Abstract
Ion channel biogenesis involves an intricate interplay between subunit folding and assembly. Channel stoichiometries vary and give rise to diverse functions, which impacts on neuronal signalling. AMPA glutamate receptor (AMPAR) assembly is modulated by RNA processing. Here, we provide mechanistic insight into this process. First, we show that a single alternatively spliced residue within the ligand-binding domain alters AMPAR secretion from the ER. Local contacts differ between Leu758 of the GluR2-flop splice form as compared with the flip-specific Val758, which is transmitted globally to alter resensitization kinetics. Detailed biochemical and functional analysis of mutants suggest that AMPARs sample the gating cascade prior to ER export. Irreversibly locking the receptor within various states of the cascade severely attenuates ER transit. Alternative RNA processing by contrast, shifts equilibria between transition states reversibly and thereby modulates secretion kinetics. These data reveal how RNA processing tunes AMPAR biogenesis, and imply that gating transitions in the ER determine iGluR secretory traffic.
Keywords: AMPA receptor gating, endoplasmic reticulum, RNA editing
Introduction
Fast excitatory neurotransmission in vertebrate brains is mediated by three subfamilies of ionotropic glutamate receptors (iGluRs)—AMPA (α-amino-3-hydroxy-5-methyl-4-isoxazolepropionate receptors), NMDA (N-methyl-D-aspartic acid) and kainate receptors (Dingledine et al, 1999). Each family comprises a set of subunits, which assemble in various combinations, giving rise to a spectrum of functionally distinct receptor tetramers (Dingledine et al, 1999). During brain development neurons engage different sets of iGluR heteromers, which is crucial for establishing and refining functional connections (Monyer et al, 1991; Cull-Candy et al, 2001). In the adult, specific tetramers are confined to certain neuronal circuits (Gardner et al, 2001), and can be selectively targeted to dendritic subdomains (Tóth and McBain, 2000). Moreover, synapses can employ channels with altered stoichiometries depending on the type and pattern of excitation received (Cull-Candy et al, 2006; Liu and Zukin, 2007). Signalling through diverse receptor heteromers endows neurons with dynamic response properties that shape the postsynaptic response, and ultimately control action potential output.
AMPARs mediate the fast component of iGluR signalling. They are composed of four subunits, GluR1–4 (or GluRA–D), which are diversified further by RNA processing—flip/flop alternative splicing and adenosine-to-inosine editing (Dingledine et al, 1999; Reenan, 2001; Seeburg, 2002; Seeburg and Hartner, 2003). Re-coding at the Q/R-editing site within the pore loop of the GluR2 subunit is highly efficient and alters ion conductance (Isaac et al, 2007). By contrast, R/G editing and flip/flop splicing within the ligand-binding domain (LBD) is developmentally regulated and modulates desensitization properties (Sommer et al, 1990; Lomeli et al, 1994). Apart from altering channel function, these RNA processing sites also impact on AMPAR assembly (Greger and Esteban, 2007; Greger et al, 2007).
Assembly of membrane proteins, which occurs at the endoplasmic reticulum (ER) membrane, poses a central cell biological question: how does the cell achieve preferential heteromerization? As individual subunit mRNAs are translated by polysomes (approximately 30 ribosomes could decorate the length of a GluR2 mRNA; Staehlin et al, 1964), a high concentration of a given subunit will accumulate in the ER in a spatiotemporal manner, a scenario expected to favour homomer formation. Subunit associations could, however, be reversible and preferential heteromerization might be facilitated by tighter or thermodynamically more favourable interactions between heteromeric assembly partners.
The folding of membrane proteins is tightly regulated, acquisition of a native quaternary structure signals ER export competence (Ellgaard and Helenius, 2003). Studies in yeast demonstrated a correlation between the stability of the native fold and secretion efficiency, with higher conformational stability promoting ER export (Kowalski et al, 1998). The biogensesis of oligomers involves concerted folding and assembly steps, which are monitored by the ER quality control (QC) machinery at multiple stages (Wanamaker et al, 2003; Schwappach, 2008). Details of the pathways leading to natively folded and export-competent oligomers are complex and largely elusive (Deutsch, 2003). The kinetics of these processes determine ER export rates and ultimately the density of surface-expressed receptors.
In response to glutamate, synaptic iGluRs undergo a spectrum of large-scale conformational alterations. The gating cascade is initiated by glutamate docking to the upper lobe of the LBD clamshell (Figure 7A; Abele et al, 2000), resulting in cleft closure and channel opening (Mayer, 2005). In the current model, the open-channel closed-clamshell state is of high energy, and is resolved by rearrangement of adjacent LBDs into the desensitized state (Sun et al, 2002).
Here, we demonstrate that conformations resembling steps in the AMPAR gating cascade are detected in the ER. Mutations, which irreversibly lock the channel at a given state within the cascade, severely attenuate ER transit. This suggests that channel function is monitored prior to secretion and determines ER export kinetics. Guided by high-resolution structures, mutants were targeted to three strategic locations within the LBD: the helix J/K kink, the LBD dimer interface and the mouth of the binding cleft (Figure 8). Detailed biochemical and functional analyses reveal that these mutations affect AMPARs at various stages of their biogenesis––in addition to altering channel formation they also modulate the quarternary conformation of assembled tetramers. In contrast to mutation, alterations introduced into the LBD by RNA editing and alternative splicing modulate these parameters reversibly. By altering the ER dwell time they impact on assembly competence and thus the tetrameric nature of the receptor.
These findings have been reported recently in abstract format (Greger and Williams, 2007).
Results
A-to-I RNA editing provides a (potentially regulatable) genetic mechanism that alters AMPAR assembly properties (Greger and Esteban, 2007). We reported recently that R/G editing within the LBD attenuates assembly and trafficking of GluR2 (R2; Greger et al, 2006). In the course of these experiments, we realized that alternative flip/flop splicing adjacent to the R/G site (Figure 1A; Sommer et al, 1990) is also a major determinant of AMPAR biogenesis. To reveal the mechanism by which these modules operate, we analysed alternatively processed R2 isoforms, as well as a panel of structure-guided gating mutants. Three key regions of the LBD—the helix J/K kink, the LBD dimer interface and the mouth of the clamshell—were mutated. We provide below detailed mechanistic insight into how these LBD segments shape AMPAR biogenesis in the ER.
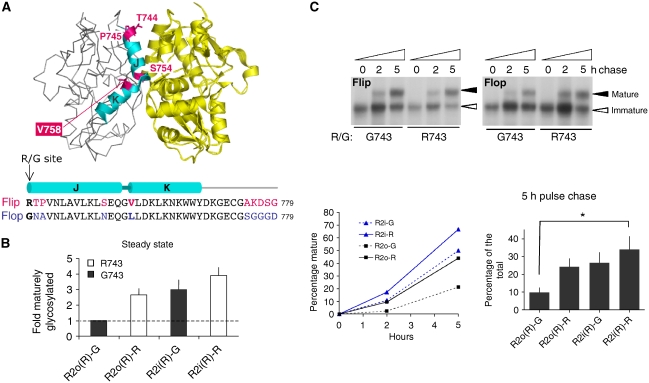
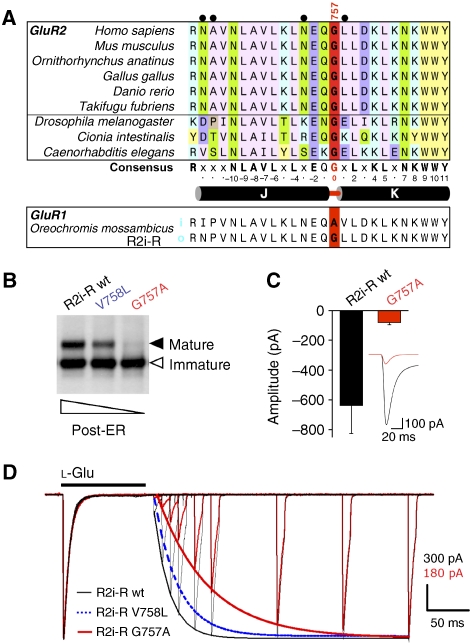
Figure 1.
Flip/flop splicing alters R2 assembly and ER exit kinetics. (A) Crystal structure of the R2i LBD (pdb: 2UXA), the two protomers are colour-coded (grey and yellow). On the left protomer, helices J and K are highlighted in cyan, with alternative residues indicated in magenta. The sequence alignment below encompasses helices J and K plus downstream sequence, which is altered by i/o splicing. Flip-specific residues are in magenta, flop residues are in blue. The R/G editing site is denoted in bold. The figure was drawn with PyMol (DeLano, 2002). (B) R2 post-ER receptor isoforms at steady state. Levels of maturely glycosylated receptor relative to total receptor was determined. Results were normalized to levels of maturely glycosylated R2o(R)-G receptor, to compensate for experimental variations. Means of fold difference±s.e.m. are presented (n=7). (C) [35S]Methionine/cysteine pulse-chase analysis. HEK293 cells were pulsed for 10 min and chased for up to 5 h. Samples were immunoprecipitated, EndoH digested and visualized by flurography. Flip R/G isoforms are shown on the left, flop varieties on the right. Immaturely glycosylated (ER) receptor is denoted by an empty arrowhead, mature receptor by a filled arrowhead. This experiment is shown quantified below (left). Radioactive signals were detected in a Phosphoimager (Typhoon) and analysed with ImageQuant software. Means±s.e.m. from pooled data (5 h time points) are shown bottom right (n=6; Kruskal–Wallis test; *P<0.05).
Flip/flop splicing alters GluR2 oligomerization and ER secretion kinetics
Flip/flop alternative splicing changes residues along helices J and K within the LBD dimer interface (Figure 1A). Together with R/G editing at position 743, four alternative GluR2 (R2) isoforms are generated, which coexist in neurons. Editing Arg743 to Gly743 drastically attenuated R2 ER exit in HEK293 cells, reducing levels of endoglycosidase H-resistant, and thus maturely glycosylated (post-ER) receptors at steady state (i.e. 36 h post-transfection; Figure 1B). This editing effect was subtle in flip receptors (1.3±0.4-fold, n=7) but pronounced in flop isoforms (2.7±0.4-fold, n=7), resembling trafficking patterns observed in neurons (Greger et al, 2006; for a key to isoform abbreviations, see Materials and methods). Interestingly, the splice-form per se exhibited altered R2 trafficking properties. Greater levels of post-ER receptors accumulated for flip isoforms, when compared with their flop counterparts (Figure 1B). Of the four R2 isoforms, R2i(R)-R matures most efficiently (3.9±0.5-fold, n=7), relative to R2o(R)-G. More detailed kinetic data, using a [35S]methionine/cysteine pulse-chase protocol (Greger et al, 2002), produced a similar pattern and revealed accelerated maturation kinetics of flip receptors within 5 h of chase (Figure 1C). Splice-form-specific trafficking properties have also been observed recently for GluR4 (Coleman et al, 2006), and together with our findings they reveal a central role for alternative RNA processing in AMPAR secretory traffic (Greger and Esteban, 2007).
As the biogenesis of oligomeric polypeptides encompasses a poorly understood interplay between subunit folding and assembly prior to ER exit (Deutsch, 2003), we tested whether i/o splicing also alters AMPAR assembly. As Q/R-edited R2(R) subunits give rise to unstable tetramers (Greger et al, 2003), we analysed oligomerization of Q/R-unedited R2(Q) isoforms. Glycerol velocity sedimentation indeed demonstrated greater levels of flip receptors in a peak harbouring AMPAR tetramers (P2; Supplementary Figure S1A; Greger et al, 2003). Furthermore, blue-native PAGE analysis (BN–PAGE) of total cell lysates revealed extensive smearing of flop receptors above the tetrameric range (T; Supplementary Figure S1B). This heterogeneity, which was less evident in flip, may reflect folding intermediates and/or receptor–chaperone complexes, and indicates slower ER processing of R2o. In summary, in addition to RNA editing, alternative splicing in the LBD provides an endogenous switch that modulates assembly and forward traffic of the functionally critical R2 subunit.
The Leu758/Val758 switch
To reveal the determinants underlying this regulation, flip residues were altered to flop, and levels of post-ER receptor were assessed. In R2, nine amino-acid residues differ between the flip and flop exons (Figure 1A; Sommer et al, 1990); three locate to helix J, which forms the heart of the LBD dimer interface (Armstrong and Gouaux, 2000), a Val to Leu change occurs at the start of helix K (position 758), and five consecutive alterations take place outside the LBD core, close to the start of transmembrane segment 4. Mutation to triple glycines (776–778) located outside the LBD core into R2i did not significantly impact on ER traffic (not shown). Of the three side chains projecting from helix J mutation of Ser754 only subtly reduced R2i trafficking (1.3±0.6-fold, n=5), whereas residues 744 and 745 adjacent to the R/G site had no visible effect even when mutated together (Figure 2A). However, the conservative and seemingly minor alteration from Val758 to Leu at the helix J/K kink, substantially reduced ER exit of R2(R)i-R (2.3±0.3-fold, n=7; Figure 2A). In the reverse experiment, secretion efficiency of the L758V flop mutant increased by ∼2-fold (1.9±0.9-fold, n=6) relative to flop wt at steady state (Figure 2A), and after 5 h of chase (Figure 2B). This point mutation essentially reversed the flip/flop trafficking phenotype. The impact of the 758 switch was observed in both R2 isoforms, R/G edited and unedited (Supplementary Figure S2A). In addition to trafficking in HEK293 cells, Leu758 also lowered levels of maturely glycosylated R2i(R)-R in hippocampal neurons (not shown). Therefore, extension of the 758 side chain by a single methylene group largely underlies the different trafficking properties of the flip and flop isoforms. The Val/Leu780 position has recently been shown to determine splice-form specific trafficking of GluR4 (Coleman et al, 2006).
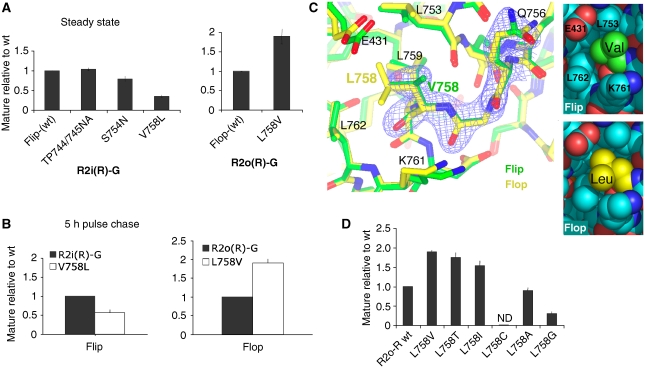
Figure 2.
The Leu/Val758 switch determines the flip/flop trafficking phenotype. (A) Position 758 reverses the i/o trafficking phenotype. Mutagenesis of alternative residues within the R2i(R)-G LBD core (left) and mutation of L758 to Val in R2o(R)-G (right). Levels of maturely glycosylated receptor, relative to total receptor, was determined as described in Figure 1B. (B) Residue 758 alters ER export kinetics. The 5 h chase time points of [35S]met/cys-labelled R2i(R)-G wt, and V758L mutant (left), and the R2o(R)-G, L758V pair (right) are shown. Mutant values were normalized and expressed relative to wt; means±s.e.m. (n=6). (C) Superposition of the glutamate-bound LBDs, R2i-R (green; pdb 2UXA) and R2o-G (yellow; pdb 1FTJ). Only helix K backbone coordinates were used for the alignment, electron density is shown for R2i-R from Q756 to V758 (0.17e/Å3 contour level). Residues within 4 Å of position 758 are indicated. Local contacts of position 758 are shown in space filling representation on the right. Closer contacts between Leu758 and E431, L753, K761 and L762 relative to Val758 are evident (bottom panel). Figure 2C was drawn with CCP4mg. (D) Mutagenesis of position 758 in the R2o-R background. Levels of maturely glycosylated mutants, relative to wt, was determined as described in Figure 1B. Levels of maturely glycosylated L758C were too low to be determined accurately (ND, not determined).
Glycerol velocity sedimentation indicated that the Leu/Val758 switch also impacts on subunit assembly. Altered sedimentation properties were observed in HEK293 cells and in neurons (Supplementary Figure S2B). For R2i-R V758L, stable tetramers formed and sedimented in P2, albeit to a lesser extent than R2i-R wt. In addition, increased appearance of receptor running at ∼250 kDa was observed, which likely reflects SDS-resistant assembly intermediates (Greger et al, 2006). This assembly preference may underlie the functional dominance of flip receptors reported earlier (Brorson et al, 2004). Therefore, a seemingly subtle change in the helix J/K kink region introduced by alternative splicing alters R2 assembly and secretion from the ER.
The atomic environment of the Leu758/Val758 switch
How can a relatively minor side chain alteration have a profound impact on receptor biogenesis? Structural information exists for glutamate-bound R2i and R2o LBDs (Armstrong and Gouaux, 2000; Greger et al, 2006), enabling us to examine the atomic environment of the Val/Leu758 switch. We considered that the Val to Leu change is transmitted along the protein backbone, and impacts on conformational alterations within the LBD. Position 758 locates to the joint of helices J and K (Figure 1A), adjacent to a highly conserved glycine (Gly757; Figure 5A). Helix J contributes to the LBD dimer interface, which rearranges during desensitization (Sun et al, 2002; Mayer, 2005). Helix K engages in intersubunit interactions on desensitization (Armstrong et al, 2006). The C terminus of helix K is disulphide bonded to helix I, which itself undergoes extensive backbone motions during gating (Fenwick and Oswald, 2008). Helix K thus constitutes a third link between the two clamshell lobes.
Local alignments of helix K coordinates indicate subtle changes at the C terminus of the helix (0.7 Å), close to the disulphide (Cys773), which connects the two lobes and which undergoes dynamic changes in solution (Fenwick and Oswald, 2008). At the level of the 758 side chains, differences inherent to the stereochemistry of Val and Leu residues are apparent (Figure 2C). First, the longer, more flexible leucine reaches deeper towards the protein interior than the shorter flip-specific Val758. Space filling representations illustrate how the δ2 moiety of Leu758 engages Glu431, Leu753 and Leu762 in van der Waals contacts, and that the back of this pocket is formed by Leu759 (Figure 2C). Another key difference concerns the Cβ branch of Val758, with the γ2 group projecting within 4 Å of Gln756 Cβ, which might contribute to local steric hindrance at the helix J/K joint.
To test size and structural constraints for this key residue, we generated a series of mutations at position 758 (Figure 2D). Replacing Leu758 in R2o-R with threonine, which resembles the structure of Val and is therefore expected to populate similar rotamer space (Supplementary Figure S3A), accelerated R2o-R ER exit similar to valine (1.7±0.7-fold, n=4). Further truncation of the 758 side chain to Ala did not improve forward traffic, and in the cases of L758C and L758G was even inhibitory, suggesting a specific spatial requirement such as a β-branch rather than simply reduced chain volume. Indeed mutation to isoleucine, L758I, facilitated R2o-R traffic to levels approaching the L758V mutant (1.5±0.6-fold, n=9). Isoleucine is expected to fill the space occupied by the Val758 γ2 group (Supplementary Figure S3B), and occurs naturally at the analogous position in R3 flip (Sommer et al, 1990). Conversely, when introduced into R2i-R, V758I slightly reduced ER exit, which was severely reduced for the truncation mutants, V758A and V758C (not shown). Therefore, a specific structural requirement at position 758 mediates accelerated assembly and forward traffic. To determine whether the Val/Leu switch does contribute to conformational alterations, we next assayed its impact on AMPAR gating.
Alterations in the helix J/K kink region modulate AMPAR resensitization
We first assayed whether residue 758 affected ligand binding. However, saturation binding experiments using the agonist [3H]flurowillardiine ([3H]FW) (Kessler and Arai, 2006) revealed a similar Kd for R2o-R and the L758V mutant (Supplementary Figure S4). Similar results were obtained with the R2i pair (R2i-R and V758L mutant; not shown). To determine whether mutation of the helical kink impacts on receptor conformation, we initially analysed gross functional properties, by recording whole-cell AMPAR currents from transiently transfected HEK293 cells. As residues in helices J/K determine desensitization properties, we performed paired-pulse protocols, which measure recovery from desensitization and so probe the stability of the desensitized state (two 10 ms pulses of 1 mM glutamate separated by 0.1–3 s in 0.1-s intervals). R2i-R recovered from desensitization visibly faster than R2o-R (Figure 3A), as has been reported earlier (Koike et al, 2000; Quirk et al, 2004). R2i-R V758L mutant currents, however, recovered markedly slower than R2i-R, approaching a recovery profile seen with R2o-R (Figure 3A and B). Interestingly, the reverse was observed for the flop pair, where L758V recovered from desensitization 1.75-fold faster than R2o-R, resembling the R2i-R profile (Figure 3B and C). This indicates that Leu758 stabilizes the desensitized state and suggests a link between ER trafficking and recovery rates, with slowed recovery correlating with reduced ER exit. Indeed, speeding of recovery was also observed with the β-branch mutants, R2o-R L758T and L758I (Supplementary Figure S5; see below), both of which facilitate ER exit (Figure 2D). This relationship between the stability of the desensitized conformation and secretory traffic was dissected further.
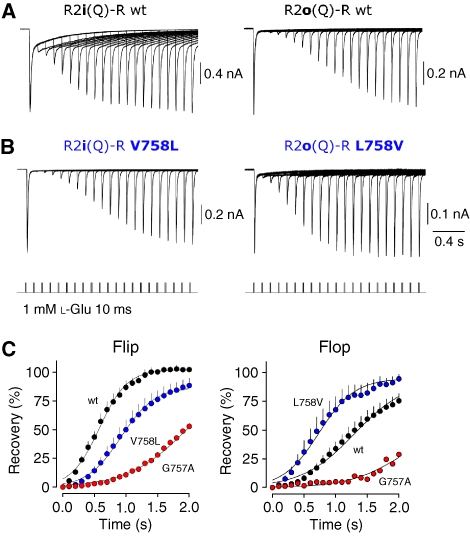
Figure 3.
Recovery from desensitization is shaped by the 758 side chain. (A) Recovery from desensitization in response to a 10 ms pulse of 1 mM glutamate is probed by the subsequent application of a second glutamate pulse delivered between 0.1 and 2 s later (lower traces). Overlain whole-cell currents show that recovery from desensitization is faster for R2i(Q)-R (left) than for R2o(Q)-R (right). (B) Position 758 switches recovery profiles bidirectionally. V758L slows recovery relative to flip wt (left), whereas L758V speeds recovery relative to flop wt (right). (C) Summary data describing the time course of recovery from desensitization for R2i wt plus corresponding 757 and 758 mutants (left), and R2o wt (right). Lines represent fits with a single Boltzmann function.
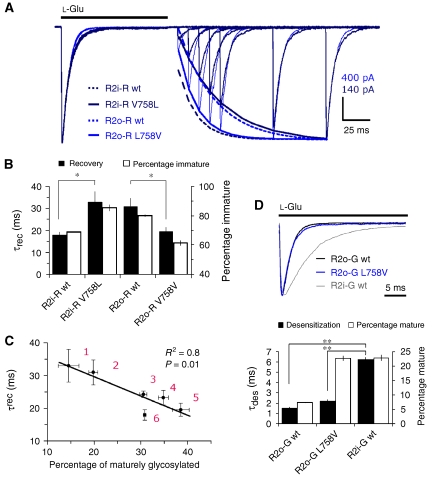
First, to assess these findings quantitatively, we determined absolute gating kinetics utilizing fast solution exchange to membrane patches (∼200 μs). Rate constants for deactivation (not shown), desensitization and recovery of R2 wt receptors were similar to earlier reports (Table I; Supplementary Figure S6; Koike et al, 2000; Quirk et al, 2004). However, as observed with whole-cell recording, switching Val to Leu slowed recovery of R2i-R V758 L (∼1.8-fold) relative to R2i-R (Figure 4A; Table I). The opposite was observed for flop receptors where R2o-R L758V recovered faster than R2o-R (∼1.6-fold; Table I), and approached values of R2i-R (Figure 4A). These bidirectional shifts in recovery almost precisely matched trafficking phenotypes, where fast recovery correlated with increased ER export (Figure 4B and C). Moreover, this relationship could be extended to the additional β-branch 758 mutants (Table I; Figure 4C). In contrast to recovery, there was no obvious match between trafficking and entry into desensitization for these kink mutants (Table I; Supplementary Figure S7). This is illustrated for the R2o-G pair where desensitization decays of wt and L758V mutant largely superimpose and are clearly faster than R2i-G (Figure 4D). Trafficking rates of R2i-G, however, are almost identical to R2o-G L758V. These results reveal an unexpected relationship between the desensitized conformation and ER export, with more stably desensitized receptors displaying attenuated forward traffic.
Table 1.
Kinetic properties of R2 gating mutants
| τrec | τdes | %des | N | |
|---|---|---|---|---|
| R2o(Q)-R | 30.9 (±3.8) | 3.2 (±0.3) | 99.8 | 6 |
| R2o(Q)-R L758T | 23.2 (±2.2) | 3.6 (±0.3) | 99.3 | 6 |
| R2o(Q)-R L758I | 24.2 (±1.0) | 3.7 (±0.4) | 98.9 | 4 |
| R2o(Q)-R L758V | 19.5 (±2.0) | 5.0 (±0.9) | 99.5 | 5 |
| R2i(Q)-R | 17.9 (±1.6) | 5.9 (±0.6) | 99.0 | 5 |
| R2i(Q)-R V758L | 32.9 (±5.0) | 5.0 (±0.6) | 99.5 | 5 |
| R2i(Q)-R G757A | 82.4 (±11.0) | 6.6 (±0.3) | 99.3 | 5 |
| R2i(Q)-G | 13.8 (±0.5) | 6.2 (±0.3) | 96.7 | 4 |
| R2o(Q)-G | 20.3 (±3.4) | 1.5 (±0.1) | 99.5 | 4 |
| R2o(Q)-G L758V | 15.5 (±1.0) | 2.2 (±0.2) | 98.8 | 5 |
| R2o(Q)-G L483A | 21.3 (±0.8) | 4.6 (±0.4) | 97.3 | 7 |
Figure 4.
Correlation between ER traffic and recovery from desensitization. (A) L-Glu (1 mM) was applied to membrane patches for 100 ms to desensitize receptors, consecutive 100 ms applications were typically delivered at 10-ms intervals, to map the time course of recovery (>90%). Representative recovery traces of 758 mutants are normalized and superimposed. R2i(Q) V758L displays marked slowing of recovery relative to the corresponding flop mutant, R2o(Q) L758V (fits are denoted by solid lines), whereas desensitization decays largely overlay. The bidirectional switch in kinetics mediated by 758 is emphasized by recovery fits for the corresponding wt receptors, indicated by stippled lines. In order of appearance in the legend (top to bottom), recovery time constants (τrec, ms) for the fits were 16.4, 35.7, 34.2 and 17.3. Current amplitude scales are indicated for the 758 mutant traces. (B) Relationship between recovery kinetics and trafficking. Because of the inverse relationship between the two parameters, percentage of immaturely glycosylated receptor is plotted (right axis). Recovery kinetics were significantly different between the wt and corresponding 758 mutant receptors *P<0.05; ANOVA. Error bars represent s.e.m. (C) Correlation (Pearson's) between recovery kinetics (τrec) and levels of maturely glycosylated kink mutants. Data were fitted by linear regression (R2=0.8). Sample size for each data point is listed in brackets for τrec and glycosylation measurements, respectively: 1: R2i-R V758L (n=5 and 7); 2: R2o-R wt (n=6 and 7); 3: R2o-R L758I (n=4 and 4); 4: R2o-R L758T (n=6 and 4); 5: R2o-R L758V (n=5 and 6); 6: R2i-R wt (n=5 and 7). Error bars represent s.e.m. (D) Desensitization of kink mutants does not correlate with trafficking properties. Representative desensitization decays to a 100 ms application of L--Glu (1 mM) are normalized to illustrate almost perfect superposition of R2o-G and the corresponding L758V mutant (top panel). However, their trafficking properties vary >2-fold (bottom panel). Trafficking is almost identical for R2o-G L758V and R2i-G, which display marked differences in desensitization kinetics (black bars). In order of appearance in the legend (top to bottom), current amplitude (nA) and τdes (ms) for traces in the top panel were −0.72, −0.86 and −1.35, and 1.4, 1.9 and 6.1, respectively. Error bars in the bottom panel represent s.e.m.; **P=0.001; ANOVA. From left to right, sample size for τdes and glycosylation measurements were 4, 5 and 4, and 7, 4 and 7, respectively.
To gain further insight into this regulation, we generated an additional mutant. Adjacent to position 758 lies Gly757, which is highly conserved within AMPAR orthologues (Figure 5A). This Gly residue also occurs in prokaryotic LBD analogues, which exhibit a conserved tertiary structure (Supplementary Figure S8A; Quiocho and Ledvina, 1996). Interestingly, a Gly is switched to Ala at the helical joint in GluR1i of Oreochromis mossambicus, as a result of i/o splicing (Figure 5A). As glycines give rise to greater main chain flexibility, mutation to Ala is expected to alter conformational freedom of this segment, and may thereby transmit conformational changes analogous to the adjacent position 758. G757A reduced levels of post-ER R2i-R receptors to a greater extent than V758L, in neurons (not shown) and in HEK293 cells (Figure 5B). Reduced surface expression of G757A was also evident by smaller current amplitudes when compared with R2i-R (peak amplitude in whole-cell mode: R2o=−650±184 pA, n=6; R2o G757A=−80±15 pA, n=6; Student's two-tailed t-test; P<0.01; Figure 5C). Importantly, in line with the correlation observed above, this mutation profoundly slowed recovery kinetics (∼4.6-fold; Table I). In fact, G757A resulted in a larger attenuation of recovery than V758L, closely matching the gradual reduction in ER traffic observed for these kink mutants (Figures 3C and 5D). Again, desensitization rates of G757A were almost identical to R2i-R corroborating that the helix J/K kink modulates aspects of recovery (Figure 5D; Table I). Taken together, these results reveal that the helix K kink shapes AMPAR resensitization, and suggest that conformational alterations associated with this state are sensed by ER QC. Natural alterations (V758 and L758), modulate ER exit rates—they likely produce reversible shifts in thermodynamic equilibria between conformational states (Figure 8). The engineered G757A mutation, however, shifts the equilibrium farther towards a conformation resembling the desensitized state, which signals ER retention.
Figure 5.
Mutation of Gly757 to Ala stabilizes the desensitized state. (A) ClustalW alignment of R2 orthologues. Alignment of protein sequence encompassing helices J and K highlights the high level of sequence conservation within vertebrates (top six alignments), and between vertebrates and invertebrates (D. melanogaster, C. intestinalis and C. elegans). Gly757 (red) is surrounded by a scaffold of highly conserved residues (‘consensus'). Residues altered by i/o splicing (grey spheres) are slotted into this conserved scaffold (G757=0); L758 forms van der Waals contacts with some conserved side chains - L753 (−4), L759 (+2), K761 (+4) and L762 (+5). (B) The Gly757 to Ala mutation results in ER retention. EH glycosylation profiles of R2i(R)-R and the corresponding, V758L and G757A mutants. The graded reduction in ER export competence is indicated below the gel. (C) Pooled data showing the peak amplitude of ensemble glutamate-activated whole-cell currents generated by R2i(Q)-R wt and G757A (red) mutant receptors. The inset illustrates the ensemble average of responses generated in each cell expressing R2i(Q)-R wt or R2i(Q)-R G757A (red). (D) Normalized recovery traces emphasize the slowing of resensitization for the R2i-R G757 mutant (red trace). The fit for a representative R2i-R V758L trace is indicated by a stippled blue line. This profile matches levels of post-ER receptor shown in (B). Desensitizing application of 1 mM L--Glu for 100 ms was followed by 10 ms applications to map the time course of recovery. Note the almost perfect overlay of the first response, indicating similar rates of desensitization between R2i-R (black) and the R2i-R G757A mutant (red). In order of appearance in the legend (top to bottom), recovery time constants (τrec, ms) for the fits were 18.5, 28.3 and 60.9. Current amplitude scales are indicated for the R2i-R and R2i-R G757A traces. See Table I.
Reversible stabilization of the LBD dimer interface promotes secretion
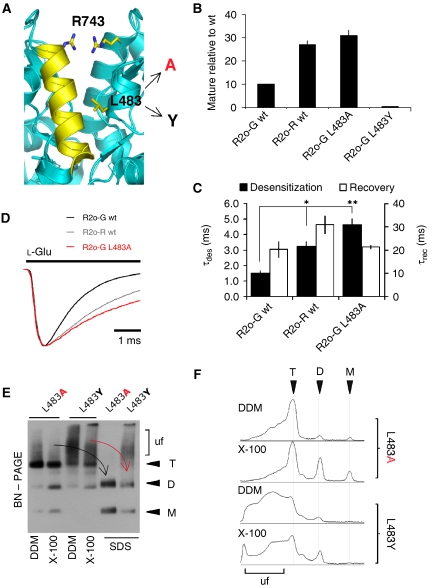
The finding that trapping the receptor within a transition state produces ER-retained conformations was encountered recently with mutants that localize to another LBD region, the LBD dimer interface (Greger et al, 2006). Stability of this interface determines the rate of entry into the desensitized state (Sun et al, 2002; Mayer, 2005). We reported that irreversibly locking the R2 interface through mutation (L483Y) severely attenuates ER exit (Greger et al, 2006). L483Y stabilizes LBD dimers in solution (Sun et al, 2002), and produces non-desensitizing receptors (Stern-Bach et al, 1998; Sun et al, 2002). In contrast to the profound attenuation of desensitization conferred by L483Y, Arg743 at the R/G site (Figure 6A) slows desensitization rates of R2o only by approximately 2-to 3-fold relative to the edited Gly743 isoform (Figure 6C and D; Grosskreutz et al, 2003), and facilitates assembly and forward traffic (Figure 6B; Greger et al, 2006). Slowing of desensitization to a similar extent has been observed with another interface mutant, R2i-G L483A (Horning and Mayer, 2004), and is seen when L483A is introduced into the R2o-G background (Figure 6D). Interestingly, L483A enhanced forward transport to an extent observed with Arg743 (3.1±0.8-fold, n=4), whereas irreversibly stabilizing dimer contacts through L483Y resulted in robust ER retention, as observed in neurons (Figure 6B; Greger et al, 2006). Therefore, in analogy to the helical kink region, shifting the equilibrium of this (‘upstream') gating step beyond an optimal range produces ER-retained receptors.
Figure 6.
Relationship between LBD interface stability and ER traffic. (A) Structure of the R2i LBD interface (pdb: 2UXA), showing R743 (the unedited R/G site) projecting from helix J, and L483 (projecting from helix D). L483 has been mutated to Ala and Tyr (arrows). (B) Levels of maturely glycosylated mutants, relative to flop-(wt), was determined as described in Figure 1B. Levels of mature L483Y mutant were below detection limit and are not determined (ND). (C) Bar graph plotting rates of desensitization and of recovery. The gradual slowing of desensitization matches the gradual increase in trafficking of R2o-R and R2o-G L483A, relative to R2o-G (Figure 6B). Desensitization kinetics were significantly different from R2o-G. *P<0.01, **P<0.001; ANOVA; from left to right sample size was n=4, 6 and 7. (D) Representative desensitization traces of R2o-G (black), R2o-R (grey) and R2o-G L483A (red). Increased trafficking of R2o-R and the L483A mutant correlate with reduced rates of desensitization. In order of appearance in the legend (top to bottom), current amplitude (nA) and τdes (ms) for traces were −0.70, −0.45 and −0.41, and 1.4, 2.9 and 4.8, respectively. (E) BN–PAGE analysis of L483 mutants. HEK293 lysates were extracted in dodecyl maltoside (DDM; lanes 1 and 3) or in Triton X-100 (X-100; lanes 2 and 4). Triton samples were supplemented with 1% SDS (for 10 min) prior to loading (SDS; lanes 5 and 6). Assembly intermediates (M, D and T) are denoted on the right, ‘uf' (‘unfolded'). (F) Line scans (NIH Image) of the first four lanes are shown on the right; they highlight the pronounced heterogeneity seen for the L483Y mutant relative to L483A. Note the absence of monomeric L483Y subunits.
A more detailed BN–PAGE analysis revealed drastic differences between trafficking competent and retained mutants. L483A migrated as sharp, clearly defined assembly intermediates, when extracted in either dodecyl maltoside or Triton X-100. The receptor tetramer collapsed into mono- and dimeric intermediates on addition of 1% SDS, whereas for the retained tyrosine mutant smearing above the tetrameric range was pronounced, and even persisted in the presence of SDS (Figure 6E and F). Receptor migrating in this high-molecular weight range may resemble assembly intermediates and/or subunit chaperone complexes. Taken together, these findings strongly suggest that a reversible stabilization of the LBD dimer interface modulates AMPAR ER processing.
Motions associated with clamshell closure are sensed in the ER
The above data reveal a tight link between various functional states and AMPAR secretory traffic. Recent findings implicated actual ligand binding in this process (Mah et al, 2005; Valluru et al, 2005; Fleck, 2006), which, together with our current results, suggest that conformations underlying the AMPAR gating cascade are sensed in the ER. To investigate this intriguing possibility further, we next dissected this initial large-scale conformational step in greater detail (Figure 8, step 1). Gating is triggered by ligand binding, which induces clamshell closure (Mayer, 2005; Oswald, 2007). The closed conformation is stabilized by hydrogen bonds across the cleft, such as the E402–T686 pair, the side chains of which approach within 2.7 Å on cleft closure (Figure 7A; Armstrong and Gouaux, 2000). Recent dose–response data showed that straining H-bond formation, by shortening the H-donor side chain (T686S), reduces agonist affinity, which is diminished further by breaking the H-bond (T686A; Robert et al, 2005). These mutations shift the equilibrium away from the closed-cleft conformation.
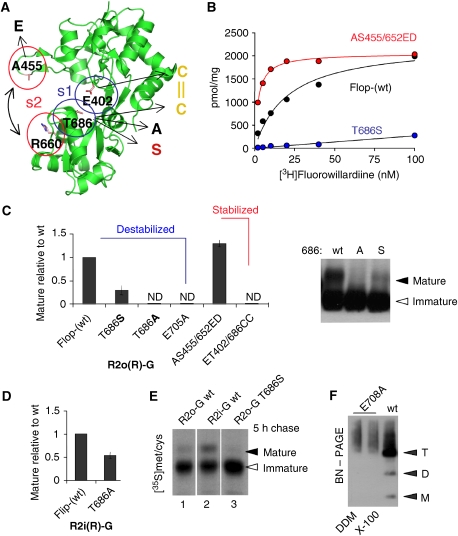
Figure 7.
Clamshell motions are sensed in the ER lumen. (A) Apo R2o-G LBD structure (pdb: 1FTO). The two lobes of the clamshell are shown in the open state. Residues that determine closed-cleft stability are shown as stick: site 1 (s1) contains E402 in the upper lobe and T686 in the lower lobe. Mutation to other residues are indicated in single letter code (arrows). Site 2 (s2) contains A455, which when mutated to Glu interacts with R660, and stabilizes the closed cleft (Weston et al, 2006) S652, another critical residue of s2 is not shown. (B) Saturation binding of [3H]FW to HEK293 cell membranes expressing R2o(R)-G (black circles), R2o(R)-G/T686S (blue circles) and R2o(R)-G/AS455/652ED (red circles). The binding Kd was 17.8 nM for R2o wt, 5 nM for AS455/652ED and >90 nM for the T686S mutant. The graph was generated with Axograph software. (C) Levels of maturely glycosylated mutants, relative to R2o-G wt, was determined as described in Figure 1B. Levels of mature T686A, E705A and ET402/686CC were too low to be determined accurately (ND, not determined). The right panel shows an overexposed blot (probed with anti-R2C; Chemicon) to emphasize the graded loss of ER export competence of R2o-G mutants with destabilization of site 1. (D) Reduction of maturely glycosylated T686S mutant, relative to R2i-G wt. (E) As described in Figure 1C,5 h [35S]met/cys pulse chase is shown. Lane 1 (R2o(R)-G), lane 2 (R2i(R)-G) and lane 3 (R2o(R)-G/T686S). Mature T686S (lane 3) was barely detected after 5 h of chase. (F) BN–PAGE analysis of the E705A ligand-binding mutant. E705A migrated as a broad band above the tetrameric range (T), no defined assembly intermediates (M, D and T) were detected.
This dose–response pattern (Robert et al, 2005) translated directly into ER exit efficiency, where lowered agonist potency conferred reduced trafficking (Figure 7A, site 1 (s1)). Straining H-bond formation (T686S), which lowers agonist Kd in binding assays approximately five-fold (Figure 7B), reduced ER exit relative to R2o-G by 2.9±0.7-fold, n=5 (Figure 7C). Strikingly, abolishing this H-bond, T686A, attenuated forward traffic further, indicating that relatively subtle changes in cleft stability are sensed by ER QC (Figure 7C). T686A also reduced ER export of R2i-G, albeit to a lesser extent (1.8±0.7, n=4; Figure 7D). Moreover, [35S] pulse/chase assays revealed >2-fold reduced ER export rates for R2o-G T6868S (Figure 7E). These data are in line with the findings that ligand binding is required for iGluR traffic (Fleck, 2006; Greger et al, 2007). Ligand docking triggers clamshell closure, destabilization of ligand shifts the equilibrium towards the open-cleft conformation (Figure 8, step 1). Indeed, Figure 7C shows that the E705A mutation, which abolishes anchorage of the agonists' α-amino group, results in ER retention. Analysis of E705A by BN–PAGE (Figure 7F) and by velocity sedimentation (not shown) suggests folding defects, evidenced by high molecular weight smearing on BN–PAGE and a broad distribution on glycerol gradients. Taken together, cleft closure and stability of the closed state appear to be critical for AMPA receptor biogenesis.
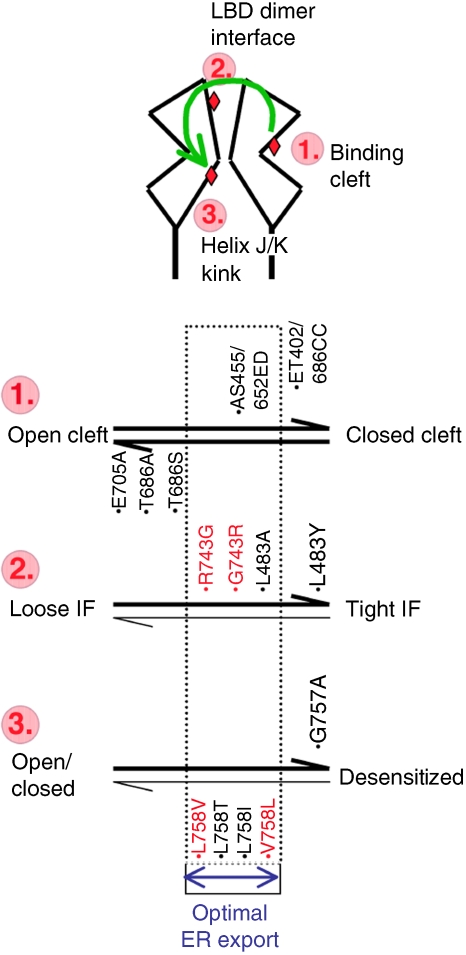
Figure 8.
Summary diagram. Top: schematic of an LBD dimer in the open-cleft state. Steps 1–3 refer to the three segments that were targeted by mutations (red diamonds), and which sequentially undergo conformational transitions (green arrow) in response to ligand docking. Bottom: equilibria within states 1–3 are affected by mutation (denoted in black) and by alternative residues (red). Step 1: the open–closed clamshell thermodynamic equilibrium is altered by mutations targeted to the mouth of the cleft (see main text). Mutations that shift the equilibrium to either extreme (i.e. open or closed, horizontal arrows) result in R2 ER retention. Reversibly stabilizing the closed-cleft state facilitates secretion. This mutation falls into the window of ‘optimal ER export' (stippled rectangle). This theme is repeated in steps 2 and 3. Naturally introduced alterations (R/G743 and V/L758, red) are within the window of optimal export, these changes modulate receptor assembly properties. Note that to date no mutants have been described that map to the left side of steps 2 and 3 (thin horizontal arrows).
To extend these observations, we examined the effect of shifting the equilibrium to the other extreme, to a constitutively closed cleft. Cleft closure brings Glu402 and Thr686 within 2.7 Å of one another, the ET402/686CC double mutant is thus expected to oxidize, form a disulphide bridge and lock the cleft in a closed conformation (Figure 7A). Surprisingly, ET402/686CC was retained in the ER, suggesting that the cleft closure motion needs to be reversible. Together with the data above, this result underlines a common theme—the importance of reversibility of transition states. Indeed, reversibly stabilizing the closed-cleft conformation did not restrict ER exit—the AS455/652ED double mutation, which shifts the equilibrium towards a closed-cleft state (Weston et al, 2006), facilitates the rate (not shown) and extent (1.3±0.5-fold, n=4; Figure 7C) of ER secretion. As expected, this mutant binds [3H]FW with higher affinity than R2o wt (∼8-fold; Figure 7B). In summary, clamshell closure, which triggers the gating cascade, is under ER QC. As observed with other gating rearrangements, this step needs to be reversible to allow consecutive gating transitions to occur, ultimately resulting in AMPAR traffic from the ER.
Discussion
This study implicates the existence of a sophisticated ER QC machinery capable of detecting a spectrum of conformations associated with AMPAR gating transitions. We suggest that this mechanism stringently selects only those receptors, capable of successfully sampling the gating cascade, which ensures employment of only fully functional receptors at synaptic sites. Our findings provide novel insight into the pivotal role of RNA processing in AMPAR formation. They reveal conformational spectra, which are sampled in the ER towards acquisition of an export-competent structure. They underline a role for ligand binding in iGluR biogenesis, and imply the presence of glutamate in the ER lumen. Moreover, as a wide array of AMPAR modulators targets certain LBD conformers (Lynch, 2002), a secondary role of these compounds in AMPAR secretory traffic can be considered.
Clamshell motions and ER exit
The LBD belongs to the group II periplasmic-binding protein (PBP) family, which undergoes substantial hinge motions on ligand binding (Gerstein et al, 1994; Quiocho and Ledvina, 1996). In bacterial PBPs, cleft closure also occurs in the absence of ligand (Oh et al, 1993). In iGluRs, motions of the unliganded LBD might be constricted by surrounding N- and C-terminal domains, and by the Glu705–Lys730 link, which is broken on ligand binding (Armstrong and Gouaux, 2000). Therefore in AMPARs, formation of the closed-cleft state appears to require ligand. Mutations that abolish agonist binding shift the thermodynamic equilibrium towards the open-cleft conformation. These mutants give rise to ER-retained receptors (Fleck, 2006), which have folding deficits (Greger et al, 2006). We cannot rule out that E705A causes gross misfolding. However, the charge conservative R485K mutant, which prevents ligand docking (Lampinen et al, 1998; Armstrong and Gouaux, 2000), produced a similar folding phenotype (Greger et al, 2006). Both (Arg485 and Glu705) side chains are not buried in the protein core (of the apo form) and are thus less likely to affect tertiary structure.
Recent molecular dynamic simulations divided cleft closure into distinct substrates (Lau and Roux, 2007). State 1 involves ligand docking to the upper lobe, which is followed by cleft closure and H-bond formation between the ligand and the lower lobe (including Glu705; state 2). Subsequently, Glu402 and Thr686 associate (Figure 7A) and stabilize the closed-cleft conformation through an additional H-bond (state 3; Lau and Roux, 2007). Interfering with step 3 lowers agonist efficacy (Robert et al, 2005; Zhang et al, 2008), and affinity (Figure 7B). State 3 mutants are ER-retained (Figure 7C). Retention correlates with the severity of the mutation (Figure 8, step 1), suggesting that subtle conformational alterations associated with cleft closure (Zhang et al, 2008) are monitored in the ER. By contrast, stabilization of the closed cleft, AS455/652ED (Figure 7A, site 2) results in enhancement of ER traffic, which is lost when the equilibrium is shifted far towards the closed-cleft state (ET402/686CC). We suggest that irreversibly shifting the LBD to either open (E705A), or closed (ET402/686CC) states hinders subsequent downstream transition states and acquisition of an ER export-competent quarternary structure. This theme of reversibility is also encountered with downstream gating motions.
The LBD interface
Cleft closure gives rise to a high-energy conformation, which collapses into a non-conducting, desensitized state. Structurally, desensitization involves a rearrangement of adjacent LBDs, thereby uncoupling cleft closure from the channel gate (Sun et al, 2002; Armstrong et al, 2006). As a result, the stability of the dimer interface determines the rate of entry into desensitization. Locking this interface through the L483Y mutation blocks desensitization (Stern-Bach et al, 1998; Sun et al, 2002; Horning and Mayer, 2004; Armstrong et al, 2006).
L483Y also impacts on receptor biogenesis—it facilitates R2 tetramerization but renders mutant tetramers ER-retained (Greger et al, 2006; Greger and Esteban, 2007). An analogous observation was made recently with kainate receptors (Priel et al, 2006). iGluRs have evolved different mechanisms to modulate this interface, including R/G RNA editing in AMPARs (Seeburg and Hartner, 2003), and anion binding in kainate receptors (Plested and Mayer, 2007). R2o-R assembles and trafficks more efficiently than the edited Gly743 counterpart (Greger et al, 2006). In contrast to L483Y, Arg743 slows desensitization rates of R2o by only ∼2- to 3-fold (Figure 6C and D; Grosskreutz et al, 2003). Arg743 forms an unconventional Arg bridge across the interface (Figure 6A; Greger et al, 2006), whereas Tyr at position 483 engages the opposite protomer in cation–pi interactions (Sun et al, 2002). The importance of reversibility is validated with the L483A mutant, which attenuates desensitization to a similar level as Arg743. The graded slowing of desensitization illustrated in Figure 6D, almost exactly matched increased ER traffic (Figure 6B).
The helix J/K kink region
AMPARs open and desensitize in parallel on agonist binding (Raman and Trussell, 1995; Robert and Howe, 2003). The desensitized state is energetically favoured and long lived (Raman and Trussell, 1995). Recent work has identified the interface that likely forms on desensitization, which is centred around the helix J/K kink (Armstrong et al, 2006). Mutations in this region alter recovery kinetics and thus the stability of the desensitized state (Figures 3, 4 and 5). In AMPARs, this segment is remodelled by i/o splicing, which adds/removes a single methyl group at residue 758. How this subtle alteration impacts on receptor conformation structurally is currently unclear, although LBD structures provide some hints.
First, in contrast to Val758 the more flexible Leu side chain is buried between helices B and J, where it engages surrounding residues in van der Waals contacts (Figure 2C). These interactions might be facilitated by ligand binding, which is apparent when superimposing the upper lobes from agonist-bound R2 LBD structures with the apo form (Supplementary Figure S9A and B). Ligand-induced rotation of Leu758 is accompanied by side chain movements of Glu431 and Leu753 (Supplementary Figure S9C). It should be noted that these observations do not hold for all structures inspected. In contrast to Leu, the β-branched side chain of Val is more rigid, and projects its γ2 group within 3.9 Å of Gln756 Cβ. By restraining conformational freedom of Gln756 at the end of helix J, Val758 could alter coupling between helices J and K. Although helices J and K perfectly overlay when superimposing upper lobes of the open and closed forms, alignment of the lower lobes of R2i with R2o structures indicates a relative shift between J and K in some instances (pdb codes 2UXA/1FTJ; chains A and B). Helix K connects the two lobes through a disulphide bond, which itself undergoes changes in solution (Fenwick and Oswald, 2008). The helix J/K segment might thereby sense and transmit motions associated with clamshell closure.
Regardless of its exact structural underpinning, the 758 side chain is the main determinant for recovery kinetics and trafficking properties of the R2 i/o splice forms. The correlation between these two parameters is striking and could be confirmed by other β-branched side chains introduced at 758 (Ile and Thr), and by the G757A mutant (Table I). This dataset suggests that the desensitized state is not the ER export-competent conformation (see also Vivithanaporn et al, 2007), and that the receptor needs to escape this state within a certain time window. Just as with the two preceding gating transitions, reversibility appears to hold key (Figure 8). Thus, AMPARs might not adopt a certain quarternary conformation prior to budding from the ER. Rather once the conformational spectrum is successfully sampled within a certain timeframe, forward traffic ensues from any transition state. However, the question of glutamate in the ER has to be considered in this scenario. The data described herein, as well as recent work (Fleck, 2006) strongly (albeit indirectly) imply intracellular ligand binding, and thus the presence of glutamate in the ER. Although ambient glutamate would shift the equilibrium towards desensitization, even at very low concentrations (Robert and Howe, 2003), the receptor will transiently enter unbound states. Moreover, glutamate may not be evenly distributed, and be concentrated in intralumenal pockets, depending on its route of entry; by passive diffusion from the cytosol (Le Gall et al, 2004), through specific transporters (Jong et al, 2005; Csala et al, 2006) or by leaking through the translocon channel (Simon et al, 1989). In the latter case, it would be at the right place during receptor formation.
Whereas mutations targeted to the three LBD segments shift transition-state equilibria irreversibly (Figure 8), regions two and three are modulated reversibly by RNA editing and alternative splicing. By altering the assembly competence and the relative stability of tetramers, as well as their ER export kinetics (see also Mu et al, 2003), these modules will contribute to AMPAR heterogeneity during development and potentially in response to altered network activity. As the editing machinery recodes predominantly polypeptides involved in fast neurotransmission (Jepson and Reenan, 2008), other editing targets may similarly undergo dynamic remodelling and thereby provide a so far underappreciated substrate for the changes induced by synaptic plasticity.
Materials and methods
Abbreviations
Alternatively processed R2 isoforms were abbreviated as described earlier (Greger et al, 2006), with lowercase letters indicating the splice form (i, o), bracketed letters denote the Q/R-editing state and letters separated by a dash denote the R/G state. For example R2i(Q)-R, refers to fully unedited (i.e. Q/R, R/G) GluR2 flip. To simplify this nomenclature, assignment of the Q/R site is mostly omitted in the text; all recordings and sedimentation experiments utilized (Q) forms, all trafficking assays utilized the (R) form.
Plasmids, viruses and mutagenesis
All mutants were generated by long-range PCR using the quick-change procedure (Stratagene). Primer sequences are available on request. R2 varieties were expressed from pCDNA3 vectors (Invitrogen) in HEK293 cells, and from sindbis virus vectors in primary hippocampal neurons (DIV12-14), as described (Greger et al, 2002).
HEK293 transfections
HEK293 cells, grown in poly-L-lysine-coated six-well plates, were transiently transfected with 0.4 μg pcDNA3-R2 varieties using Effectene (Qiagen). Expression was conducted routinely for ∼36 h (±3 h) at 37°C (5% CO2). To identify transfected cells for recording, pCDNA3-R2 constructs were co-transfected with pEGFP (Clontech) at a 5:1 ratio.
EndoH digestions, BN–PAGE and glycerol velocity sedimentation
EH assays were essentially as described in Greger et al (2006). Levels of maturely glycosylated mutants (relative to total receptor) were normalized to levels of maturely glycosylated R2o(R)-G, to compensate for experimental variations. Western blots were routinely probed with anti-GluR2/3 polyclonal ab (Chemicon).
Total cell lysates for BN–PAGE were prepared by washing cells with cold PBS and scraping into hypotonic buffer (HTB; in mM: 50 NaCl, 1.5 MgCl2, 1 EDTA, 20 HEPES, pH 7.2). After 10 min incubation in HTB on ice, cell suspensions were homogenized by passing 20 times through a 23-gauge needle, unbroken cells and nuclei were removed by centrifugation at 400 g for 10 min, and remaining membranes were pelleted by centrifugation at 50 000 gav for 20 min. Membrane pellets were either snap frozen and stored at −80°C or processed for BN–PAGE as described (Greger et al, 2003). Glycerol velocity gradient (10–50% glycerol) centrifugations were as described (Greger et al, 2006).
Ligand binding
Transiently transfected HEK293T cells (36–48 h) were washed with cold PBS, and scraped and osmotically ruptured in HTB buffer (as described above in the BN–PAGE section). Membrane pellets from the first 50K spin were resuspended in ligand-binding buffer (in mM: 50 KSCN, 2.5 Ca2Cl, 30 Tris–HCl, pH 7.3), and the high-speed step was repeated twice to remove endogenous glutamate. Membranes (∼50 μg) were incubated with 2–300 nM [3H] (S)-5-fluorowillardiine (30 Ci/mmol; American Radiolabeled Chemicals) for 1 h on ice before filtration onto 0.3% PEI-treated GF/C filters (Whatman). Signals were obtained by liquid scintillation counting in a Beckman LS6000 SC counter. Data were fit with an expression of the form y=(a × x)/(b+x) using Axograph (Molecular Devices, CA).
Electrophysiology
HEK293 cells were maintained in an extracellular solution (mM): NaCl (145), KCl (3), CaCl (2), MgCl (1), HEPES (10) and glucose (10), pH 7.4, in the recording chamber of an upright microscope (BX-51; Olympus, UK) at room temperature (24–26°C). Whole-cell recordings were made from isolated transiently transfected cells identified under fluorescent microscopy by the co-expression of EGFP. Recording pipettes were filled with (mM): CsCl (120), EGTA (10), CaCl (1), MgCl (2) and HEPES (10), pH adjusted to 7.2–7.3 with CsOH. Low series resistance (<15 MΩ) recordings were made with a dedicated patch-clamp amplifier (Axopatch 200B; Molecular Devices), the series resistance was electronically compensated for by >80% (compensation and predication, lag <10 μs). The brief pressure application (10 ms, Picospritzer III; Parker Instruments) of 1 mM glutamate dissolved in the extracellular fluid from a delivery pipette positioned under visual guidance close to the recorded cell (3–10 μm) resulted in the generation of fast inward currents from a holding potential of between −40 and −60 mV. During paired-pulse experiments, two 10 ms pulses of glutamate were applied with a delay of between 0.1 and 3 s (0.1-s intervals), every 20 s, whereas longer applications of glutamate (1 s) were delivered every 30 or 60 s. Control experiments showed that 10 ms pulses of glutamate evoked maximal inward currents in each AMPAR mutant examined and that lifting the recorded HEK293 cell from the cover slip did not alter the kinetics of single responses or the time course of recovery from desensitization. In each cell, the time course of recovery from desensitization was fit by a single Boltzmann function of the form: y=1/[1+ex1/2−x)/K] where x1/2 represents the half recovery time.
For fast solution exchange, EGFP positive cells were visualized with an inverted Diaphot 200 (Nikon) microscope using infrared differential interference contrast. Voltage clamp recordings were performed at room temperature (20–26°C) on outside-out patches using an Axopatch-1D (Molecular Devices). Glass pipettes had an open tip resistance of 2–5 MΩ when filled with intracellular solution (in mM): 140 CsCl, 10 EGTA, 10 HEPES, 2 MgCl2, 2 ATP (sodium salt), pH 7.3 with CsOH. Glutamate (1 mM) in external solution was applied with a θ-pipette mounted on a piezoelectric stack (PZS-200; Burleigh) powered by a PZ-150M amplifier (Burleigh). Patches were held at −60 mV and were blown off following recordings to measure solution exchange of the open tip (Supplementary Figure 6A). Agonist-containing barrel was supplemented with 0.1% NaCl to increase the amplitude of the open tip response (∼100 pA) and 0.2% sucrose to improve visibility of the solution interface. Filtered current recordings (10 kHz; Bessel Filter) were acquired at 20 kHz using pClamp 9 (Molecular Devices). A paired-pulse protocol was used to measure recovery from desensitization: a 100 ms desensitizing pulse was followed by 10 or 100 ms test pulses at intervals increasing by 10 ms. Sweeps were separated by 2–5 s. Recordings were discarded when agonist-evoked currents randown >15%. Traces were filtered offline at 3 kHz and analysed in Axograph (Molecular Devices). Desensitization (80–20% of the peak) and recovery were fitted with a single exponential.
Supplementary Material
Supplementary Figures S1
Supplementary Figures S2
Supplementary Figures S3
Supplementary Figures S4
Supplementary Figures S5
Supplementary Figures S6
Supplementary Figures S7
Supplementary Figures S8
Supplementary Figures S9
Supplementary Figure Legends
Acknowledgments
We thank Carol Deutsch, Ruth Murrell-Lagnado and Zsofia Nemoda for critically reading the paper. We are grateful to Annette Lenton for help with figure design. We also thank the MRC LMB workshop for skillfully crafting various gadgets for the e-physiology rig. This study was supported by the Royal Society (IHG), and the MRC (ACP, SRW and IHG).
References
- Abele R, Keinanen K, Madden DR (2000) Agonist-induced isomerization in a glutamate receptor ligand-binding domain. A kinetic and mutagenetic analysis. J Biol Chem 275: 21355–21363 [DOI] [PubMed] [Google Scholar]
- Armstrong N, Gouaux E (2000) Mechanisms for activation and antagonism of an AMPA-sensitive glutamate receptor: crystal structures of the GluR2 ligand binding core. Neuron 28: 165–181 [DOI] [PubMed] [Google Scholar]
- Armstrong N, Jasti J, Beich-Frandsen M, Gouaux E (2006) Measurement of conformational changes accompanying desensitization in an ionotropic glutamate receptor. Cell 127: 85–97 [DOI] [PubMed] [Google Scholar]
- Brorson JR, Li D, Suzuki T (2004) Selective expression of heteromeric AMPA receptors driven by flip–flop differences. J Neurosci 24: 3461–3470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman SK, Möykkynen T, Cai C, von Ossowski L, Kuismanen E, Korpi ER, Keinänen K (2006) Isoform-specific early trafficking of AMPA receptor flip and flop variants. J Neurosci 26: 11220–11229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csala M, Bánhegyi G, Benedetti A (2006) Endoplasmic reticulum: a metabolic compartment. FEBS Lett 580: 2160–2165 [DOI] [PubMed] [Google Scholar]
- Cull-Candy S, Brickley S, Farrant M (2001) NMDA receptor subunits: diversity, development and disease. Curr Opin Neurobiol 11: 327–335 [DOI] [PubMed] [Google Scholar]
- Cull-Candy S, Kelly L, Farrant M (2006) Regulation of Ca2+-permeable AMPA receptors: synaptic plasticity and beyond. Curr Opin Neurobiol 16: 288–297 [DOI] [PubMed] [Google Scholar]
- DeLano WL (2002) The PyMOL Molecular Graphics System. Palo Alto, CA, USA: DeLano Scientific [Google Scholar]
- Deutsch C (2003) The birth of a channel. Neuron 40: 265–276 [DOI] [PubMed] [Google Scholar]
- Dingledine R, Borges K, Bowie D, Traynelis SF (1999) The glutamate receptor ion channels. Pharmacol Rev 51: 7–61 [PubMed] [Google Scholar]
- Ellgaard L, Helenius A (2003) Quality control in the endoplasmic reticulum. Nat Rev Mol Cell Biol 4: 181–191 [DOI] [PubMed] [Google Scholar]
- Fenwick MK, Oswald RE (2008) NMR spectroscopy of the ligand-binding core of ionotropic glutamate receptor 2 bound to 5-substituted willardiine partial agonists. J Mol Biol 378: 673–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleck MW (2006) Glutamate receptors and endoplasmic reticulum quality control: looking beneath the surface. Neuroscientist 12: 232–244 [DOI] [PubMed] [Google Scholar]
- Gardner SM, Trussell LO, Oertel D (2001) Correlation of AMPA receptor subunit composition with synaptic input in the mammalian cochlear nuclei. J Neurosci 21: 7428–7437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein M, Lesk AM, Chothia C (1994) Structural mechanisms for domain movements in proteins. Biochemistry 33: 6739–6749 [DOI] [PubMed] [Google Scholar]
- Greger IH, Akamine P, Khatri L, Ziff EB (2006) Developmentally regulated, combinatorial RNA processing modulates AMPA receptor biogenesis. Neuron 51: 85–97 [DOI] [PubMed] [Google Scholar]
- Greger IH, Esteban JA (2007) AMPA receptor biogenesis and trafficking. Curr Opin Neurobiol 17: 289–297 [DOI] [PubMed] [Google Scholar]
- Greger IH, Khatri L, Kong X, Ziff EB (2003) AMPA receptor tetramerization is mediated by Q/R editing. Neuron 40: 763–774 [DOI] [PubMed] [Google Scholar]
- Greger IH, Khatri L, Ziff EB (2002) RNA editing at Arg607 controls AMPA receptor exit from the endoplasmic reticulum. Neuron 34: 759–772 [DOI] [PubMed] [Google Scholar]
- Greger IH, Williams SR (2007) AMPA receptor gating in the endoplasmic reticulum. Soc Neurosci Abstr 40: 7 [Google Scholar]
- Greger IH, Ziff EB, Penn AC (2007) Molecular determinants of AMPA receptor subunit assembly. Trends Neurosci 30: 407–416 [DOI] [PubMed] [Google Scholar]
- Grosskreutz J, Zoerner A, Schlesinger F, Krampfl K, Dengler R, Bufler J (2003) Kinetic properties of human AMPA-type glutamate receptors expressed in HEK293 cells. Eur J Neurosci 17: 1173–1178 [DOI] [PubMed] [Google Scholar]
- Horning MS, Mayer ML (2004) Regulation of AMPA receptor gating by ligand binding core dimers. Neuron 41: 379–388 [DOI] [PubMed] [Google Scholar]
- Isaac JT, Ashby M, McBain CJ (2007) The role of the GluR2 subunit in AMPA receptor function and synaptic plasticity. Neuron 54: 859–871 [DOI] [PubMed] [Google Scholar]
- Jepson JE, Reenan RA (2008) RNA editing in regulating gene expression in the brain. Biochim Biophys Acta 1779: 459–470 [DOI] [PubMed] [Google Scholar]
- Jong YJ, Kumar V, Kingston AE, Romano C, O'Malley KL (2005) Functional metabotropic glutamate receptors on nuclei from brain and primary cultured striatal neurons. Role of transporters in delivering ligand. J Biol Chem 280: 30469–30480 [DOI] [PubMed] [Google Scholar]
- Kessler M, Arai A (2006) Use of [3H] fluorowillardiine to study properties of AMPA receptor allosteric modulators. Brain Res 1076: 25–41 [DOI] [PubMed] [Google Scholar]
- Koike M, Tsukada S, Tsuzuki K, Kijima H, Ozawa S (2000) Regulation of kinetic properties of GluR2 AMPA receptor channels by alternative splicing. J Neurosci 20: 2166–2174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalski JM, Parekh RN, Mao J, Wittrup KD (1998) Protein folding stability can determine the efficiency of escape from endoplasmic reticulum quality control. J Biol Chem 273: 19453–19458 [DOI] [PubMed] [Google Scholar]
- Lampinen M, Pentikainen O, Johnson MS, Keinanen K (1998) AMPA receptors and bacterial periplasmic amino acid-binding proteins share the ionic mechanism of ligand recognition. EMBO J 17: 4704–4711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau AY, Roux B (2007) The free energy landscapes governing conformational changes in a glutamate receptor ligand-binding domain. Structure 15: 1203–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Gall S, Neuhof A, Rapoport T (2004) The endoplasmic reticulum membrane is permeable to small molecules. Mol Biol Cell 15: 447–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SJ, Zukin RS (2007) Ca2+-permeable AMPA receptors in synaptic plasticity and neuronal death. Trends Neurosci 30: 126–134 [DOI] [PubMed] [Google Scholar]
- Lomeli H, Mosbacher J, Melcher T, Hoger T, Geiger JR, Kuner T, Monyer H, Higuchi M, Bach A, Seeburg PH (1994) Control of kinetic properties of AMPA receptor channels by nuclear RNA editing. Science 266: 1709–1713 [DOI] [PubMed] [Google Scholar]
- Lynch G (2002) AMPA receptor modulators as cognitive enhancers. Curr Opin Pharmacol 4: 4–11 [DOI] [PubMed] [Google Scholar]
- Mah SJ, Cornell E, Mitchell NA, Fleck MW (2005) Glutamate receptor trafficking: endoplasmic reticulum quality control involves ligand binding and receptor function. J Neurosci 25: 2215–2225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer ML (2005) Glutamate receptor ion channels. Curr Opin Neurobiol 15: 282–288 [DOI] [PubMed] [Google Scholar]
- Monyer H, Seeburg PH, Wisden W (1991) Glutamate-operated channels: developmentally early and mature forms arise by alternative splicing. Neuron 6: 799–810 [DOI] [PubMed] [Google Scholar]
- Mu Y, Otsuka T, Horton AC, Scott DB, Ehlers MD (2003) Activity-dependent mRNA splicing controls ER export and synaptic delivery of NMDA receptors. Neuron 40: 581–594 [DOI] [PubMed] [Google Scholar]
- Oh BH, Pandit J, Kang CH, Nikaido K, Gokcen S, Ames GF, Kim SH (1993) Three-dimensional structures of the periplasmic lysine/arginine/ornithine-binding protein with and without a ligand. J Biol Chem 268: 11348–11355 [PubMed] [Google Scholar]
- Oswald RE (2007) Flexibility of a glutamate-binding domain. Structure 15: 1157–1158 [DOI] [PubMed] [Google Scholar]
- Plested AJ, Mayer ML (2007) Structure and mechanism of kainate receptor modulation by anions. Neuron 53: 829–841 [DOI] [PubMed] [Google Scholar]
- Priel A, Selak S, Lerma J, Stern-Bach Y (2006) Block of kainate receptor desensitization uncovers a key trafficking checkpoint. Neuron 52: 1037–1046 [DOI] [PubMed] [Google Scholar]
- Quiocho FA, Ledvina PS (1996) Atomic structure and specificity of bacterial periplasmic receptors for active transport and chemotaxis: variation of common themes. Mol Microbiol 20: 17–25 [DOI] [PubMed] [Google Scholar]
- Quirk JC, Siuda ER, Nisenbaum ES (2004) Molecular determinants responsible for differences in desensitization kinetics of AMPA receptor splice variants. J Neurosci 24: 11416–11420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman IM, Trussell LO (1995) The mechanism of alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionate receptor desensitization after removal of glutamate. Biophys J 68: 137–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reenan RA (2001) The RNA world meets behavior: A—>I pre-mRNA editing in animals. Trends Genet 17: 53–56 [DOI] [PubMed] [Google Scholar]
- Robert A, Armstrong N, Gouaux JE, Howe JR (2005) AMPA receptor binding cleft mutations that alter affinity, efficacy, and recovery from desensitization. J Neurosci 25: 3752–3762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert A, Howe JR (2003) How AMPA receptor desensitization depends on receptor occupancy. J Neurosci 23: 847–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwappach B (2008) An overview of trafficking and assembly of neurotransmitter receptors and ion channels. Review Mol Membr Biol 25: 270–278 [DOI] [PubMed] [Google Scholar]
- Seeburg PH (2002) A-to-I editing: new and old sites, functions and speculations. Neuron 35: 17–20 [DOI] [PubMed] [Google Scholar]
- Seeburg PH, Hartner J (2003) Regulation of ion channel/neurotransmitter receptor function by RNA editing. Curr Opin Neurobiol 13: 279–283 [DOI] [PubMed] [Google Scholar]
- Simon SM, Blobel G, Zimmerberg J (1989) Large aqueous channels in membrane vesicles derived from the rough endoplasmic reticulum of canine pancreas or the plasma membrane of E. coli. Proc Natl Acad Sci USA 86: 6176–6178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer B, Keinanen K, Verdoorn TA, Wisden W, Burnashev N, Herb A, Kohler M, Takagi T, Sakmann B, Seeburg PH (1990) Flip and flop: a cell-specific functional switch in glutamate-operated channels of the CNS. Science 249: 1580–1585 [DOI] [PubMed] [Google Scholar]
- Staehlin T, Wettstein FO, Oura H, Noll H (1964) Determination of the coding ratio based on molecular weight of messenger ribonucleic acid associated with ergosomes of different aggregate size. Nature 201: 264–270 [DOI] [PubMed] [Google Scholar]
- Stern-Bach Y, Russo S, Neuman M, Rosenmund C (1998) A point mutation in the glutamate binding site blocks desensitization of AMPA receptors. Neuron 21: 907–918 [DOI] [PubMed] [Google Scholar]
- Sun Y, Olson R, Horning M, Armstrong N, Mayer M, Gouaux E (2002) Mechanism of glutamate receptor desensitization. Nature 417: 245–253 [DOI] [PubMed] [Google Scholar]
- Tóth K, McBain CJ (2000) Target-specific expression of pre- and postsynaptic mechanisms. J Physiol 15: 41–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valluru L, Xu J, Zhu Y, Yan S, Contractor A, Swanson GT (2005) Ligand binding is a critical requirement for plasma membrane expression of heteromeric kainate receptors. J Biol Chem 280: 6085–6093 [DOI] [PubMed] [Google Scholar]
- Vivithanaporn P, Lash LL, Marszalec W, Swanson GT (2007) Critical roles for the M3–S2 transduction linker domain in kainate receptor assembly and postassembly trafficking. J Neurosci 27: 10423–10433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanamaker CP, Christianson JC, Green WN (2003) Regulation of nicotinic acetylcholine receptor assembly. Ann NY Acad Sci 998: 66–80 [DOI] [PubMed] [Google Scholar]
- Weston MC, Gertler C, Mayer ML, Rosenmund C (2006) Interdomain interactions in AMPA and kainate receptors regulate affinity for glutamate. J Neurosci 26: 7650–7658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Cho Y Jr, Lolis E, Howe JR (2008) Structural and single-channel results indicate that the rates of ligand binding domain closing and opening directly impact AMPA receptor gating. J Neurosci 28: 932–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures S1
Supplementary Figures S2
Supplementary Figures S3
Supplementary Figures S4
Supplementary Figures S5
Supplementary Figures S6
Supplementary Figures S7
Supplementary Figures S8
Supplementary Figures S9
Supplementary Figure Legends