Table 2, Refs. [54,55]. Interestingly, we have characterized the effects of each of these proteins on ERα activity and found that each of these proteins influences expression of estrogen-responsive genes [38-40,42,43,45] and unpublished data).
Figure 4.
ERα-associated proteins form interconnected networks. A. Interactions between ERα and the DNA repair proteins MPG, PCNA, FEN1, and APE1. B. Interactions between ERα and the SET complex proteins TAF-Iβ, pp32, NM23-H1, HMG-2, and APE1.
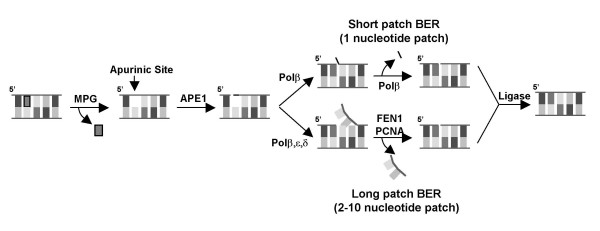
The ERα-associated proteins we isolated are each endowed with specific activities that collectively alter basic cellular processes. As shown in Fig. 5, the DNA glycosylase MPG catalyzes the removal of a damaged or modified base and the formation of an apurinic site [56,57]. APE1 recognizes this apurinic site and initiates strand incision. DNA repair is then completed by polymerase-induced insertion of a single nucleotide and ligation. This process of replacing a single base is referred to as short patch BER. Alternatively, the DNA can be repaired through long patch BER in which FEN1 removes a short flap of nucleotides. PCNA serves as a platform for FEN1, stabilizes the interaction of FEN1 with the DNA flap, and enhances FEN1 cleavage efficiency [58]. The interaction of these DNA repair proteins with ERa is both physical and functional, but more importantly, their identification led to the discovery of an integrated protein network associated with the DNA-bound receptor that is involved in DNA repair (Figs. 4A and 5 and Table 2).
Figure 5.
ERα-associated proteins are involved in base excision repair. MPG, PCNA, FEN1, and APE1 form an interconnected network of proteins involved in short- and long-patch BER.
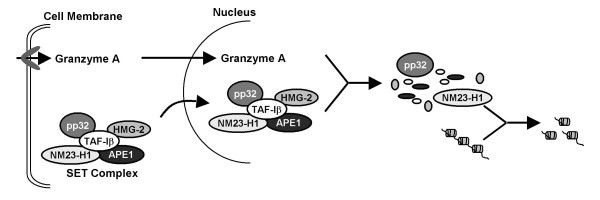
The SET complex is likewise comprised of an interactive group of proteins involved in regulating cellular processes, which has been described in detail by Lieberman and coworkers [52,54,59-61]. In normal cells, NM23-H1 assists in maintaining DNA integrity by nicking DNA and initiating DNA repair. In these cells, NM23-H1's DNase I activity is limited by its inhibitor, TAF-Iβ. However, a different scenario ensues when cytotoxic T lymphocytes detect a virally infected or tumor cell. In this instance, the cytotoxic T lymphocytes release Granzyme A, which enters the target cell and cleaves the inhibitor of NM23-H1, TAF-Iβ, as well as HMG-2 and APE1 (Fig. 6). With its inhibitor destroyed, NM23H1-induced DNA nicking is increased and caspase-independent apoptosis is initiated. The destruction of APE1 in these cells further hobbles the DNA repair machinery and helps to ensure that the cells undergo apoptosis. We isolated all of the SET complex proteins (pp32, TAF-Iβ, NM23-H1, HMG-2, and APE1) in our protein-DNA complexes. We and others showed previously that HMG proteins interact with ERa and other nuclear receptors, enhance receptor-DNA interaction, and increase receptor-mediated transactivation [50,62-64].
Figure 6.
ERα-associated proteins are in the SET complex. TAF-Iβ, pp32, HMG-2, NM23-H1, and APE1 form an interconnected network of proteins involved in DNA repair and/or apoptosis. (Adapted from Refs. [54,55].
We are intrigued by the fact that the proteins we isolated interact with ERα and with each other (Fig. 4 and Table 2) providing evidence that these proteins belong to interactive networks of proteins with discrete cellular functions. The interaction of the protein networks may be fostered by the association of a protein with more than one network. For example, APE1, which interacts with ERα, components of long- and short-patch BER complexes, and SET complex proteins, may help to coordinate the actions of these protein networks and link DNA repair and transcription.
The interaction of ERα with its associated proteins may not only be physical, but may have functional consequences for both proteins. We know that MPG influences ERα-mediated transactivation and that, in turn, ERα enhances the association of MPG with modified DNA and promotes base excision [65]. Thus, by recruiting protein complexes involved in DNA repair, ERα may help to preferentially maintain the integrity of transcriptionally-active, estrogen-responsive genes.
Taken together, our findings suggest that the ERE-bound ERα serves as a nucleating factor to recruit a cohort of proteins with a variety of cellular functions that influence estrogen-responsive gene expression and that ERα may in turn enhance DNA repair and ultimately help to determine cell fate.
Conclusion
The electrophoretic agarose fractionation protocol that we have developed provides a method to isolate interrelated networks of ERα-associated proteins involved in regulating estrogen-responsive gene expression. These studies have provided a fascinating glimpse of the complexity involved in regulating estrogen-responsive genes. This agarose gel fractionation method should be readily adaptable to a variety of cultured cell lines, DNA sequences, and transcription factors and help to define how proteins associated with DNA-bound transcription factors influence gene expression and other critical cellular processes.
Methods
Small scale characterization of protein-DNA complex formation
HeLa nuclear extracts and baculovirus-expressed, purified ERα were prepared as previously described [3,66]. Oligos containing the Xenopus laevis vitellogenin A2 estrogen response element flanked by the native DNA sequence (ERE, (5'-GAT TAA CTG TCC AAA GTC AGG TCA CAG TGA CCT GAT CAA AGT TAA TGT AA-3' and 5'-TTA CAT TAA CTT TGA TCA GGT CAC TGT GAC CTG ACT TTG GAC AGT TAA TC-3') were annealed and end labeled with 32γP-ATP. Radiolabeled oligos (10 pmol) were incubated with 400 fmol purified ERα in binding buffer (15 mM Tris, 0.2 mM EDTA, 80 mM KCl, 50 μM ZnCl, 5 mM MgOAc, 10% glycerol, 4 mM DTT) with 1 μg of poly dI/dC, 1 μg salmon sperm DNA, 1 μg BSA, and 10 μM 17 β-estradiol (E2) for ten minutes at room temperature. HeLa nuclear extracts (10 μg) were then added and incubated at room temperature for an additional 20 minutes. Reactions lacking ERα were run in parallel with additional BSA added to maintain constant protein concentrations. 200 ng of antibody directed against YY1 (control antibody) or ERα (sc-7341 or sc-8005, Santa Cruz Biotechnology, Santa Cruz, CA) or 10 pmol of unlabeled double-stranded oligos containing an ERE or nonspecific DNA sequence (NS, 5'-CTA GAT TAC TTC TCA TGT TAG ACA TAC TCA-3', and 5'GAT CTG AGT ATG TCT AAC ATG AGA AGT AAT CTA G-3') were included in the binding reactions as indicated. The complexes and the free DNA were separated on a horizontal 1.25% low melt agarose gel (BioRad, Hercules, CA) in a modified TBE buffer (0.45 mM Tris pH 7.9, 4.5 mM boric acid, 2 mM EDTA) containing 5 mM MgOAc at 100 volts for two hours at 4°C. The gel was dried on DE81 ion exchange cellulose acetate (Whatman, Florham Park, NJ) at 65 C for 30 minutes under vacuum and visualized by autoradiography.
Large scale complex formation
For large scale isolation of protein-ERα-ERE complexes, DNA oligos containing the Xenopus laevis A2 ERE and surrounding DNA sequence were annealed and the binding reactions were incubated as described above except that they were increased to include 50 pmol DNA, 260 μg of HeLa nuclear extract with or without 18 pmol of ERα in a total volume of 200 μl. 3.2 μg of an ERα-specific antibody (sc-8002, Santa Cruz Biotechnology, Santa Cruz, CA) was added to help stabilize the ERα-containing complexes. All samples were loaded onto 10 cm × 15 cm horizontal 1.25% agarose gels prepared with molecular biology grade agarose (BioRad, Hercules, CA) and modified TBE buffer. Unlabeled ERE-containing oligos were utilized in all samples submitted for mass spectrometry analysis. Marker lanes, which contained radiolabeled oligos, 3 pmol ERα, 50 μg HeLa nuclear extract, and 0.6 μg of ERα-specific antibody in 40 μl total volumes were run in parallel at 100 V for 2 h to indicate the position of the complexes. After fractionation, the wet gel was subjected to autoradiography overnight at room temperature and the regions containing the unlabeled protein-ERα-ERE complexes were excised. Proteins were isolated with the Montage gel extraction kit (Millipore, Billerica, MA) according to manufacturer's directions. The extracted proteins were concentrated using Microcon YM-10 size exclusion columns (Millipore, Billerica, MA) with a molecular weight cutoff of 10 kDa and then subjected to mass spectrometry analysis as previously described [38]. Peptide fragments found in multiple proteins were excluded from the data analysis.
Western analysis of ERα-associated proteins
Proteins isolated from large-scale agarose gels were fractionated on denaturing SDS-PAGE, transferred to nitrocellulose, and subjected to Western analysis. Blots were probed with antibodies specific to ERα, p300, RNA polymerase II (sc-8005, sc-585, or sc-899, respectively, Santa Cruz Biotechnologies, Santa Cruz, CA) or AIB-1 (A79920, BD Transduction Labs) and a horseradish peroxidase-conjugated secondary antibody. Proteins were visualized using a chemiluminescent detection system as previously described [4].
Abbreviations
ERα: estrogen receptor α; ERE: estrogen response element; NCoR: nuclear receptor corepressor; SMRT: silencing mediator for RXR and TR; TIF-2: transcription intermediary factor 2; AIB1: amplified in breast cancer 1; Pol II: RNA polymerase II; MPG: 3-methyladenine DNA glycosylase; APE1: apurinic endonuclease 1; PCNA: proliferating cell nuclear antigen; FEN1: flap endonuclease 1; BER: base excision repair; TAF-Iβ: template activating factor Iβ; HMG-2: high mobility group protein 2; NM23-H1: nonmetastatic protein 23 homolog 1.
Authors' contributions
VSL developed the initial approach of isolating protein-receptor-DNA complexes using agarose gel fractionation. The method was further refined by JRS-N and then by YSZ, who isolated the majority of the proteins identified. JRY and coworkers identified each of the receptor-associated proteins using mass spectrometry analysis and AMN guided the overall project. All authors have read and approved the final manuscript.
Supplementary Material
ERα-interacting proteins isolated from agarose gel complexes. HeLa nuclear extracts were incubated with ERα-specific antibody and radiolabeled ERE-containing oligos in the absence or presence of purified ERα. Isolated proteins were subjected to trypsin digestion and mass spectrometry analysis. Peptide sequences were blasted against the SEQUEST database to identify the isolated proteins. Proteins identified in more than one independent experiment are grouped by cellular function and listed by gene name, accession number, and protein description. The number of independent experiments in which each protein was identified, the number of peptides isolated, and the percent of amino acid sequence identified are indicated. Data are compiled from 4 independent experiments.
Acknowledgments
Acknowledgements
We are grateful to members of the Yates laboratory, who identified the proteins isolated in our large-scale agarose gels. This work was supported by NIH grant R01 DK 53884 (to AMN) and NIH P41 RR11823-10 (to JRY). JRS-N was supported by NIH Reproductive Biology T32 HD07028.
Contributor Information
Jennifer R Schultz-Norton, Email: jschultznorton@mail.millikin.edu.
Yvonne S Ziegler, Email: yziegler@uiuc.edu.
Varsha S Likhite, Email: varsha.likhite@gmail.com.
John R Yates, Email: jyates@scripps.edu.
Ann M Nardulli, Email: anardull@life.uiuc.edu.
References
- Beato M, Herrlich P, Schutz G. Steroid hormone receptors: many actors in search of a plot. Cell. 1995;83:851–857. doi: 10.1016/0092-8674(95)90201-5. [DOI] [PubMed] [Google Scholar]
- Wood JR, Greene GL, Nardulli AM. Estrogen response elements function as allosteric modulators of estrogen receptor conformation. Mol Cell Biol. 1998;18:1927–1934. doi: 10.1128/mcb.18.4.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood JR, Likhite VS, Loven MA, Nardulli AM. Allosteric modulation of estrogen receptor conformation by different estrogen response elements. Mol Endocrinol. 2001;15:1114–1126. doi: 10.1210/me.15.7.1114. [DOI] [PubMed] [Google Scholar]
- Loven MA, Likhite VS, Choi I, Nardulli AM. Estrogen response elements alter coactivator recruitment through allosteric modulation of estrogen receptor beta conformation. J Biol Chem. 2001;276:45282–45288. doi: 10.1074/jbc.M106211200. [DOI] [PubMed] [Google Scholar]
- Loven MA, Wood JA, Nardulli AM. Interaction of estrogen receptors alpha and beta with estrogen response elements. Mol Cell Endocrinol. 2001;181:151–163. doi: 10.1016/S0303-7207(01)00491-9. [DOI] [PubMed] [Google Scholar]
- Klinge CM. Estrogen receptor interaction with estrogen response elements. Nucleic Acids Res. 2001;29:2905–2919. doi: 10.1093/nar/29.14.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall JM, McDonnell DP, Korach KS. Allosteric regulation of estrogen receptor structure, function, and coactivator recruitment by different estrogen response elements. Mol Endocrinol. 2002;16:469–486. doi: 10.1210/me.16.3.469. [DOI] [PubMed] [Google Scholar]
- Robyr D, Wolffe AP, Wahli W. Nuclear hormone receptor coregulators in action: diversity for shared tasks. Mol Endocrinol. 2000;14:329–347. doi: 10.1210/me.14.3.329. [DOI] [PubMed] [Google Scholar]
- Torchia J, Glass C, Rosenfeld MG. Co-activators and co-repressors in the integration of transcriptional responses. Curr Opin Cell Biol. 1998;10:373–383. doi: 10.1016/S0955-0674(98)80014-8. [DOI] [PubMed] [Google Scholar]
- Halachmi S, Marden E, Martin G, MacKay H, Abbondanza C, Brown M. Estrogen receptor-associated proteins: possible mediators of hormone-induced transcription. Science. 1994;264:1455–1458. doi: 10.1126/science.8197458. [DOI] [PubMed] [Google Scholar]
- Oñate SA, Tsai SY, Tsai M-J, O'Malley BW. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science. 1995;270:1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- Anzick SL, Kononen J, Walker RL, Azorsa DO, Tanner MM, Guan X-Y, Sauter G, Kallioniemi O-P, Trent JM, Meltzer PS. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science. 1997;277:965–968. doi: 10.1126/science.277.5328.965. [DOI] [PubMed] [Google Scholar]
- Chen H, Lin RJ, Schiltz RL, Chakravarti D, Nash A, Nagy L, Privalsky ML, Nakatani Y, Evans RM. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell. 1997;90:569–580. doi: 10.1016/S0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- Li H, Gomes PJ, Chen JD. RAC3, a steroid/nuclear receptor-associated coactivator that is related to SRC-1 and TIF2. Proc Natl Acad Sci USA. 1997;94:8479–8484. doi: 10.1073/pnas.94.16.8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voegel JJ, Heine MJS, Zechel C, Chambon P, Gronemeyer H. TIF 2, a 160 kDa transcriptional mediator for the ligand-dependent activation function AF-2 of nuclear receptors. EMBO J. 1996;15:101–108. [PMC free article] [PubMed] [Google Scholar]
- Hong H, Kulwant K, Trivedi A, Johnson DL, Stallcup MR. GRIP1, a novel mouse protein that serves as a transcriptional coactivator in yeast for the hormone binding domains of steroid receptors. Proc Natl Acad Sci USA. 1996;93:4948–4952. doi: 10.1073/pnas.93.10.4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong H, Kohli K, Garabedian MJ, Stallcup MR. GRIP1, a transciptional coactivator for the AF-2 transactivation domain of steroid, thyroid, retinoid, and vitamin D receptors. Mol Cell Biol. 1997;17:2735–2744. doi: 10.1128/mcb.17.5.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XJ, Ogryzko VV, Nishikawa J, Howard BH, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- Ogryzko VV, Schiltz RL, Russanova V, Howard BH, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/S0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- Spencer TE, Jenster G, Burcin MM, Allis CD, Zhou J, Mizzen CA, McKenna NJ, Onate SA, Tsai SY, Tsai M-J, O'Malley BW. Steroid receptor coactivator-1 is a histone acetyltransferase. Nature. 1997;389:194–198. doi: 10.1038/38304. [DOI] [PubMed] [Google Scholar]
- Eckner R, Ewen ME, Newsome D, Gerdes M, Decaprio JA, Lawrence JB, Livingston DM. Molecular cloning and functional analysis of the adenovirus E1A-associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes Dev. 1994;8:869–884. doi: 10.1101/gad.8.8.869. [DOI] [PubMed] [Google Scholar]
- Smith CL, Oñate SA, Tsai M-J, O'Malley BW. CREB binding protein acts synergistically with steroid receptor coactivator-1 to enhance steroid receptor-dependent transcription. Proc Natl Acad Sci USA. 1996;93:8884–8888. doi: 10.1073/pnas.93.17.8884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarti D, LaMorte VJ, Nelson MC, Nakajima T, Schulman IG, Juguilon H, Montminy M, Evans RM. Role of CBP/P300 in nuclear receptor signaling. Nature. 1996;383:99–103. doi: 10.1038/383099a0. [DOI] [PubMed] [Google Scholar]
- Hanstein B, Liu H, Yancisin MC, Brown M. Functional analysis of a novel estrogen receptor beta isoform. Mol Endocrinol. 1999;13:129–137. doi: 10.1210/me.13.1.129. [DOI] [PubMed] [Google Scholar]
- Fondell JD, Ge H, Roeder RG. Ligand induction of a transcriptionally active thyroid hormone receptor coactivator complex. Proc Natl Acad Sci USA. 1996;93:8329–8333. doi: 10.1073/pnas.93.16.8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachez C, Suldan Z, Ward J, Chang CP, Burakov D, Erdjument-Bromage H, Tempst P, Freedman LP. A novel protein complex that interacts with the vitamin D3 receptor in a ligand-dependent manner and enhances VDR transactivation in a cell-free system. Genes Dev. 1998;12:1787–1800. doi: 10.1101/gad.12.12.1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachez C, Lemon BD, Suldan Z, Bromleigh V, Gamble M, Naar AM, Erdjument-Bromage H, Tempst P, Freedman LP. Ligand-dependent transcription activation by nuclear receptors requires the DRIP complex. Nature. 1999;398:824–828. doi: 10.1038/19783. [DOI] [PubMed] [Google Scholar]
- Yuan CX, Ito M, Fondell JD, Fu ZY, Roeder RG. The TRAP220 component of a thyroid hormone receptor-associated protein (TRAP) coactivator complex interacts directly with nuclear receptors in a ligand-dependent fashion. Proc Natl Acad Sci USA. 1998;95:7939–7944. doi: 10.1073/pnas.95.14.7939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burakov D, Wong CW, Rachez C, Cheskis BJ, Freedman LP. Functional interactions between the estrogen receptor and DRIP205, a subunit of the heteromeric DRIP coactivator complex. J Biol Chem. 2000;275:20928–20934. doi: 10.1074/jbc.M002013200. [DOI] [PubMed] [Google Scholar]
- Zamir I, Harding HP, Atkins GB, Horlein A, Glass CK, Rosenfeld MG, Lazar MA. A nuclear hormone receptor corepressor mediates transcriptional silencing by receptors with distinct repression domains. Mol Cell Biol. 1996;16:5458–5465. doi: 10.1128/mcb.16.10.5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JD, Evans RM. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature. 1995;377:454–457. doi: 10.1038/377454a0. [DOI] [PubMed] [Google Scholar]
- Smith C, Nawaz Z, O'Malley B. Coactivator and corepressor regulation of agonist/antagonist activity of the mixed antiestrogen, 4-hydroxytamoxifen. Mol Endocrinol. 1997;11:657–666. doi: 10.1210/me.11.6.657. [DOI] [PubMed] [Google Scholar]
- Heinzel T, Lavinsky RM, Mullen TM, Soderstrom M, Laherty CD, Torchia J, Yang WM, Brard G, Ngo SD, Eisenman RN, Rose DW, Glass CK, Rosenfeld MG. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature. 1997;386:43–48. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- Nagy L, Kao H-Y, Chakravarti D, Lin RJ, Hassig CA, Ayer DE, Schreiber SL, Evans RM. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell. 1997;89:373–380. doi: 10.1016/S0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- Hanstein B, Eckner R, DiRenzo J, Halachmi S, Liu H, Searcy B, Kurokawa R, Brown M. p300 is a component of an estrogen receptor coactivator complex. Proc Natl Acad Sci USA. 1996;93:11540–11545. doi: 10.1073/pnas.93.21.11540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranish JA, Yudkovsky N, Hahn S. Intermediates in formation and activity of the RNA polymerase II preinitiation complex: holoenzyme recruitment and a postrecruitment role for the TATA box and TFIIB. Genes Dev. 1999;13:49–63. doi: 10.1101/gad.13.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutoglou E, Talianidis I. Coordination of PIC assembly and chromatin remodeling during differentiation-induced gene activation. Science. 2002;295:1901–1904. doi: 10.1126/science.1068356. [DOI] [PubMed] [Google Scholar]
- Loven MA, Muster N, Yates JR, Nardulli AM. A novel estrogen receptor alpha associated protein, template activating factor I beta, inhibits acetylation and transactivation. Mol Endocrinol. 2003;17:67–78. doi: 10.1210/me.2002-0280. [DOI] [PubMed] [Google Scholar]
- Loven MA, Davis RE, Curtis CD, Muster N, Yates JR, Nardulli AM. A novel estrogen receptor alpha-associated protein alters receptor-deoxyribonucleic acid interactions and represses receptor-mediated transcription. Mol Endocrinol. 2004;18:2649–2659. doi: 10.1210/me.2003-0195. [DOI] [PubMed] [Google Scholar]
- Likhite VS, Cass EI, Anderson SD, Yates JR, Nardulli AM. Interaction of estrogen receptor alpha with 3-methyladenine DNA glycosylase modulates transcription and DNA repair. J Biol Chem. 2004;279:16875–16882. doi: 10.1074/jbc.M313155200. [DOI] [PubMed] [Google Scholar]
- Schultz-Norton JR, McDonald WH, Yates JR, Nardulli AM. Protein disulfide isomerase serves as a molecular chaperone to maintain estrogen receptor {alpha} structure and function. Mol Endocrinol. 2006;20:1982–1995. doi: 10.1210/me.2006-0006. [DOI] [PubMed] [Google Scholar]
- Schultz-Norton JR, Walt KA, Ziegler YS, McLeod IX, Yates JR, Raetzman LT, Nardulli AM. The deoxyribonucleic acid repair protein flap endonuclease-1 modulates estrogen-responsive gene expression. Mol Endocrinol. 2007;21:1569–1580. doi: 10.1210/me.2006-0519. [DOI] [PubMed] [Google Scholar]
- Schultz-Norton JR, Gabisi VA, Ziegler YS, McLeod IX, Yates JR, Nardulli AM. Interaction of estrogen receptor alpha with proliferating cell nuclear antigen. Nucleic Acids Res. 2007;35:5028–5038. doi: 10.1093/nar/gkm533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Marzouk S, Schultz-Norton JR, Likhite VS, McLeod IX, Yates JR, Nardulli AM. Rho GDP dissociation inhibitor alpha interacts with estrogen receptor alpha and influences estrogen responsiveness. J Mol Endocrinol. 2007;39:249–259. doi: 10.1677/JME-07-0055. [DOI] [PubMed] [Google Scholar]
- Curtis CD, Likhite VS, McLeod IX, Yates JR, Nardulli AM. Interaction of the tumor metastasis suppressor nonmetastatic protein 23 homologue H1 and estrogen receptor alpha alters estrogen-responsive gene expression. Cancer Res. 2007;67:10600–10607. doi: 10.1158/0008-5472.CAN-07-0055. [DOI] [PubMed] [Google Scholar]
- Rao AK, Ziegler YS, McLeod IX, Yates JR, Nardulli AM. Effects of cu/zn superoxide dismutase on estrogen responsiveness and oxidative stress in human breast cancer cells. Mol Endocrinol. 2008;22:1113–1124. doi: 10.1210/me.2007-0381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creekmore AL, Walt KA, Schultz-Norton JR, Ziegler YS, McLeod IX, Yates JR, Nardulli AM. The role of retinoblastoma associated proteins 46 and 48 in estrogen receptor alpha mediated gene expression. Mol Cell Endocrinol. 2008;291:79–86. doi: 10.1016/j.mce.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landel CC, Kushner PJ, Greene GL. The interaction of human estrogen receptor with DNA is modulated by receptor-associated proteins. Mol Endocrinol. 1994;8:1407–1419. doi: 10.1210/me.8.10.1407. [DOI] [PubMed] [Google Scholar]
- Boonyaratanakornkit V, Melvin V, Prendergast P, Altmann M, Ronfani L, Bianchi ME, Taraseviciene L, Nordeen SK, Allegretto EA, Edwards DP. High-mobility group chromatin proteins 1 and 2 functionally interact with steroid hormone receptors to enhance their DNA binding in vitro and transcriptional activity in mammalian cells. Mol Cell Biol. 1998;18:4471–4487. doi: 10.1128/mcb.18.8.4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romine L, Wood J, Lamia L, Prendergast P, Edwards D, Nardulli A. The high mobility group protein 1 enhances binding of the estrogen receptor DNA binding domain to the estrogen response element. Mol Endocrinol. 1998;12:664–674. doi: 10.1210/me.12.5.664. [DOI] [PubMed] [Google Scholar]
- Vaesen M, Barnikol-Watanabe S, Gotz H, Awni LA, Cole T, Zimmerman B, Kratzin HD, Hilschmann N. Purification and characterization of two putative HLA class II associated proteins: PHAPI and PHAPII. Biol Chem Hoppe-Seyler. 1994;375:113–126. doi: 10.1515/bchm3.1994.375.2.113. [DOI] [PubMed] [Google Scholar]
- Beresford PJ, Kam CM, Powers JC, Lieberman J. Recombinant human granzyme A binds to two putative HLA-associated proteins and cleaves one of them. Proc Natl Acad Sci USA. 1997;94:9285–9290. doi: 10.1073/pnas.94.17.9285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo SB, McNamara P, Heo S, Turner A, Lane WS, Chakravarti D. Regulation of histone acetylation and transcription by INHAT, a human cellular complex containing the set oncoprotein. Cell. 2001;104:119–130. doi: 10.1016/S0092-8674(01)00196-9. [DOI] [PubMed] [Google Scholar]
- Lieberman J, Fan Z. Nuclear war: the granzyme A-bomb. Curr Opin Immunol. 2003;15:553–559. doi: 10.1016/S0952-7915(03)00108-0. [DOI] [PubMed] [Google Scholar]
- Chowdhury D, Beresford PJ, Zhu P, Zhang D, Sung JS, Demple B, Perrino FW, Lieberman J. The exonuclease TREX1 is in the SET complex and acts in concert with NM23-H1 to degrade DNA during granzyme A-mediated cell death. Mol Cell. 2006;23:133–142. doi: 10.1016/j.molcel.2006.06.005. [DOI] [PubMed] [Google Scholar]
- Lindahl T, Sedgwick B, Sekiguchi M, Nakabeppu Y. Regulation and expression of the adaptive response to alkylating agents. Annu Rev Biochem. 1988;57:133–157. doi: 10.1146/annurev.bi.57.070188.001025. [DOI] [PubMed] [Google Scholar]
- Wood RD. DNA repair in eukaryotes. Annu Rev Biochem. 1996;65:135–167. doi: 10.1146/annurev.bi.65.070196.001031. [DOI] [PubMed] [Google Scholar]
- Tom S, Henricksen LA, Bambara RA. Mechanism whereby proliferating cell nuclear antigen stimulates flap endonuclease 1. J Biol Chem. 2000;275:10498–10505. doi: 10.1074/jbc.275.14.10498. [DOI] [PubMed] [Google Scholar]
- Beresford PJ, Zhang D, Oh DY, Fan Z, Greer EL, Russo ML, Jaju M, Lieberman J. Granzyme A activates an endoplasmic reticulum-associated caspase-independent nuclease to induce single-stranded DNA nicks. J Biol Chem. 2001;276:43285–43293. doi: 10.1074/jbc.M108137200. [DOI] [PubMed] [Google Scholar]
- Fan Z, Beresford PJ, Oh DY, Zhang D, Lieberman J. Tumor suppressor NM23-H1 is a granzyme A-activated DNase during CTL-mediated apoptosis, and the nucleosome assembly protein SET is its inhibitor. Cell. 2003;112:659–672. doi: 10.1016/S0092-8674(03)00150-8. [DOI] [PubMed] [Google Scholar]
- Fan Z, Beresford PJ, Zhang D, Xu Z, Novina CD, Yoshida A, Pommier Y, Lieberman J. Cleaving the oxidative repair protein Ape1 enhances cell death mediated by granzyme A. Nat Immunol. 2003;4:145–153. doi: 10.1038/ni885. [DOI] [PubMed] [Google Scholar]
- Melvin VS, Edwards DP. Coregulatory proteins in steroid hormone receptor action: the role of chromatin high mobility group proteins HMG-1 and -2. Steroids. 1999;64:576–586. doi: 10.1016/S0039-128X(99)00036-7. [DOI] [PubMed] [Google Scholar]
- Verrier CS, Roodi N, Yee CJ, Bailey R, Jensen RA, Bustin M, Parl FF. High-mobility group (HMG) protein HMG-1 and TATA-binding protein-associated factor TAFII30 affect estrogen receptor-mediated transcriptional activation. Mol Endocrinol. 1997;11:1009–1019. doi: 10.1210/me.11.8.1009. [DOI] [PubMed] [Google Scholar]
- Zhang CC, Krieg S, Shapiro DJ. HMG-1 stimulates estrogen response element binding by estrogen receptor from stably transfected HeLa cells. Mol Endocrinol. 1999;13:632–643. doi: 10.1210/me.13.4.632. [DOI] [PubMed] [Google Scholar]
- Aaltomaa S, Lipponen P, Syrjanen K. Proliferating cell nuclear antigen (PCNA) immunolabeling as a prognostic factor in axillary lymph node negative breast cancer. Anticancer Res. 1993;13:533–538. [PubMed] [Google Scholar]
- Kim J, Petz LN, Ziegler YS, Wood JR, Potthoff SJ, Nardulli AM. Regulation of the estrogen-responsive pS2 gene in MCF-7 human breast cancer cells. J Steroid Biochem Mol Biol. 2000;74:157–168. doi: 10.1016/S0960-0760(00)00119-9. [DOI] [PubMed] [Google Scholar]
- Sakurai S, Kitano K, Okada K, Hamada K, Morioka H, Hakoshima T. Preparation and crystallization of human flap endonuclease FEN-1 in complex with proliferating-cell nuclear antigen, PCNA. Acta Crystallogr D Biol Crystallogr. 2003;59:933–935. doi: 10.1107/S0907444903004815. [DOI] [PubMed] [Google Scholar]
- Dianova II, Bohr VA, Dianov GL. Interaction of human AP endonuclease 1 with flap endonuclease 1 and proliferating cell nuclear antigen involved in long-patch base excision repair. Biochemistry. 2001;40:12639–12644. doi: 10.1021/bi011117i. [DOI] [PubMed] [Google Scholar]
- Fan Z, Beresford PJ, Zhang D, Lieberman J. HMG2 interacts with the nucleosome assembly protein SET and is a target of the cytotoxic T-lymphocyte protease granzyme A. Mol Cell Biol. 2002;22:2810–2820. doi: 10.1128/MCB.22.8.2810-2820.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia L, Zheng L, Lee HW, Bates SE, Federico L, Shen B, O'Connor TR. Human 3-methyladenine-DNA glycosylase: effect of sequence context on excision, association with PCNA, and stimulation by AP endonuclease. J Mol Biol. 2005;346:1259–1274. doi: 10.1016/j.jmb.2005.01.014. [DOI] [PubMed] [Google Scholar]
- Li X, Li J, Harrington J, Lieber MR, Burgers PM. Lagging strand DNA synthesis at the eukaryotic replication fork involves binding and stimulation of FEN-1 by proliferating cell nuclear antigen. J Biol Chem. 1995;270:22109–22112. doi: 10.1074/jbc.270.38.22109. [DOI] [PubMed] [Google Scholar]
- Wu X, Li J, Li X, Hsieh CL, Burgers PM, Lieber MR. Processing of branched DNA intermediates by a complex of human FEN-1 and PCNA. Nucleic Acids Res. 1996;24:2036–2043. doi: 10.1093/nar/24.11.2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosfield DJ, Mol CD, Shen B, Tainer JA. Structure of the DNA repair and replication endonuclease and exonuclease FEN-1: coupling DNA and PCNA binding to FEN-1 activity. Cell. 1998;95:135–146. doi: 10.1016/S0092-8674(00)81789-4. [DOI] [PubMed] [Google Scholar]
- Scharer OD, Jiricny J. Recent progress in the biology, chemistry and structural biology of DNA glycosylases. Bioessays. 2001;23:270–281. doi: 10.1002/1521-1878(200103)23:3<270::AID-BIES1037>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Das D, Peterson RC, Scovell WM. High mobility group B proteins facilitate strong estrogen receptor binding to classical and half-site estrogen response elements and relax binding selectivity. Mol Endocrinol. 2004;18:2616–2632. doi: 10.1210/me.2004-0125. [DOI] [PubMed] [Google Scholar]
- Hong R, Chakravarti D. The human proliferating cell nuclear antigen regulates transcriptional coactivator p300 activity and promotes transcriptional repression. J Biol Chem. 2003;278:44505–44513. doi: 10.1074/jbc.M303138200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
ERα-interacting proteins isolated from agarose gel complexes. HeLa nuclear extracts were incubated with ERα-specific antibody and radiolabeled ERE-containing oligos in the absence or presence of purified ERα. Isolated proteins were subjected to trypsin digestion and mass spectrometry analysis. Peptide sequences were blasted against the SEQUEST database to identify the isolated proteins. Proteins identified in more than one independent experiment are grouped by cellular function and listed by gene name, accession number, and protein description. The number of independent experiments in which each protein was identified, the number of peptides isolated, and the percent of amino acid sequence identified are indicated. Data are compiled from 4 independent experiments.