Abstract
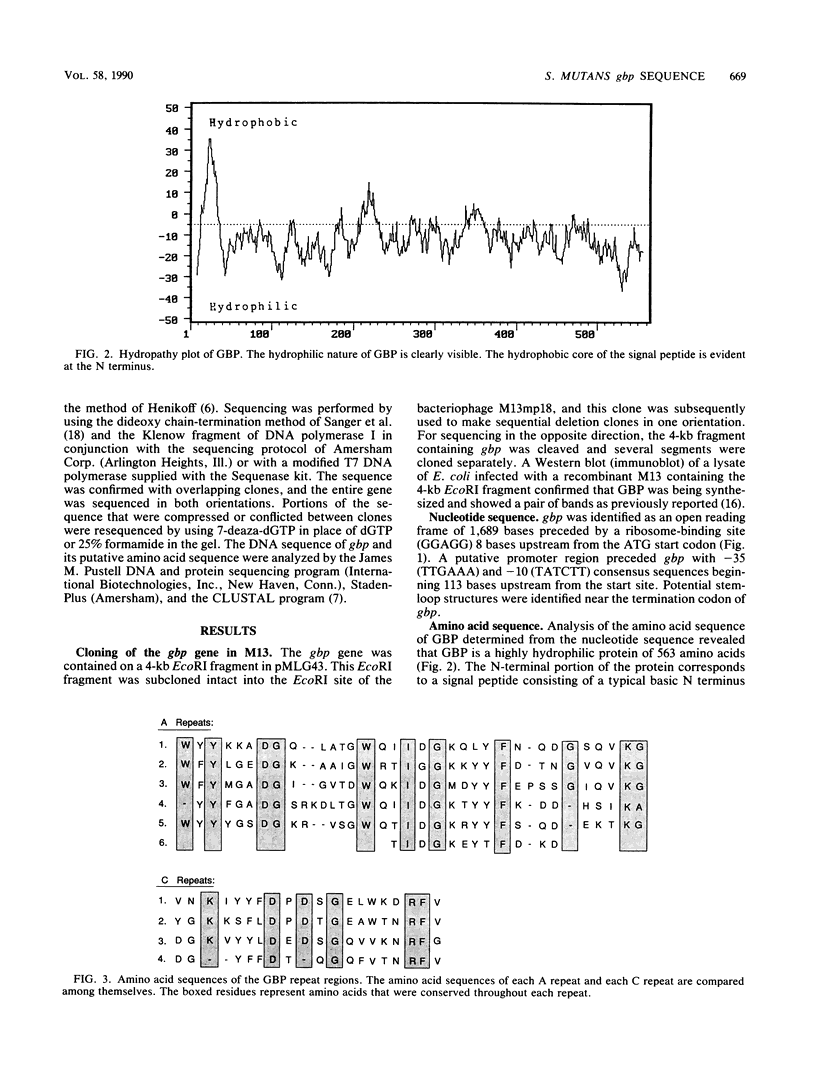
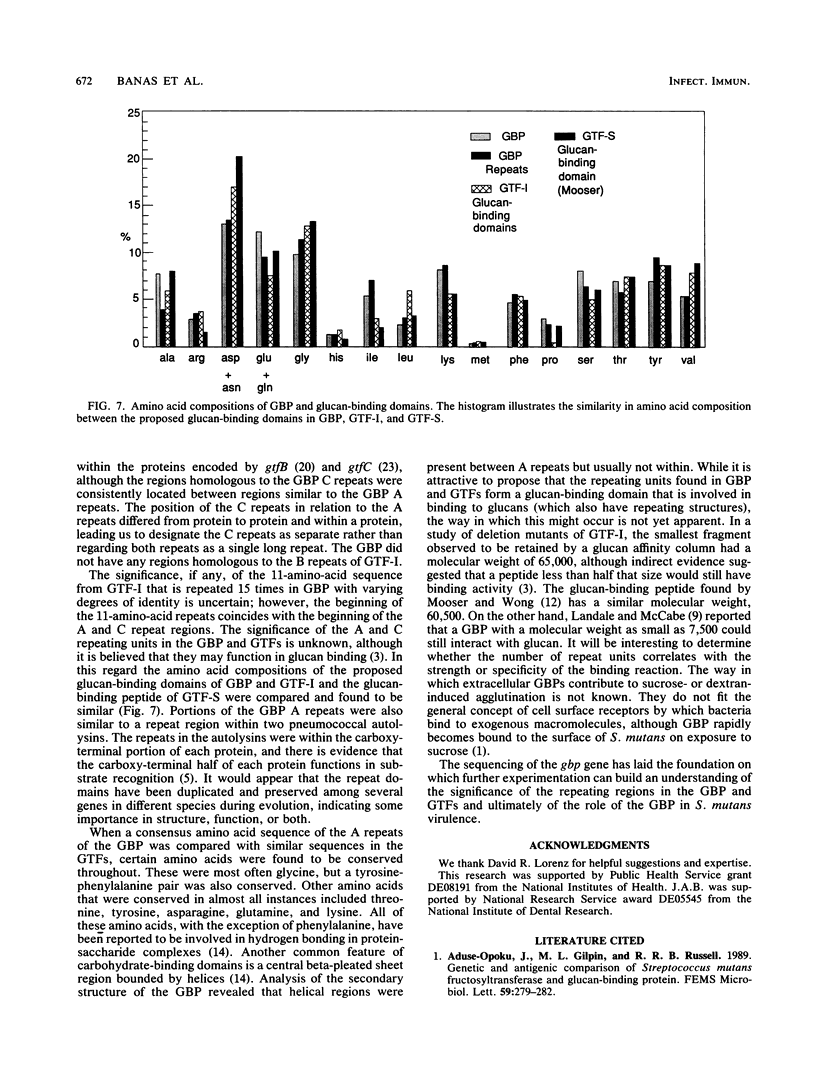
The nucleotide sequence of the gbp gene, which encodes the glucan-binding protein (GBP) of Streptococcus mutans, was determined. The reading frame for gbp was 1,689 bases. A ribosome-binding site and putative promoter preceded the start codon, and potential stem-loop structures were identified downstream from the termination codon. The deduced amino acid sequence of the GBP revealed the presence of a signal peptide of 35 amino acids. The molecular weight of the processed protein was calculated to be 59,039. Two series of repeats spanned three-quarters of the carboxy-terminal end of the protein. The repeats were 32 to 34 and 17 to 20 amino acids in length and shared partial identity within each series. The repeats were found to be homologous to sequences hypothesized to be involved in glucan binding in the GTF-I of S. downei and to sequences within the protein products encoded by gtfB and gtfC of S. mutans. The repeated sequences may represent peptide segments that are important to glucan binding and may be distributed among GBPs from other bacterial inhabitants of plaque or the oral cavity.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aduse-Opoku J., Gilpin M. L., Russell R. R. Genetic and antigenic comparison of Streptococcus mutans fructosyltransferase and glucan-binding protein. FEMS Microbiol Lett. 1989 Jun;50(3):279–282. doi: 10.1016/0378-1097(89)90432-1. [DOI] [PubMed] [Google Scholar]
- Douglas C. W., Russell R. R. Effect of specific antisera on adherence properties of the oral bacterium Streptococcus mutans. Arch Oral Biol. 1982;27(12):1039–1045. doi: 10.1016/0003-9969(82)90009-7. [DOI] [PubMed] [Google Scholar]
- Drake D., Taylor K. G., Bleiweis A. S., Doyle R. J. Specificity of the glucan-binding lectin of Streptococcus cricetus. Infect Immun. 1988 Aug;56(8):1864–1872. doi: 10.1128/iai.56.8.1864-1872.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferretti J. J., Gilpin M. L., Russell R. R. Nucleotide sequence of a glucosyltransferase gene from Streptococcus sobrinus MFe28. J Bacteriol. 1987 Sep;169(9):4271–4278. doi: 10.1128/jb.169.9.4271-4278.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferretti J. J., Russell R. R., Dao M. L. Sequence analysis of the wall-associated protein precursor of Streptococcus mutans antigen A. Mol Microbiol. 1989 Apr;3(4):469–478. doi: 10.1111/j.1365-2958.1989.tb00193.x. [DOI] [PubMed] [Google Scholar]
- García E., García J. L., García P., Arrarás A., Sánchez-Puelles J. M., López R. Molecular evolution of lytic enzymes of Streptococcus pneumoniae and its bacteriophages. Proc Natl Acad Sci U S A. 1988 Feb;85(3):914–918. doi: 10.1073/pnas.85.3.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Higgins D. G., Sharp P. M. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene. 1988 Dec 15;73(1):237–244. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- Kühn S., Fritz H. J., Starlinger P. Close vicinity of IS1 integration sites in the leader sequence of the gal operon of E. coli. Mol Gen Genet. 1979 Jan 2;167(3):235–241. doi: 10.1007/BF00267414. [DOI] [PubMed] [Google Scholar]
- Landale E. C., McCabe M. M. Characterization by affinity electrophoresis of an alpha-1,6-glucan-binding protein from Streptococcus sobrinus. Infect Immun. 1987 Dec;55(12):3011–3016. doi: 10.1128/iai.55.12.3011-3016.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipman D. J., Pearson W. R. Rapid and sensitive protein similarity searches. Science. 1985 Mar 22;227(4693):1435–1441. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- Loesche W. J. Role of Streptococcus mutans in human dental decay. Microbiol Rev. 1986 Dec;50(4):353–380. doi: 10.1128/mr.50.4.353-380.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooser G., Wong C. Isolation of a glucan-binding domain of glucosyltransferase (1,6-alpha-glucan synthase) from Streptococcus sobrinus. Infect Immun. 1988 Apr;56(4):880–884. doi: 10.1128/iai.56.4.880-884.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller-Hill B., Crapo L., Gilbert W. Mutants that make more lac repressor. Proc Natl Acad Sci U S A. 1968 Apr;59(4):1259–1264. doi: 10.1073/pnas.59.4.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiocho F. A. Carbohydrate-binding proteins: tertiary structures and protein-sugar interactions. Annu Rev Biochem. 1986;55:287–315. doi: 10.1146/annurev.bi.55.070186.001443. [DOI] [PubMed] [Google Scholar]
- Russell R. R., Coleman D., Dougan G. Expression of a gene for glucan-binding protein from Streptococcus mutans in Escherichia coli. J Gen Microbiol. 1985 Feb;131(2):295–299. doi: 10.1099/00221287-131-2-295. [DOI] [PubMed] [Google Scholar]
- Russell R. R., Donald A. C., Douglas C. W. Fructosyltransferase activity of a glucan-binding protein from Streptococcus mutans. J Gen Microbiol. 1983 Oct;129(10):3243–3250. doi: 10.1099/00221287-129-10-3243. [DOI] [PubMed] [Google Scholar]
- Russell R. R. Glucan-binding proteins of Streptococcus mutans serotype c. J Gen Microbiol. 1979 May;112(1):197–201. doi: 10.1099/00221287-112-1-197. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiroza T., Kuramitsu H. K. Sequence analysis of the Streptococcus mutans fructosyltransferase gene and flanking regions. J Bacteriol. 1988 Feb;170(2):810–816. doi: 10.1128/jb.170.2.810-816.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiroza T., Ueda S., Kuramitsu H. K. Sequence analysis of the gtfB gene from Streptococcus mutans. J Bacteriol. 1987 Sep;169(9):4263–4270. doi: 10.1128/jb.169.9.4263-4270.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoker N. G., Fairweather N. F., Spratt B. G. Versatile low-copy-number plasmid vectors for cloning in Escherichia coli. Gene. 1982 Jun;18(3):335–341. doi: 10.1016/0378-1119(82)90172-x. [DOI] [PubMed] [Google Scholar]
- Ueda S., Shiroza T., Kuramitsu H. K. Sequence analysis of the gtfC gene from Streptococcus mutans GS-5. Gene. 1988 Sep 15;69(1):101–109. doi: 10.1016/0378-1119(88)90382-4. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- von Heijne G. Patterns of amino acids near signal-sequence cleavage sites. Eur J Biochem. 1983 Jun 1;133(1):17–21. doi: 10.1111/j.1432-1033.1983.tb07424.x. [DOI] [PubMed] [Google Scholar]


