Abstract
RECQ helicase protein-like 4 (RECQL4) is a member of the human RECQ family of DNA helicases. Two-thirds of patients with Rothmund-Thomson syndrome (RTS) carry biallelic inactivating mutations in the RECQL4 gene. RTS is an autosomal recessive disorder characterized by poikiloderma, sparse hair, small stature, skeletal abnormalities, cataracts, and an increased risk of cancer. Mutations in two other RECQ helicases, BLM and WRN, are responsible for the cancer predisposition conditions Bloom and Werner syndromes, respectively. Previous studies have shown that BLM and WRN-deficient cells demonstrate increased sensitivity to hydroxyurea (HU), camptothecin (CPT), and 4-nitroquinoline 1-oxide (4NQO). Little is known about the sensitivity of RECQL4-deficient cells to these and other genotoxic agents. The purpose of this study was to determine if RTS cells display any distinct cellular phenotypes in response to DNA damaging agents or replication blocks that could provide insight into the molecular function of the RECQL4 protein. Our results show that primary fibroblasts from RTS patients carrying two deleterious RECQL4 mutations, compared to wild type (WT) fibroblasts, have increased sensitivity to HU, CPT, and doxorubicin (DOX), modest sensitivity to other DNA damaging agents including ultraviolet (UV) irradiation, ionizing radiation (IR), and cisplatin (CDDP), and relative resistance to 4NQO. The RECQ family of DNA helicases has been implicated in the regulation of DNA replication, recombination, and repair. Because HU, CPT and DOX exert their effects primarily during S phase, these results support a greater role for the RECQL4 protein in DNA replication as opposed to repair of exogenous damage.
Keywords: Rothmund-Thomson Syndrome, Bloom syndrome, Werner syndrome, RECQL4, Cellular sensitivity, Colony survival assay, Genotoxic stress
Introduction
Rothmund-Thomson syndrome (RTS) is a rare autosomal recessive disorder characterized by a poikilodermatous rash starting in infancy. Other clinical features include growth retardation, skeletal abnormalities, hair loss, gastrointestinal disturbances, juvenile cataracts, and a high incidence of malignancy, particularly osteosarcoma (OS) (Larizza et al., 2006; Wang et al., 2003). Cytogenetic analyses of cells from RTS patients demonstrate mosaic chromosomal abnormalities and genomic instability (Lindor et al., 2000; Miozzo et al., 1998; Wang et al., 2001). In 1999, Kitao and coworkers linked a subset of RTS cases to mutations in the human RECQ helicase protein-like 4 gene, RECQL4 (Kitao et al., 1999). Sequence analysis of RECQL4 in a cohort of RTS patients revealed that two-thirds carried truncating mutations, and the majority of these were compound heterozygotes (Wang et al., 2003). Genotype-phenotype analysis of this cohort showed that patients with mutations were at a significantly higher risk of developing osteosarcoma, a tumor originating in bone, compared to those RTS patients who did not carry RECQL4 mutations (Wang et al., 2003). In addition to RTS, mutations in RECQL4 have also been found in subgroups of patients with two other genetic disorders, RAPADILINO syndrome (Dietschy et al., 2007; Siitonen et al., 2003) and Baller-Gerold syndrome (BGS) (Van Maldergem et al., 2006). Unlike mostly truncating mutations seen in RTS and BGS, the most common RECQL4 mutation in RAPADILINO is an intronic deletion that results in in-frame skipping of exon 7 and mislocalization of the protein. This mutation is not found in RTS patients. Two of the three BGS mutations described so far have also been detected in RTS. Similarly, there is overlap in some, but not all, clinical features between these three disorders, including small stature, skeletal abnormalities, and increased cancer risk in RAPADILINO, and together they form a spectrum of RECQL4 diseases.
Mutations in two other members of the RECQ family, BLM and WRN, are responsible for Bloom syndrome (BS) (Ellis et al., 1995) and Werner syndrome (WS) (Yu et al., 1996), respectively, both of which are also cancer predisposition syndromes. Cells from both BLM and WRN mutated patients demonstrate increased sensitivity to several genotoxic agents, including hydroxyurea (HU) (Blank et al., 2004; Davies et al., 2004; Dhillon et al., 2007), camptothecin (CPT) (Blank et al., 2004; Lebel and Leder, 1998; Poot et al., 2002b; Rodriguez-Lopez et al., 2007), and 4-nitroquinoline 1-oxide (4NQO) (Hisama et al., 2000; Miao et al., 2006; Poot et al., 2002b; Prince et al., 1999; Rodriguez-Lopez et al., 2007). These agents cause DNA damage through a variety of mechanisms. HU is a ribonucleotide reductase inhibitor that inhibits DNA synthesis, producing cell death in S phase and synchronization of the fraction of surviving cells. CPT is a topoisomerase I inhibitor which causes DNA strand breaks and S phase arrest. 4NQO is a carcinogen that produces multiple DNA lesions, including oxidative adducts and alkylated purines (Poot et al., 2002a). Further studies of BLM and WRN have revealed some of their biochemical and cellular properties. BLM acts as a DNA structure-specific helicase in DNA replication, and can also catalyze DNA strand annealing and resolution of recombination intermediates. BLM interacts with other proteins that are involved in DNA repair and recombination. In addition to its helicase function in DNA replication, WRN also possesses 3′→5′ exonuclease function, and it may have a role in dissociating alternate or secondary structures to allow proper replication and repair at the telomere (Bachrati and Hickson, 2008; Hanada and Hickson, 2007). Both BLM and WRN play important roles in the resumption of synthesis after DNA damage or in the maintenance of fork progression after DNA damage (Davies et al., 2007; Sidorova et al., 2008).
In contrast to BLM and WRN, much less is known about the functional role of RECQL4 or the sensitivity of RECQL4 deficient cells to genotoxic agents. RECQL4 has recently been shown in a Xenopus model and in mouse embryonic fibroblasts (MEFs) to be important for initiation of replication (Sangrithi et al., 2005). Hypersensitivity of RECQL4-deficient cells to oxidative stress, manifested as decreased cell growth, decreased S-phase cells in the cell cycle, and reduced DNA synthesis after recovery from H2O2-induced damage has been reported (Werner et al., 2006). RTS cells have also been shown to display abnormal responses after treatment with other agents such as HU, ultraviolet (UV) and ionizing (IR) radiation, but results have been inconsistent (Grant et al., 2000; Hoki et al., 2003; Park et al., 2006; Shinya et al., 1993; Smith and Paterson, 1981; Smith and Paterson, 1982; Vasseur et al., 1999). One possibility for the inconsistencies could be the use of different experimental systems. There has to date been no systematic study of the response of RECQL4-deficient RTS patient cells to a wide range of genotoxic agents. A summary of results from previous publications as well as this study are summarized in Table 1.
Table 1.
Summary of studies of RECQL4-deficient cellular response to genotoxic agents
| Previous publications | Cell type and number of strains used | RECQL4 mutation status known | Assay | Genotoxic Agent | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| UV-C | IR | CDDP | HU | CPT | DOX | 4NQO | H2O2 | ||||
| Smith et al. 1981 | RTS patient fibroblasts (n=2) | No | CSA | Normal sensitivity |
-- | -- | -- | -- | -- | -- | -- |
| Smith et al. 1982 | RTS patient fibroblasts (n=4)* | No | CSA; DNA repair assay |
-- | Increased sensitivity in 2 cases; defective DNA repair | -- | -- | -- | -- | -- | -- |
| Shinya et al. 1993 | RTS patient fibroblasts (n=1) | No | CSA | Slight sensitivity |
-- | -- | -- | -- | -- | -- | -- |
| Vasseur et al. 1999 | RTS patient lymphocytes and fibroblasts (n=2) | No | Unscheduled DNA synthesis |
Decreased nucleotide excision repair | -- | -- | -- | -- | -- | -- | -- |
| Grant et al. 2000 | RTS patient lymphocytes (n=1) | No | Unscheduled DNA synthesis |
Normal nucleotide excision repair | -- | -- | -- | -- | -- | -- | -- |
| Hoki et al. 2003 | Recql4 KO MEF (n=1) | Yes | CSA | Normal sensitivity |
Normal sensitivity |
-- | -- | -- | -- | -- | -- |
| Park et al. 2006 | RTS patient fibroblasts (n=1); T-Rex-293 cells/RECQL4 shRNA (n=1) | Yes | FACS | Defect in S-phase arrest |
-- | -- | Defect in S-phase arrest |
-- | -- | -- | -- |
| Werner et al. 2006 | RTS patient fibroblasts (n=1) | Yes | Proliferation assay; BrdU incorporation; FACS; IF | -- | -- | -- | -- | -- | -- | -- | Growth arrest; reduced DNA synthesis and S-phase cells; nuclear translocation and foci formation |
| This report | |||||||||||
| Jin et al. 2008 | RTS patient fibroblasts (n=10) | Yes | CSA | Modest sensitivity |
Modest sensitivity |
Modest sensitivity |
Increased sensitivity |
Increased sensitivity |
Increased sensitivity |
Decreased Sensitivity |
-- |
Abbreviations: RTS, Rothmund-Thomson syndrome; CSA, colony survival assay; KO, knockout; MEF, mouse embryonic fibroblasts; GFP, green fluorescent protein; IF, immunofluorescence
Two of these patients are the same as in Smith et al., 1981 publication
In this report, we studied the clonogenic survival of RECQL4-deficient RTS primary fibroblasts after treatment with DNA damaging agents or inhibitors of replication. Since one-third of patients with RTS do not carry RECQL4 mutations, there may be one or more other genes responsible for the RTS phenotype which have not yet been identified (Wang et al., 2003). Thus, we focused our study on subjects with defined inactivating mutations in RECQL4 in order not to confound our findings with potential effects of other gene products on cellular sensitivity. In addition to studying response to HU, CPT, and 4NQO, we also examined survival after treatment with UV, IR, doxorubicin (DOX), and cisplatin (CDDP) in RECQL4-deficient RTS fibroblasts. UV causes DNA inter- and intra-strand links and DNA adducts, while IR results predominantly in double strand breaks. DOX is an anthracycline anticancer agent that acts as a topoisomerase II inhibitor but can also form formaldehyde mediated DNA adducts, and this is an S-phases specific event (Aubel-Sadron and Londos-Gagliardi, 1984). CDDP resembles an alkylating agent and causes DNA cross-links (Martin et al., 2008). Both DOX and CDDP are active chemotherapy agents used to treat osteosarcoma. The purpose of this study was to determine whether RECQL4-deficient RTS cells display any distinct cellular phenotypes in response to DNA damaging agents or replication blocks that could provide insight into the molecular function of the RECQL4 protein. Our results show that RECQL4-deficient RTS cells are hypersensitive to agents that interfere with DNA replication such as HU, CPT, and DOX and are less sensitive to those agents (UV, IR, CDDP) that predominantly cause damage requiring double strand break repair or nucleotide excision repair (NER) mechanisms. Interestingly, RTS cells were found to be relatively resistant to the agent 4NQO, unlike the related WRN and BLM cells.
Materials and methods
Subjects and primary fibroblast cultures
All subjects included in this study met the clinical diagnostic criteria for RTS (Wang et al., 2001) and were RECQL4-deficient; i.e., they all carried mutations in both alleles of RECQL4. Primary RTS skin fibroblasts and wild type (WT) control skin fibroblasts were isolated at Texas Children's Cancer Center, Baylor College of Medicine (BCM, Houston, TX, USA) through the BCM Mental Retardation and Developmental Disabilities Research Center Tissue Culture Core. All subjects or their parents provided informed written consent to participate in a research protocol approved by the Institutional Review Board for Human Subjects Research at BCM.
Primary untransformed fibroblasts from ten RTS subjects were used for this study and are listed along with RECQL4 mutation status in Table 2. For subjects 1-5, mutation analysis of the RECQL4 gene was performed by the BCM Medical Genetics Laboratory (http://www.bcm.edu/geneticlabs/). The RECQL4 mutation status of the remaining subjects has been previously reported (Wang et al., 2003). Mutation nomenclature followed the Nomenclature Working Group guidelines (den Dunnen and Antonarakis, 2001) and used the human RECQL4 cDNA (NM_004260) as a reference. “Sensitive” control cells (defined as those cells known to be hypersensitive to an agent compared to WT cells) were obtained from the Coriell Institute Cell Repositories (Camden, NJ, USA), and included BS (AG06040) and WS fibroblasts (AG03141) for HU, CPT and 4NQO experiments, xeroderma pigmentosum complementation group D (XP-D) fibroblasts (GM00434) for UV experiments (Auerbach and Verlander, 1997), and ataxia-telangiectasia (AT) fibroblasts (GM02052) for IR and DOX experiments (Auerbach and Verlander, 1997;Tamminga et al., 2002). All cells were grown as adherent monolayers in Minimum Essential Medium Alpha Medium (Gibco/Invitrogen, Grand Island, NY, USA) supplemented with 10% or 15% (v/v) fetal bovine serum (ATCC, Manassas, VA, USA) in a humidified, 5% CO2, 37°C incubator. RTS fibroblasts were of low (5-9) passage number, and passage matched cells were used in each experiment.
Table 2. RECQL4 Genotypes of RTS Subjects.
| Subject No. | Allele I mutation | Position* | Mutation type |
|---|---|---|---|
| Allele II mutation | |||
| 1‡ | c.2637_2638insC | Exon 15 | Frameshift |
|
|
|||
| c.1397C>T/P466L | Exon 8 | Missense | |
| c.1772C>T/P591L | Exon 11 | Missense | |
| c.3313G>A/G1105S | Exon 19 | Missense | |
|
| |||
| 2 | c.1048_1049delAG | Exon 5 | Frameshift |
|
|
|||
| c.2269C>T/Q757X | Exon 14 | Nonsense | |
|
| |||
| 3 | IVS2+27_51del25 | Intron 2 | Splicing |
|
|
|||
| IVS16-2A>T | Intron 16 | Splicing | |
|
| |||
| 4 | c.1015_1016insC | Exon 5 | Frameshift |
|
|
|||
| c.2269C>T/Q757X | Exon 14 | Nonsense | |
|
| |||
| 5 | c.2269C>T/Q757X | Exon 14 | Nonsense |
|
|
|||
| c.2662C>T/Q888X | Exon 15 | Nonsense | |
|
| |||
| 6† | c.2269C>T/Q757X | Exon 14 | Nonsense |
|
|
|||
| c.3072_3073delAG | Exon 18 | Frameshift | |
|
| |||
| 7† | c.3072_3073delAG | Exon 18 | Frameshift |
|
|
|||
| c.3276delG | Exon 19 | Frameshift | |
|
| |||
| 8† | IVS7-1G>A | Intron 7 | Splicing |
|
|
|||
| c.1573delT | Exon 9 | Frameshift | |
|
| |||
| 9† | c.2476C>T/ R826X | Exon 15 | Nonsense |
|
|
|||
| IVS11+5G>A | Intron 11 | Splicing | |
|
| |||
| 10† | IVS8+17del11 | Intron 8 | Splicing |
|
|
|||
| IVS8+17del11 | Intron 8 | Splicing | |
Position of mutation in the RECQL4 gene
In addition to a frameshift mutation, this subject had three missense mutations in relatively well-conserved residues; it is not known whether these lie on the same allele.
Previously published subjects/mutations (Wang, 2003)
DNA damaging and replication block agents
All chemicals were obtained from Sigma (St. Louis, MO, USA). Stock solutions of HU (1 M in phosphate-buffered saline, PBS), CPT (10 mM in DMSO), DOX (3.4 mM in 0.9% NaCl), 4NQO (100 mM in DMSO), and CDDP (3.3 mM in 0.9% NaCl) were stored at -80°C and protected from light exposure. Drugs were diluted in PBS immediately before adding to cell culture dishes. Cells were treated with CPT for 72 hours, HU, 4NQO and CDDP for 24 hours, or DOX for 30 minutes in complete medium. UV light of 254 nm was administered at a dose rate of 0.5 J/M2 per second at 13 inches (Model UVGL-55 MineraLight lamp, UVP, Upland, CA, USA). For IR treatments, the dose rate was 8.33 Gy/minute using a 137Cs source irradiator (GammaCell 40 Exactor, Nordion International, Kanata, Canada).
Colony survival assays
Clonogenic survival of unexposed cells was determined by plating 300 or 600 cells in 10 cm cell culture plates (Falcon, Becton-Dickinson Labware, Franklin Lakes, NJ, USA). Higher cell numbers (up to 3600 cells) were used in the culture plates when testing survival after treatment. 24 hours after plating, cells were treated with corresponding doses of agents. For UV irradiation, cells were washed once with PBS, irradiated, and complete medium was added thereafter. For all other treatments, cells were maintained in culture medium and were treated with drugs or IR for the indicated length of time, rinsed twice with PBS, and then cultured in complete medium. Following treatment, cells were incubated in a humidified 5% CO2 incubator at 37°C for 10 to 14 days. Plates were then rinsed with PBS, fixed and stained with crystal violet (0.35%, EMD Chemicals Inc., Gibbstown, NJ, USA). Colonies consisting of more than 50 cells were counted to determine survival (except for subjects 9 and 10 which were slow growing, and colonies with greater than 16 cells were counted).
Statistical Analysis
Descriptive statistics including mean and standard deviation were provided for summarizing the experimental data. For each type of cell, empirical survival percentages were plotted against the dosages of the genotoxic agent. In Figs. 1-3, the survival curves for WT represent an average of three WT controls, and those for RTS represent an average of six RTS subjects (10 subjects for DOX and CDDP) in at least three independent experiments. Dose-response curves for studying the sensitivity of each cell type were fitted by the generalized linear regression model (GLM) for the probability of colony survival as a function of dose levels (Prince et al., 1999). The differences in cell sensitivities were compared by the shapes of estimated dose-response curves. We calculated the corresponding dose-survival fraction curves from the fitted dose-response curves (Supplemental Fig. 2). LD10 was defined as the dose at which 10% survival fraction was achieved (Franken et al., 2006), and was calculated from the estimated dose-response curves. A 95% confidence interval (CI) for the estimated LD10 for each cell line was calculated by the delta method and is represented by error bars in LD10 plots. Statistical analyses used the statistical packages R (version 2.6.1) and STATA (version 10.0). The estimated dose-response curves and dose-survival fraction curves were plotted by the statistical package R.
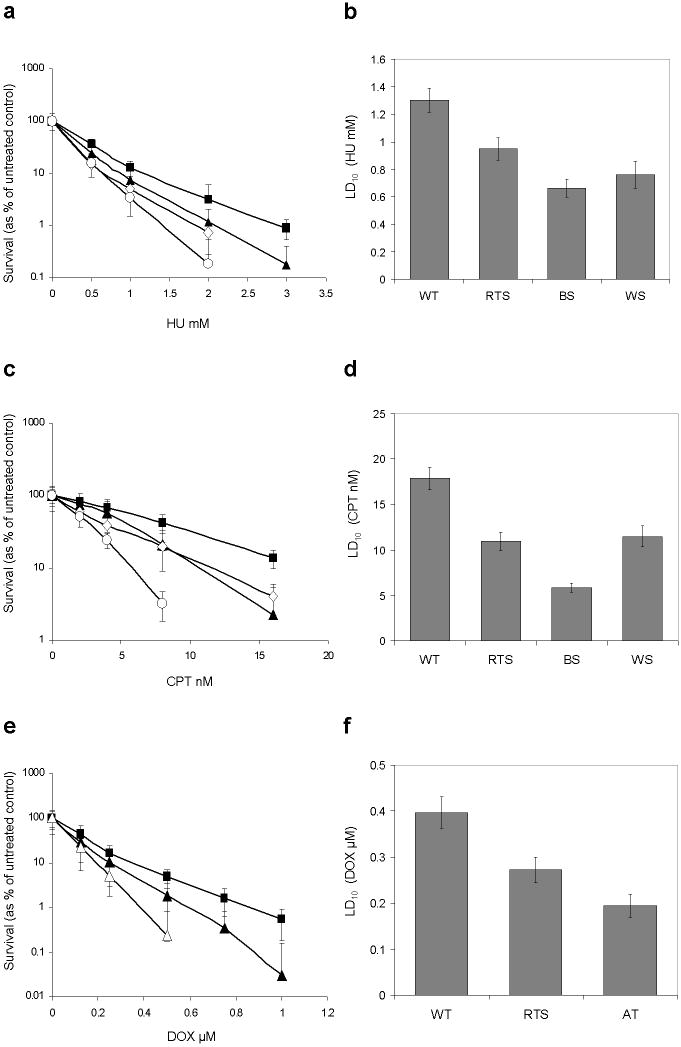
Fig. 1. RTS fibroblasts show hypersensitivity to HU, CPT, and DOX compared to WT controls.
a, c, e Mean log percentage survival curves and standard deviations for three independent WT controls (closed square) or six (10 for DOX) independent RTS subjects (closed triangle) after treatment with HU, CPT, or DOX. BS (open circle) and WS (open diamond), or AT (open triangle) fibroblasts were used as sensitive (positive) controls.
b, d, f Estimated LD10 values and 95% CIs for indicated fibroblasts in each treatment
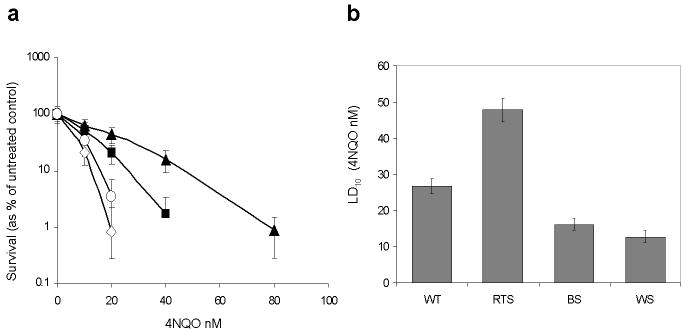
Fig. 3. RTS fibroblasts show resistance to 4NQO compared to WT controls.
a Mean log percentage survival curves and standard deviations for three independent WT controls (closed square) or six independent RTS subjects (closed triangle) after treatment with 4NQO. BS (open circle) and WS (open diamond) fibroblasts were used as sensitive controls.
b Estimated LD10 values and 95% CIs for indicated fibroblasts
Results
We studied the sensitivity of RECQL4-deficient primary fibroblasts from six RTS subjects (10 for DOX and CDDP) to various genotoxic agents and compared them to three different WT control fibroblasts, as well as fibroblasts with known hypersensitivity to specific agents. Survival curves for RTS and WT controls represent the average for the respective number of samples. Supplemental Fig. 1 shows an example of individual survival curves after CPT treatment to demonstrate the variation between individual WT and RTS samples, and is representative of results of treatment with other genotoxic agents used in this study (data not shown).
RECQL4-deficient RTS fibroblasts are hypersensitive to hydroxyurea, camptothecin, and doxorubicin compared to wild type fibroblasts
Hydroxyurea
Logarithmic survival curves are shown in Fig. 1a. Compared to WT controls, RTS fibroblasts were more sensitive to HU, but less so than BS and WS sensitive controls at concentrations between 0.5 and 3 mM. The estimated LD10 values along their 95% confidence intervals are shown in Fig. 1b. The LD10 for WT cells treated with HU was 1.32 mM (95% CI 1.27-1.37 mM), whereas for RTS fibroblasts it was 0.94 mM (95% CI 0.90-0.98 mM) compared to 0.67 mM for BS (95% CI 0.61-0.72 mM) and 0.78 mM for WS (95% CI 0.68-0.88 mM). These represented a 1.4 fold decrease in LD10 for RTS, a 2.0 fold decrease for BS, and a 1.7 fold decrease for WS cells compared to WT controls.
Camptothecin
Compared with WT controls, RTS fibroblasts were more sensitive to CPT, similar to WS, but less sensitive than BS fibroblasts at concentrations between 2 and 16 nM, as shown in the survival curves in Fig. 1c. The estimated LD10 for WT controls was 17.87 nM (95% CI 17.10-18.64 nM), whereas for RTS fibroblasts it was 10.95 nM (95% CI 10.58-11.32 nM) compared to 5.86 nM for BLM (95% CI 5.36-6.36 nM) and 11.47 nM for WS (95% CI 9.67-13.28 nM) as shown in Fig. 1d. These represented a 1.6 fold decrease in LD10 for both RTS and WS and a 3.0 fold decrease for BS cells compared to WT controls.
Doxorubicin
RTS fibroblasts were more sensitive to DOX compared to WT controls, but less so than AT sensitive control cells at concentrations between 0.125 and 1 μM, as shown in the survival curves in Fig. 1e. The LD10 for WT controls was 0.40 μM (95% CI 0.38-0.41 μM), whereas for RTS fibroblasts it was 0.27 μM (95% CI 0.26-0.28 μM) compared to 0.19 μM for AT fibroblasts (95% CI 0.18-0.21 μM) as shown in Fig. 1f. These represented a decrease in LD10 of 1.5 fold and 2.0 fold for RTS and AT cells, respectively, compared to WT controls.
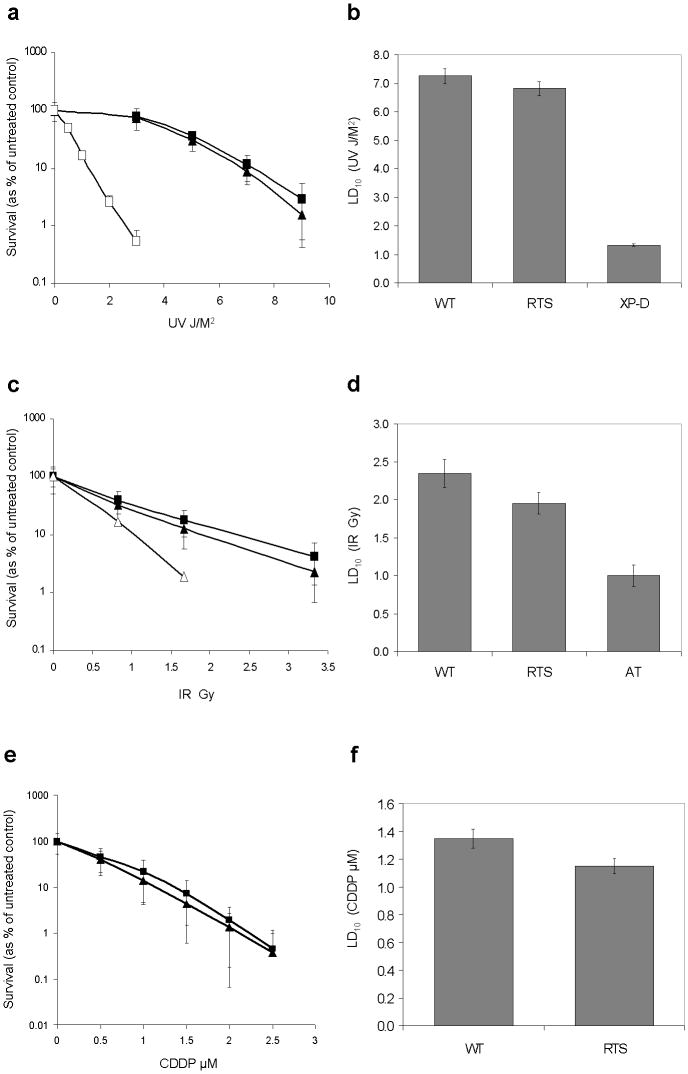
RECQL4-deficient RTS cells have modest sensitivity to UV, ionizing radiation, and cisplatin compared to wild type fibroblasts
UV radiation
The survival curve for RTS fibroblasts treated with UV at doses between 3-9 J/M2 was closer to WT control fibroblasts compared to XP-D sensitive controls (Fig. 2a). The estimated LD10 for WT controls was 7.25 J/M2 (95% CI 7.08-7.71 J/M2), while for RTS it was 6.82 J/M2 (95% CI 6.69-6.94 J/M2), and for XP-D it was 0.19 J/M2 (95% CI 0.18-0.21 J/M2) as shown in Fig. 2b. These represented a 1.2 fold decrease in LD10 for RTS and a 38.2 fold decrease for XP-D cells compared to WT controls.
Fig. 2. RTS fibroblasts show modest sensitivity to UV, IR, and CDDP compared to WT controls.
a, c, e Mean log percentage survival curves and standard deviations for three independent WT controls (closed square) or six (10 for CDDP) independent RTS subjects (closed triangle) after treatment with UV, IR, or CDDP. XP (open square) or AT (open triangle) fibroblasts were used as sensitive controls for UV and IR experiments, respectively.
b, d, f Estimated LD10 values and 95% CIs for indicated fibroblasts in each treatment
Ionizing radiation
Similar to UV, the survival curve for RTS fibroblasts treated with IR at doses between 0.83-3.33 Gy was closer to that of WT control fibroblasts compared to AT sensitive controls (Fig. 2c). The estimated LD10 value of WT controls was 2.35 Gy (95% CI 2.17-2.54 Gy), while for RTS it was 1.95 Gy (95% CI 1.80-2.09 Gy), and for AT it was 1.00 Gy (95% CI 0.86-1.14 Gy) as shown in Fig. 2d. These represented a 1.2 fold decrease in LD10 for RTS and a 2.4 fold decrease for AT cells compared to WT controls.
Cisplatin
For CDDP treatment, although we did not have sensitive control cells, we observed that RTS fibroblasts had survival curves close to WT control fibroblasts at concentrations between 0.5-2.5 μM (Fig. 2e). The estimated LD10 value of WT controls was 1.35 μM (95% CI 1.28-1.41 μM), while for RTS fibroblasts it was 1.15 μM (95% CI 1.09-1.21 μM) as shown in Fig. 2f. This represented a 1.2 fold decrease in LD10 for RTS compared to WT controls.
RECQL4-deficient RTS fibroblasts are resistant to 4-nitroquinoline 1-oxide compared to wild type fibroblasts
RTS fibroblasts were relatively resistant to 4NQO compared to WT control fibroblasts, while BS and WS positive controls were more sensitive at concentrations between 10 and 80 nM, as shown in the survival curves in Fig. 3a. The estimated LD10 value of WT controls was 26.73 nM (95% CI 24.58-28.89 nM), and for RTS fibroblasts it was 47.83 nM (95% CI 44.58-51.07 nM). The LD10 for BS was 16.11 nM (95% CI 14.54-17.69 nM), and for WS it was 12.71 nM (95% CI 11.12-14.31 nM) as shown in Fig. 3b. These represented an increase in LD10 of 1.8 fold for RTS and a decrease of 1.7 fold and 2.1 fold for BS and WS cells, respectively, compared to WT controls.
Discussion
In summary, we have shown that RECQL4-deficient primary fibroblasts from RTS patients demonstrate hypersensitivity to HU, CPT, and DOX, modest sensitivity to UV, IR and CDDP, and relative resistance to 4NQO compared to WT controls. While there have been a few other studies of the cellular sensitivity of RECQL4-deficient cells to genotoxic agents, this is to our knowledge the first systematic study of the sensitivity of multiple RTS primary fibroblasts derived from patients with known deleterious mutations in the RECQL4 gene to a panel of agents that inhibit replication or cause DNA damage using a standardized colony survival assay. This approach offers the advantage of avoiding confounding effects resulting from immortalization of cells or from other genetic loci causing the RTS phenotype in addition to RECQL4, minimizing any aberrant phenomenon stemming from any single RTS patient, and perhaps more accurately reflecting what may be occurring in vivo in humans with RTS.
The RecQ helicases as a group have been shown to play a general role in replication. More specific functions have been delineated for BLM and WRN, including replication repair to ensure the faithful resolution of structural abnormalities that arise during replication, such as those that occur when replication forks encounter DNA lesions. Recent studies of RECQL4 show that it also plays a role in the initiation of DNA replication as well as cell cycle progression (Bachrati and Hickson, 2008; Hanada and Hickson, 2007; Sangrithi et al., 2005). Thus, sensitivity of RECQ-deficient cells to agents that inhibit replication is consistent with these described functions. Similar to WS and BS-deficient fibroblasts, RECQL4-deficient RTS fibroblasts in our study showed increased sensitivity to HU, CPT and DOX. Cell cycle profiles of untreated RECQL4-deficient RTS fibroblasts by flow cytometry, however, appeared similar to WT controls (data not shown). The sensitivity of RTS fibroblasts to these agents was to a lesser degree than that seen in BS or WS fibroblasts. This may be attributable to differences between RECQL4 and the other RECQ members in terms of structure and function. Although it possesses the conserved helicase domain and possesses ATPase activity, the RECQL4 protein appears to lack helicase activity (Macris et al., 2006; Sangrithi et al., 2005), unlike WRN and BLM. In addition, the RECQL4 gene's NLS is located in its amino terminus in contrast to the carboxy terminus in WRN and BLM (Burks et al., 2007; Woo et al., 2006). Therefore differences in response to inhibitors of replication between RTS-deficient cells and BLM- or WRN-deficient cells may be attributable to these differences in helicase activity or gene structure. Another explanation for the intermediate effect in RTS cells is that the cells from RTS patients possess residual RECQL4 activity. Based on studies in both mouse (reviewed in Dietschy et al., 2007) and Xenopus (Sangrithi et al., 2005), it appears that complete loss of RECQL4 function is lethal, while mutant proteins derived from truncating mutations are partially active due to retention of the Sld2 homology region near the N-terminus of the protein in the Xenopus model. This is corroborated by the fact that in none of the RTS patients studied thus far do both alleles carry far 5′ mutations, and no whole gene deletions have been identified. Thus the potential for expression of truncated or missense proteins to convey hypomorphic effects in the RTS cells used in our experiments may account for the intermediate response to replication inhibitors compared to BLM and WRN cells, but this remains to be experimentally verified. Thus far no biochemical or genetic evidence exists to prove that truncated proteins are expressed. Western blots on RTS subjects with RECQL4 mutations on both alleles showed no bands (data not shown), but these were performed using a C-terminal RECQL4 antibody, so truncated proteins may not have been detected.
The resistance of RTS cells to 4NQO is a unique and unanticipated finding. The exact mechanism of action of 4NQO is not clear, but it is known to be metabolically activated to 4-hydroxyaminoquinoline-1-oxide. The latter reacts with DNA to form purine adducts. Similar to UV photolesions, 4NQO DNA adducts are removed by the NER pathway in normal human cells (Zelle and Bootsma, 1980). 4NQO also generates substantial amounts of reactive oxygen species in the cell, there by producing DNA strand breaks and alkali-labile sites (Nunoshiba and Demple, 1993). In addition, it induces formation of irreversible topoisomerase cleavage complexes (Top1cc) that are converted into DNA breaks, similar to CPT (Miao et al., 2006). Interestingly, this is the only drug tested to which RTS cells had an opposite response compared to WRN and BLM cells, which may provide some direction into study of the functional differences between RECQL4 and the other RECQ helicases. Previous papers have shown that BLM and WRN are hypersensitive to 4NQO (Hisama et al., 2000; Miao et al., 2006; Poot et al., 2002a; Prince et al., 1999; Rodriguez-Lopez et al., 2007), except for one report by Honma et al. demonstrating that BLM cells were resistant to this agent (Honma et al., 2002). However, these researchers used EBV transformed lymphoblastoid cell lines and indirectly demonstrated cytotoxicity through relative proliferation, calculated as cell density of the treated versus the non-treated culture, whereas our study used primary untransformed fibroblasts and clonogenic survival assays. Why defects in RECQL4 result in “resistance” to this carcinogen remains a question. RTS cells are not hypersensitive to UV compared to XP-D cells, indicating that RECQL4 is not likely to play an important role in NER pathways, which is also suggested by the fact that RTS cells are not hypersensitive to CDDP. Perhaps the response of RTS cells to 4NQO relates to this drug's ability to generate reactive oxygen species, although RTS cells would then be predicted to be more sensitive to this agent based on previous studies by other groups showing sensitivity of RECQL4-deficient cells to oxidative stress (Werner et al., 2006; Woo et al., 2006). Similarly, if RTS cells were prone to forming Top1cc complexes, then they would be predicted to be more sensitive to 4NQO given their increased sensitivity to CPT (Miao et al., 2006). Alternatively, the relative resistance of RTS cells to 4NQO but not to UV, which also induces the NER pathway for repair, may indicate a defect in the uptake or metabolic activation of 4NQO in RECQL4-deficient cells.
Previous case reports of UV sensitivity in RTS cells (Park et al., 2006; Shinya et al., 1993) along with the consideration of RTS as a DNA damage repair disease similar to ataxia-telangiectasia, have impacted the clinical management of patients with RTS. For example, some patients have been advised major lifestyle alterations based on sun avoidance, while others diagnosed with cancer have received decreased doses of chemotherapy based on the assumption that RTS patients would not be able to tolerate DNA damaging chemotherapeutic agents at standard doses. Our results show that RTS-deficient fibroblasts do not demonstrate hypersensitivity to ionizing or UV radiation of the magnitude displayed by AT or XP-D cells, respectively, compared to WT cells. Although the difference in sensitivity of RTS cells to UV, IR and CDDP compared to WT cells was statistically significant based on LD10 values, the magnitude of the difference compared to AT and XP sensitive controls cells is much lower. Similarly the magnitude of difference in RTS vs. WT cells with these agents is less than that seen with HU, CPT and DOX treatments. These findings are consistent with observations by Hoki et al. who tested the sensitivity to IR and UV of MEFs derived from Recql4 mutant mice that had skipping of exon 13. No statistically significant difference in sensitivity to either form of radiation was found between WT, heterozygous or homozygous mutant Recql4 MEF lines. The current findings are also consistent with clinical data from our cohort of RTS patients who do not demonstrate a significant or persistent history of erythema, blistering, or peeling after sun exposure (data not shown). Thus, patients with RTS may not need to practice complete sun avoidance, but would benefit from sensible use of sunscreens and reasonable UV exposure.
Another clinical issue that has been debated is the potential risk of radiation exposure from radiologic screening for osteosarcoma in RECQL4 mutation positive RTS patients, who are at significantly increased risk of developing this tumor (Wang et al., 2003). While our results do not argue for or against screening, they do provide some evidence that RTS patients may not be much more sensitive to IR than the general population, unlike AT patients, and that IR hypersensitivity would not serve as a strong argument against radiographic screening. Two of the main chemotherapy agents used for the treatment of osteosarcoma are DOX and CDDP. In our studies, RTS cells demonstrated hypersensitivity to DOX but less sensitivity to CDDP. This is consistent with clinical studies of toxicities of cancer therapy in RTS patients treated for osteosarcoma, in whom side effects in the form of mucositis after DOX treatment, but no apparent increased toxicities to CDDP were observed (Hicks et al., 2007).
While our results show that RECQL4-deficient cells are more sensitive to agents that block replication compared to agents that predominantly cause damage requiring double strand break or nucleotide excision repair mechanisms, these data do not preclude the involvement of RECQL4 in these repair processes. Other groups have shown in different experimental systems that RECQL4 may play a role in the repair of double-stranded DNA breaks and suppression of homologous recombination (Bagherieh-Najjar et al., 2005; Kumata et al., 2007; Petkovic et al., 2005). It is likely that RECQL4, similar to the other RECQ proteins, carries out multiple functions in the cell. The molecular mechanisms underlying the differences in response of RECQL4-deficient cells to different genotoxic agents remain to be elucidated. The results of our studies may provide additional insight into the functional differences between the various RECQ helicase family members, and may also find application to future studies aimed at understanding the biologic and clinical differences between members of the RECQL4 spectrum of disorders.
Supplementary Material
Sensitivity of individual RTS fibroblast samples to CPT compared to WT, BS, and WS controls
a Log percentage survival curves and standard deviations for three independent WT controls and one sample each for RTS, WS, and BS fibroblasts
b Log percentage survival curves and standard deviations for three independent RTS samples and one WT control and one BS control
c Same as b except using three other independent RTS samples
Plots of survival fraction curves as a function of various doses of genotoxic agent for indicated fibroblasts. Curves were estimated by the generalized linear regression models fitted for the probability of surviving colonies. LD10 levels for each agent are indicated by the horizontal lines. Fibroblasts were treated with the following agents: a HU; b CPT; c DOX; d UV; e IR; f CDDP; g 4NQO
Acknowledgments
The authors thank the patients and families for their participation in this research. We thank the following ongoing collaborators in our research study: Moise Levy, MD and Richard Lewis, MD. We gratefully acknowledge the Baylor College of Medicine Mental Retardation Developmental Disabilities Research Center, Tissue Culture Core for technical assistance, Stephen M. Gottschalk, MD for scientific advice, and Alison Bertuch, MD, PhD for critical reading of the manuscript and helpful discussions.
Funding: National Institutes of Health (K08 HD42136-05 to L.W., K12 HD41648-01, HD24064); Doris Duke Charitable Foundation; V-Foundation for Cancer Research
Footnotes
Conflicts of Interest Statement: The authors indicated no potential conflicts of interest.
Contributor Information
Weidong Jin, Department of Pediatrics, Section of Hematology/Oncology, Texas Children's Cancer Center, Baylor College of Medicine, Houston, TX 77030, USA.
Hao Liu, Division of Biostatistics, Dan L. Duncan Cancer Center, Baylor College of Medicine, Houston, TX, 77030, USA.
Yiqun Zhang, Division of Biostatistics, Dan L. Duncan Cancer Center, Baylor College of Medicine, Houston, TX, 77030, USA.
Subhendu K. Otta, Department of Pediatrics, Section of Hematology/Oncology, Texas Children's Cancer Center, Baylor College of Medicine, Houston, TX 77030, USA
Sharon E. Plon, Departments of Human and Molecular Genetics and Pediatrics, Section of Hematology/Oncology, Texas Children's Cancer Center, Baylor College of Medicine, Houston, TX 77030, USA
Lisa L. Wang, Department of Pediatrics, Section of Hematology/Oncology, Texas Children's Cancer Center, Baylor College of Medicine, Houston, TX 77030, USA
References
- Aubel-Sadron G, Londos-Gagliardi D. Daunorubicin and doxorubicin, anthracycline antibiotics, a physicochemical and biological review. Biochimie. 1984;66:333–352. doi: 10.1016/0300-9084(84)90018-x. [DOI] [PubMed] [Google Scholar]
- Auerbach AD, Verlander PC. Disorders of DNA replication and repair. Curr Opin Pediatr. 1997;9:600–616. doi: 10.1097/00008480-199712000-00010. [DOI] [PubMed] [Google Scholar]
- Bachrati CZ, Hickson ID. RecQ helicases: guardian angels of the DNA replication fork. Chromosoma. 2008 doi: 10.1007/s00412-007-0142-4. in press. [DOI] [PubMed] [Google Scholar]
- Bagherieh-Najjar MB, de Vries OM, Hille J, Dijkwel PP. Arabidopsis RecQI4A suppresses homologous recombination and modulates DNA damage responses. Plant J. 2005;43:789–798. doi: 10.1111/j.1365-313X.2005.02501.x. [DOI] [PubMed] [Google Scholar]
- Blank A, Bobola MS, Gold B, Varadarajan S, Kolstoe D, Meade EH, Rabinovitch PS, Loeb LA, Silber JR. The Werner syndrome protein confers resistance to the DNA lesions N3-methyladenine and O6-methylguanine: implications for WRN function. DNA Repair (Amst) 2004;3:629–638. doi: 10.1016/j.dnarep.2004.02.003. [DOI] [PubMed] [Google Scholar]
- Burks LM, Yin J, Plon SE. Nuclear import and retention domains in the amino terminus of RECQL4. Gene. 2007;391:26–38. doi: 10.1016/j.gene.2006.11.019. [DOI] [PubMed] [Google Scholar]
- Davies SL, North PS, Dart A, Lakin ND, Hickson ID. Phosphorylation of the Bloom's syndrome helicase and its role in recovery from S-phase arrest. Mol Cell Biol. 2004;24:1279–1291. doi: 10.1128/MCB.24.3.1279-1291.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies SL, North PS, Hickson ID. Role for BLM in replication-fork restart and suppression of origin firing after replicative stress. Nat Struct Mol Biol. 2007;14:677–679. doi: 10.1038/nsmb1267. [DOI] [PubMed] [Google Scholar]
- den Dunnen JT, Antonarakis SE. Nomenclature for the description of human sequence variations. Hum Genet. 2001;109:121–124. doi: 10.1007/s004390100505. [DOI] [PubMed] [Google Scholar]
- Dhillon KK, Sidorova J, Saintigny Y, Poot M, Gollahon K, Rabinovitch PS, Monnat RJ., Jr Functional role of the Werner syndrome RecQ helicase in human fibroblasts. Aging Cell. 2007;6:53–61. doi: 10.1111/j.1474-9726.2006.00260.x. [DOI] [PubMed] [Google Scholar]
- Dietschy T, Shevelev I, Stagljar I. The molecular role of the Rothmund-Thomson-, RAPADILINO- and Baller-Gerold-gene product, RECQL4: recent progress. Cell Mol Life Sci. 2007;64:796–802. doi: 10.1007/s00018-007-6468-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis NA, Groden J, Ye TZ, Straughen J, Lennon DJ, Ciocci S, Proytcheva M, German J. The Bloom's syndrome gene product is homologous to RecQ helicases. Cell. 1995;83:655–666. doi: 10.1016/0092-8674(95)90105-1. [DOI] [PubMed] [Google Scholar]
- Franken NA, Rodermond HM, Stap J, Haveman J, van Bree C. Clonogenic assay of cells in vitro. Nat Protoc. 2006;1:2315–2319. doi: 10.1038/nprot.2006.339. [DOI] [PubMed] [Google Scholar]
- Grant SG, Wenger SL, Latimer JJ, Thull D, Burke LW. Analysis of genomic instability using multiple assays in a patient with Rothmund-Thomson syndrome. Clin Genet. 2000;58:209–215. doi: 10.1034/j.1399-0004.2000.580308.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada K, Hickson ID. Molecular genetics of RecQ helicase disorders. Cell Mol Life Sci. 2007;64:2306–2322. doi: 10.1007/s00018-007-7121-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks MJ, Roth JR, Kozinetz CA, Wang LL. Clinicopathologic features of osteosarcoma in patients with Rothmund-Thomson syndrome. J Clin Oncol. 2007;25:370–375. doi: 10.1200/JCO.2006.08.4558. [DOI] [PubMed] [Google Scholar]
- Hisama FM, Chen YH, Meyn MS, Oshima J, Weissman SM. WRN or telomerase constructs reverse 4-nitroquinoline 1-oxide sensitivity in transformed Werner syndrome fibroblasts. Cancer Res. 2000;60:2372–2376. [PubMed] [Google Scholar]
- Hoki Y, Araki R, Fujimori A, Ohhata T, Koseki H, Fukumura R, Nakamura M, Takahashi H, Noda Y, Kito S, Abe M. Growth retardation and skin abnormalities of the Recql4-deficient mouse. Hum Mol Genet. 2003;12:2293–2299. doi: 10.1093/hmg/ddg254. [DOI] [PubMed] [Google Scholar]
- Honma M, Tadokoro S, Sakamoto H, Tanabe H, Sugimoto M, Furuichi Y, Satoh T, Sofuni T, Goto M, Hayashi M. Chromosomal instability in B-lymphoblasotoid cell lines from Werner and Bloom syndrome patients. Mutat Res. 2002;520:15–24. doi: 10.1016/s1383-5718(02)00144-4. [DOI] [PubMed] [Google Scholar]
- Kitao S, Shimamoto A, Goto M, Miller RW, Smithson WA, Lindor NM, Furuichi Y. Mutations in RECQL4 cause a subset of cases of Rothmund-Thomson syndrome. Nat Genet. 1999;22:82–84. doi: 10.1038/8788. [DOI] [PubMed] [Google Scholar]
- Kumata Y, Tada S, Yamanada Y, Tsuyama T, Kobayashi T, Dong YP, Ikegami K, Murofushi H, Seki M, Enomoto T. Possible involvement of RecQL4 in the repair of double-strand DNA breaks in Xenopus egg extracts. Biochim Biophys Acta. 2007;1773:556–564. doi: 10.1016/j.bbamcr.2007.01.005. [DOI] [PubMed] [Google Scholar]
- Larizza L, Magnani I, Roversi G. Rothmund-Thomson syndrome and RECQL4 defect: splitting and lumping. Cancer Lett. 2006;232:107–120. doi: 10.1016/j.canlet.2005.07.042. [DOI] [PubMed] [Google Scholar]
- Lebel M, Leder P. A deletion within the murine Werner syndrome helicase induces sensitivity to inhibitors of topoisomerase and loss of cellular proliferative capacity. Proc Natl Acad Sci U S A. 1998;95:13097–13102. doi: 10.1073/pnas.95.22.13097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindor NM, Furuichi Y, Kitao S, Shimamoto A, Arndt C, Jalal S. Rothmund-Thomson syndrome due to RECQ4 helicase mutations: report and clinical and molecular comparisons with Bloom syndrome and Werner syndrome. Am J Med Genet. 2000;90:223–228. doi: 10.1002/(sici)1096-8628(20000131)90:3<223::aid-ajmg7>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- Macris MA, Krejci L, Bussen W, Shimamoto A, Sung P. Biochemical characterization of the RECQ4 protein, mutated in Rothmund-Thomson syndrome. DNA Repair (Amst) 2006;5:172–180. doi: 10.1016/j.dnarep.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Martin LP, Hamilton TC, Schilder RJ. Platinum resistance: the role of DNA repair pathways. Clin Cancer Res. 2008;14:1291–1295. doi: 10.1158/1078-0432.CCR-07-2238. [DOI] [PubMed] [Google Scholar]
- Miao ZH, Rao VA, Agama K, Antony S, Kohn KW, Pommier Y. 4-nitroquinoline-1-oxide induces the formation of cellular topoisomerase I-DNA cleavage complexes. Cancer Res. 2006;66:6540–6545. doi: 10.1158/0008-5472.CAN-05-4471. [DOI] [PubMed] [Google Scholar]
- Miozzo M, Castorina P, Riva P, Dalpra L, Fuhrman Conti AM, Volpi L, Hoe TS, Khoo A, Wiegant J, Rosenberg C, Larizza L. Chromosomal instability in fibroblasts and mesenchymal tumors from 2 sibs with Rothmund-Thomson syndrome. Int J Cancer. 1998;77:504–510. doi: 10.1002/(sici)1097-0215(19980812)77:4<504::aid-ijc5>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- Nunoshiba T, Demple B. Potent intracellular oxidative stress exerted by the carcinogen 4-nitroquinoline-N-oxide. Cancer Res. 1993;53:3250–3252. [PubMed] [Google Scholar]
- Park SJ, Lee YJ, Beck BD, Lee SH. A positive involvement of RecQL4 in UV-induced S-phase arrest. DNA Cell Biol. 2006;25:696–703. doi: 10.1089/dna.2006.25.696. [DOI] [PubMed] [Google Scholar]
- Petkovic M, Dietschy T, Freire R, Jiao R, Stagljar I. The human Rothmund-Thomson syndrome gene product, RECQL4, localizes to distinct nuclear foci that coincide with proteins involved in the maintenance of genome stability. J Cell Sci. 2005;118:4261–4269. doi: 10.1242/jcs.02556. [DOI] [PubMed] [Google Scholar]
- Poot M, Gollahon KA, Emond MJ, Silber JR, Rabinovitch PS. Werner syndrome diploid fibroblasts are sensitive to 4-nitroquinoline-N-oxide and 8-methoxypsoralen: implications for the disease phenotype. FASEB J. 2002a;16:757–758. doi: 10.1096/fj.01-0906fje. [DOI] [PubMed] [Google Scholar]
- Poot M, Silber JR, Rabinovitch PS. A novel flow cytometric technique for drug cytotoxicity gives results comparable to colony-forming assays. Cytometry. 2002b;48:1–5. doi: 10.1002/cyto.10101. [DOI] [PubMed] [Google Scholar]
- Prince PR, Ogburn CE, Moser MJ, Emond MJ, Martin GM, Monnat RJ., Jr Cell fusion corrects the 4-nitroquinoline 1-oxide sensitivity of Werner syndrome fibroblast cell lines. Hum Genet. 1999;105:132–138. doi: 10.1007/s004399900078. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Lopez AM, Whitby MC, Borer CM, Bachler MA, Cox LS. Correction of proliferation and drug sensitivity defects in the progeroid Werner's Syndrome by Holliday junction resolution. Rejuvenation Res. 2007;10:27–40. doi: 10.1089/rej.2006.0503. [DOI] [PubMed] [Google Scholar]
- Sangrithi MN, Bernal JA, Madine M, Philpott A, Lee J, Dunphy WG, Venkitaraman AR. Initiation of DNA replication requires the RECQL4 protein mutated in Rothmund-Thomson syndrome. Cell. 2005;121:887–898. doi: 10.1016/j.cell.2005.05.015. [DOI] [PubMed] [Google Scholar]
- Shinya A, Nishigori C, Moriwaki S, Takebe H, Kubota M, Ogino A, Imamura S. A case of Rothmund-Thomson syndrome with reduced DNA repair capacity. Arch Dermatol. 1993;129:332–336. [PubMed] [Google Scholar]
- Sidorova JM, Li N, Folch A, Monnat RJ., Jr The RecQ helicase WRN is required for normal replication fork progression after DNA damage or replication fork arrest. Cell Cycle. 2008;7:796–807. doi: 10.4161/cc.7.6.5566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siitonen HA, Kopra O, Kaariainen H, Haravuori H, Winter RM, Saamanen AM, Peltonen L, Kestila M. Molecular defect of RAPADILINO syndrome expands the phenotype spectrum of RECQL diseases. Hum Mol Genet. 2003;12:2837–2844. doi: 10.1093/hmg/ddg306. [DOI] [PubMed] [Google Scholar]
- Smith PJ, Paterson MC. Abnormal responses to mid-ultraviolet light of cultured fibroblasts from patients with disorders featuring sunlight sensitivity. Cancer Res. 1981;41:511–518. [PubMed] [Google Scholar]
- Smith PJ, Paterson MC. Enhanced radiosensitivity and defective DNA repair in cultured fibroblasts derived from Rothmund Thomson syndrome patients. Mutat Res. 1982;94:213–228. doi: 10.1016/0027-5107(82)90183-x. [DOI] [PubMed] [Google Scholar]
- Tamminga RY, Dolsma WV, Leeuw JA, Kampinga HH. Chemo- and radiosensitivity testing in a patient with ataxia telangiectasia and Hodgkin disease. Pediatr Hematol Oncol. 2002;19:163–171. doi: 10.1080/088800102753541314. [DOI] [PubMed] [Google Scholar]
- Van Maldergem L, Siitonen HA, Jalkh N, Chouery E, De RM, Delague V, Muenke M, Jabs EW, Cai J, Wang LL, Plon SE, Fourneau C, Kestila M, Gillerot Y, Megarbane A, Verloes A. Revisiting the craniosynostosis-radial ray hypoplasia association: Baller-Gerold syndrome caused by mutations in the RECQL4 gene. J Med Genet. 2006;43:148–152. doi: 10.1136/jmg.2005.031781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasseur F, Delaporte E, Zabot MT, Sturque MN, Barrut D, Savary JB, Thomas L, Thomas P. Excision repair defect in Rothmund Thomson syndrome. Acta Derm Venereol. 1999;79:150–152. doi: 10.1080/000155599750011417. [DOI] [PubMed] [Google Scholar]
- Wang LL, Gannavarapu A, Kozinetz CA, Levy ML, Lewis RA, Chintagumpala MM, Ruiz-Maldanado R, Contreras-Ruiz J, Cunniff C, Erickson RP, Lev D, Rogers M, Zackai EH, Plon SE. Association between osteosarcoma and deleterious mutations in the RECQL4 gene in Rothmund-Thomson syndrome. J Natl Cancer Inst. 2003;95:669–674. doi: 10.1093/jnci/95.9.669. [DOI] [PubMed] [Google Scholar]
- Wang LL, Levy ML, Lewis RA, Chintagumpala MM, Lev D, Rogers M, Plon SE. Clinical manifestations in a cohort of 41 Rothmund-Thomson syndrome patients. Am J Med Genet. 2001;102:11–17. doi: 10.1002/1096-8628(20010722)102:1<11::aid-ajmg1413>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- Werner SR, Prahalad AK, Yang J, Hock JM. RECQL4-deficient cells are hypersensitive to oxidative stress/damage: Insights for osteosarcoma prevalence and heterogeneity in Rothmund-Thomson syndrome. Biochem Biophys Res Commun. 2006;345:403–409. doi: 10.1016/j.bbrc.2006.04.093. [DOI] [PubMed] [Google Scholar]
- Woo LL, Futami K, Shimamoto A, Furuichi Y, Frank KM. The Rothmund-Thomson gene product RECQL4 localizes to the nucleolus in response to oxidative stress. Exp Cell Res. 2006;312:3443–3457. doi: 10.1016/j.yexcr.2006.07.023. [DOI] [PubMed] [Google Scholar]
- Yu CE, Oshima J, Fu YH, Wijsman EM, Hisama F, Alisch R, Matthews S, Nakura J, Miki T, Ouais S, Martin GM, Mulligan J, Schellenberg GD. Positional cloning of the Werner's syndrome gene. Science. 1996;272:258–262. doi: 10.1126/science.272.5259.258. [DOI] [PubMed] [Google Scholar]
- Zelle B, Bootsma D. Repair of DNA damage after exposure to 4-nitroquinoline-1-oxide in heterokaryons derived from xeroderma pigmentosum cells. Mutat Res. 1980;70:373–381. doi: 10.1016/0027-5107(80)90027-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sensitivity of individual RTS fibroblast samples to CPT compared to WT, BS, and WS controls
a Log percentage survival curves and standard deviations for three independent WT controls and one sample each for RTS, WS, and BS fibroblasts
b Log percentage survival curves and standard deviations for three independent RTS samples and one WT control and one BS control
c Same as b except using three other independent RTS samples
Plots of survival fraction curves as a function of various doses of genotoxic agent for indicated fibroblasts. Curves were estimated by the generalized linear regression models fitted for the probability of surviving colonies. LD10 levels for each agent are indicated by the horizontal lines. Fibroblasts were treated with the following agents: a HU; b CPT; c DOX; d UV; e IR; f CDDP; g 4NQO