Abstract
Centromere that plays a pivotal role in chromosome segregation is composed of repetitive elements in many eukaryotes. Although chromosomal regions containing repeats are the hotspots of rearrangements, little is known about the stability of centromere repeats. Here, by using a minichromosome that has a complete set of centromere sequences, we have developed a fission yeast system to detect gross chromosomal rearrangements (GCRs) that occur spontaneously. Southern and comprehensive genome hybridization analyses of rearranged chromosomes show two types of GCRs: translocation between homologous chromosomes and formation of isochromosomes in which a chromosome arm is replaced by a copy of the other. Remarkably, all the examined isochromosomes contain the breakpoint in centromere repeats, showing that isochromosomes are produced by centromere rearrangement. Mutations in the Rad3 checkpoint kinase increase both types of GCRs. In contrast, the deletion of Rad51 recombinase preferentially elevates isochromosome formation. Chromatin immunoprecipitation analysis shows that Rad51 localizes at centromere around S phase. These data suggest that Rad51 suppresses rearrangements of centromere repeats that result in isochromosome formation.
Keywords: centromere, fission yeast, gross chromosomal rearrangement, homologous recombination, isochromosome
Introduction
Maintaining genetic information through cell divisions is crucial for cell proliferation and for preventing genetic diseases in multicellular organisms. However, external stress such as ultraviolet irradiation or internal stress such as reactive oxygen species introduces various kinds of insults into genomic DNA. When aberrant DNA structures (e.g. DNA double-strand breaks (DSBs)) are produced, a DNA checkpoint is activated that coordinates the repair process and cell cycle progression (Carr, 2002; Nyberg et al, 2002). Mammalian ataxia telangiectasia mutated (ATM) and ataxia telangiectasia and Rad3 related (ATR) are the central checkpoint kinases that are activated in response to aberrant DNA structures. DSBs are a serious threat to genome integrity (Myung and Kolodner, 2003; Prudden et al, 2003), and they can be repaired by non-homologous end joining or by homologous recombination when the template DNA strand is available (Paques and Haber, 1999; Branzei and Foiani, 2008). Although homologous recombination is a relatively conservative mechanism of DSB repair, it leads to gross chromosomal rearrangement (GCR) when associated with crossing over or break-induced replication (BIR) (Myung et al, 2001; Lambert et al, 2005). Many types of homologous recombination, including gene conversion (GC), require the Rad51 protein that promotes DNA-strand exchange and Holliday junction formation (Shinohara et al, 1993; Bianco et al, 1998; Murayama et al, 2008). However, a subset of recombination including BIR occurs even in the absence of Rad51 (Malkova et al, 1996). To maintain genome integrity, it is important to process aberrant DNA structures in an appropriate way.
Genome instability such as GCR is one of the hallmarks of cancer cells. GCR is associated with loss of heterogeneity (LOH) between two alleles and/or the alteration of gene dosage, either of which is implicated in tumorigenic process of neoplasms (Lengauer et al, 1998). BRCA2 that regulates Rad51 recombinase is frequently mutated in breast cancer cells (Thorslund and West, 2007), suggesting the involvement of homologous recombination in GCR events. Among the various types of GCR events (e.g. translocation, deletion, and so on) observed in cancer cells, isochromosome is unique in that it is the chromosome with homologous arms that are mirror images of one another (Mertens et al, 1994). Although DNA cleavage around the centromere is proposed to lead to isochromosome formation (Jin et al, 2000), the sequence complexity of the centromere hampers the relationship between the centromere and isochromosome formation.
Centromere is functionally divided into two regions: kinetochore where mitotic spindles are attached to chromosomes and pericentromeric heterochromatin where a pair of sister chromatids are held together until the onset of anaphase (Bernard et al, 2001; Pidoux and Allshire, 2005; Schueler and Sullivan, 2006). Most centromeres of animals and plants consist of arrays of DNA repeats. In humans, centromere spans 300–5000 kb and contains extensive copies of repetitive elements called α-satellite (171 bp) (Grewal and Jia, 2007). α-satellites are arranged homogeneously in the centromere core, whereas in pericentromeric heterochromatin, the stretches of α-satellites are not uniformly oriented and frequently interrupted by interspersed elements such as LINE, SINE, and LTR (Schueler and Sullivan, 2006). In the fission yeast Schizosaccharomyces pombe, centromere is relatively short (40–110 kb), but it contains repetitive elements that surround a unique central core (Grewal and Jia, 2007). DNA rearrangement or recombination occurs in chromosomal regions that contain repetitive elements such as rDNA and telomere (Kobayashi, 2006; Longhese, 2008). GC tracts can enter and extend through centromere in budding yeast (Liebman et al, 1988), showing that recombination can happen in centromere. However, little is known about the stability of centromere repeats.
Here, we have developed a system to detect GCRs using a fission yeast minichromosome ChL that is derived from chromosome III (ChIII) and contains an intact centromere 3 (cen3). Analyses of the GCR products show that translocation between homologous chromosomes, ChL and ChIII, and isochromosome are detected in our system. We found that the isochromosome is produced by the rearrangement of centromere repeats. Mutations in the Rad3 checkpoint kinase (Bentley et al, 1996) increase both types of GCRs, suggesting that DSBs accumulated in the mutant cells induce both translocation and isochromosome formation. Interestingly, the deletion of Rad51 (also called Rhp51 in fission yeast) recombinase strongly elevates the rate of isochromosome formation. Chromatin immunoprecipitation (ChIP) analysis shows the Rad51 localization at centromere around S phase. We propose that Rad51 suppresses the formation of isochromosome that is produced by the rearrangement of centromere repeats.
Results
Novel system to detect spontaneous GCRs in fission yeast
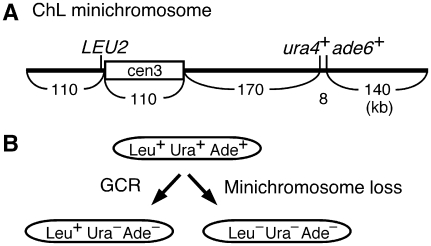
As GCRs can lead to cell death and hamper the analysis of rearranged chromosomes, a minichromosome that is dispensable for cell proliferation was used to detect the GCRs. Minichromosome Ch16 that is derived from ChIII seems to be useful for this purpose, as it contains an intact cen3 and telomere sequences, and it is transmitted to daughter cells with high fidelity (Niwa et al, 1986; Matsumoto et al, 1987). To monitor chromosome stability, we introduced three genetic markers onto the minichromosome (Figure 1A). The LEU2 gene was introduced on the left arm proximal to cen3, whereas the ura4+ and ade6+ genes were introduced around the middle of the right arm. Hereafter, we call this modified minichromosome as ChL. In a leu1 ura4 ade6 strain background, the presence of ChL is manifested as Leu+ Ura+ Ade+. When GCR associated with a specific loss of the region encompassing the ura4+ and ade6+ genes takes place, the cell becomes Leu+ Ura− Ade− (Figure 1B). The cell that has lost ChL can be detected as Leu− Ura−. To allow GCR or ChL loss to occur spontaneously, we incubated the yeast strain harbouring the ChL on non-selective media that contains Leu, Ura, and Ade. By means of a fluctuation test, it has been found that GCR and minichromosome loss occur at 2.9 × 10−5 and 3.4 × 10−5 per cell division, respectively (Table I; Wild type). This loss rate is comparable to that of Ch16 (Niwa et al, 1986), showing that the introduction of markers does not interfere with chromosome stability.
Figure 1.
System to monitor chromosome stability using ChL minichromosome. (A) Positions of the three integrated markers and the centromere are indicated. ChL was created by the introduction of LEU2, ura4+, and ade6+ markers onto Ch16 (Niwa et al, 1986). LEU2 replaced ubcp4 and chk1 genes; ura4+ replaced spcc1322.09; ade6-M216 at the original locus was converted to ade6+. (B) From the yeast strain harbouring ChL (Leu+ Ura+ Ade+), GCR associated with the specific loss of the ura4+ and ade6+ genes results in Leu+ Ura− Ade− (left arrow), whereas the loss of ChL leads to Leu− Ura− Ade− (right arrow).
Table 1.
Rates of GCR and minichromosome loss in the wild-type, rad3 and rad51 strains
| Strain | Sample no. | Genotype | GCR | Minichromosome loss | ||
|---|---|---|---|---|---|---|
| Rate ( × 10−5) | Relative | Rate ( × 10−5) | Relative | |||
| TNF1610 | 45 | Wild type | 2.9 (2.5–3.2) | 1 | 3.4 (2.6–4.1) | 1 |
| TNF1700 | 15 | rad3Δ | 24.2 (16.9–32.5) | 8 | 33.8 (23.9–61.4) | 10 |
| TNF1728 | 15 | rad3D2230A | 32.0 (28.1–38.2) | 11 | 72.0 (60.5–218.0) | 21 |
| TNF1730 | 15 | rad3N2235K | 57.6 (37.3–82.0) | 20 | 150.0 (94.0–287.0) | 44 |
| TNF1732 | 15 | rad3D2249E | 19.2 (16.2–29.3) | 7 | 41.7 (30.5–64.9) | 12 |
| TNF2094 | 35 | rad51Δ | 556 (445–641) | 192 | 1750 (1450–1980) | 515 |
| The number of independent colonies examined is shown under sample no. The mean value of the rates is indicated, and 95% confidence interval is shown within parentheses. Relative rates compared to that of the wild type are indicated. | ||||||
Two different types of GCR products are detected using ChL minichromosome
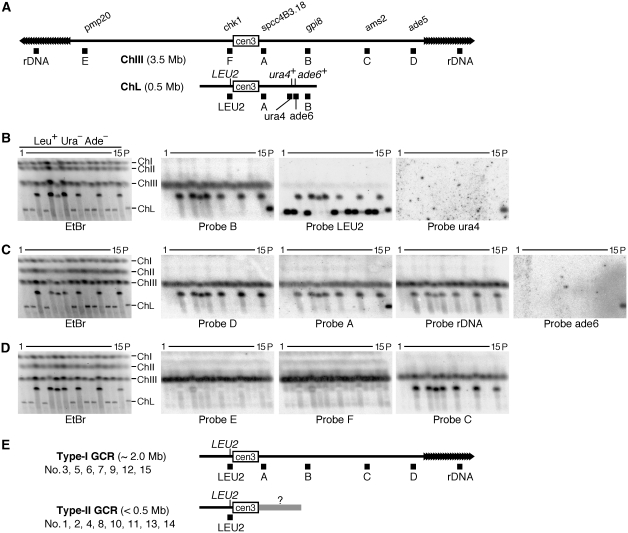
To determine the kind of chromosomal rearrangement occurring in this system, chromosomal DNA was prepared from 15 independent clones of Leu+ Ura− Ade− and the parental strain, separated by pulse field gel electrophoresis (PFGE) and stained with ethidium bromide (EtBr) (Figure 2B–D, left panels). The lengths of the minichromosomes in Leu+ Ura− Ade− clones were different from that of the parental ChL, indicating that GCRs rather than simple GCs or point mutations have occurred in these clones. To characterize the GCR products, the separated DNA was transferred onto a nylon membrane and hybridized with specific probes shown in Figure 2A (Figure 2A–D). All the minichromosomes were detected using probe LEU2 (Figure 2B), showing that they are derived from ChL. However, only the parental ChL was detected using probe ura4 or ade6 (Figure 2B and C), showing loss of the two markers in the GCR clones. It was found that half of the rearranged minichromosomes contained regions A, B, C, and D, as well as rDNA that is originally present at the ends of ChIII, and were longer than ChL (Figure 2E, type-I GCR). On the other hand, the others had lost regions A and B, and were smaller than ChL (Figure 2E, type-II GCR). None of the minichromosomes contained region E or F that is specific to the ChIII left arm. These results show that two different types of GCRs are detected in this system.
Figure 2.
Analysis of chromosomes by PFGE. (A) Positions of the probes used in Southern hybridization are indicated as filled boxes under ChIII and ChL. The name of the gene or ORF that is overlapping or nearby the probes A–F is shown above ChIII. (B–D) Chromosomal DNA was prepared from the parental strain (TNF1610) and 15 Leu+ Ura− Ade− GCR clones that were derived from different colonies on YE3S, separated by PFGE (pulse time: 1800 s at 2 V/cm for 72 h) using 0.7% agarose gel in 1 × TAE buffer and stained with EtBr (left panels). Positions of ChI, ChII, ChIII, and ChL (5.7, 4.6, 3.5, and 0.5 Mb, respectively) in the parental strain are indicated on the right of the EtBr panel. DNA was transferred onto a nylon membrane and hybridized sequentially with the specific probes indicated under each panel. (E) Type-I and type-II GCR products. For each type, the probes that hybridized to the GCR products are indicated. The size of the GCR products and the clone number are also shown.
Translocation between homologous chromosomes ChL and ChIII
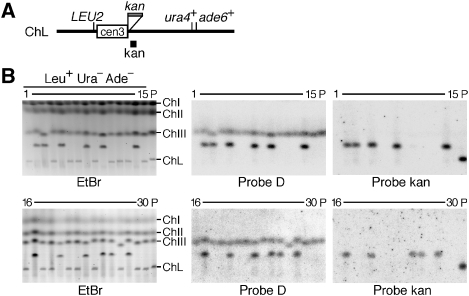
To determine the position where the translocation occurred in type-I GCR products, we introduced an additional marker, the kan gene, on the right side of cen3 (Figure 3A). The introduction of kan did not affect the chromosome stability (Supplementary Table I), and around half of the GCR products were type-I determined on the basis of the minichromosome length and the presence of region D (Figures 2 and 3B). Re-hybridization with a probe specific to kan showed that 10 of the 12 type-I products retained the kan marker. These results suggest that in most cases type-I products are formed by translocation within the region flanking cen3 and ura4+ (∼170 kb, see Figure 1A), but in some cases the translocation occurs in the centromere sequence. In contrast to type-I, none of the type-II products were detected by the kan probe, suggesting that the original right arm has been completely lost in this type of GCR products.
Figure 3.
Type-I is produced by translocation between ChIII and ChL chromosomes. (A) A ChL derivative that contains the kan gene on the right side of cen3. The kan gene was introduced between irc3R and spcc4B3.18 (see Materials and methods). (B) Chromosomal DNA was prepared from the parental strain (TNF2076) and 30 Leu+ Ura− Ade− clones that were derived from the different colonies on YE3S, separated by PFGE (2 V/cm for 72 h) in 1 × TAE buffer and stained with EtBr (left panel). The top panel: pulse time 1800 s, 0.8% agarose gel; bottom panel: pulse time 2400 s, 0.7% agarose gel. DNA was transferred onto a nylon membrane and hybridized sequentially with probe D (see Figure 2) and probe kan as indicated under each panel.
Formation of isochromosome produced around the centromere
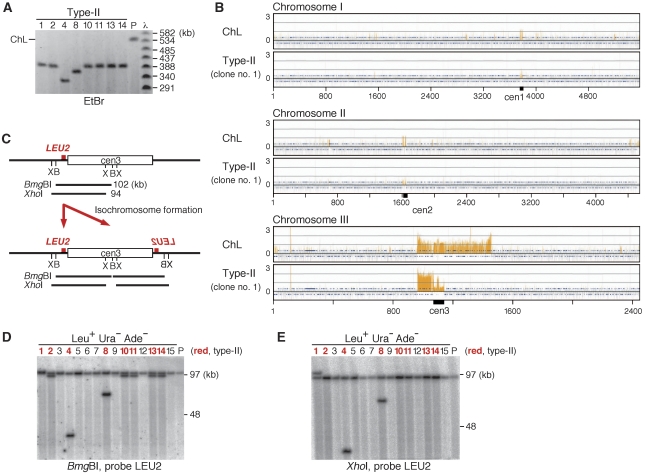
To define the length of the type-II GCR products, PFGE was carried out under the condition where 50–800 kb DNA can be resolved. Assuming λ DNA ladder as a standard, it was determined that type-II products were 330–400 kb, whereas ChL was ∼540 kb (Figure 4A). As the length of the ChL left arm plus the centromere is 220 kb (Figure 1A), type-II products seem to have acquired some DNA sequences of 110–180 kb. In an attempt to identify the sequence, the minichromosomes were recovered from the gel and subjected to comprehensive genome hybridization (CGH) using oligonucleotide arrays. When ChL was used as a probe, ∼500 kb around cen3 was detected, as expected (Figure 4B, ChL; Supplementary Figure 1). However, other than the original left arm and cen3, no consecutive sequences of >100 kb were detected using a type-II product (clone no. 1) (Figure 4B, type-II; Supplementary Figure 2). Similar results were obtained using several other type-II products (clone no. 4, 8, and 11, see Supplementary Figure 3) that are different in the length from each other and from clone no. 1. Detection of the portions of cen1 and cen2 either by ChL or type-II is due to the partial sequence identity among cen1, 2, and 3. The apparent discrepancy between the length and CGH profile can be solved if type-II product were an isochromosome in which the original right arm has been replaced by a copy of the left arm. To test this possibility, chromosome DNA was treated with the restriction enzyme BmgBI or XhoI, separated by PFGE, and the fragment containing LEU2 was detected using Southern blot. An additional copy of LEU2 would be created if an isochromosome were produced around the cen3 of ChL (Figure 4C). Using BmgBI, two separate bands were detected for seven out of eight type-II clones (indicated by the red colour), whereas only one band was detected for all of the type-I products and ChL (Figure 4D). The type-II clone that gave a single band using BmgBI (clone no. 1) produced two bands using XhoI (Figure 4E). These results show that the type-II GCR product is an isochromosome. The difference in the lengths among the isochromosomes may be due to the copy number of otr3s in cen3 (see also below).
Figure 4.
Type-II is the isochromosome produced around the centromere. (A) Chromosomal DNA prepared from the parental strain (TNF1610) and type-II GCR clones shown in Figure 2 were separated by PFGE (pulse time: 5–120 s linear at 4.5 V/cm for 48 h) using 1.0% agarose gel in 0.5 × TBE buffer and stained with EtBr. The position of ChL is indicated on the left of the panel. Sizes of the λ DNA ladder (New England Biolabs) are indicated on the right. The clone numbers are shown on the top of the panel. (B) By a DNA microarray technique, CGH was carried out for the parental ChL and a type-II GCR product. The signals obtained from the comparison of the minichromosome DNA to the total genomic DNA of TNF740 (h−, ade6Δ-D, ura4-D18, and leu1-32) are presented. Each bar represents the mean of 11 oligonucleotide probes within adjacent 250-bp windows. Orange bars represent significant binding. The chromosome position corresponds to the fully assembled virtual contigs in Sanger Center S. pombe Genome Database. The Y axis scale is log2. (C) LEU2 is duplicated when an isochromosome is produced around the centromere. Positions of BmgBI and XhoI restriction sites around cen3 are indicated as B and X, respectively. (D) Chromosomal DNA was prepared from the parental strain (TNF1610) and the same 15 Leu+ Ura− Ade− clones that are shown in Figure 2 were digested with BmgBI, separated by PFGE (pulse time: 1–6 s at 6 V/cm for 22 h in 0.5 × TBE buffer), transferred onto a nylon membrane, and hybridized with probe LEU2. (E) Chromosomal DNA was digested with XhoI instead of BmgBI; 0.8 and 0.7% agarose gel was used in the case of BmgBI and XhoI, respectively. Type-II clones are indicated by the red colour. It is estimated that clone nos. 2, 10, 11, 13, and 14 contain the same number (i.e. ∼12 copies) of otr3s on both sides of cnt3, whereas clone nos. 1, 4, and 8 contain 13, 2, or 7 copies of otr3s, respectively, on the right side (see also Figure 5).
Isochromosome is produced by rearrangement of centromere repeats
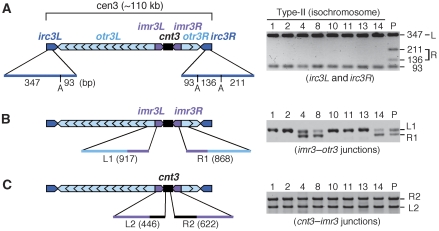
Cen3 consists of a central core, cnt3 (4.9 kb), surrounded by inverted repeats, imr3 (5.4 kb), otr3 (6.3 kb), and irc3 (2.2 kb) (Grewal and Jia, 2007) (Figure 5A). There are ∼12 and 2 tandem copies of otr3s on the left and right sides of cnt3 on ChL, respectively (Figure 4; Supplementary Figure 4). The DNA sequences of imr3L and imr3R are completely identical (Takahashi et al, 1992), whereas small differences are observed for pairs of otr3s or irc3s. To examine the sequence integrity in the centromere of type-II products (i.e. isochromosomes), PCR was carried out using the minichromosomes recovered from agarose gel. A portion of irc3 was amplified using a pair of primers, digested with ApoI, and separated by conventional agarose gel electrophoresis (Figure 5A). The fragment amplified from the left (irc3L) contains a single ApoI site, whereas that from the right (irc3R) has an additional ApoI site. Using ChL, four restriction fragments were detected as expected. However, using type-II products, irc3R-specific fragments (Figure 5A, fragment R) were not detected, indicating an alteration of irc3R in isochromosomes. We extended a similar analysis to imr3–otr3 junctions (Figure 5B). Because of the difference between otr3L and otr3R, two fragments of different sizes (L1 and R1) were amplified using a pair of primers. However, only L1 was amplified using 5 of the 8 type-II products, indicating an alteration of otr3R as well as irc3R in some isochromosomes. When we used a set of three primers to amplify both sides of cnt3–imr3 junctions, two fragments, namely, L2 and R2, were amplified in all the examined cases (Figure 5C), indicating no changes at the cnt3–imr3 junctions in the isochromosomes. In combination, these data show that the breakpoints producing the isochromosomes are located in the centromere repeats but not in a unique region, namely, cnt3. We examined 10 more isochromosomes and found that they also showed an alteration of irc3R and no changes at the cnt3–imr3 junctions (Supplementary Figure 5). Around half of the isochromosomes (11 out of 18 in total) exhibited an alteration of otr3R, demonstrating that the breakpoints are created almost equally in imr3 and in otr3–irc3 regions in wild type.
Figure 5.
Rearrangement of centromere repeats in isochromosomes. (A) Fission yeast cen3 consists of cnt3 (black) surrounded by inverted repeats of three different sequences imr3 (magenta), otr3 (cyan), and irc3 (blue). The minichromosomes shown in Figure 4A were recovered from gel and used as templates of PCR. irc3L and irc3R were amplified using irc3-R and irc3-F primers, digested with ApoI, separated by 2.0% agarose gel electrophoresis, and stained with EtBr. The length of ApoI restriction fragments is indicated. (B) Both sides of the imr3–otr3 junctions were amplified using imr3-out and dh primers, separated by 2.0% agarose gel electrophoresis. Because of small deletions on the right side, 917-bp (L1) and 868-bp (R1) fragments are produced from the left and right, respectively. (C) Both sides of the cnt3–imr3 junctions were amplified using three primers, namely, cnt3-L, cnt3-R, and imr3-in, separated by 1.2% agarose gel electrophoresis. Here, 446-bp (L2) and 622-bp (R2) fragments are produced. The clone number is shown at the top of each panel.
Rad3 checkpoint kinase suppresses translocation and isochromosome formation
To see whether the GCRs detected in this system are affected by the DNA checkpoint activated in response to DNA damage and replication stress, we constructed a deletion mutant of Rad3, the fission yeast homologue of ATR, and measured the rate of GCR. It was observed that rad3Δ significantly increased the rate of GCR (Table I). Analysis of the GCR products showed that both type-I and type-II GCRs were increased by rad3Δ (Supplementary Figure 6). Thus, it appears that translocation, as well as isochromosome formation, is suppressed by the DNA checkpoint. The introduction of kinase-dead mutations (rad3D2230A, rad3N2235K, or rad3D2249E; Bentley et al, 1996) resulted in essentially the same phenotype as that caused by rad3Δ (Table I), suggesting the requirement of the kinase activity of Rad3 for the suppression of GCRs. Consistent with the elevated rates of GCRs, the loss rate of ChL was also increased by the rad3 mutations.
Rad51 homologous recombination protein suppresses isochromosome formation
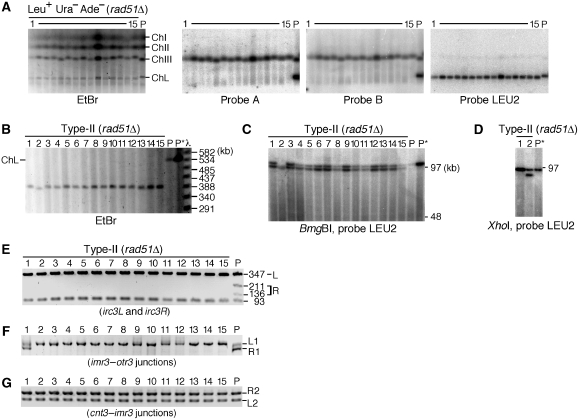
Either the translocation or isochromosome formation seen in this study seems to be mediated by homologous sequences. To explore a possible role of homologous recombination in the GCRs, we constructed a deletion mutant of rad51 (Murayama et al, 2008). Interestingly, rad51Δ strongly increased the rate of GCR as well as ChL loss (Table I). Remarkably, PFGE analysis revealed that all the 15 independent GCR products formed in rad51Δ cells have lost the original right arm and have attained a length of ∼390 kb (Figure 6A and B). Restriction digestion followed by LEU2 hybridization (Figure 6C and D) showed that LEU2 is duplicated in all the products. These data show that Rad51 suppresses isochromosome formation, although the effect of rad51Δ on translocation is unclear because of the limited number of samples. PCR analysis of the centromere of isochromosomes (Figure 6E–G) showed the localization of the breakpoint in the centromere repeats in rad51Δ as that in wild type (Figure 5). However, in contrast to the wild type (Figure 5B; Supplementary Figure 5), the right imr3–otr3 junction (R1) was not amplified in most cases of rad51Δ (Figure 6F) (11 of 18 in wild type; 14 of 15 in rad51Δ; P<0.02 in the χ2 test), suggesting that the isochromosomes are produced by rearrangement preferentially at the innermost repeat, imr3, in the absence of Rad51.
Figure 6.
Rad51 suppresses isochromosome formation. (A) Chromosomal DNA was prepared from the parental rad51Δ strain (TNF2094) and 15 Leu+ Ura− Ade− GCR clones that were derived from different colonies on YE3S, separated by PFGE as described in Figure 2, and stained with EtBr (left panel). DNA in the agarose gel was transferred onto a nylon membrane and hybridized sequentially with specific probes A, B, and LEU2, shown in Figure 2A. (B) Chromosomal DNA was separated under the condition where 50–800 kb DNA can be resolved and stained with EtBr. P and P* indicate the parental rad51Δ (TNF2094) and wild-type (TNF1610) strains, respectively. (C, D) DNA was digested with BmgBI (C) or XhoI (D) and the fragment containing LEU2 was detected as described in Figure 4. (E–G) PCR analysis of centromere was carried out as described in Figure 5.
Rad51 localization at centromere around S phase
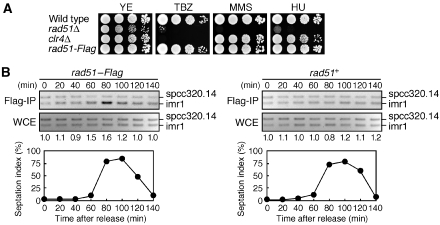
The elevated rate of the isochromosome formation suggests that Rad51 may play some role in centromere. The methyltransferase Clr4 is required for the methylation of histone H3 lysine 9 in pericentromeric heterochromatin and supports the establishment of CENP-A chromatin in kinetochore (Nakayama et al, 2001; Folco et al, 2008). The mutations that impair the centromere structure and function such as clr4Δ cause hypersensitivity to thiabendazole (TBZ), a microtubule-destabilizing reagent (Figure 7A; Ekwall et al, 1996). We found that rad51Δ caused TBZ hypersensitivity, consistent with the idea that Rad51 plays an important role for the centromere structure and function.
Figure 7.
Localization of Rad51 at centromere. (A) The rad51 mutant exhibits TBZ hypersensitivity. Exponentially growing cells of wild type, rad51Δ, clr4Δ, and rad51–Flag strains (TNF34, 1618, 2483, and 2315, respectively) were 10-folds serially diluted and spotted onto YE plates supplemented with 15 μg/ml TBZ, 0.005% MMS, or 4 mM HU, and incubated for 4–6 days at 30°C. (B) ChIP analysis of Rad51–Flag. Synchronous cultures in EMM medium were prepared using rad51–Flag cdc25-22 (TNF2437) and rad51+ cdc25-22 (TNF701) strains. PCR was carried out in the presence of spcc320.14F, spcc320.14R, imr1-HF, and imr1-HR primers. The PCR products (spcc320.14, 170 bp; imr1, 126 bp) were separated by 2.5% agarose gel electrophoresis in 1 × TBE buffer and stained with EtBr. Relative fold enrichments depicting the ratios of the signals at imr1 relative to spcc320.14, between Flag-IP and WCE, are shown beneath each lane. The percentage of the cells containing the septum is also shown. At each time point, >200 cells stained with calcofluor were counted under the microscope. The yeast strains used here contain no minichromosomes.
To see whether Rad51 localizes at centromere, we tagged Rad51 with the Flag epitope at the C terminus and performed ChIP assay using anti-Flag monoclonal antibody. The Rad51–Flag protein appears to be fully functional because the replacement of the rad51+ gene with the rad51–Flag gene did not change the sensitivity to TBZ as well as to a DNA-damaging reagent methyl methanesulphonate (MMS) and a replication inhibitor hydroxyurea (HU) (Figure 7A). Log phase cultures of the cdc25-22 background stains were temporally incubated at 36°C for 3 h to induce G2 arrest and then returned to 25°C, allowing synchronous progression of the cell cycle thereafter. We prepared DNA either from Flag immunoprecipitate (Flag-IP) or from whole cell extract (WCE), and carried out PCR by using the primers specific to a site close to a boundary of pericentromeric heterochromatin in imr1 (Scott et al, 2006) and those to a site near spcc320.14 on a chromosome arm. Using Flag-IP, a specific amplification of imr1 over spcc320.14 was observed at around 80 min after the release from G2 where the cells containing the septum were accumulated (Figure 7B, left panel). However, similar levels of amplification for imr1 and spcc320.14 were observed using WCE. The specific amplification of imr1 is dependent on Rad51–Flag, because no such enrichment was observed when we used rad51+ cells (Figure 7B, right panel). Although further analyses are required to address all the Rad51-binding sites in centromere, these results show that the Rad51 protein is localized at centromere around S phase.
Discussion
Using an extra minichromosome ChL, we developed a fission yeast system to detect spontaneous GCRs. Analyses of the rearranged chromosomes revealed two types of GCRs: translocation between homologous chromosomes and isochromosome formation. We found that the isochromosome was produced by the rearrangement of centromere repeats. Both types of GCRs were increased by the mutations of Rad3 checkpoint kinase. In contrast, the deletion of Rad51 preferentially increased the isochromosome formation.
As GCR can lead to cell death, a dispensable chromosome ChL was constructed and used to detect GCR in this study. The rate of GCR was determined to be 2.9 × 10−5 per cell division, which is even higher than that of allelic GC (∼1–4 × 10−6 per cell division) (Virgin et al, 2001). The high incidence of GCR is not surprising, because it has been observed that GCR occurs at comparable rates when a dispensable chromosome is monitored in budding yeast (Huang and Koshland, 2003). Although an affect of an extra chromosome on the GCR rate cannot be excluded, it is possible that GCR occurs at a high rate in normal cells. In the wild type, two different types of GCR products were observed. Around half of the GCR products (type-I) acquired the right arm of ChIII and became larger than ChL. It appears that type-I products are formed by translocation between homologous chromosomes ChL and ChIII and contain the ChIII right arm to its end, as detected by a probe specific to rDNA originally present at the ChIII ends. The length of the type-I products (∼2.0 Mb) is consistent with this idea. In most cases, the breakpoint appears to be located in a 170-kb arm region flanking cen3 and ura4+. A similar type of translocation was induced by a site-specific DSB, and a pedigree analysis revealed that the translocation occurs either by reciprocal crossing over or nonreciprocal BIR mechanism (Prudden et al, 2003; Cullen et al, 2007). The translocation observed in this study may also be initiated by a DSB produced at fragile sites on the chromosomes (Cha and Kleckner, 2002). The tRNA gene is one of the fragile sites (Ivessa et al, 2003; Admire et al, 2006), and ChL has three such genes on the right arm at ∼70, 110, and 220 kb from cen3. It is also possible that extensive degradation of DNA (>140 kb) from a chromosome end that passes the ura4+ and ade6+ genes leads to translocation. The deletion of a chromosome end combined with de novo addition of telomere sequences has been reported to occur in haploid cells (Chen et al, 1998). However, such kind of GCR products was not seen in this study, probably because the rearrangement between homologous sequences is prevailing when homologous chromosomes or sequences are present in the same nucleus. The translocation between the cen3 sequences of ChL and ChIII was observed, although that between cen3 and other centromeres, was not detected. Different sequences of cnt, imr, and irc and different arrangements of otr repeats may prevent GCR between different centromeres. It should be noted that the rate of type-I GCR seems to be underestimated, as the cells harbouring type-I products grew less effectively as compared with those containing the parental ChL or isochromosomes (type-II). The slow growth may be due to the increased copy number of the rDNA and/or the other genes present on the right arm of ChIII (Chikashige et al, 2007).
The second type of GCR products (type-II) has lost the original right arm and they became smaller than ChL. CGH and Southern blotting revealed that the original right arm was replaced by a copy of the left arm, resulting in the formation of the isochromosome that has homologous arms of the mirror image. Fission yeast centromere is composed of a unique central core, cnt, surrounded by pairs of inverted repeats, namely, imr, otr, and irc, except cen2 where no irc is recognized. In all the examined isochromosomes, the right edge of cen3 (i.e. irc3R) had been altered, whereas the central cnt3–imr3 junctions were preserved, indicating that the isochromosome is produced by the rearrangement of centromere repeats. As isochromosome formation occurs at a rate comparable to that of the translocation between homologous chromosomes, centromere seems to be more prone to rearrangement than expected. Consistent with this idea, it has been reported recently that in mammalian cells mitotic recombination occurs at centromere at a higher frequency than those at chromosome arm or telomere (Jaco et al, 2008). Our data show that centromere is one of the fragile sites where deleterious chromosome rearrangements such as isochromosome formation and translocation can happen.
With regard to the structure and function, centromere can be divided into two regions: kinetochore and pericentromeric heterochromatin. The kinetochore-specific histone H3 variant (i.e. CENP-A) is incorporated around cnt to create kinetochore, whereas specific modifications of histones (e.g. histone H3 K9 methylation) take place in pericentromeric heterochromatin. Chromatin boundaries of pericentromeric heterochromatin are present in imr and irc (Cam et al, 2005; Scott et al, 2006). tRNA genes are also embedded in imr. Cruciform DNA has been detected by two-dimensional gel electrophoresis in the centromere of fission yeast (Smith et al, 1995). Pausing of the replication forks has been seen in the centromere of budding yeast (Greenfeder and Newlon, 1992). Thus, it is possible that aberrant DNA structures produced in centromere triggers isochromosome formation. This type of GCR (i.e. isochromosome formation) may occur for any chromosome in fission yeast. This is because isochromosome is produced by the rearrangement of inverted repeats surrounding a central core, cnt, and all the centromeres are essentially mirror images. Interestingly, an intramolecular loop structure around the centromere (C-loop) that brings both sides of the centromere-flanking regions close has been observed in budding yeast (Yeh et al, 2008). Higher order structures of the centromere such as the C-loop might facilitate the interaction between centromere repeats.
Rad3 is the central player of the DNA structure checkpoint, and it is activated when DNA damage or replication stress is induced. We found that amino-acid substitutions in the kinase domain, as well as a rad3 deletion (rad3Δ), increased the rate of GCR. Thus, it is likely that the kinase function of Rad3 is important to suppress spontaneous GCR. Analyses of the GCR products formed in a rad3Δ mutant suggest that both translocation and isochromosome formation were increased. Alteration of the size of ChIII observed in the rad3 mutant suggests that Rad3 may also affect the rDNA repeats on ChIII. As it has been shown that DSBs are accumulated in the absence of the checkpoint kinase (Brown and Baltimore, 2000; de Klein et al, 2000; Cha and Kleckner, 2002), our data suggest that translocation, as well as isochromosome formation, is initiated by DSBs. Consistent with this notion, chronic exposure of wild-type cells with MMS or HU that may induce DSBs increased the rate of GCR as well as minichromosome loss (Supplementary Figure 7).
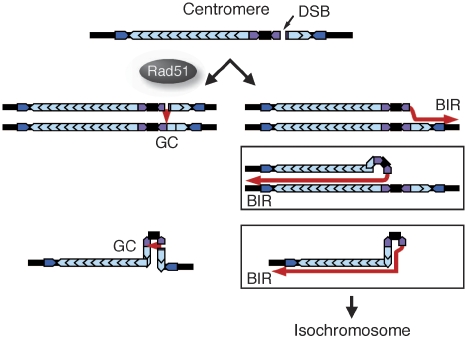
In contrast to rad3Δ, the rate of isochromosome formation was preferentially and strongly increased by rad51Δ, indicating that Rad51 suppresses isochromosome formation. It should be noted that rad51Δ might also increase other types of GCR that cause extreme slow growth or cell death in the rad51Δ background. Although the breakpoints are equally found in imr3 and in otr3–irc3 in wild-type isochromosomes, they are significantly enriched in imr3 in rad51Δ isochromosomes, suggesting that Rad51 may have some specific function in the imr region of centromere. The high incidence of minichromosome loss and the hypersensitivity to the microtubule-destabilizing drug TBZ suggest that Rad51 has a function in the centromere structure and/or function. The observation that rad51Δ increases missegregation of chromosomes following HU treatment (Bailis et al, 2008) supports this idea. We have found that Rad51 localizes at centromere around S phase, suggesting that Rad51-mediated recombination occurs at centromere around S phase. Figure 8 shows our current model for the isochromosome formation and its suppression by Rad51. In wild type, aberrant DNA structures such as DSBs created at centromere may be repaired either by the conservative GC or by BIR mechanism (Figure 8, the left and right arrows respectively). GC between sister chromatids or between the centromere repeats from a chromatid would not cause the isochromosome formation. However, in the absence of Rad51, the aberrant DNA may be repaired mainly by BIR. Because, it has been shown in budding yeast that BIR can occur in a Rad51-independent manner, whereas GC is severely impaired by a rad51 deletion (Malkova et al, 1996; Fasullo et al, 2001). It appears that Rad51 suppresses BIR by facilitating GC. BIR between sisters may or may not result in the isochromosome formation, but BIR between the centromere repeats from a chromatid always lead to an isochromosome (Figure 8, the boxes). On the basis of the sizes of the two restriction fragments containing LEU2, it appears that most of the isochromosomes that have the breakpoint in imr3 possess the same number of the outer repeats on either side of centromere. However, we found three exceptions where different numbers of the outer repeats are present, although the breakpoint is in imr3 (clone no. 1 and 17, wild type; clone no. 2, rad51Δ). It is possible that they have been produced by several rounds of strand invasion, DNA synthesis and dissociation. Because template switching has been found to occur during BIR (Smith et al, 2007), these data further support the idea that isochromosomes are produced by BIR. Although further studies are required to understand the molecular mechanism by which isochromosomes are produced, our data provide the first evidence for a role of Rad51 in the suppression of rearrangements of centromere repeats that results in isochromosome formation.
Figure 8.
A model for the isochromosome formation at centromere and its suppression by Rad51. GC, gene conversion; BIR, break-induced replication.
Materials and methods
Yeast strains and media
The yeast strains used in this study are listed in Supplementary Table II. To create a rad3∷kan strain, the PCR product obtained by using rad3-k5 and rad3-k3 primers and pFA6a-kanMX6 (Bahler et al, 1998) was introduced into yeast cells by lithium acetate transformation. To create a rad51∷kan strain, pTN471 was used for yeast transformation after digestion with XbaI and XhoI. A rad51-6His-3Flag:kan strain was created as follows: 0.5- and 0.4-kb regions flanking the rad51+ stop codon were amplified using rad51-1 and rad51-2ter primers and rad51-3His and rad51-4 primers, respectively. The second PCR was carried out in the presence of the amplified fragments, pU6H3Flag plasmid (De Antoni and Gallwitz, 2000; Katou et al, 2003), and rad51-1 and rad51-4 primers. The PCR products of 2.4 kb was introduced into the yeast strain TNF34, creating TNF2315. The transformants were selected using a medium supplemented with 50 μg/ml of G418 (Nacalai Tesque). Correct integration was confirmed by PCR. Cells were grown in complete (YE) or minimum (EMM) medium supplemented with the indicated amino acid at a final concentration of 225 μg/ml (Moreno et al, 1991). Solid medium contains 1.5% agar (Nacalai Tesque). 5FOA medium contains 56 μg/ml Ura, 7.0 g/l of yeast nitrogen base supplemented with ammonium sulphate (BD Biosciences), 2% glucose, and 1% 5FOA (Wako).
Construction of the ChL minichromosome
To construct the ChL chromosome, LEU2, ura4+, and ade6+ genes were integrated into Ch16 (Niwa et al, 1986) as follows: a 1.0-kb region flanking the ubcp4+ gene was amplified using III_1057F and III_1057R primers by PCR and digested with MscI and HindIII. The resulting 0.5-kb MscI–HindIII fragment was introduced between the HincII and HindIII sites of pBluescript II SK+ (Stratagene), creating the plasmid pTN593. A 1.8-kb region flanking the chk1+ gene was amplified using III_1062F and III_1062R primers, digested with SacI and HindIII, and the resulting 0.4-kb SacI–HindIII fragment was introduced between the SacI and HindIII sites of pTN593, creating pTN594. A 2.2-kb HindIII fragment containing LEU2 from pREP81 (Maundrell, 1993) was introduced into the HindIII site of pTN594, creating pTN595. A 3.1-kb SacI–XhoI fragment containing the ubcp4∷LEU2∷chk1 construct from pTN595 was introduced into a yeast strain harbouring Ch16, and the transformants were selected using a medium lacking Leu, creating the yeast strain TNF848. A 1.7-kb PvuII–HindIII fragment containing 5′-half of ade6+ from pTN435 was introduced into the TNF848 strain, and the transformants were selected using a medium lacking Ade, creating the TNF1383 strain. Further, 0.3- and 0.5-kb regions flanking the ORF, spcc1322.09, were amplified using III_1310A and III_1310B+ura4AN5 primers and III_1310C+ura4AN3 and III_1310D primers, respectively. The second PCR was carried out in the presence of the amplified fragments, pTN445 plasmid containing ura4+, and III_1310A and III_1310D primers. The PCR product of 2.5 kb was introduced into the TNF1383 strain, and the transformants were selected using a medium lacking Ura, creating the TNF1610 strain.
A ChL derivative that contains the kan gene on the right arm proximal to cen3 was created as follows: 0.2-kb regions flanking the ORF, spcc4B3.18, were amplified using irc3R7 and irc3R8+kanAN3 primers and irc3R9+kanAN5 and irc3R10 primers. The second PCR was carried out in the presence of the amplified fragments, pFA6a-kanMX6 plasmid, and the irc3R7 and irc3R10 primers. The PCR product of 1.8 kb was introduced into TNF1610, creating the TNF2076 strain. The sequence of the primers used in this study is listed in Supplementary Table III. Correct integration was confirmed by PCR or by Southern blotting.
PFGE and Southern hybridization
Chromosomal DNA was prepared essentially as described before (Smith et al, 1987). For details about PFGE, see Supplementary data. The probe for Southern hybridization was prepared using Megaprime DNA Labeling System (GE Healthcare) with α-32P-dATP. Radioactive signals were detected using a BAS2500 phosphorimager (Fuji Film).
Rates of GCR and minichromosome loss
The yeast strain harbouring ChL was grown for 3–4 days on the complete media supplemented with Leu, Ura, and Ade (YE3S). A single colony was picked, suspended in distilled water, and plated onto YE3S and 5FOA media supplemented with Leu and Ade (5FOA+LA). After incubation for 4–5 days, the colonies were counted to determine the total number of colony-forming cells and the number of Ura− cells. The Ura− colonies were replica plated onto the minimum media supplemented with Ura (EMM+U) and the media with Ura and Ade (EMM+UA). After further incubation for 3–4 days, the colonies were counted to determine the numbers of Leu+ Ura− Ade+ and Leu+ Ura− cells. The number of Leu+ Ura− Ade− cells indicative of GCR was obtained by subtracting the number of Leu+ Ura− Ade+ cells from that of Leu+ Ura− cells, whereas the number of Leu− Ura− cells indicative of minichromosome loss was obtained by subtracting the number of Leu+ Ura− cells from that of Ura− cells. Because rad51Δ cells grow slowly, they were incubated for 2 days longer than the others on YE3S and 5FOA+AL. Yeast cells were incubated at 30°C. Rates of GCR and minichromosome loss were determined by means of a fluctuation analysis using the method of medians (Lea and Coulson, 1949; Lin et al, 1996). We determined 95% confidential interval as described (Dixon and Massey, 1969).
PCR analysis of the centromere region of ChL
ChL chromosomes separated by PFGE were recovered from the gel using the QIAquick gel extraction kit (Qiagen). The PCR products were separated by agarose gel electrophoresis in 1 × TAE buffer. The oligonucleotide primers used are listed in Supplementary Table III.
CGH analysis
DNA microarray analysis was carried out as described (Hayashi et al, 2007) using minichromosomes recovered from pulse-field agarose gel and purified using the QIAquick extraction kit (Qiagen).
ChIP analysis
ChIP analysis was carried out as described (Nitani et al, 2008), using anti-Flag M2 mouse monoclonal antibody (Sigma) and the magnetic beads conjugated to sheep anti-mouse IgG (Dynabeads M-280; Invitrogen). The signals of the PCR products were quantified by using Image Gauge v.3.3 software (Fuji Film).
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Figure 5
Supplementary Figure 6
Supplementary Figure 7
Supplementary Figure Legeds
Supplementary Tables I–III
Supplementary Data
Acknowledgments
We are grateful to Osami Niwa, Mitsuhiro Yanagida, and Antony M Carr for providing the yeast strains. This study was supported by a Grant-in-Aid for Cancer Research from MEXT and funding from the Sumitomo Foundation and the Naito Foundation to TN.
References
- Admire A, Shanks L, Danzl N, Wang M, Weier U, Stevens W, Hunt E, Weinert T (2006) Cycles of chromosome instability are associated with a fragile site and are increased by defects in DNA replication and checkpoint controls in yeast. Genes Dev 20: 159–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A III, Steever AB, Wach A, Philippsen P, Pringle JR (1998) Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14: 943–951 [DOI] [PubMed] [Google Scholar]
- Bailis JM, Luche DD, Hunter T, Forsburg SL (2008) Minichromosome maintenance proteins interact with checkpoint and recombination proteins to promote s-phase genome stability. Mol Cell Biol 28: 1724–1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley NJ, Holtzman DA, Flaggs G, Keegan KS, DeMaggio A, Ford JC, Hoekstra M, Carr AM (1996) The Schizosaccharomyces pombe rad3 checkpoint gene. EMBO J 15: 6641–6651 [PMC free article] [PubMed] [Google Scholar]
- Bernard P, Maure JF, Partridge JF, Genier S, Javerzat JP, Allshire RC (2001) Requirement of heterochromatin for cohesion at centromeres. Science 294: 2539–2542 [DOI] [PubMed] [Google Scholar]
- Bianco PR, Tracy RB, Kowalczykowski SC (1998) DNA strand exchange proteins: a biochemical and physical comparison. Front Biosci 3: 570–603 [DOI] [PubMed] [Google Scholar]
- Branzei D, Foiani M (2008) Regulation of DNA repair throughout the cell cycle. Nat Rev Mol Cell Biol 9: 297–308 [DOI] [PubMed] [Google Scholar]
- Brown EJ, Baltimore D (2000) ATR disruption leads to chromosomal fragmentation and early embryonic lethality. Genes Dev 14: 397–402 [PMC free article] [PubMed] [Google Scholar]
- Cam HP, Sugiyama T, Chen ES, Chen X, FitzGerald PC, Grewal SI (2005) Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nat Genet 37: 809–819 [DOI] [PubMed] [Google Scholar]
- Carr AM (2002) DNA structure dependent checkpoints as regulators of DNA repair. DNA Repair (Amst) 1: 983–994 [DOI] [PubMed] [Google Scholar]
- Cha RS, Kleckner N (2002) ATR homolog Mec1 promotes fork progression, thus averting breaks in replication slow zones. Science 297: 602–606 [DOI] [PubMed] [Google Scholar]
- Chen C, Umezu K, Kolodner RD (1998) Chromosomal rearrangements occur in S. cerevisiae rfa1 mutator mutants due to mutagenic lesions processed by double-strand-break repair. Mol Cell 2: 9–22 [DOI] [PubMed] [Google Scholar]
- Chikashige Y, Tsutsumi C, Okamasa K, Yamane M, Nakayama J, Niwa O, Haraguchi T, Hiraoka Y (2007) Gene expression and distribution of Swi6 in partial aneuploids of the fission yeast Schizosaccharomyces pombe. Cell Struct Funct 32: 149–161 [DOI] [PubMed] [Google Scholar]
- Cullen JK, Hussey SP, Walker C, Prudden J, Wee BY, Dave A, Findlay JS, Savory AP, Humphrey TC (2007) Break-induced loss of heterozygosity in fission yeast: dual roles for homologous recombination in promoting translocations and preventing de novo telomere addition. Mol Cell Biol 27: 7745–7757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Antoni A, Gallwitz D (2000) A novel multi-purpose cassette for repeated integrative epitope tagging of genes in Saccharomyces cerevisiae. Gene 246: 179–185 [DOI] [PubMed] [Google Scholar]
- de Klein A, Muijtjens M, van Os R, Verhoeven Y, Smit B, Carr AM, Lehmann AR, Hoeijmakers JH (2000) Targeted disruption of the cell-cycle checkpoint gene ATR leads to early embryonic lethality in mice. Curr Biol 10: 479–482 [DOI] [PubMed] [Google Scholar]
- Dixon WJ, Massey FJ Jr (1969) Introduction to Statistical Analysis. New York: McGraw-Hill Book Company [Google Scholar]
- Ekwall K, Nimmo ER, Javerzat JP, Borgstrom B, Egel R, Cranston G, Allshire R (1996) Mutations in the fission yeast silencing factors clr4+ and rik1+ disrupt the localisation of the chromo domain protein Swi6p and impair centromere function. J Cell Sci 109: 2637–2648 [DOI] [PubMed] [Google Scholar]
- Fasullo M, Giallanza P, Dong Z, Cera C, Bennett T (2001) Saccharomyces cerevisiae rad51 mutants are defective in DNA damage-associated sister chromatid exchanges but exhibit increased rates of homology-directed translocations. Genetics 158: 959–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folco HD, Pidoux AL, Urano T, Allshire RC (2008) Heterochromatin and RNAi are required to establish CENP-A chromatin at centromeres. Science 319: 94–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenfeder SA, Newlon CS (1992) Replication forks pause at yeast centromeres. Mol Cell Biol 12: 4056–4066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal SI, Jia S (2007) Heterochromatin revisited. Nat Rev Genet 8: 35–46 [DOI] [PubMed] [Google Scholar]
- Hayashi M, Katou Y, Itoh T, Tazumi A, Yamada Y, Takahashi T, Nakagawa T, Shirahige K, Masukata H (2007) Genome-wide localization of pre-RC sites and identification of replication origins in fission yeast. EMBO J 26: 1327–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D, Koshland D (2003) Chromosome integrity in Saccharomyces cerevisiae: the interplay of DNA replication initiation factors, elongation factors, and origins. Genes Dev 17: 1741–1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivessa AS, Lenzmeier BA, Bessler JB, Goudsouzian LK, Schnakenberg SL, Zakian VA (2003) The Saccharomyces cerevisiae helicase Rrm3p facilitates replication past nonhistone protein–DNA complexes. Mol Cell 12: 1525–1536 [DOI] [PubMed] [Google Scholar]
- Jaco I, Canela A, Vera E, Blasco MA (2008) Centromere mitotic recombination in mammalian cells. J Cell Biol 181: 885–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Jin C, Salemark L, Martins C, Wennerberg J, Mertens F (2000) Centromere cleavage is a mechanism underlying isochromosome formation in skin and head and neck carcinomas. Chromosoma 109: 476–481 [DOI] [PubMed] [Google Scholar]
- Katou Y, Kanoh Y, Bando M, Noguchi H, Tanaka H, Ashikari T, Sugimoto K, Shirahige K (2003) S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature 424: 1078–1083 [DOI] [PubMed] [Google Scholar]
- Kobayashi T (2006) Strategies to maintain the stability of the ribosomal RNA gene repeats–collaboration of recombination, cohesion, and condensation–. Genes Genet Syst 81: 155–161 [DOI] [PubMed] [Google Scholar]
- Lambert S, Watson A, Sheedy DM, Martin B, Carr AM (2005) Gross chromosomal rearrangements and elevated recombination at an inducible site-specific replication fork barrier. Cell 121: 689–702 [DOI] [PubMed] [Google Scholar]
- Lea DE, Coulson CA (1949) The distribution of the numbers of mutants in bacterial populations. J Genet 49: 264–285 [DOI] [PubMed] [Google Scholar]
- Lengauer C, Kinzler KW, Vogelstein B (1998) Genetic instabilities in human cancers. Nature 396: 643–649 [DOI] [PubMed] [Google Scholar]
- Liebman SW, Symington LS, Petes TD (1988) Mitotic recombination within the centromere of a yeast chromosome. Science 241: 1074–1077 [DOI] [PubMed] [Google Scholar]
- Lin M, Chang CJ, Green NS (1996) A new method for estimating high mutation rates in cultured cells. Mutat Res 351: 105–116 [DOI] [PubMed] [Google Scholar]
- Longhese MP (2008) DNA damage response at functional and dysfunctional telomeres. Genes Dev 22: 125–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkova A, Ivanov EL, Haber JE (1996) Double-strand break repair in the absence of RAD51 in yeast: a possible role for break-induced DNA replication. Proc Natl Acad Sci USA 93: 7131–7136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto T, Fukui K, Niwa O, Sugawara N, Szostak JW, Yanagida M (1987) Identification of healed terminal DNA fragments in linear minichromosomes of Schizosaccharomyces pombe. Mol Cell Biol 7: 4424–4430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maundrell K (1993) Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene 123: 127–130 [DOI] [PubMed] [Google Scholar]
- Mertens F, Johansson B, Mitelman F (1994) Isochromosomes in neoplasia. Genes Chromosomes Cancer 10: 221–230 [DOI] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P (1991) Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol 194: 795–823 [DOI] [PubMed] [Google Scholar]
- Murayama Y, Kurokawa Y, Mayanagi K, Iwasaki H (2008) Formation and branch migration of Holliday junctions mediated by eukaryotic recombinases. Nature 451: 1018–1021 [DOI] [PubMed] [Google Scholar]
- Myung K, Chen C, Kolodner RD (2001) Multiple pathways cooperate in the suppression of genome instability in Saccharomyces cerevisiae. Nature 411: 1073–1076 [DOI] [PubMed] [Google Scholar]
- Myung K, Kolodner RD (2003) Induction of genome instability by DNA damage in Saccharomyces cerevisiae. DNA Repair (Amst) 2: 243–258 [DOI] [PubMed] [Google Scholar]
- Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI (2001) Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292: 110–113 [DOI] [PubMed] [Google Scholar]
- Nitani N, Yadani C, Yabuuchi H, Masukata H, Nakagawa T (2008) Mcm4 C-terminal domain of MCM helicase prevents excessive formation of single-stranded DNA at stalled replication forks. Proc Natl Acad Sci USA 105: 12973–12978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa O, Matsumoto T, Yanagida M (1986) Construction of a mini-chromosome by deletion and its mitotic and meiotic behaviour in fission yeast. Mol Gen Genet 203: 397–405 [Google Scholar]
- Nyberg KA, Michelson RJ, Putnam CW, Weinert TA (2002) Toward maintaining the genome: DNA damage and replication checkpoints. Annu Rev Genet 36: 617–656 [DOI] [PubMed] [Google Scholar]
- Paques F, Haber JE (1999) Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev 63: 349–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidoux AL, Allshire RC (2005) The role of heterochromatin in centromere function. Philos Trans R Soc Lond B Biol Sci 360: 569–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prudden J, Evans JS, Hussey SP, Deans B, O'Neill P, Thacker J, Humphrey T (2003) Pathway utilization in response to a site-specific DNA double-strand break in fission yeast. EMBO J 22: 1419–1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schueler MG, Sullivan BA (2006) Structural and functional dynamics of human centromeric chromatin. Annu Rev Genomics Hum Genet 7: 301–313 [DOI] [PubMed] [Google Scholar]
- Scott KC, Merrett SL, Willard HF (2006) A heterochromatin barrier partitions the fission yeast centromere into discrete chromatin domains. Curr Biol 16: 119–129 [DOI] [PubMed] [Google Scholar]
- Shinohara A, Ogawa H, Matsuda Y, Ushio N, Ikeo K, Ogawa T (1993) Cloning of human, mouse and fission yeast recombination genes homologous to RAD51 and recA. Nat Genet 4: 239–243 [DOI] [PubMed] [Google Scholar]
- Smith CE, Llorente B, Symington LS (2007) Template switching during break-induced replication. Nature 447: 102–105 [DOI] [PubMed] [Google Scholar]
- Smith CL, Matsumoto T, Niwa O, Klco S, Fan JB, Yanagida M, Cantor CR (1987) An electrophoretic karyotype for Schizosaccharomyces pombe by pulsed field gel electrophoresis. Nucleic Acids Res 15: 4481–4489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JG, Caddle MS, Bulboaca GH, Wohlgemuth JG, Baum M, Clarke L, Calos MP (1995) Replication of centromere II of Schizosaccharomyces pombe. Mol Cell Biol 15: 5165–5172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Murakami S, Chikashige Y, Funabiki H, Niwa O, Yanagida M (1992) A low copy number central sequence with strict symmetry and unusual chromatin structure in fission yeast centromere. Mol Biol Cell 3: 819–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorslund T, West SC (2007) BRCA2: a universal recombinase regulator. Oncogene 26: 7720–7730 [DOI] [PubMed] [Google Scholar]
- Virgin JB, Bailey JP, Hasteh F, Neville J, Cole A, Tromp G (2001) Crossing over is rarely associated with mitotic intragenic recombination in Schizosaccharomyces pombe. Genetics 157: 63–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E, Haase J, Paliulis LV, Joglekar A, Bond L, Bouck D, Salmon ED, Bloom KS (2008) Pericentric chromatin is organized into an intramolecular loop in mitosis. Curr Biol 18: 81–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Figure 5
Supplementary Figure 6
Supplementary Figure 7
Supplementary Figure Legeds
Supplementary Tables I–III
Supplementary Data