Abstract
Background
Despite a plethora of functional genomic efforts, the function of many genes in sequenced genomes remains unknown. The increasing amount of microarray data for many species allows employing the guilt-by-association principle to predict function on a large scale: genes exhibiting similar expression patterns are more likely to participate in shared biological processes.
Results
We developed Prosecutor, an application that enables researchers to rapidly infer gene function based on available gene expression data and functional annotations. Our parameter-free functional prediction method uses a sensitive algorithm to achieve a high association rate of linking genes with unknown function to annotated genes. Furthermore, Prosecutor utilizes additional biological information such as genomic context and known regulatory mechanisms that are specific for prokaryotes. We analyzed publicly available transcriptome data sets and used literature sources to validate putative functions suggested by Prosecutor. We supply the complete results of our analysis for 11 prokaryotic organisms on a dedicated website.
Conclusion
The Prosecutor software and supplementary datasets available at http://www.prosecutor.nl allow researchers working on any of the analyzed organisms to quickly identify the putative functions of their genes of interest. A de novo analysis allows new organisms to be studied.
Background
One of the central challenges in computational biology is the prediction of gene function [1]. The inference of gene function typically starts with DNA sequence analysis based on ortholog information [2-5]. Although this method has proven to be successful in many cases, considerable numbers of genes (20–50%) in current genome annotations still are of unknown function. Complementary approaches are therefore required to characterize the function of these genes.
Since the start of the DNA microarray era, the "guilt-by-association" (GBA) methodology has been used to infer gene function [6-9]. This concept is based on the assumption that genes involved in similar cellular functions are likely to display correlated expression behavior [10-12]. In addition, this correlated behavior might identify common regulatory mechanisms.
Ultimately, to understand the function of a new gene, one should exploit all available experimental data sources (e.g., transcriptomics, proteomics, protein-protein interactions and metabolomics) [13,14] or even by the joint efforts of many scientists in a community annotation [15]. Previous work on gene function prediction has mainly been focused on higher organisms using multiple high-throughput data sources [16-18]. On the other hand, genome organizational principles that are unique for prokaryotes supply valuable additional information about gene function.
However, it is expected that the GBA method is particularly powerful for prokaryotes, due to their tight coupling of transcription and translation [19]. In addition, for many prokaryotes, the available gene expression datasets greatly outnumber other experimental data sources.
To improve the analysis of the predictions, Prosecutor provides additional information for each annotated gene, most notably in its genomic context, which is particularly useful for operons. The occurrence of adjacent divergent co-expressed genes is also highlighted since these are expected to be co-regulated [20]. Finally, putative new members of transcriptional modules are examined for the presence of the same regulatory motif that is already known for the module.
Our Prosecutor software imposes no constraints on the biological annotations used; it generates hypotheses based on large variety of annotation sources e.g., Gene Ontology, metabolic pathways, UniProt keywords, etc. This is in contrast to most other methods [11,12,16-18,21-24] which, with few exceptions [8,10], are focused on coupling genes to Gene Ontology sources only.
We discuss some of the functional assignments obtained by Prosecutor, as well as a number of mining capabilities provided by the software. We find that the increasing variety of experimental conditions used in DNA microarray experiments has greatly improved the ability to identify the function of unknown genes using GBA principles.
Results and discussion
Prosecutor software
Prosecutor is a standalone application developed in Java and shares its functional database structure with the FIVA software [25]. It features an iterative implementation of the GBA method which is based on iterative Group Analysis algorithm (iGA) [26]. Several characteristics of the software analysis modules are described below.
The Iterative Guilt-By-Association (iGBA) method
The iGBA method requires DNA microarray datasets and functional categories from annotation sources to infer putative gene functions. The rationale for our approach is the GBA principle, i.e., genes that are functionally involved in, or linked to, the same function will in general show higher expression correlations than genes that are not functionally related. The prediction algorithm of Prosecutor calculates the significance of association for all pairs of genes and functional categories. For n genes, expression profiles from DNA microarrays (Fig. 1A) are used to create an n × n correlation matrix M (Fig. 1B). Each row j of this matrix represents the (Pearson or Spearman) expression correlation between gene gj and all other genes. To annotate each gene gj, we sort all other genes by their correlation with gene gj, and subject the resulting sorted gene list to iGA (Fig. 1C). This results in a list of functional categories that are over-represented among the genes that are highly correlated with gene gj, with associated p-values. The iGA algorithm works iteratively and therefore does not require a fixed cutoff of the sorted correlation list, no minimum correlation has to be defined. Instead, iGA determines the appropriate cutoff that yields the lowest p-value for each individual analysis of a gene to a functional category. As a consequence, the function assignment by iGA is very sensitive [26] compared to methods which use a predefined correlation cut-off.
Figure 1.
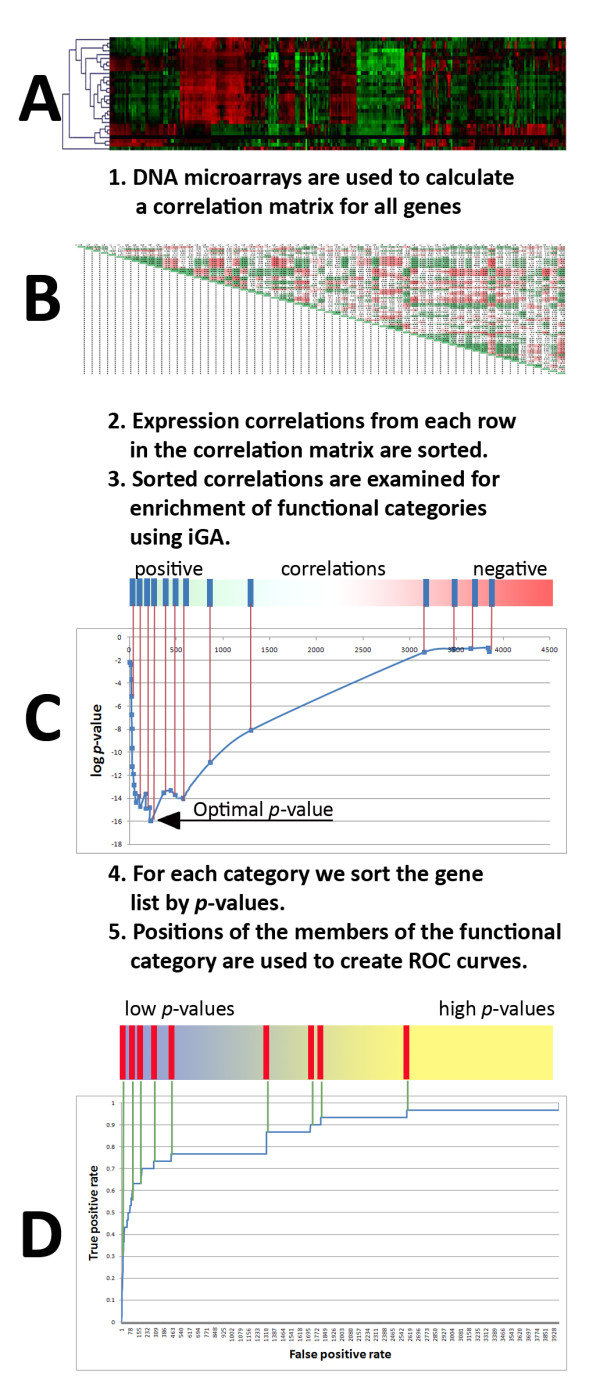
Flowchart of Prosecutor. Flowchart of the functional prediction process in Prosecutor. First, the expression profiles from DNA microarrays (1A) are used to create a correlation matrix (1B). For every gene, the correlations with the remaining genes are retrieved from the correlation matrix and sorted (1B2). The sorted gene list is used to perform an iterative Group Analysis for every functional category (1B3). The resulting p-value is indicative for the prediction of a gene as a member of a functional category (1C). At this step, the regular iGBA process ends. However, to also assess the reliability of each prediction, the following steps are added. The complete list of p-values for every functional category is sorted (1C4), after which the positions of the members of the functional category are determined (1C5). These positions are used to create ROC curves (1D; see Results section for more information concerning ROC curves). The corresponding Area Under the ROC Curve (AUC) is then used as a measure of expression coherence value of a functional category.
Performance of functional categories
Receiver Operating Characteristic curves
The performance on well-annotated genes was assessed to evaluate the sensitivity of the iGBA method. This evaluation has to be specific for each functional category, because for some of them we expect that all members show close correlation, while others are so general that their members will not correlate and iGBA is expected to fail. The category specific evaluation of expression coherence is done as follows: Our iGBA algorithm yields a p-value for every pair of gene-functional category pair (Fig. 1C). This p-value is indicative of the confidence of the assignment of a gene to a functional category. For each category we sort the gene list by p-values and examine the positions of the p-values of its known members in this sorted list. We are then able to calculate an "expression coherence value" for each functional category by plotting the true and false positive rates on Receiver Operating Characteristic (ROC) curves (Fig. 1D) [27]. The corresponding Area Under the ROC Curve (AUC) is a quantitative measure of the expression coherence of the genes of a functional category. A functional category in which all known members show strong co-expression will have an AUC close to 1.0, whereas a randomly predicting functional category (i.e., a category that does not show coexpression of its members) would yield AUC values around 0.5. Using the AUC measure, we are now able to select the most promising functional categories for further analysis.
Parameter free approach
Various methods have been developed that specifically employ data from microarrays studies [21-24]. Some of these methods are designed for temporal gene expression profiles [23,24] or calculate a functional enrichment for each dataset [22]. Other approaches require preprocessing of the annotation data, e.g., generating a set of validated and highly unlikely associations (see [28] for more information) used for training of the prediction model [21]. Our Prosecutor application improves on previous methods by providing a parameter free approach for the inferral of gene function. No trusted set of functional associations between proteins is required since Prosecutor treats every functional category individually, thereby circumventing preselection toward particular processes.
Additional layers of information
The strength of Prosecutor comes also from its additional prokaryote-specific layers of information combined with a convenient visualization of the functional predictions. This prioritizing of the results allows for the rapid identification of the most promising function predictions.
Genomic context analysis
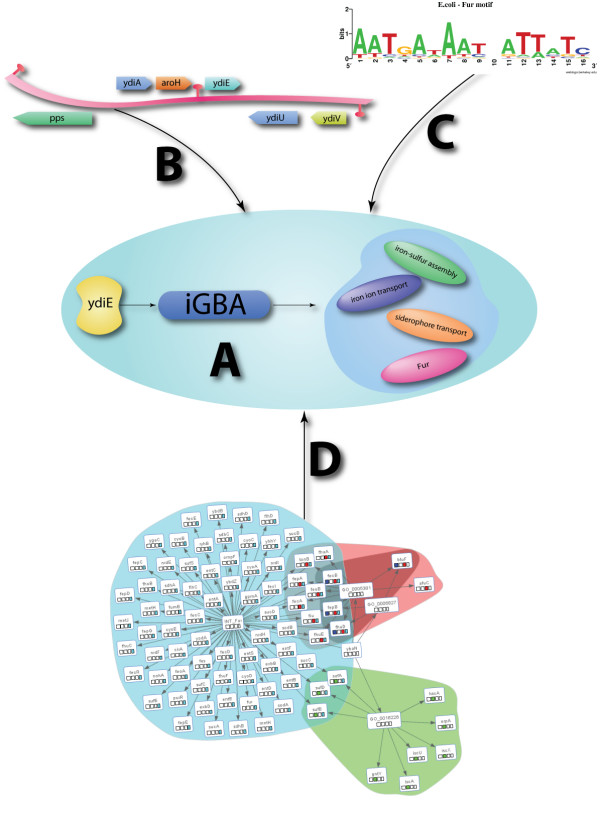
The function predictions generated by Prosecutor are provided for individual genes. Genes co-transcribed to a polycistronic messenger RNA are known as operons whose members typically share biological function. Predictions for genes of which other member(s) of the same operon were already linked to the predicted function are highlighted in the visualization of the results. The same procedure is applied to divergent genes which share the same upstream region (Fig. 2B). This layer of information that is based on the genomic context of genes provides additional, and in some case cases vital, information concerning putative function predictions.
Figure 2.
Schematic overview of the additional information provided by Prosecutor. Various layers of information are supplied for the iGBA results (2A) from Prosecutor. Predicted functional assignments for genes whose operon members are already linked to the predicted function are indicated in the results (2B). In addition, this protocol is also followed for divergent genes that share the same upstream region (in this example pps and ydiA). The operon information that is used for the genomic context analysis is also used to detect known regulatory sequences for transcriptional modules (2C). Lastly, graph visualization is used to visualize the gene redundancy of the different functional assignments of Prosecutor (2D). Nodes in the graph represent functional categories and genes. Arrows represent membership of gene nodes to a functional category node as well as the putative functional prediction of the studied gene. The members of individual categories are placed in colored aggregates. In addition to the aggregates, a colored square is placed in each gene member of a category. The squares are colored using the colors of their matching aggregates. Members of different categories can easily be distinguished using the colored squares. An example of a functional prediction found by Prosecutor for ydiE from E. coli is shown. The expression of this gene was correlated with members of various functional categories involved in the uptake of iron. In addition to the functional association with the transcriptional module Fur, the upstream region of ydiE also contains a putative Fur DNA binding site.
Regulatory mechanism analysis
Transcriptional modules represent genes that are regulated by a common regulator. The regulatory mechanisms underlying the co-expression of members of a transcriptional module are used as additional evidence to prioritize the Prosecutor results. For some organisms, functional annotations based on curated knowledge of transcriptional modules are available [29,30]. Motif instances from all members of a transcriptional module are used to create a position specific scoring matrix. This matrix is used to search for additional hits in the upstream and coding regions from the first gene of the operon as well as the gene of interest (in case of residing in an operon). Using this approach, we are able to predict putative new targets for transcriptional modules that exhibit significant co-expression with known members of the transcriptional module and a putative regulatory motif in their upstream regions (Fig. 2C).
Graph visualization
Functional predictions are represented by Prosecutor as graphs using the Prefuse toolkit [31] to visualize the gene redundancy and overlap between the functional categories of different functional predictions. This method allows to visually determine the uniqueness of each of the function predictions. A force-directed layout from the Prefuse visualization framework is used to position the different nodes (genes) in the network (Fig. 2D).
Performance compared to random microarray data
The performance of different annotation sources (e.g., Gene Ontology terms) was investigated by comparing AUC results for real and random data using a two-sample Kolmogorov-Smirnov test. This method was used to compare the distribution of AUC values of our algorithm based on 305 microarrays from E. coli (Fig. 3A) as compared to results for which the genes were randomized (the link between expression and annotation is expected to be lost) (Fig. 3B). The null hypothesis that the true data do not significantly deviate from the random distribution is rejected with a p-value of 2e-16. The real data yield significantly higher AUC values than expected by chance. This confirms that the coexpression enrichment of many functional categories is predictive of gene function. Additional analysis of the AUC distribution across the annotation sources shows that the transcription module annotation source contains a large number of high scoring functional categories (i.e., categories exceeding an AUC value of 0.9). Moreover, we found that applying a Pearson correlation measure for calculating the correlation matrix outperforms Spearman correlations, generating 16% more functional categories with an AUC value of 0.8 or higher (data not shown).
Figure 3.
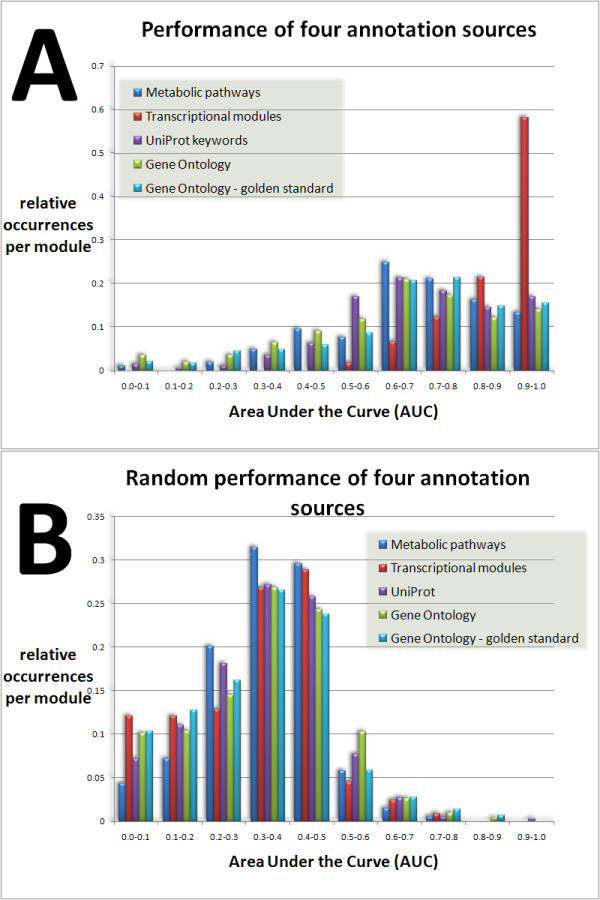
Prediction ability of four annotation sources. Histograms of ROC areas (Area Under the Curve) for four annotation sources for E. coli based on 305 microarrays (3A) compared to randomized results (3B). The real data reveal a large amount of categories with AUC values larger than 0.8, which are almost absent in randomized results. These categories are the most promising candidates for which the iGBA approach will enable confident gene assignments functional predictions. Analysis of the AUC distribution across the annotation sources shows that the "transcription module" annotation source is the most informative, i.e., contains the largest amount of categories exceeding an AUC value of 0.9 (3A). This is intuitively very convincing as shared transcriptional regulation is the basis of coexpression. In addition to ROC areas for all GO terms, we have also analyzed the distribution of ROC areas for the GO annotation source using the "gold standard" [28]. This proposed "gold standard" (GS) consists of a specific trusted set of biological processes that maps proteins to well-defined functional classes to evaluate predictions. The authors supply a set of biological processes that is based on selection by a panel of biology experts. We have included AUC results for the GO annotation for E. coli using the GS. Analysis of the AUC distributions shows that the distribution of relative occurrences of the GS analysis and the analysis using a fixed member cutoff is comparable.
Prosecutor test-cases
Most genome annotations deposited to GenBank are rarely if ever updated [32]. As research progresses, knowledge of many previously uncharacterized genes improves. This annotation gap enables us to analyze results obtained by Prosecutor by manual literature mining of genes for which no function was available in the original genome annotation. For this validation, only functional categories exhibiting strong predictive properties, with AUC values higher than 0.7, were taken into account.
First test-case: validating results of Prosecutor
The first analysis deals with results obtained from Prosecutor for all tested organisms and was based on data from dual-dye microarrays. Prosecutor predicted a large number of gene functions for previously unannotated genes which could be validated using literature information (Table 1). The complete results of this analysis is available on the supplemental website. Analysis of the results for the model organisms E. coli and B. subtilis was facilitated by the large diversity of microarray perturbation studies available. A detailed analysis for B. subtilis revealed that for 25% of the best 160 predictions sufficient literature data was available to positively confirm the predictions (data not shown).
Table 1.
Confirmed results from Prosecutor
| Organism | gene | functional category | rank | auc | reference |
| Campylobacter jejuni | Cj0391c | Pathway flagellar assembly | 2 | 0.76 | [41,42] |
| Cj1242 | GO:0003774 motor activity | 3 | 0.75 | [42] | |
| Cj1316c | GO:0019861 agellum | 7 | 0.72 | [43] | |
| Escherichia coli | yncE | GO:0015343 siderophore-iron transmembrane transporter activity | 7 | 0.96 | [44] |
| ybiX | UP:Enterobactin biosynthesis | 1 | 0.99 | [44] | |
| cho | GO:0009432 SOS response | 17 | 0.92 | [45] | |
| ybeD | GO:0051082 unfolded protein binding | 1 | 0.78 | [46] | |
| ulaC | GO:0019852 L-ascorbic acid metabolic process | 1 | 0.76 | [47] | |
| yciW | GO:0006534 cysteine metabolic process | 10 | 0.82 | [48] | |
| Bacillus subtilis | ypbG | transcriptional module SigM | 18 | 0.81 | [49] |
| ykuO | transcriptional module Fur | 8 | 0.75 | [50] | |
| yviF | GO:0006935 chemotaxis | 37 | 0.90 | [51] | |
| ylxF | Pathway Flagellar assembly | 7 | 0.89 | [52] | |
| yfnE | transcriptional module GerE | 52 | 0.81 | [53] | |
| Streptomyces coelicolor | SCBAC28G1.05 | PW:Biosynthesis of type II polyketide back- bone | 1 | 0.99 | [54] |
| SCBAC28G1.07 | PW:Biosynthesis of type II polyketide products | 8 | 0.76 | [54] | |
| Vibroi cholera | VC1688 | GO:0006826 iron ion transport | 2 | 0.87 | [55] |
| VCA0216 | GO:0019290 siderophore biosynthetic process | 8 | 0.98 | [55] | |
| VC1267 | GO:0019290 siderophore biosynthetic process | 6 | 0.98 | [55] | |
Functional predictions identified by Prosecutor for several organisms that are confirmed using literature information. Gene: the gene for which a validated functional prediction with a functional category (column three) was found. Rank: the position of a gene in the prioritized list based on p-values. These p-values describe the functional prediction significance for every individual gene with a specific functional category. AUC: the expression coherence value for a functional category with respect to its own members. Notice the examples of genes for which a particular annotation is assigned rank 1. This means that this gene is more close associated with this functional category then any of the original known members of the category, indicating a very high confidence in the prediction.
Second test-case: extending transcriptional modules in E. coli
The second analysis dealt with the detection of putative new members of existing transcriptional modules in E. coli (Table 2). We used gene expression data from 305 Affymetrix genechips [33] combined with functional annotations based on curated regulatory network information from RegulonDB [30]. The results of Prosecutor were supplemented with data obtained from the position specific scoring matrices. These matrices were based on aligned motif sequences of the known DNA binding sites from the members of every transcriptional module. We found that some of the newly identified putative transcriptional module members had been confirmed in the literature, but are not yet catalogued in RegulonDB. The remainder of the putative transcriptional module members which could not be verified using literature information are marked "putative" in Table 2. Due to the exceptional predictive performance (almost 60% of the transcriptional modules shows an AUC value above 0.9) and the additional analysis of the results using known regulatory mechanisms, we were able to reliably predict a large number of putative and validated members for transcription modules.
Table 2.
Extending transcriptional modules of E. coli
| transcriptional module |
gene |
prosecutor rank |
motif rank | motif sequence in the intergenic region of either the gene or its operon | literature reference |
| ArgR Amino acid biosynthesis: Arginine. AUC 0.92 | artJ | 21 | 8 | TGCATAACATTGCG | [56] |
| aroP | 58 | 39 | TGATTTTTAATTCA | [57] | |
| artI | 131 | 50 | TGCATAATTATTCT | [56] | |
| hisL | 16 | 4 | TGAATAAACATTCA | putative | |
| pyrL | 32 | 61 | TGACTTTTAATTCA | putative | |
| metH | 36 | 76 | TGAATTTTTATTAA | putative | |
| ydcS | 43 | 63 | TGAATAAATTTTCT | putative | |
| stpA | 132 | 21 | TGCATTTTTATTCA | putative | |
| hisG | 141 | 8 | TGAATAAACATTCA | putative | |
| hisJ | 144 | 27 | TGCATTGAAATGCA | putative | |
| hisC | 145 | 13 | TGAATAAACATTCA | putative | |
| hisA | 147 | 14 | TGAATAAACATTCA | putative | |
| potF | 162 | 46 | TGCATAAAAATTTG | putative | |
| CysB Amino acid biosynthesis: Cysteine AUC 0.91 | sbp | 12 | 0 | CGCAAGTTATAGCCAATCTTTTTTTATTCTT | [48,58] |
| nlpA | 36 | 17 | CAGACTTTATATTCCACTTTTATTCCTTTTT | [48] | |
| mmuP | 28 | 40 | AACGCGGTATAACAAACCTTCTTTGGATGTT | putative | |
| Fur iron regulatory gene AUC 0.84 | yncD | 74 | 32 | GGGAATGGTAATCATTATT | [44] |
| ybaN | 37 | 5 | GAAAATGATAATTGTTATG | putative | |
| folE | 101 | 29 | GGCAATTACAATAATTATC | putative | |
| LexA major regulator of DNA repair AUC 0.87 | yebG | 0 | 21 | CTGTATAAAATCACAG | [59,60] |
| dinI | 2 | 6 | CTGTATAAATAACCAG | [61,62] | |
| dinB | 6 | 51 | CTGTATACTTTACCAG | [63] | |
| dinD | 19 | 0 | CTGTATATAAATACAG | [45] | |
| yjiW | 39 | 20 | CTGATGATATATACAG | [45] | |
| ybfE | 120 | 31 | CTGATTAAAAACCCAG | [45] | |
| sbmC | 125 | 4 | CTGTATATAAAAACAG | [64] | |
| MetJ Amino acid biosynthesis: Methionine AUC 0.88 | ybdH | 10 | 6 | AGACGTTTAGATGTCT | [65] |
| ybdL | 106 | 0 | AGACATCTAAACGTCT | [65] | |
| ycbK | 198 | 17 | AGTCATCTTGACGTCT | [65] | |
| mmuP | 14 | 15 | GGATGTTTAGATGTCC | putative | |
Transcriptional module member predictions identified by Prosecutor applied to well-known transcriptional modules for E. coli. Gene: the gene for which a validated functional prediction with a transcriptional module (column three) was found. Rank: represents the position of a gene in the sorted list with p-values. These p-values describe the functional prediction significance for every individual gene with a specific functional category. More significant p-values are matched with lower ranks. AUC: describes the association efficiency for a transcriptional module with respect to its own members. In addition to the rank information provided by Prosecutor, supplemental motif information is provided. This data is obtained by applying a position specific scoring matrix (PSSM) to the upstream sequences of all genes in the genome. The PSSM is derived from aggregating all known consensus target sequences (DNA regulatory binding sites). The additional motif information allows users to concentrate on genes that exhibit coexpression with a transcriptional module as well as possessing a predicted consensus sequence. This additional evidence contributes to the confidence in assigning a gene to a particular transcriptional module. Motif rank: based on the results for a PSSM when matched to every upstream sequence in the genome. For example, based on the PSSM of the regulator LexA, the upstream region of gene dinD contains the best ranking motif (rank 1).
Third test-case: performance of annotation sources for Saccharomyces cerevisiae
The genome annotation of S. cerevisae is available in Genbank as well as EMBL format, allowing our Prosecutor software to perform an iGBA analysis. For this third analysis we used two annotation sources (metabolic pathways and Gene Ontology). The gene expression data was obtained from the Stanford microarray database [34]. The distribution of AUC values of our algorithm (Fig. 4A) is compared to results for which the genes were randomized (Fig. 4B) The results based on the real data yield more large AUC values than expected by chance. The categories with high AUC values will presumably allow our iGBA method to assign reliable functional predictions. This demonstrates that Prosecutor, while being specifically optimized for prokaryotes, will also be a useful tool for the general biologist community.
Figure 4.
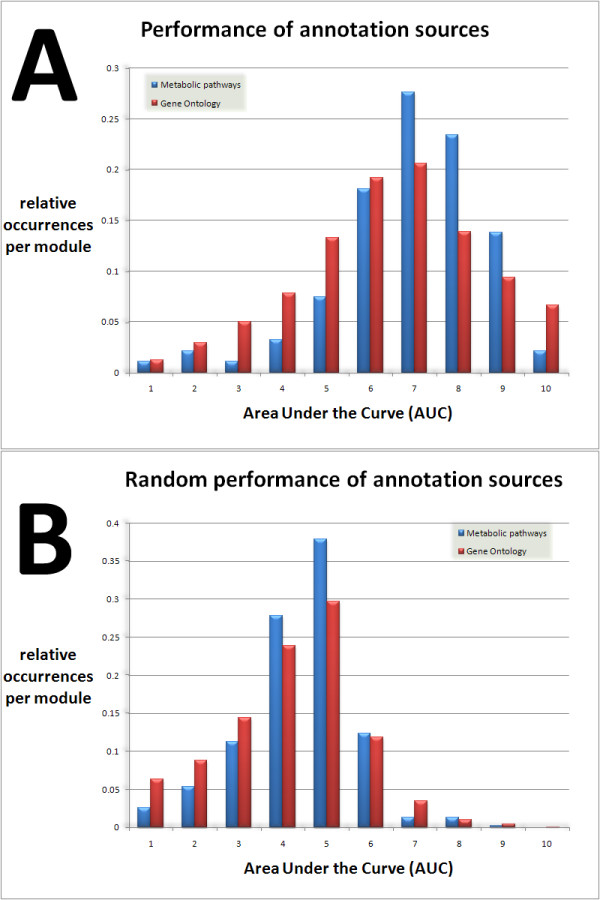
Prediction ability of two annotation sources for yeast. Histograms of ROC areas (Area Under the Curve) for two annotation sources (Gene Ontology and metabolic pathways) for S. cerevisae based on 1079 datasets from Stanford microarray database (4A) compared to randomized results (4B). The real data reveal a large number of categories with AUC values larger than 0.8, which are almost absent in randomized results. These categories are the most promising candidates for which the iGBA approach will enable confident gene assignments of functional predictions.
Community resource
The complete results of the annotation efforts from our software for twelve organisms are available on the supplemental website [35]. On this dedicated web-site functional couplings can be mined in three ways: 1) through a list of the best functional couplings for each functional category; this allows "browsing" through the most promising associations, 2) a sorted list of functional categories and their predictive power (AUC); in case that one is interested in the genes that are associated with a specific functional category, and 3) a sorted list of genes; allows to identify to which functional categories a gene of interest is associated. All data sources used for analysis are available, allowing researchers studying any of the analyzed organisms to perform a functional analysis for their expression dataset and/or functional categories.
Conclusion
Prosecutor uses DNA microarray data combined with functional annotations to infer putative gene functions. We show that multiple annotation sources are informative and non-redundant and allow maximizing the use of all available DNA microarray data. For B. subtilis, we were able to confirm 40 out of the 160 best functional predictions generated by Prosecutor, using published literature. We therefore believe that the other functional assignments based on our analysis are also likely to be informative and reliable. Combined with regulatory motif information for the species B. subtilis and E. coli, Prosecutor allows the identification of new transcriptional module members. Prosecutor can thus serve as a generic tool for a genome-wide (re)annotation of gene functions in prokaryotes. The results of such a re-annotation effort, for 11 widely studied bacterial species, is supplied as a community resource at the associated website [35].
Methods
Implementation & Availability
Prosecutor was programmed as a multithreaded standalone application in Java using the Eclipse framework http://www.eclipse.org/ as a Rich Client Platform (source code is available upon request). Prosecutor runs on all Java-supporting operating systems (MS Windows, Linux and Mac OS). The Prosecutor was developed from a bacterial perspective and therefore supports the two major prokaryotic genome annotation formats (Genbank and EMBL). A simplified tabulated genome annotation format can also be used, enabling organisms for which no Genbank or EMBL file is available to be studied.
Data sources
The basic requirements of an analysis consist of a genome annotation (i.e., Genbank or EMBL) and a collection of microarray data. Currently, six different annotation sources are implemented: (i) transcriptional modules, (ii) gene ontologies (GO) [36], (iii) metabolic pathways from the KEGG database [37] (iv) UniProt keywords [38], (v) InterPro domains [39] and (vi) user-defined categories.
DNA microarray datasets
DNA microarray data used in this study consisted of dual dye arrays for 11 prokaryotic organisms and yeast from the KEGG expression database [40] and the Stanford microarray database [34]. For E. coli, an additional 305 Affymetrix expression arrays were obtained from the M3D Database [33].
Multiple testing correction
A typical problem in genome-wide statistical analysis is the occurrence of many false positives (i.e., a functional prediction that is mistakenly found significant due to multiple testing). The incidence of false positives is roughly proportional to the number of tests performed. Since a typical search in Prosecutor may consist of thousands of tests, the chance of obtaining false positive predictions is large. We have used a strict Bonferroni multiple testing correction method to correct the raw p-values from the iGA results to minimize this problem.
Authors' contributions
EJB conceived the study and programmed the software. EJB and RB devised the iGBA algorithm. KJH designed and programmed the analysis interface. EJB and SAFTH wrote the manuscript. JBTMR and OPK guided and coordinated the project. All authors read, corrected and approved the final manuscript.
Acknowledgments
Acknowledgements
This study was supported by a grant from the Netherlands Organisation for Scientific Research and industrial partners in the NWO-BMI project number 050.50.206 on Computational Genomics of Prokaryotes and by the Center IOP Genomics. Work of SvH was in part supported by the BMBF within the framework of the transnational SysMO initiative in the project BaCell-SysMO. This work was in part supported by an EU program in FW6: Bacell Health, European Union Grant LSHG-CT-2004-503468. We thank Grayson H. Kleine C.B. de Miranda for creating a multi-threaded implementation of the iGBA algorithm.
Contributor Information
Evert Jan Blom, Email: e.j.blom@rug.nl.
Rainer Breitling, Email: R.Breitling@rug.nl.
Klaas Jan Hofstede, Email: klaasjanhofstede@home.nl.
Jos BTM Roerdink, Email: j.b.t.m.roerdink@rug.nl.
Sacha AFT van Hijum, Email: sacha.vanhijum@nizo.nl.
Oscar P Kuipers, Email: o.p.kuipers@rug.nl.
References
- Friedberg I. Automated protein function prediction-the genomic challenge. Brief Bioinform. 2006;7:225–242. doi: 10.1093/bib/bbl004. [DOI] [PubMed] [Google Scholar]
- Overbeek R, Begley T, Butler RM, Choudhuri JV, Chuang HY, Cohoon M, de Crécy-Lagard V, Diaz N, Disz T, Edwards R, Fonstein M, Frank ED, Gerdes S, Glass EM, Goesmann A, Hanson A, Iwata-Reuyl D, Jensen R, Jamshidi N, Krause L, Kubal M, Larsen N, Linke B, McHardy AC, Meyer F, Neuweger H, Olsen G, Olson R, Osterman A, Portnoy V, Pusch GD, Rodionov DA, Rückert C, Steiner J, Stevens R, Thiele I, Vassieva O, Ye Y, Zagnitko O, Vonstein V. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 2005;33:5691–5702. doi: 10.1093/nar/gki866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynen M, Snel B, Lathe W, Bork P. Predicting protein function by genomic context: quantitative evaluation and qualitative inferences. Genome Res. 2000;10:1204–1210. doi: 10.1101/gr.10.8.1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Hu Z, DeLisi C. Gene annotation and network inference by phylogenetic profiling. BMC Bioinformatics. 2006;7:80. doi: 10.1186/1471-2105-7-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Su Z, Mao F, Olman V, Xu Y. Prediction of functional modules based on comparative genome analysis and Gene Ontology application. Nucleic Acids Res. 2005;33:2822–2837. doi: 10.1093/nar/gki573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brent R. Functional genomics: learning to think about gene expression data. Curr Biol. 1999;9:R338–R341. doi: 10.1016/S0960-9822(99)80208-5. [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aerts S, Lambrechts D, Maity S, Loo PV, Coessens B, Smet FD, Tranchevent LC, Moor BD, Marynen P, Hassan B, Carmeliet P, Moreau Y. Gene prioritization through genomic data fusion. Nat Biotechnol. 2006;24:537–544. doi: 10.1038/nbt1203. [DOI] [PubMed] [Google Scholar]
- Quackenbush J. Computational analysis of microarray data. Nat Rev Genet. 2001;2:418–427. doi: 10.1038/35076576. [DOI] [PubMed] [Google Scholar]
- Hughes TR, Marton MJ, Jones AR, Roberts CJ, Stoughton R, Armour CD, Bennett HA, Coffey E, Dai H, He YD, Kidd MJ, King AM, Meyer MR, Slade D, Lum PY, Stepaniants SB, Shoemaker DD, Gachotte D, Chakraburtty K, Simon J, Bard M, Friend SH. Functional discovery via a compendium of expression profiles. Cell. 2000;102:109–126. doi: 10.1016/S0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- Wolfe CJ, Kohane IS, Butte AJ. Systematic survey reveals general applicability of "guilt-by-association" within gene coexpression networks. BMC Bioinformatics. 2005;6:227. doi: 10.1186/1471-2105-6-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotte EM, Pellegrini M, Thompson MJ, Yeates TO, Eisenberg D. A combined algorithm for genome-wide prediction of protein function. Nature. 1999;402:83–86. doi: 10.1038/47048. [DOI] [PubMed] [Google Scholar]
- Noordewier MO, Warren PV. Gene expression microarrays and the integration of biological knowledge. Trends Biotechnol. 2001;19:412–415. doi: 10.1016/S0167-7799(01)01735-8. [DOI] [PubMed] [Google Scholar]
- Espadaler J, Eswar N, Querol E, Aviles FX, Sali A, Marti-Renom M, Oliva B. Prediction of enzyme function by combining sequence similarity and protein interactions. BMC Bioinformatics. 2008;9:249. doi: 10.1186/1471-2105-9-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mons B, Ashburner M, Chichester C, van Mulligen E, Weeber M, den Dunnen JT, Musen M, Cockerill M, Hermjakob H, Packer A, Pacheco R, Lewis S, Berkeley A, Melton W, Barris N, Borner K, Meijssen G, Moeller E, Roes PJ, Mons A, van Ommen GJ, Wales J, Bairoch A. Calling on a million minds for community annotation in WikiProteins. Genome Biol. 2008;9:R89. doi: 10.1186/gb-2008-9-5-r89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlidis P, Weston J, Cai J, Noble WS. Learning gene functional classifications from multiple data types. J Comput Biol. 2002;9:401–411. doi: 10.1089/10665270252935539. [DOI] [PubMed] [Google Scholar]
- Troyanskaya OG, Dolinski K, Owen AB, Altman RB, Botstein D. A Bayesian framework for combining heterogeneous data sources for gene function prediction (in Saccharomyces cerevisiae) Proc Natl Acad Sci USA. 2003;100:8348–8353. doi: 10.1073/pnas.0832373100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I, Li Z, Marcotte EM. An improved, bias-reduced probabilistic functional gene network of baker's yeast, Saccharomyces cerevisiae. PLoS ONE. 2007;2:e988. doi: 10.1371/journal.pone.0000988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller OL, Hamkalo BA, Thomas CA. Visualization of bacterial genes in action. Science. 1970;169:392–395. doi: 10.1126/science.169.3943.392. [DOI] [PubMed] [Google Scholar]
- Korbel JO, Jensen LJ, von Mering C, Bork P. Analysis of genomic context: prediction of functional associations from conserved bidirectionally transcribed gene pairs. Nat Biotechnol. 2004;22:911–917. doi: 10.1038/nbt988. [DOI] [PubMed] [Google Scholar]
- Huttenhower C, Hibbs M, Myers C, Troyanskaya OG. A scalable method for integration and functional analysis of multiple microarray datasets. Bioinformatics. 2006;22:2890–2897. doi: 10.1093/bioinformatics/btl492. [DOI] [PubMed] [Google Scholar]
- Hibbs MA, Hess DC, Myers CL, Huttenhower C, Li K, Troyanskaya OG. Exploring the functional landscape of gene expression: directed search of large microarray compendia. Bioinformatics. 2007;23:2692–2699. doi: 10.1093/bioinformatics/btm403. [DOI] [PubMed] [Google Scholar]
- Lagreid A, Hvidsten TR, Midelfart H, Komorowski J, Sandvik AK. Predicting gene ontology biological process from temporal gene expression patterns. Genome Res. 2003;13:965–979. doi: 10.1101/gr.1144503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Zou S, Song J. Term-tissue specific models for prediction of gene ontology biological processes using transcriptional profiles of aging in Drosophila melanogaster. BMC Bioinformatics. 2008;9:129. doi: 10.1186/1471-2105-9-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blom EJ, Bosman DWJ, van Hijum SA, Breitling R, Tijsma L, Silvis R, Roerdink JB, Kuipers OP. FIVA: Functional Information Viewer and Analyzer extracting biological knowledge from transcriptome data of prokaryotes. Bioinformatics. 2007;23:1161–1163. doi: 10.1093/bioinformatics/btl658. [DOI] [PubMed] [Google Scholar]
- Breitling R, Amtmann A, Herzyk P. Iterative Group Analysis (iGA): a simple tool to enhance sensitivity and facilitate interpretation of microarray experiments. BMC Bioinformatics. 2004;5:34. doi: 10.1186/1471-2105-5-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasko TA, Bhagwat JG, Zou KH, Ohno-Machado L. The use of receiver operating characteristic curves in biomedical informatics. J Biomed Inform. 2005;38:404–415. doi: 10.1016/j.jbi.2005.02.008. [DOI] [PubMed] [Google Scholar]
- Myers CL, Barrett DR, Hibbs MA, Huttenhower C, Troyanskaya OG. Finding function: evaluation methods for functional genomic data. BMC Genomics. 2006;7:187. doi: 10.1186/1471-2164-7-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierro N, Makita Y, de Hoon M, Nakai K. DBTBS: a database of transcriptional regulation in Bacillus subtilis containing upstream intergenic conservation information. Nucleic Acids Res. 2008;36:D93–D96. doi: 10.1093/nar/gkm910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salgado H, Gama-Castro S, Peralta-Gil M, Díaz-Peredo E, Sánchez-Solano F, Santos-Zavaleta A, Martínez-Flores I, Jiménez-Jacinto V, Bonavides-Martínez C, Segura-Salazar J, Martínez-Antonio A, Collado-Vides J. RegulonDB (version 5.0): Escherichia coli K-12 transcriptional regulatory network, operon organization, and growth conditions. Nucleic Acids Res. 2006;34:D394–D397. doi: 10.1093/nar/gkj156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heer J, Card SK, Landay JA. Prefuse: a toolkit for interactive information visualization. CHI 05: Proc SIGCHI conf Human factors in computing systems 2005, April 2–7, Portland, OR.
- Salzberg SL. Genome re-annotation: a wiki solution? Genome Biol. 2007;8:102. doi: 10.1186/gb-2007-8-6-r102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faith JJ, Driscoll ME, Fusaro VA, Cosgrove EJ, Hayete B, Juhn FS, Schneider SJ, Gardner TS. Many Microbe Microarrays Database: uniformly normalized Affymetrix compendia with structured experimental metadata. Nucleic Acids Res. 2007 doi: 10.1093/nar/gkm815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demeter J, Beauheim C, Gollub J, Hernandez-Boussard T, Jin H, Maier D, Matese JC, Nitzberg M, Wymore F, Zachariah ZK, Brown PO, Sherlock G, Ball CA. The Stanford Microarray Database: implementation of new analysis tools and open source release of software. Nucleic Acids Res. 2007;35:D766–D770. doi: 10.1093/nar/gkl1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosecutor website http://www.prosecutor.nl
- GeneOntology http://www.geneontology.org/GO.downloads.ontology.shtml
- Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, Yamanishi Y. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uniprot keywords http://www.expasy.org/cgi-bin/keywlist.pl
- Interpro domains http://www.ebi.ac.uk/interpro/
- KEGG Expression database http://www.genome.jp/kegg/expression/
- Kalmokoff M, Lanthier P, Tremblay TL, Foss M, Lau PC, Sanders G, Austin J, Kelly J, Szymanski CM. Proteomic analysis of Campylobacter jejuni 11168 biofilms reveals a role for the motility complex in biofilm formation. J Bacteriol. 2006;188:4312–4320. doi: 10.1128/JB.01975-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo CD, Taboada E, Nash JHE, Lanthier P, Kelly J, Lau PC, Verhulp R, Mykytczuk O, Sy J, Findlay WA, Amoako K, Gomis S, Willson P, Austin JW, Potter A, Babiuk L, Allan B, Szymanski CM. Genome-wide expression analyses of Campylobacter jejuni NCTC11168 reveals coordinate regulation of motility and virulence by flhA. J Biol Chem. 2004;279:20327–20338. doi: 10.1074/jbc.M401134200. [DOI] [PubMed] [Google Scholar]
- Guerry P, Ewing CP, Schirm M, Lorenzo M, Kelly J, Pattarini D, Majam G, Thibault P, Logan S. Changes in flagellin glycosylation affect Campylobacter autoagglutination and virulence. Mol Microbiol. 2006;60:299–311. doi: 10.1111/j.1365-2958.2006.05100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHugh JP, Rodríguez-Quinoñes F, Abdul-Tehrani H, Svistunenko DA, Poole RK, Cooper CE, Andrews SC. Global iron-dependent gene regulation in Escherichia coli. A new mechanism for iron homeostasis. J Biol Chem. 2003;278:29478–29486. doi: 10.1074/jbc.M303381200. [DOI] [PubMed] [Google Scholar]
- Fernández De Henestrosa AR, Ogi T, Aoyagi S, Chafin D, Hayes JJ, Ohmori H, Woodgate R. Identification of additional genes belonging to the LexA regulon in Escherichia coli. Mol Microbiol. 2000;35:1560–1572. doi: 10.1046/j.1365-2958.2000.01826.x. [DOI] [PubMed] [Google Scholar]
- Lesley SA, Graziano J, Cho CY, Knuth MW, Klock HE. Gene expression response to misfolded protein as a screen for soluble recombinant protein. Protein Eng. 2002;15:153–160. doi: 10.1093/protein/15.2.153. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Aboulwafa M, Smith MH, Saier MH. The ascorbate transporter of Escherichia coli. J Bacteriol. 2003;185:2243–2250. doi: 10.1128/JB.185.7.2243-2250.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyaneshwar P, Paliy O, McAuliffe J, Popham DL, Jordan MI, Kustu S. Sulfur and nitrogen limitation in Escherichia coli K-12: specific homeostatic responses. J Bacteriol. 2005;187:1074–1090. doi: 10.1128/JB.187.3.1074-1090.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jervis AJ, Thackray PD, Houston CW, Horsburgh MJ, Moir A. SigM-responsive genes of Bacillus subtilis and their promoters. J Bacteriol. 2007;189:4534–4538. doi: 10.1128/JB.00130-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore CM, Nakano MM, Wang T, Ye RW, Helmann JD. Response of Bacillus subtilis to nitric oxide and the nitrosating agent sodium nitroprusside. J Bacteriol. 2004;186:4655–4664. doi: 10.1128/JB.186.14.4655-4664.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrell DS, Thompson LJ, Kim CC, Mitchell H, Tompkins LS, Lee A, Falkow S. Growth phase-dependent response of Helicobacter pylori to iron starvation. Infect Immun. 2003;71:6510–6525. doi: 10.1128/IAI.71.11.6510-6525.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amati G, Bisicchia P, Galizzi A. DegU-P represses expression of the motility fla-che operon in Bacillus subtilis. J Bacteriol. 2004;186:6003–6014. doi: 10.1128/JB.186.18.6003-6014.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichenberger P, Fujita M, Jensen ST, Conlon EM, Rudner DZ, Wang ST, Ferguson C, Haga K, Sato T, Liu JS, Losick R. The program of gene transcription for a single differentiating cell type during sporulation in Bacillus subtilis. PLoS Biol. 2004;2:e328. doi: 10.1371/journal.pbio.0020328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YJ, Song JY, Moon MH, Smith CP, Hong SK, Chang YK. pH shock induces overexpression of regulatory and biosynthetic genes for actinorhodin productionin Streptomyces coelicolor A3 (2) Appl Microbiol Biotechnol. 2007;76:1119–1130. doi: 10.1007/s00253-007-1083-9. [DOI] [PubMed] [Google Scholar]
- Mey AR, Wyckoff EE, Kanukurthy V, Fisher CR, Payne SM. Iron and fur regulation in Vibrio cholerae and the role of fur in virulence. Infect Immun. 2005;73:8167–8178. doi: 10.1128/IAI.73.12.8167-8178.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldara M, Minh PNL, Bostoen S, Massant J, Charlier D. ArgR-dependent repression of arginine and histidine transport genes in Escherichia coli K-12. J Mol Biol. 2007;373:251–267. doi: 10.1016/j.jmb.2007.08.013. [DOI] [PubMed] [Google Scholar]
- Bulyk ML, McGuire AM, Masuda N, Church GM. A motif co-occurrence approach for genome-wide prediction of transcription-factor-binding sites in Escherichia coli. Genome Res. 2004;14:201–208. doi: 10.1101/gr.1448004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploeg JR van der, Eichhorn E, Leisinger T. Sulfonate-sulfur metabolism and its regulation in Escherichia coli. Arch Microbiol. 2001;176:1–8. doi: 10.1007/s002030100298. [DOI] [PubMed] [Google Scholar]
- Lomba MR, Vasconcelos AT, Pacheco AB, de Almeida DF. Identification of yebG as a DNA damage-inducible Escherichia coli gene. FEMS Microbiol Lett. 1997;156:119–122. doi: 10.1111/j.1574-6968.1997.tb12715.x. [DOI] [PubMed] [Google Scholar]
- Courcelle J, Khodursky A, Peter B, Brown PO, Hanawalt PC. Comparative gene expression profiles following UV exposure in wild-type and SOS-deficient Escherichia coli. Genetics. 2001;158:41–64. doi: 10.1093/genetics/158.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faith JJ, Hayete B, Thaden JT, Mogno I, Wierzbowski J, Cottarel G, Kasif S, Collins JJ, Gardner TS. Large-scale mapping and validation of Escherichia coli transcriptional regulation from a compendium of expression profiles. PLoS Biol. 2007;5:e8. doi: 10.1371/journal.pbio.0050008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda T, Morimatsu K, Horii T, Nagata T, Ohmori H. Inhibition of Escherichia coli RecA coprotease activities by DinI. EMBO J. 1998;17:3207–3216. doi: 10.1093/emboj/17.11.3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Capilla T, Baquero MR, Gómez-Gómez JM, Ionel A, Martín S, Blázquez J. SOS-independent induction of dinB transcription by beta-lactam-mediated inhibition of cell wall synthesis in Escherichia coli. J Bacteriol. 2005;187:1515–1518. doi: 10.1128/JB.187.4.1515-1518.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baquero MR, Bouzon M, Varea J, Moreno F. sbmC, a stationary-phase induced SOS Escherichia coli gene, whose product protects cells from the DNA replication inhibitor microcin B17. Mol Microbiol. 1995;18:301–311. doi: 10.1111/j.1365-2958.1995.mmi_18020301.x. [DOI] [PubMed] [Google Scholar]
- Liu R, Blackwell TW, States DJ. Conformational model for binding site recognition by the E. coli MetJ transcription factor. Bioinformatics. 2001;17:622–633. doi: 10.1093/bioinformatics/17.7.622. [DOI] [PubMed] [Google Scholar]