Abstract
Oxidative maturation of secretory and membrane proteins in the endoplasmic reticulum (ER) is powered by Ero1 oxidases. To prevent cellular hyperoxidation, Ero1 activity can be regulated by intramolecular disulphide switches. Here, we determine the redox-driven shutdown mechanism of Ero1α, the housekeeping Ero1 enzyme in human cells. We show that functional silencing of Ero1α in cells arises from the formation of a disulphide bond—identified by mass spectrometry—between the active-site Cys94 (connected to Cys99 in the active enzyme) and Cys131. Competition between substrate thiols and Cys131 creates a feedback loop where activation of Ero1α is linked to the availability of its substrate, reduced protein disulphide isomerase (PDI). Overexpression of Ero1α-Cys131Ala or the isoform Ero1β, which does not have an equivalent disulphide switch, leads to augmented ER oxidation. These data reveal a novel regulatory feedback system where PDI emerges as a central regulator of ER redox homoeostasis.
Keywords: disulphide-bond formation, endoplasmic reticulum, ER oxidoreductin 1, protein disulphide isomerase, redox homoeostasis
Introduction
The endoplasmic reticulum (ER) maintains redox conditions that are optimal for the essential process of native disulphide-bond formation in proteins of the secretory pathway. ER redox homoeostasis is achieved by an interplay between the ER-localized disulphide-generating oxidases of the Ero1 family (Sevier and Kaiser, 2008) and reduced glutathione (GSH) that transfers reducing equivalents from the cytosol into the ER (Cuozzo and Kaiser, 1999; Molteni et al, 2004). By competing with nascent polypeptides for Ero1-derived oxidizing equivalents, GSH is oxidized to glutathione disulphide (GSSG) in the ER lumen. Together, GSH and GSSG form a redox buffer that helps to maintain the ER redox homoeostasis. Thus, redox-dependent processes such as oxidative protein folding are hampered under conditions of glutathione depletion (Cuozzo and Kaiser, 1999; Chakravarthi and Bulleid, 2004; Molteni et al, 2004).
In principle, Ero1-catalysed oxidation can lead to undesirable, constant consumption of cellular GSH (Thorpe and Coppock, 2007). Moreover, potentially cytotoxic reactive oxygen species (ROS) accumulate as a side product of Ero1 activity that generates stoichiometric amounts of H2O2 per newly formed disulphide (Gross et al, 2006). ER oxidation therefore requires control mechanisms that link the process of disulphide-bond generation to the overall redox conditions in the ER. The recent detection of two regulatory disulphide bridges in Saccharomyces cerevisiae Ero1 (Ero1p) that must be reduced for catalytic activity has provided the first direct evidence for such a feedback system (Sevier et al, 2007). The question of the physiological reductant(s) of the Ero1p disulphide switches, that is, the redox sensing mechanism of the enzyme, has not yet been resolved.
The oxidases of the Ero1 family are flavoproteins. They harbour two di-cysteine active-site motifs that cooperate in the transfer of electrons from substrate to the flavin adenine dinucleotide (FAD) cofactor: an outer active site (also termed the ‘shuttle cysteines') interacting with substrate, and an inner active site that is FAD adjacent (Sevier and Kaiser, 2008). The shuttle cysteines are located in a flexible polypeptide segment that can reach into the helical core domain and shuttle electrons to the inner active site (Gross et al, 2004). In Ero1p, the regulatory disulphides (Cys90–Cys349 and Cys150–Cys295) link the flexible loop region to the core domain.
In human cells, two Ero1 isoforms exist that are both transcriptionally upregulated under conditions of ER stress (Pagani et al, 2000; Marciniak et al, 2004). Whereas the Ero1α protein is expressed in most cell types, detectable protein levels of Ero1β are found only in select tissues (Dias-Gunasekara et al, 2005). Ero1α exists as two oxidized redox forms in vivo, OX1 and OX2, that differ in their electrophoretic mobility on non-reducing gels (Benham et al, 2000). As the faster-migrating OX2 form is favoured under oxidizing conditions and disfavoured upon treatment with a reducing agent (Anelli et al, 2002; Molteni et al, 2004), it appears to comprise at least one additional long-range disulphide bond as compared with the slower-migrating OX1 species. For an overview of the cysteines in Ero1α and Ero1p, see Figure 2B.
Apart from the monomeric OX1 and OX2 forms of Ero1α, a band of ∼130 kDa representing a covalent complex between Ero1α and protein disulphide isomerase (PDI) linked by an inter-chain disulphide has been identified by non-reducing SDS–PAGE (Benham et al, 2000; Mezghrani et al, 2001). PDI is the only conserved substrate in mammalian cells and yeast (Frand and Kaiser, 2000; Tu et al, 2000) and is generally believed to be the primary acceptor of disulphide bonds from Ero1. Thus, one function of PDI is to oxidize nascent polypeptides (Appenzeller-Herzog and Ellgaard, 2008b). Similar to its close homologue ERp57, PDI has two redox-active domains—a and a′—that each feature a Cys-Gly-His-Cys active-site sequence for the catalysis of thiol–disulphide exchange. In HEK293 cells, PDI is predominantly reduced (∼50% of all molecules) at steady state, but fully oxidized (∼16%) and semi-oxidized (oxidized in one active site only, ∼33%) species are also present (Appenzeller-Herzog and Ellgaard, 2008a).
Here, we describe the molecular basis of the OX1–OX2 redox switch in Ero1α that directly regulates enzyme activity in vivo. Furthermore, we show that the activation state of Ero1α is coupled to the availability of reduced PDI, demonstrating the existence of a regulatory feedback loop in ER redox control.
Results
Overexpressed Ero1 variants are active, but elicit only minimal ER stress
Our studies of ER redox regulation by Ero1 necessitated a well-controlled system for ectopic inducible expression of human Ero1α and Ero1β. For this purpose, we used a recombinase-mediated integration system to generate four HEK293-based cell lines. In addition to cells encoding myc- and His-tagged wild-type Ero1α and Ero1β, we created cells expressing the two Ero1α mutants Cys131Ala (C131A) and Cys394Ala (C394A). As described in further detail below, the former cannot build a critical regulatory disulphide, whereas the latter carries a mutation in the inner active site and has been suggested to be a dominant-negative mutant (Mezghrani et al, 2001). Western blot analysis of cells untreated or induced with doxycyclin for 24 and 48 h confirmed doxycyclin-dependent expression of all constructs (Figure 1A). As expected for stable cell clones generated using a system for gene integration at a single genomic location (O'Gorman et al, 1991), the recombinant Ero1 proteins were expressed at similar levels. Compared with the levels of endogenous Ero1α, induction of Ero1α cells for 24 or 48 h led to approximately 20-fold or 30-fold overexpression, respectively (Supplementary Figure S1).
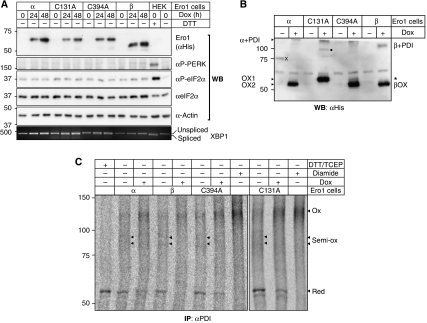
Figure 1.
Characterization of inducible Ero1 cell lines. (A) Ero1α (α), Ero1α-C131A (C131A), Ero1α-C394A (C394A) and Ero1β (β) cells were induced with doxycyclin (Dox) for 0, 24 or 48 h, the cell lysates normalized for protein content, separated by SDS–PAGE under reducing conditions and analysed by western blotting using the indicated antibodies. The distribution of spliced and unspliced XBP1 mRNA was assessed by RT–PCR and analysed on an agarose gel. As positive and negative controls for the phosphorylation of PERK and eIF2α, and the splicing of XBP1 mRNA, HEK293 cells (HEK) were treated or not with the known ER stress inducer DTT (2 mM, 1 h). (B) Ero1 cells were incubated with or without doxycyclin (Dox), treated with NEM to quench post-lysis thiol–disulphide exchange reactions, and the oxidation state of exogenous Ero1 variants was visualized by western blotting using αHis after non-reducing SDS–PAGE. The gel mobilities of monomeric Ero1α variants and Ero1β are indicated as OX1, OX2 and βOX, respectively. α+PDI and β+PDI denote the mixed disulphides between Ero1α/Ero1β and PDI. Note that compared with the endogenous protein (see, e.g., Figure 3A), the epitope-tagged Ero1α proteins migrated slower by SDS–PAGE. Asterisk, background band; X, His-tagged 75 kDa molecular weight marker (leakage from neighbouring lane); filled circle, unidentified mixed-disulphide complex preferentially formed with Ero1α-C131A (our unpublished data). (C) The in vivo redox state of the two active sites in PDI (a and a′) in uninduced (−Dox) versus doxycyclin-induced (+Dox) Ero1 cells (α, β, C394A, C131A) was determined as described previously (Appenzeller-Herzog and Ellgaard, 2008a). After in situ modification of free cysteines with NEM, metabolically labelled PDI was immunoprecipitated, and disulphide-bonded cysteines were reduced and then modified with mPEG-mal. Differentially alkylated PDI was analysed by SDS–PAGE and phosphorimaging. Samples reduced with DTT and TCEP, or oxidized with diamide ahead of NEM-modification, served as mobility markers. Red, both active sites reduced; Ox, both active sites oxidized; Semi-ox, semi-oxidized PDI (owing to the faint appearance of the two semi-oxidized forms mainly present in the uninduced lanes, arrowheads mark their mobility in the autoradiograms). Note that mPEG-mal-modified bands are not sharp in appearance because of the large size dispersity of the reagent.
Overexpression of Ero1 holds the potential to increase the cellular load of ROS (Harding et al, 2003; Haynes et al, 2004). In higher eukaryotes, such ER-derived oxidative stress is counteracted by activation of the ER kinase PERK that triggers two signalling pathways—Nrf2 (Cullinan and Diehl, 2004) and eIF2α (Harding et al, 2003)—both leading to an increase in the cellular antioxidant capacity through increased glutathione synthesis. We therefore asked if overexpression of Ero1 in our cell lines activated PERK signalling. Lysates of induced Ero1 cells were analysed by western blotting using antibodies specific for activated (phosphorylated) PERK and eIF2α. In none of the Ero1-expressing cell lines were we able to detect increased phosphorylation of PERK or eIF2α (Figure 1A). Consistent with this, we did not measure altered glutathione levels in Ero1-overexpressing cells (see below, Figure 4D). As the splicing of XBP1 mRNA, another hallmark of ER stress signalling, was only weakly observed (Figure 1A), the overexpression of Ero1 in our cell lines just slightly triggered ER stress. Thus, the doxycyclin-inducible system provided a good tool to analyse the effects of Ero1 overexpression.
Similar to transient expression of Ero1 in HeLa cells (Benham et al, 2000; Bertoli et al, 2004; Dias-Gunasekara et al, 2005), the stably transfected Ero1 constructs showed characteristic gel mobilities when analysed under non-reducing conditions (Figure 1B). The wild-type Ero1α and Ero1α-C394A proteins were predominantly confined to the OX2 form (Figure 1B). This was unlike the case for endogenous Ero1α where the relative abundance of Ero1α redox forms varied significantly, or HeLa cells where a considerable fraction of overexpressed Ero1α was found in the OX1 configuration (Supplementary Figure S2; Anelli et al, 2002; Bertoli et al, 2004). As observed previously (Bertoli et al, 2004), Ero1α-C131A was unable to form OX2 and migrated as OX1. In addition to these monomeric forms, all four exogenous Ero1 proteins were found in a mixed-disulphide complex with endogenous PDI (Figure 1B). Importantly, the Ero1 variants clearly demonstrated oxidase activity as judged by their ability to increase the oxidation level of both active sites in PDI (Figure 1C). In a mock cell line stably transfected with empty vector, the redox state of PDI was not affected by doxycyclin treatment (data not shown). The finding that PDI became oxidized by the presumed dominant-negative Ero1α-C394A was surprising. Furthermore, overexpression of Ero1α-C394A showed the same effect as wild-type Ero1α on the redox state of endogenous Ero1α (see below, Figure 3A). These data indicated that Ero1α-C394A does not exert an effect through a dominant-negative, but rather a competitive mechanism (see Supplementary data).
A disulphide bond between Cys94 and Cys131 defines the OX2 configuration of Ero1α
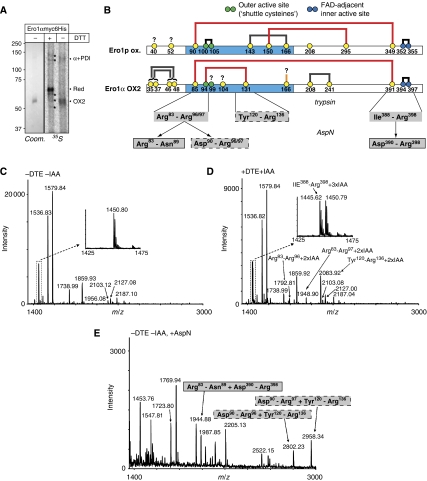
To explore the molecular basis for the OX1–OX2 interconversion in Ero1α, we set out to determine the disulphide-bridge pattern of the protein. We first treated doxycyclin-induced Ero1α cells with the cell-permeable alkylating agent N-ethylmaleimide (NEM) to prevent post-lysis disulphide exchange and then purified His-tagged Ero1α under denaturing conditions. Consistent with the results in Figure 1B, overexpressed Ero1α was trapped predominantly in the OX2 form. Purified Ero1α OX2—some of which had been obtained from cells labelled with 35S-Cys (Figure 2A)—was digested with trypsin, and the resulting peptides were separated by reverse-phase HPLC. Analysis by mass spectrometry (MS) of the HPLC fractions A, B and C containing 35S-labelled material allowed the identification of disulphides in Ero1α OX2 (Figure 2B and Table I).
Figure 2.
The OX2 redox species of Ero1α is molecularly defined by the regulatory Cys94–Cys131 disulphide. (A) Ero1αmyc6His was two-step purified from unlabelled or 35S-Cys-labelled, doxycyclin-induced and NEM-treated Ero1α cells. A Coomassie-stained gel (Coom.) and a phosphorimager scan (35S) from the unlabelled and labelled protein batch, respectively, are shown. Ero1α OX2 was shifted to the reduced (Red) form upon treatment of the sample with DTT. α+PDI, mixed-disulphide complex between Ero1α and PDI; asterisks, background bands. (B) Schematic representation of the cysteine connectivities in oxidized (ox.) Ero1p (Gross et al, 2004) and Ero1α OX2 (this study). The cysteines are shown as yellow, green (outer active site) or blue (inner active site) circles with amino-acid numbering, and disulphides as thick grey (likely structural), black (active site) or red (reported or inferred regulatory function; Sevier et al, 2007 and this study) lines. The thick orange line at Cys166 indicates the connection to a likely (but unidentified) disulphide partner. The question marks denote cysteines with unresolved oxidation state. The flexible loop regions are coloured in light blue. The braces for Cys35/37 and Cys46/48 indicate the two possible connections that could not be distinguished from our data. The three disulphide-linked peptides after trypsin cleavage and their trimming products upon further digestion with AspN are shown in grey boxes where the solid and dashed borders reflect their connectivity. Note that the alternative tryptic cleavage at Arg96 or Arg97 gave rise to two peptides that were both disulphide-linked to the Tyr120–Arg136 peptide (see panel E). (C, D) Mass spectra of HPLC fraction B before (C) and after (D) reduction and alkylation with DTE and IAA (±DTE±IAA), respectively. The monoisotropic masses at m/z 1445.62 (insets), 1792.81, 1948.90 and 2083.92 appear only in the reduced and alkylated sample and correspond to the four IAA-modified peptides labelled in the spectrum (see also Table I). (E) The monoisotropic masses at m/z 1944.88, 2802.83 and 2958.34 upon AspN digestion of fraction B correspond to the indicated disulphide-linked di-peptides (see also Table I). Bordering patterns of boxes match those in panel B.
Table 1.
Mapping of disulphide bonds in Ero1α
| Fractionsa | Treatmentsb | Peptidesc | Cysteinesd | Observed masse (MH+) | Mass differencef (MH+) | |
|---|---|---|---|---|---|---|
| DTE+IAA | Second digest | |||||
| A | − | − | Cys35–Arg55 | 35, 37, 46, 48 | 2380.99 | −3.95 (2 S-S) |
| + | − | Cys35–Arg55 | 35, 37, 46, 48 | 2613.10 | 228.16 (4 IAA) | |
| − | AspN | Cys35–Asp44+Asp45–Cys48 | 35, 37, 46, 48 | 1570.54 | −4.01 (2 S-S) | |
| B | + | − | Arg83–Arg96 | 85, 94 | 1792.81 | 114.06 (2 IAA) |
| Arg83–Arg97 | 85, 94 | 1948.90 | 114.05 (2 IAA) | |||
| Tyr120–Arg136 | 131 | 2083.92 | 57.06 (1 IAA) | |||
| Ile388–Arg398 | 391, 394, 397 | 1445.62 | 171.08 (3 IAA) | |||
| − | AspN | Arg83–Asn89+Asp390–Arg398 | 85, 391, 394, 397 | 1944.88 | −3.96 (2 S-S) | |
| Asp90–Arg96+Tyr120–Arg136 | 94, 131 | 2802.23 | −1.97 (1 S-S) | |||
| Asp90–Arg97+Tyr120–Arg136 | 94, 131 | 2958.34 | −1.97 (1 S-S) | |||
| C | + | − | His158–Arg187 | 166 | 3493.53 | 57.06 (1 IAA) |
| Ile199–Lys215 | 208 | 2182.12 | 57.05 (1 IAA) | |||
| Arg216–Lys244 | 241 | 3282.52 | 56.98 (1 IAA) | |||
| Arg216–Arg245 | 241 | 3438.50 | 56.85 (1 IAA) | |||
| − | GluC | Ala176–Arg187 | 1431.74 | |||
| Tyr178–Arg187 | 1231.68 | |||||
| Ile199–Glu206 | 1065.48 | |||||
| Arg216–Glu230 | 1555.70 | |||||
| Asn231–Glu238 | 1059.48 | |||||
| Asn207–Lys215+Gly239–Glu243 | 208, 241 | 1595.75 | −2.05 (1 S-S) | |||
| aFractions from the tryptic digest of Ero1α identified to contain disulphide-linked peptides. | ||||||
| bThe fractions were analysed by MS before and after reduction with DTE followed by alkylation with IAA. In some cases, the samples were further digested with endoproteinase AspN or endoproteinase GluC. | ||||||
| cPeptides identified by MALDI-TOF-MS. | ||||||
| dPositions of cysteines contained in the identified peptides. | ||||||
| eObserved monoisotopic molecular masses determined by MALDI-TOF-MS. | ||||||
| fDifference between observed and calculated masses. The number and type of modifications corresponding to the mass difference are given in parentheses: disulphide bridge (S–S) and alkylation with iodoacetamide (IAA). | ||||||
Reduction and alkylation of the peptides in fraction B revealed peaks with masses corresponding to the peptides Arg83–Arg96/97 (containing Cys85 and Cys94), Ile388–Arg398 (containing Cys391, Cys394 and Cys397) and Tyr120–Arg136 (containing Cys131) (Figure 2D), which were not detected in the non-reduced fraction (Figure 2C). This suggested the existence of a disulphide-linked tri-peptide. Indeed, further digestion of the non-reduced fraction with endopeptidase AspN, which separated Cys85 and Cys94 by cleaving at Asp90, followed by MS confirmed this notion (Table I). Specifically, under these conditions, we detected three peaks (Figure 2E) that demonstrated the presence of disulphides between the peptides comprising Cys85 and Cys391/Cys394/Cys397, and Cys94 and Cys131, respectively (Figure 2B). In agreement with the presence of the Cys94–Cys131 disulphide in Ero1α OX2, both Ero1α mutants C131A (Figure 1B) and C94S are unable to form OX2 (Bertoli et al, 2004). By comparison with Ero1p where one regulatory disulphide is formed by Cys90 and Cys349 (Figure 2B), the long-range disulphide bridge involving Cys85 was most likely to Cys391, one of three cysteines in the Ile388–Arg398 peptide. The internal disulphide detected in this peptide (Table I) would then be between Cys394 and Cys397 and represent the oxidized state of the inner active site. Additional disulphides were identified between Cys35 and Cys46 or Cys48, Cys37 and Cys46 or Cys48, and Cys208 and Cys241 (fractions A+C, Table I). Finally, the peptide comprising Cys166 was not detected in an NEM-modified state, but only upon reduction and alkylation of fraction C (Table I). Although this suggested it to be disulphide-bonded, we did not identify a potential partner. No additional Cys-containing Ero1α peptides were detected in the MS analysis, suggesting that the disulphide pattern depicted in Figure 2B represents the situation in the large majority of protein molecules.
Cys131 competes with PDI for disulphide exchange with Cys94–Cys99
Ero1-catalysed oxidation of PDI is initiated through the nucleophilic attack by a deprotonated active-site cysteine in PDI on the disulphide bond between the Ero1 shuttle cysteines (Cys94–Cys99 in Ero1α; Figure 2B). Consequently, a transient mixed-disulphide intermediate is formed between PDI and Cys94. The present identification of the intramolecular Cys94–Cys131 disulphide bond that defines Ero1α OX2 suggested an alternative reaction pathway. Here, a Cys131-derived thiolate anion would react with the Cys94–Cys99 active-site disulphide to form Cys94–Cys131 and thus inactivate the enzyme.
To investigate the hypothesis that the conserved Cys131 (Supplementary Figure S3) competes with reduced PDI, we examined if manipulation of substrate availability influenced the relative levels of OX1 and OX2. First, we reasoned that overexpression of Ero1 would decrease the abundance of reduced PDI (see Figure 1C) and thereby lower the substrate level for endogenous Ero1α. We therefore studied the redox state of endogenous Ero1α upon overexpression of exogenous Ero1. In this experiment, exogenous Ero1α variants were depleted through the His-tag after cell lysis (Supplementary Figure S4), allowing detection of the endogenous protein only by subsequent western blotting. Indeed, the mixed-disulphide complex between endogenous Ero1α and PDI became less abundant when Ero1 was overexpressed (Figure 3A). Concomitantly, we observed a moderate, but clear and reproducible (see Supplementary Figure S5) decrease in the OX1/OX2 ratio of endogenous Ero1α. These effects were seen for all four constructs. Importantly, the OX2 shifts observed upon overexpression of Ero1 variants were not a result of general ER hyperoxidation, as the overexpression of Ero1α did not perturb the GSSG/GSH ratio (see below, Figure 4).
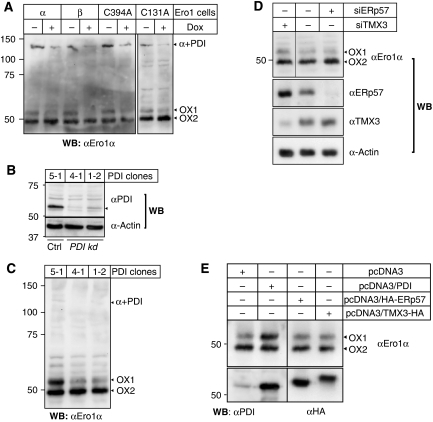
Figure 3.
The redox state of Cys94–Cys131 is modulated by the availability of PDI. (A) Uninduced (−Dox) or induced (+Dox) Ero1 cells (α, C131A, C394A, β) were treated with NEM, solubilized in lysis buffer and the lysates were depleted of exogenous, His-tagged Ero1α using TALON beads. The supernatant was subjected to precipitation with ConA-sepharose, and the glycoprotein fraction was analysed by western botting using αEro1α following non-reducing SDS–PAGE. Stripping and reprobing the same membrane with αPDI identified the ∼130 kDa band as the mixed-disulphide complex between endogenous Ero1α and PDI (α+PDI, data not shown). (B) Lysates of a control (Ctrl) shRNA clone (5-1) and two PDI knockdown (kd) clones (4-1 and 1-2) (Ou and Silver, 2006) were analysed by western blotting using αPDI to monitor knockdown efficiency and α-Actin as loading control. (C) 5-1, 4-1 and 1-2 cells were treated with NEM, lysed and the ConA-sepharose-precipitated glycoprotein fraction was subjected to αEro1α western blot analysis. The three independent samples were not normalized for protein content, meaning that no conclusions about the relative abundance of Ero1α between lanes can be made. (D) HeLa cells left untreated or treated with siRNAs against TMX3 (siTMX3) or ERp57 (siERp57) were harvested and lysed in the presence of NEM. Lysates were directly analysed either by western blotting with the indicated antibodies or, for the detection of Ero1α, after ConA-precipitation to concentrate the protein. Similar results were obtained with HEK293 cells (not shown). The hairline indicates where a lane has been removed. (E) HEK293 cells were transfected with the indicated cDNAs, harvested and lysed in the presence of NEM. Lysates were either directly subjected to western blot analysis using αPDI and αHA (lower panels) or incubated with ConA-sepharose to concentrate the glycoprotein fraction ahead of western blot detection of endogenous Ero1α (upper panel).
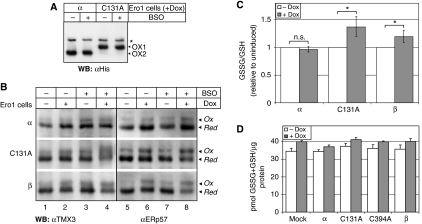
Figure 4.
Increased in vivo activity of Ero1α-C131A and Ero1β is buffered by GSH. (A) Ero1α and C131A cells were induced (+Dox), treated with or without BSO and processed in the presence of NEM before αHis western blot analysis. (B) Ero1 cells were treated with doxycyclin (Dox) and/or BSO as indicated, and the in vivo redox states of TMX3 and ERp57 trapped by differential alkylation with NEM and AMS, followed by western blotting. Treatment with BSO reduced GSSG+GSH to 20% of control (data not shown). The mobilities of the reduced (Red) and oxidized (Ox) species, as verified by control samples using lysates from DTT- or diamide-treated cells (not shown), are indicated. Note that the mobility shift of the ERp57 band reflects the oxidation state of the a′ domain (Supplementary Figure S6). The results are representative of three independent experiments. (C) Cellular GSSG levels increase upon overexpression of Ero1α-C131A or Ero1β. Ero1 cells (α, C131A, β) were left untreated or induced with doxycyclin for 24 h and analysed for the intracellular levels of GSSG and GSH. The relative changes of GSSG/GSH caused by Ero1 overexpression along with 95% confidence intervals as calculated by using a linear model to correct for variation between experimental batches are plotted (n=12; for the raw data, refer to Supplementary Figure S7). Asterisks indicate statistical significance (P<0.03). n.s., not significant. (D) GSSG+GSH and total protein was determined in samples obtained from the indicated cell lines with (+Dox) or without (−Dox) doxycyclin induction for 24 h. GSSG+GSH (in pmol) was normalized to protein content (in μg) (mean±s.d., n=3).
We next used two different HeLa-derived cell lines stably transfected with a PDI-specific short hairpin RNA (shRNA) (Ou and Silver, 2006) to study a potential influence of PDI knockdown on the redox state of Ero1α (Figure 3B). Under these conditions of decreased substrate level, the OX1/OX2 ratio of endogenous Ero1α was clearly diminished and less covalent Ero1α–PDI complex was present (Figure 3C). Importantly, cells treated with siRNA against ERp57, a close homologue of PDI, or a transmembrane PDI-family member called TMX3 (Haugstetter et al, 2005) did not exhibit an OX2 shift in Ero1α (Figure 3D).
In contrast to the above-mentioned data, overexpression of PDI (but not of ERp57 or TMX3) affected the activation state of endogenous Ero1α so that the OX1/OX2 ratio was now increased (Figure 3E). The redox state of exogenous Ero1α responded to co-expression of PDI in a similar way (Otsu et al, 2006; and data not shown). We concluded that the Cys131-linked activation state of Ero1α is governed by the availability of its own substrate, PDI.
The OX2 switch in Ero1α contributes to ER redox homoeostasis
We next characterized the OX2-impaired C131A mutant of Ero1α. Yeast cells expressing the Ero1p-C150A-C295A mutant that lacks one regulatory disulphide and also does not form the other under normal cellular conditions exhibit a severe growth defect caused by Ero1p hyperactivity (Sevier et al, 2007). In contrast, induced and uninduced Ero1α-C131A cells proliferated equally well (our unpublished observation). Treatment of yeast cells with L-buthionine-sulphoximine (BSO), an inhibitor of GSH synthesis, leads to oxidative inactivation of Ero1p-C150A-C295A through the formation of the second regulatory disulphide Cys90-Cys349 (corresponding to Cys85–Cys391 in Ero1α) (Sevier et al, 2007). However, Ero1α-C131A in HEK293 cells remained in the partially oxidized OX1 configuration irrespective of BSO-mediated depletion of the cellular GSH pool (Figure 4A). These results suggested that in Ero1α-C131A, unlike the case in Ero1p-C150A-C295A, Cys85–Cys391 remained oxidized under steady-state ER redox conditions.
To more closely monitor the effect of exogenous Ero1 variants on ER redox homoeostasis, we assayed the redox distribution of ER-resident oxidoreductases and GSH in Ero1-expressing cells. We first examined the in vivo redox state of the PDI homologues ERp57 and TMX3 by means of a 4-acetamido-4′-maleimidylstilbene-2,2′-disulphonic acid (AMS) alkylation assay that results in a decreased electrophoretic mobility of oxidized species (Supplementary Figure S6). Overexpression of Ero1α did not cause any clear effect on the redox state of the two oxidoreductases (Figure 4B; top panel, compare lanes 1 and 2, and 5 and 6). Interestingly, however, overexpression of Ero1α-C131A or Ero1β, which also exhibits only one oxidized species (Dias-Gunasekara et al, 2005), caused a moderate oxidative shift in the redox state of both TMX3 and ERp57 (Figure 4B; middle and bottom panel, compare lanes 1 and 2, and lanes 5 and 6). As documented in the Supplementary data, for ERp57, the AMS assay most clearly monitors the redox state of the a′ domain (Supplementary Figure S6). Moreover, depletion of glutathione by treatment with BSO aggravated the oxidative phenotypes of Ero1α-C131A- and Ero1β-overexpressing cells (Figure 4B; middle and bottom panel, compare lanes 2 and 4, and 6 and 8). Thus, in vivo, the activity of Ero1α is counterbalanced by the glutathione redox buffer and negatively regulated by Cys131-mediated inactivation.
The oxidative effect on TMX3 and ERp57 caused by overexpression of Ero1α-C131A and Ero1β may have resulted from moderate, direct oxidation of these PDI-like proteins and/or from oxidation through the glutathione redox buffer. To test if overexpression of Ero1 variants affected the overall ER thiol–disulphide homoeostasis, we determined the ratio of oxidized to reduced glutathione (GSSG/GSH) in Ero1-expressing and control cells. As GSSG is hardly present in the cytosol, mitochondria and nuclei (Go and Jones, 2008), and as the ER by far is the largest organelle of the secretory pathway in mammalian cells (De Menezes et al, 1996), it can be assumed that most GSSG detected in an assay on whole cells is present in the ER. Treatment of Ero1α-C131A and Ero1β cells with doxycyclin significantly increased GSSG/GSH, whereas ectopic expression of wild-type Ero1α had no effect (Figure 4C and Supplementary Figure S7). These differences could potentially be a result of Ero1 construct-specific changes in the total glutathione concentration (GSSG+GSH). Although doxycyclin treatment per se produced a marginal increase in GSSG+GSH (Figure 4D, mock cells), none of the Ero1 proteins further modulated cellular glutathione levels when overexpressed (Figure 4D). These results differ from the situation in transiently transfected HeLa cells (Molteni et al, 2004). The effects on GSSG/GSH suggested that the Ero1-dependent oxidative shifts of TMX3 and ERp57 were at least in part caused by a change in the overall ER thiol–disulphide homoeostasis. Taken together, the overexpression of Ero1 variants lacking an inactivated OX2 form affected the glutathione-buffered ER redox homoeostasis in the direction of more oxidizing conditions.
Discussion
A central feedback loop for redox control in the ER of human cells
The results presented here provide critical mechanistic insight into ER redox control in human cells. The most oxidized redox form of Ero1α, OX2, represents a negatively regulated species. The silencing mechanism is straightforward and direct—the formation of the Cys94–Cys131 regulatory disulphide bridge covalently blocks the catalytically essential, first shuttle cysteine, Cys94. Owing to competition between Cys131 and substrate thiols for reaction with the active-site Cys94–Cys99 disulphide, the availability of substrate would predictably regulate the formation of Cys94–Cys131. Indeed, the manipulation of both the redox state (Figure 3A) and levels (Figure 3C and E) of the Ero1α substrate PDI affected the extent of Cys131-mediated shutdown that is given by the OX1/OX2 ratio.
Protein disulphide isomerase fulfils a central position in ER thiol–disulphide homoeostasis by passing on Ero1-generated oxidizing equivalents to the ER thiol pool. Our data suggest that Ero1α is designed to adapt its activation state to the redox distribution of PDI through a direct feedback loop (Figure 5). Consequently, Ero1α-mediated generation of disulphides will be stalled by feedback inhibition when, under oxidizing ER conditions, PDI accumulates in the oxidized form. Under reducing conditions, however, PDI active-site cysteines in the reduced state will be abundant as substrates for Ero1α. By interacting with Ero1α in the OX1 state to reduce Cys94–Cys99, reduced PDI will disfavour Cys131-mediated shutdown. At the same time, overexpression of a PDI variant devoid of all four active-site cysteines also leads to some OX2-to-OX1 conversion in Ero1α (Otsu et al, 2006), indicating that this mutant binds and stabilizes OX1 as well. We suggest that the PDI-Ero1α feedback loop relies on both thiol–disulphide exchange and a specific binding of PDI to Ero1α. Accordingly, overexpression or downregulation of the PDI-related oxidoreductases TMX3 and ERp57 did not influence the OX1/OX2 ratio in Ero1α.
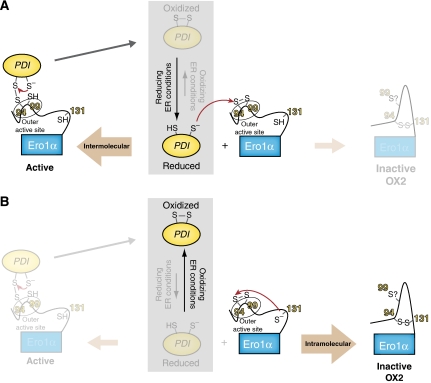
Figure 5.
Model for homoeostatic control of the PDI redox state by Cys131-dependent feedback regulation of Ero1α. The thiol–disulphide composition in the ER depends on the load of nascent polypeptides—many of which are substrates for PDI-mediated oxidation—as well as on the redox distribution of ER-localized glutathione. As both of these terms vary under physiological conditions, the redox state of the thiol–disulphide system in the ER can fluctuate (reducing versus oxidizing ER conditions). As detailed in the text, such fluctuations are likely mirrored in the steady-state distribution between oxidized and reduced PDI (grey box). Panels (A) and (B) depict the two extreme cases where the outer active-site disulphide in Ero1α is exclusively attacked by a PDI- or Cys131-derived thiolate anion. In the cell, these two reaction pathways compete with each other. (A) Under reducing ER conditions, PDI (for simplicity, only one active site in PDI is depicted) accumulates in the reduced form. Reduced PDI is a substrate for oxidation by Ero1α, which is initiated through the intermolecular nucleophilic attack by an active-site cysteine in PDI on Cys94 in the oxidized outer active site of Ero1α. The competing intramolecular reaction of Cys131 with the Cys94–Cys99 disulphide is disfavoured by the abundance of reduced PDI. A thiol–disulphide exchange reaction between Ero1α (in the active form) and PDI then results in the formation of oxidized PDI. By this mechanism, the activity of Ero1α counterbalances reducing ER conditions. (B) Under oxidizing ER conditions, PDI is predominantly oxidized and the outer active-site disulphide of Ero1α will, therefore, preferentially react with Cys131. The resulting OX2 configuration of Ero1α is inactive because the outer active Cys94 is covalently blocked by Cys131. The mechanism of reactivation of Ero1α OX2 remains unclear. In principle, it could involve a nucleophilic attack of Cys99 on Cys94 to resolve Cys94–Cys131, but the oxidation state of Cys99 in OX2 is not known (indicated by the question mark).
In most experiments, we observed OX1/OX2 ratios lower than one (Figure 3A, C and E and Supplementary Figure S2, lower panel). However, it is worth noting that the in vivo redox distribution of endogenous Ero1α is variable under normal cellular conditions, presumably reflecting physiological fluctuations in the thiol–disulphide household of the ER (Supplementary Figure S2). Still, in all our experiments and regardless of the thiol-trapping method used (NEM or rapid acidification by trichloroacetic acid), endogenous Ero1α-OX2 was prominently detected. Mammalian cells apparently maintain a considerable pool of inactive molecules that can promptly be activated upon need. Indeed, ER reoxidation after DTT treatment of cells is in the time scale of seconds and further accelerated by overexpression of Ero1α (our unpublished observations).
An intact OX2 switch is required for ER redox homoeostasis
Our results showed that despite the higher relative abundance of oxidized PDI upon overexpression of Ero1α (Figure 1C), the general ER redox state did not measurably change (Figure 4B and C). This homoeostatic stability depended on an intact OX2 switch in overexpressed Ero1α that was absent in Ero1α-C131A, and apparently also Ero1β (Figure 4B and C). The latter finding fits well with the observation that Ero1β does not form a redox form similar to OX2 (Dias-Gunasekara et al, 2005), although a cysteine at the position equivalent of Ero1α Cys131 is conserved in Ero1β. In contrast to Ero1α, however, Ero1β harbours three N-glycosylation sites proximal to that cysteine, two of which have been experimentally verified (Lewandrowski et al, 2006). Although speculative at present, it is possible that a conformation similar to OX2 in Ero1β is sterically disfavoured by these N-glycans. Ero1β also harbours an additional cysteine that is not conserved in Ero1α. Overall, a possible disulphide-based regulatory mechanism of Ero1β activity awaits further clarification.
The mechanism by which overexpression of Ero1α-C131A and Ero1β perturb the ER redox balance is unclear. However, the effect may arise from faster oxidation kinetics and an increased rate of disulphide generation that is not visible in the steady-state readout of the PDI redox state where these two proteins showed an effect similar to Ero1α (Figure 1C). In support of this, the deregulated Ero1p-C150A-C295A mutant shows increased reaction kinetics in vitro (Sevier et al, 2007).
Glutathione counterbalances the activity of Ero1
Apart from the OX2 regulatory feedback mechanism, it has previously been shown that a proper ER redox balance critically depends on GSH (Cuozzo and Kaiser, 1999; Chakravarthi and Bulleid, 2004; Molteni et al, 2004). Accordingly, the redox states of ERp57 and TMX3 showed more prominent oxidative shifts upon Ero1α-C131A or Ero1β overexpression when cellular glutathione levels were diminished by treatment with BSO (Figure 4B). Although we cannot exclude the possibility that TMX3 and/or ERp57 were also directly oxidized by Ero1α-C131A and Ero1β, the Ero1-dependent oxidative shifts in these two proteins were clearly less prominent than those observed in PDI (Figure 1C). Therefore, the shifts in TMX3 and ERp57 are probably mainly promoted by the increase in GSSG/GSH (Figure 4C). Unfortunately, our assay did not allow us to accurately determine Ero1-dependent changes in GSSG/GSH at the low levels of glutathione present in BSO-treated cells.
Unique structural and functional features of Ero1α OX2
The MS analysis of Ero1α OX2 revealed several distinguishing features compared with the oxidized form of Ero1p. Most intriguingly, a disulphide corresponding to the long-range regulatory Cys150–Cys295 in Ero1p, which connects the extended loop with the core, is missing in Ero1α OX2 that instead contains the Cys94–Cys131 regulatory disulphide (Figure 2B). This is positioned in the extended loop, with Cys131 reaching directly into the outer active site. As a consequence, the regulatory mechanisms are clearly different between the two enzymes. Structurally, the constraint placed by the Cys94–Cys131 disulphide on the backbone conformation of the extended loop in Ero1α must cause the structure of this region to differ significantly from the one in oxidized Ero1p.
The position of the second regulatory disulphide in Ero1p is conserved in Ero1α (Cys85–Cys391; Figure 2B), but it does not appear to be equally regulated—in Ero1α-C131A, unlike the case in Ero1p-C150A-C295A (Sevier et al, 2007), the disulphide resisted GSH-mediated reduction in vivo. On the basis of the sequence similarity, we consider it most likely that Cys85–Cys391 constitutes a regulatory disulphide in Ero1α as well that, however, seems to be more stable in the ER than its counterpart in Ero1p. Unfortunately, the potential functional conservation of Cys85–Cys391 will be difficult to test in vivo, as both Ero1α-C85S and Ero1α-C391A—presumably owing to folding defects when expressed in cultured cells—display reduced oxidase activity, a gel migration pattern different from wild-type Ero1α, and the formation of high-molecular weight complexes with PDI unlike the normal one at ∼130 kDa (Bertoli et al, 2004).
The data identified three additional disulphides. Two connect the two N-terminal Cys-Xaa-Cys clusters—as an N-terminally truncated version of Ero1p was used for structural studies, the oxidation state of the Cys40/Cys52 couple in Ero1p is not known (Figure 2B). On the basis of sequence alignment and homology modelling, the Cys208–Cys241 bond is expected to be structural and connect two antiparallel helices of the core structure. Although Cys208 is conserved, the equivalent bond is missing in Ero1p that does not contain a cysteine corresponding to Cys241. We did not recover the tryptic peptide containing Cys99 and Cys104. It is tempting to speculate, however, that in Ero1α OX2 where Cys131 blocks Cys94 (the partner of Cys99 in the active enzyme), Cys99 forms a disulphide with the close-by Cys104. However, at present, we cannot rule out that either Cys99 or Cys104 is connected to Cys166, the presumed disulphide partner of which was not detected in our MS analysis.
Conclusion
On the basis of the identification of a crucial regulatory disulphide bond in human Ero1α, we show the existence of a feedback mechanism where the activation state of this ER oxidase is controlled by the redox state and availability of its substrate, PDI. This places PDI as a central regulator of ER lumenal redox homoeostasis in human cells.
A principle conclusion of this study is that human Ero1α differs significantly from Ero1p. In addition, the redox distribution of PDI is significantly more oxidizing in yeast than in HEK293 cells (Frand and Kaiser, 2000; Appenzeller-Herzog and Ellgaard, 2008a). Despite these differences, it is quite possible, on the basis of the conservation of Ero1 and PDI, that yeast PDI has an important function in regulating the redox state of Ero1p. These details of the yeast system are, however, presently unknown. Clearly, more experiments are required to clarify the exact mechanisms that govern the activation of Ero1p, Ero1α and Ero1β, as well as redox differences between various cell types. Moreover, in mammalian cells, other redox enzymes such as the QSOX oxidases (Thorpe and Coppock, 2007) or ER-resident PDI peroxidases (L Ruddock, personal communication) may have important, as yet unrecognized functions in ER redox homoeostasis.
It is interesting to consider how the ER redox system has evolved to deal with the inherent danger of hyperoxidation. In vitro, PDI is a relatively poor substrate of Ero1p compared with the much more reducing thioredoxin (Sevier et al, 2007). It thus appears that the ER redox system has evolved to carefully regulate redox conditions at the expense of optimal catalytic activity.
Materials and methods
Recombinant DNA
DNA work was performed as detailed in the Supplementary data. pcDNA3/PDI was a gift from I Braakman, Utrecht, pcDNA3/HA-ERp57 from R Sitia, Milan, and pcDNA3/TMX3-HA has been described previously (Haugstetter et al, 2005).
Cell lines, transfection and antibodies
HEK293 and HeLa cells were grown in α-minimal essential medium (Invitrogen), supplemented with 10% fetal calf serum at 37°C, 5% CO2. Doxycyclin-inducible cell lines were generated by transfecting pcDNA5/Ero1αmyc6his, pcDNA5/Ero1α-C131Amyc6his, pcDNA5/Ero1α-C394Amyc6his and pcDNA5/Ero1βmyc6his into Flp-In TRex-293 cells (Invitrogen) using the CaPO4 method and selecting for stably transfected clones with 100 μg/ml hygromycin B and 15 μg/ml blasticidin (both Invitrogen). Ero1-expressing cells were grown in the presence of these antibiotics. Ero1 expression was induced for 24 h (unless otherwise stated) using 1 μg/ml doxycyclin (Sigma). BSO (1 mM) was added for 20 h. PDI shRNA clones 5-1, 4-1 and 1-2 (Ou and Silver, 2006), a gift from W Ou, Bethesda, were maintained in the presence of 100 μg/ml hygromycin B. Transient transfections were performed with Lipofectamine 2000 (Invitrogen). For the knockdown of ERp57 and TMX3, cells were transfected with the Hs_GRP58_5 HP (10 nM) and Hs_TXNDC10_7 HP (20 nM) siRNAs (both from Qiagen) using Lipofectamine RNAiMAX (Invitrogen) and analysed 72 h post-transfection.
The following mouse monoclonal antibodies were used: Tetra-His (αHis, Qiagen), HA.11 (αHA, Covance), 9E10 (αmyc, Covance), AC-15 (α-actin, Sigma). The rabbit polyclonal antisera used were as follows: D5 (αEro1α, a gift from I Braakman, Utrecht), SPA-890 (αPDI, Stressgen), αTMX3 (Haugstetter et al, 2005), αERp57 (a gift from A Helenius, Zürich), αP-PERK (Santa Cruz Biotechnology), αP-eIF2α and αeIF2α (both Cell Signaling Technology).
Purification of recombinant Ero1α
Ero1α cells grown in twenty 175 cm2 T-flasks to ∼50% confluency and induced with 1 μg/ml doxycyclin for 48 h were washed once with ice-cold PBS containing 20 mM NEM and incubated with the same buffer for 20 min at 4°C. Cells were then lysed for 1 h at 4°C in 10 ml lysis buffer (100 mM NaPO4, pH 8.0, 1% TX-100, 20 mM NEM, 0.2 mM phenylmethylsulphonylfluoride) per T-flask and the lysates cleared by centrifugation (100 000 g, 4°C, 30 min). A total of 50 μl ConA (Concanavalin A)-Sepharose 4B (Sigma) was added per 10 ml of lysate, and the bead suspension was incubated for 1 h at 4 °C under constant agitation. After precipitation, the beads were washed 4 × in lysis buffer and 1 × in the same buffer lacking TX-100. Bound (glyco)proteins were eluted by two consecutive incubations in 5 ml (per 50 μl ConA beads) denaturation buffer (50 mM NaPO4, pH 7.0, 300 mM NaCl, 20 mM NEM, 8 M urea) for 1.5 h each at room temperature. The pooled eluate was loaded onto a column packed with 0.5 ml TALON affinity resin (Clontech) and recirculated over the column overnight at room temperature using a peristaltic pump. After washing with 300 bead volumes of denaturation buffer and 20 bead volumes of a 1:1 dilution of the same buffer with H2O (0.5 × denaturation buffer), the bound protein was eluted in 3 × 0.5 ml of 0.5 × denaturation buffer containing 300 mM imidazole. The purified Ero1α protein (∼50 μg) was concentrated using a Biomax-5 Ultrafree-0.5 centrifugal filter device (Millipore) and stored at −20°C. In parallel, Ero1α cells grown in fifteen 10-cm dishes were labelled with 5 mCi 35S-Cys (PerkinElmer) for 24 h in the presence of 1 μg/ml doxycyclin and 2% fetal calf serum and the protein was purified as described above.
Disulphide-bridge mapping
The samples of labelled (∼106 cpm) and unlabelled purified Ero1α were pooled and digested with sequencing-grade modified trypsin (Promega) at 37°C overnight. Tryptic peptides were separated by reverse-phase HPLC on a μRPC C2/C18 PC 2.1/10 column connected to an ÄKTA Explorer (GE Healthcare). The peptides were separated in 0.1% trifluoroacetic acid (TFA) and eluted with a gradient of 80% acetonitrile in 0.1% TFA. Aliquots of each fraction were subjected to liquid scintillation counting to identify 35S-Cys-containing peptides.
For reduction or subdigestion of disulphide bond-containing peptides, 20 μl of HPLC fractions were dried in a vacuum centrifuge. For reduction, the samples were resuspended in 20 μl of 15 mM 1,4-dithioerythritol (DTE) in 50 mM ammonium bicarbonate and incubated at 56°C for 45 min. Free cysteines were then alkylated by addition of iodoacetamide (IAA, to 50 mM) and incubation for 30 min in the dark at room temperature. Endoproteinase digestions were performed in 20 μl of either 50 mM sodium phosphate (pH 8.0) at 37°C for 18 h (AspN, Roche Diagnostics) or 0.1 M ammonium bicarbonate at 37°C for 3.5 h (GluC, Sigma) using 120 ng of each enzyme.
For mass spectrometric analyses using a Voyager DE-PRO MALDI-TOF mass spectrometer (Applied Biosystems), peptide samples were mixed with a saturated solution of 2,5-dihydroxybenzoic acid (LaserBio Labs) in a 1:1 ratio directly on the target probe. All spectra were obtained in positive reflector-ion mode using a nitrogen laser at 337 nm and an acceleration voltage of 20 kV. Typically, 50–100 laser shots were added per spectrum and calibrated with external standards. The theoretical peptide masses were calculated using the GPMAW program (Lighthouse Data).
Sample preparation, AMS and mPEG-mal modification
N-ethylmaleimide-alkylated TX-100 lysates for ConA- or TALON-precipitation were obtained as described for the purification of Ero1α. Depletion of His-tagged Ero1 was performed by repeated incubation of the lysate with TALON-beads at 4°C on an end-over-end rotator. Reduced and oxidized control lysates were obtained from cells treated with 10 mM DTT or 5 mM diamide (both from Sigma) for 5 min at 37°C in full-growth medium. AMS (Invitrogen) and mPEG-mal (methoxy polyethylene glycol 5000 maleimide, Laysan Bio) modification protocols have been described previously (Appenzeller-Herzog and Ellgaard, 2008a).
Analysis of XBP1 splicing
The splicing of XBP1 mRNA was analysed as described by Yoshida et al (2001).
GSH and protein measurements
A total of 5 × 106 cells per dish were seeded into 10-cm dishes 1 day before treatment with or without doxycyclin for 24 h. Cells were washed twice with PBS and lysed in 400 μl of 1% sulphosalicylic acid. The lysate was cleared by centrifugation (20 000 g, 5 min) and analysed for GSSG+GSH and GSSG (expressed as GSH equivalents) as described in detail elsewhere (Minich et al, 2006). Briefly, GSSG+GSH was measured directly with a 5,5′-dithiobis(2-nitrobenzoic acid)GSH reductase recycling assay, whereas for the determination of GSSG, the lysate was treated with 2-vinylpyridine to modify GSH before analysis. Protein levels were determined using the BCA assay (Pierce).
Supplementary Material
Supplementary data
Acknowledgments
We thank Sandra Abel Nielsen and Jaco den Otter for excellent technical assistance, and the Institute of Biochemistry, ETH Zürich, for support. We also thank Roberto Sitia, Ineke Braakman, Jonathan Silver, Ulrike Kutay and Ari Helenius for providing constructs and reagents, Aron Eklund and Klaus Holst for help with statistical analysis, Pascal Benkert and Torsten Schwede for structural modelling, Neil Bulleid for sharing unpublished data and the members of the Ellgaard laboratory and Jacob R. Winther for helpful discussions and critical reading of the manuscript. Funding obtained from the Swiss National Science Foundation, the Böhringer Ingelheim Foundation, the Novartis Stiftung, Carlsbergfondet and Novo Nordisk Fonden is gratefully acknowledged.
References
- Anelli T, Alessio M, Mezghrani A, Simmen T, Talamo F, Bachi A, Sitia R (2002) ERp44, a novel endoplasmic reticulum folding assistant of the thioredoxin family. EMBO J 21: 835–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appenzeller-Herzog C, Ellgaard L (2008a) In vivo reduction–oxidation state of protein disulfide isomerase: the two active sites independently occur in the reduced and oxidized forms. Antioxid Redox Signal 10: 55–64 [DOI] [PubMed] [Google Scholar]
- Appenzeller-Herzog C, Ellgaard L (2008b) The human PDI family: versatility packed into a single fold. Biochim Biophys Acta 1783: 535–548 [DOI] [PubMed] [Google Scholar]
- Benham AM, Cabibbo A, Fassio A, Bulleid N, Sitia R, Braakman I (2000) The CXXCXXC motif determines the folding, structure and stability of human Ero1-Lalpha. EMBO J 19: 4493–4502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertoli G, Simmen T, Anelli T, Molteni SN, Fesce R, Sitia R (2004) Two conserved cysteine triads in human Ero1alpha cooperate for efficient disulfide bond formation in the endoplasmic reticulum. J Biol Chem 279: 30047–30052 [DOI] [PubMed] [Google Scholar]
- Chakravarthi S, Bulleid NJ (2004) Glutathione is required to regulate the formation of native disulfide bonds within proteins entering the secretory pathway. J Biol Chem 279: 39872–39879 [DOI] [PubMed] [Google Scholar]
- Cullinan SB, Diehl JA (2004) PERK-dependent activation of Nrf2 contributes to redox homeostasis and cell survival following endoplasmic reticulum stress. J Biol Chem 279: 20108–20117 [DOI] [PubMed] [Google Scholar]
- Cuozzo JW, Kaiser CA (1999) Competition between glutathione and protein thiols for disulphide-bond formation. Nat Cell Biol 1: 130–135 [DOI] [PubMed] [Google Scholar]
- De Menezes Y, De Faria FP, Sesso A (1996) In human hepatocellular carcinoma cells the total membrane surface area of each major organelle is a particular allometric function of the cytoplasmic volume. A morphometric study. J Submicrosc Cytol Pathol 28: 573–582 [PubMed] [Google Scholar]
- Dias-Gunasekara S, Gubbens J, van Lith M, Dunne C, Williams JA, Kataky R, Scoones D, Lapthorn A, Bulleid NJ, Benham AM (2005) Tissue-specific expression and dimerization of the endoplasmic reticulum oxidoreductase Ero1beta. J Biol Chem 280: 33066–33075 [DOI] [PubMed] [Google Scholar]
- Frand AR, Kaiser CA (2000) Two pairs of conserved cysteines are required for the oxidative activity of Ero1p in protein disulfide bond formation in the endoplasmic reticulum. Mol Biol Cell 11: 2833–2843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Go YM, Jones DP (2008) Redox compartmentalization in eukaryotic cells. Biochim Biophys Acta 1780: 1273–1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross E, Kastner DB, Kaiser CA, Fass D (2004) Structure of Ero1p, source of disulfide bonds for oxidative protein folding in the cell. Cell 117: 601–610 [DOI] [PubMed] [Google Scholar]
- Gross E, Sevier CS, Heldman N, Vitu E, Bentzur M, Kaiser CA, Thorpe C, Fass D (2006) Generating disulfides enzymatically: reaction products and electron acceptors of the endoplasmic reticulum thiol oxidase Ero1p. Proc Natl Acad Sci USA 103: 299–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, Sadri N, Yun C, Popko B, Paules R, Stojdl DF, Bell JC, Hettmann T, Leiden JM, Ron D (2003) An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell 11: 619–633 [DOI] [PubMed] [Google Scholar]
- Haugstetter J, Blicher T, Ellgaard L (2005) Identification and characterization of a novel thioredoxin-related transmembrane protein of the endoplasmic reticulum. J Biol Chem 280: 8371–8380 [DOI] [PubMed] [Google Scholar]
- Haynes CM, Titus EA, Cooper AA (2004) Degradation of misfolded proteins prevents ER-derived oxidative stress and cell death. Mol Cell 15: 767–776 [DOI] [PubMed] [Google Scholar]
- Lewandrowski U, Moebius J, Walter U, Sickmann A (2006) Elucidation of N-glycosylation sites on human platelet proteins: a glycoproteomic approach. Mol Cell Proteomics 5: 226–233 [DOI] [PubMed] [Google Scholar]
- Marciniak SJ, Yun CY, Oyadomari S, Novoa I, Zhang Y, Jungreis R, Nagata K, Harding HP, Ron D (2004) CHOP induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Genes Dev 18: 3066–3077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mezghrani A, Fassio A, Benham A, Simmen T, Braakman I, Sitia R (2001) Manipulation of oxidative protein folding and PDI redox state in mammalian cells. EMBO J 20: 6288–6296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minich T, Riemer J, Schulz JB, Wielinga P, Wijnholds J, Dringen R (2006) The multidrug resistance protein 1 (Mrp1), but not Mrp5, mediates export of glutathione and glutathione disulfide from brain astrocytes. J Neurochem 97: 373–384 [DOI] [PubMed] [Google Scholar]
- Molteni SN, Fassio A, Ciriolo MR, Filomeni G, Pasqualetto E, Fagioli C, Sitia R (2004) Glutathione limits Ero1-dependent oxidation in the endoplasmic reticulum. J Biol Chem 279: 32667–32673 [DOI] [PubMed] [Google Scholar]
- O'Gorman S, Fox DT, Wahl GM (1991) Recombinase-mediated gene activation and site-specific integration in mammalian cells. Science 251: 1351–1355 [DOI] [PubMed] [Google Scholar]
- Otsu M, Bertoli G, Fagioli C, Guerini-Rocco E, Nerini-Molteni S, Ruffato E, Sitia R (2006) Dynamic retention of Ero1alpha and Ero1beta in the endoplasmic reticulum by interactions with PDI and ERp44. Antioxid Redox Signal 8: 274–282 [DOI] [PubMed] [Google Scholar]
- Ou W, Silver J (2006) Role of protein disulfide isomerase and other thiol-reactive proteins in HIV-1 envelope protein-mediated fusion. Virology 350: 406–417 [DOI] [PubMed] [Google Scholar]
- Pagani M, Fabbri M, Benedetti C, Fassio A, Pilati S, Bulleid NJ, Cabibbo A, Sitia R (2000) Endoplasmic reticulum oxidoreductin 1-lbeta (ERO1-Lbeta), a human gene induced in the course of the unfolded protein response. J Biol Chem 275: 23685–23692 [DOI] [PubMed] [Google Scholar]
- Sevier CS, Kaiser CA (2008) Ero1 and redox homeostasis in the endoplasmic reticulum. Biochim Biophys Acta 1783: 549–556 [DOI] [PubMed] [Google Scholar]
- Sevier CS, Qu H, Heldman N, Gross E, Fass D, Kaiser CA (2007) Modulation of cellular disulfide-bond formation and the ER redox environment by feedback regulation of Ero1. Cell 129: 333–344 [DOI] [PubMed] [Google Scholar]
- Thorpe C, Coppock DL (2007) Generating disulfides in multicellular organisms: emerging roles for a new flavoprotein family. J Biol Chem 282: 13929–13933 [DOI] [PubMed] [Google Scholar]
- Tu BP, Ho-Schleyer SC, Travers KJ, Weissman JS (2000) Biochemical basis of oxidative protein folding in the endoplasmic reticulum. Science 290: 1571–1574 [DOI] [PubMed] [Google Scholar]
- Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K (2001) XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 107: 881–891 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data