Abstract
Embryonic stem (ES) cells are pluripotent cells with the potential to differentiate into cells or tissues that may be used for transplantation therapy. Parthenogenetic ES cells have been recently derived from both mouse and human oocytes and hold promise as a cell source which is histocompatible to the oocyte donor. Due to the importance of major histocompatibility complex (MHC) antigens in mediating tissue rejection or acceptance, we examined levels of mRNA and protein expression of MHC class I proteins, as well as several MHC class I antigen processing and presentation chaperones, in mouse embryonic stem cells derived from both fertilized (fES) and parthenogenetic (pES) embryos. We found that H-2K, Qa-2, TAP1, TAP2 and tapasin mRNAs were all expressed at low levels in undifferentiated and differentiating ES cells, and were significantly upregulated in response to interferon-γ (IFN-γ) treatment following 14 days of differentiation. Likewise, expression of H-2Kb and H-2Kk proteins were upregulated to detectable levels by IFN-γ after 14 days of differentiation, but Qa-2 protein expression remained low or absent. We also found that MHC class I, TAP1, TAP2, and tapasin mRNAs were all expressed at very low levels in ES cells compared to T cells, suggesting transcriptional regulation of these genes in ES cells. Calnexin, a chaperone molecule involved in other pathways than MHC expression, had mRNA levels that were similar in ES cells and T cells, and was not upregulated by IFN-γ in ES cells. Overall, embryonic stem cells derived from fertilized embryos and parthenogenetic embryos displayed remarkably similar patterns of gene expression at the mRNA and protein levels. The similarity between the fES and pES cell lines in regard to expression of MHC class I and antigen processing machinery provides evidence for the potential usefulness of parthenogenetic ES cells in transplantation therapy.
Keywords: antigen processing chaperones, embryonic stem cells, MHC class I, parthenogenesis
Introduction
Embryonic stem (ES) cells are pluripotent cells characterized by their ability to self-renew indefinitely and to differentiate into all cell types of the body. These unique properties, and the derivation of human embryonic stem cells [1], have established ES cells as a promising potential source of cells for transplantation therapy. Following differentiation into the desired cell types, ES cells may prove useful for a variety of therapies, including hematopoietic, neuronal and cardiac cell transplants [2-4].
Embryonic stem cells are typically derived from inner cell mass cells of a blastocyst stage embryo. Variations in this procedure have been used to establish embryonic stem cell lines, including the use of nuclear transfer, parthenogenesis, morula-stage embryos or single embryonic blastomeres [5-8]. To create parthenogenetic ES (pES) cells, oocytes are activated chemically (without paternal contribution from sperm), resulting in a diploid blastocyst-stage parthenote, from which ES cells are derived and can differentiate into cells from all three embryonic germ layers [5]. Parthenogenetic ES cells are an especially promising cell source for transplantation since the resulting parthenote embryos exhibit defects in genomic imprinting [5, 9] and cannot develop into live offspring without significant genetic manipulation [10], thereby eliminating some of the ethical controversy surrounding embryonic stem cell therapies.
Recently, new lines of major histocompatibility complex (MHC)-matched parthenogenetic embryonic stem (pES) cells were described by Kim et al. [9]. Using two different methods, they established several lines of mouse parthenogenetic ES cells that are completely histocompatible in the MHC region to the donor oocyte. These new “MHC-matched” pES cells are therefore heterozygous at the MHC, due to recombination during the oocyte maturation procedure [9]. There is some evidence that MHC heteroyzgosity may be advantageous for transplantation into MHC heterozygous recipients, since mouse bone marrow transplants from homozygous parents have been shown to be rejected by F1 hybrid (heterozygous) offspring, in a phenomenon known as hybrid resistance [11, 12].
Since MHC proteins are highly polymorphic, matching of MHC class I antigens between donor and recipient is crucial in cell and tissue transplants to improve graft survival. Despite the importance of MHC class I proteins in transplantation therapy, little is known about the regulation of MHC class Ia or class Ib antigen expression in normal fertilized or parthenogenetic embryonic stem cells. Human ES cells express low levels of MHC class Ia proteins in undifferentiated cells, which is upregulated with differentiation and treatment with the cytokine interferon-γ (IFN-γ) [13]. Similarly, it has been shown that mouse embryonic stem cells express little to no MHC class Ia proteins in the undifferentiated state, with detectable MHC class Ia protein expression only after differentiation and IFN-γ treatment [14-16]. MHC class Ib proteins have not been detected on the cell surface of either human or mouse embryonic stem cells [13, 15].
MHC class I protein surface expression is often regulated at the transcriptional level [17], but it is also partially regulated by a system of antigen processing and presentation machinery in which MHC proteins are properly folded, assembled with β2-microglobulin, loaded with a high affinity peptide, and transported to the cell surface [reviewed in 18]. Briefly, a series of antigen processing chaperones (including TAP1, TAP2, tapasin and calnexin) are involved in this process. Calnexin is one of the first chaperones to bind the MHC class I heavy chains and is involved in folding the newly formed protein into the native conformation. TAP1 and TAP2 form a transmembrane heterodimer in the endoplasmic reticulum (ER), which transports short endogenous peptides (derived from ubiquitinated proteins degraded by the proteosome) from the cytosol to the ER. Tapasin, an MHC class I-dedicated chaperone, facilitates the loading of high affinity peptides into the peptide-binding groove of the MHC class I molecule. Loss of dedicated MHC class I antigen processing chaperones results in significantly diminished MHC class I protein surface expression [19, 20]. Our laboratory has recently shown that mouse preimplantation embryos, from which ES cells are derived, express antigen processing chaperones and that tapasin and TAP1 are important for normal MHC class Ib surface expression in early embryos [21, 22]. Another recent study found that undifferentiated mouse embryonic stem cells express detectable TAP1 and TAP2 mRNA as well as proteosome mRNA [15]. However, a quantitative analysis of MHC class Ia and class Ib genes and their antigen processing chaperones during early mouse embryonic stem cell differentiation has not yet been described.
In order to further understand the immunological properties of ES cells and their differentiated progeny, we characterized and quantified the expression of MHC class Ia and Ib molecules and some key antigen processing chaperones (TAP1, TAP2, tapasin and calnexin) in differentiating mouse embryonic stem cells with or without IFN-γ treatment. Furthermore, we compared the expression of these MHC class I genes and their chaperones in embryonic stem cells derived from normal fertilized embryos (fES cells) to embryonic stem cells derived from MHC-matched parthenogenetic embryos (pES cells). This comparison of MHC expression in fES cells and pES cells provides important information for future use of parthenogenetic embryonic stem cells as a source of cells for transplantation therapies.
Materials and Methods
Cell lines
The three mouse embryonic stem cell lines analyzed were a kind gift from the laboratory of Dr. George Q. Daley (Children’s Hospital, Boston). The control cell line, termed fES, was derived from a normal fertilized blastocyst stage embryo (C57BL/6 × CBA hybrid) using standard embryonic stem cell derivation techniques [9]. The other two cell lines were derived using two different protocols for parthenogenetically activating oocytes (from a C57BL/6 × CBA hybrid mouse) and deriving embryonic stem cells from the resulting diploid parthenote blastocysts [9]. In the first protocol for parthenogenetic activation, meiosis I was interrupted and the extrusion of the first polar body was blocked. Parthenogenetic ES cells derived using this method were termed p(MI)ES cells. In the second protocol for parthenogenetic activation, the extrusion of the second polar body during meiosis II was blocked, creating p(MII)ES cells. Using both of these protocols for parthenogenetic activation, a significant proportion of the oocytes underwent recombination at the MHC during the maturation and activation procedure and subsequent pES cells were heterozygous at the MHC and termed “MHC-matched pES cells”. We obtained one MHC-matched p(MI)ES cell line and one MHC-matched p(MII) ES cell line from the Daley laboratory, both with normal diploid chromosome numbers.
T cells
Mouse T cells were harvested from B6.K2 mice, originally obtained from L. Flaherty and maintained in our laboratory, but now available from the Jackson Laboratory (Bar Harbor, ME, Stock 007959). These cells served as a reference cell type for analysis of MHC class I and chaperone molecule mRNA expression. Lymphocytes were isolated from the spleen via Histopaque (Sigma, St. Louis, MO) density centrifugation and T cells were enriched using a mouse T cell negative selection column (Stratagene, La Jolla, CA). This protocol for T cell enrichment typically results in 85-91% purity of CD3+ T cells [23].
Embryonic stem cell culture
Embryonic stem cells were maintained in an undifferentiated state by culture in mouse embryonic stem (ES) cell medium on a feeder layer of mitomycin-c inactivated mouse embryonic fibroblasts (Millipore, Billerica, MA). ES cell medium contained Knockout™ DMEM, 15% Knockout™ serum replacement, 2mM l-glutamine, 1× nonessential amino acids stock, 1× penicillin/streptomycin stock, 0.1 mM β-mercaptoethanol, and 1000 U/ml leukemia inhibitory factor (LIF, Millipore) (all media reagents were from Invitrogen, Carlsbad, CA, unless otherwise noted). For differentiation of ES cells into embryoid bodies (EBs) in suspension, we followed the ATCC standard protocol. ES cells were trypsinized to single cells and plated at 2 × 106 cells per 100 mm ultra low attachment dish (Corning Inc., Corning, NY) in ES cell differentiation medium lacking LIF (1× DMEM (Mediatech, Herndon, VA), 15% fetal bovine serum (Atlanta Biologicals, Lawrenceville, GA), 4 mM l-glutamine, 1 × nonessential amino acids, 1 × sodium pyruvate, 1× penicillin/streptomycin stock , 0.1 mM β-mercaptoethanol). After 1 week in suspension, embryoid bodies were plated onto gelatin-coated tissue culture flasks and allowed to attach and differentiate for an additional 7 days (14 days total). Embryonic stem cells were analyzed as undifferentiated cells (day 0), 7 day embryoid bodies (in suspension) and 14 day embryoid bodies (attached). Two days prior to the 7 day and 14 day time points, half the cells were treated with 10 ng/ml of interferon-γ (IFN-γ, Millipore). The IFN-γ dose of 10 ng/ml was recommended by the manufacturer and determined to be optimal in preliminary dose response experiments (data not shown). Embryonic stem cells and embryoid bodies were observed and imaged using the phase contrast objective of a Nikon Diaphot microscope (Melville, NY) and Spot camera and software.
RNA isolation and reverse transcription
RNA was isolated from 5×105 or 1×106 embryonic stem cells from each cell line at day 0, 7, and 14 of the differentiation protocol (with or without IFN-γ at day 7 and day 14). RNA was also isolated from 1× 106 B6.K2 T cells. The Absolutely RNA® Miniprep Kit (Stratagene, La Jolla, CA) was used for RNA isolation, and complementary DNA (cDNA) was synthesized using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA) according to the manufacturers’ protocols.
Quantitative real time RT-PCR
Quantitative real time RT-PCR was used to quantify relative MHC and chaperone molecule mRNA levels from day 0, day 7 (with or without IFN-γ) and day 14 (with or without IFN-γ) embryonic stem cells from fES, p(MI)ES and p(MII)ES cells. mRNA levels were analyzed for the MHC class I molecule H-2K, the MHC class Ib molecule Qa-2, and chaperone molecules TAP1, TAP2, tapasin and calnexin. GAPDH was utilized as an endogenous control. The exon-spanning primer sequences, their source and original reference, fluorescence detection method and thermocycling conditions are presented in Table 1. It should be noted that H-2Kb and H-2Kk alleles are identical in the sequences recognized by these primers, so the total mRNA levels are referred to simply as H-2K. All the PCR reactions utilized SYBR green fluorescent dye detection except tapasin detection, which utilized a Taqman probe and FAM detection. Thermocycling and fluorescence detection were carried out using an ABI Prism 7000 (Applied Biosystems). During optimization of amplification conditions, amplicon size was confirmed via agarose gel electrophoresis.
Real time PCR results were analyzed using the Pfaffl equation [24], which computes the relative expression of each gene to an untreated control, normalized to the expression of a reference RNA. The Pfaffl equation considers the efficiency (E) of each primer pair in the calculation. The PCR efficiencies were determined via a standard curve (repeated in triplicate) of serially diluted mouse T cell cDNA or ES cell cDNA and the average efficiency (E) is shown in Table 1. The reference gene used for normalization was GAPDH, while the target gene was each of the MHC and chaperone molecule genes. The relative mRNA expression of differentiating and IFN-treated cells (experimental) was compared to undifferentiated ES cells (control) for each individual cell line. For analysis of the effects of differentiation and IFN-γ, Q-Gene software [25] was used to analyze the normalized gene expression of triplicate samples from two different experiments (6 total values). A two-tailed, non-paired Student’s t-test (two degrees of freedom) was used for statistical analyses. Significance was considered P < 0.05. We compared relative mRNA levels in T cells to mRNA levels in undifferentiated fES cells utilizing the Pfaffl method described above. We designated the fES cells as the control for calibration (expression level = 1 in fES day 0 cells).
Flow cytometry
MHC class I protein expression was analyzed at day 14 (with or without IFN-γ) for each of the three embryonic stem cell lines. Three MHC class I proteins were analyzed by flow cytometry: H-2Kb (MHC class Ia), H-2Kk (MHC class Ia) and Qa-2 (MHC class Ib). The embryonic stem cell marker SSEA-1 was also analyzed to evaluate the differentiation status of the embryonic stem cells. ES cells and embryoid bodies were trypsinized to single-celled suspension, passed through a 70 μm cell strainer, and stained with primary antibody for 40 minutes in 1 x PBS with 1% BSA, 0.1% sodium azide (PSBAZ). Biotinylated primary antibodies were utilized for H-2Kb, H-2Kk and Qa-2 (all from BD Pharmingen, Franklin Lakes, NJ), while an unlabeled primary antibody was utilized for SSEA-1 (Millipore). The cells were then washed 3 times in PBSAZ, followed by incubation with a secondary reagent conjugated to a fluorophore for an additional 35-40 minutes. Streptavidin-conjugated PEAlexa Fluor 647 was used as a secondary reagent for the biotinylated primary antibodies, whereas goat anti-mouse IgM conjugated to Alexafluor-488 secondary antibody was used as a secondary reagent for the SSEA-1 antibody. After incubation with the secondary reagent, the cells were washed and then analyzed with a BD FACSCalibur instrument (Becton Dickinson). Data analysis was performed with Cellquest software to determine the percentage of MHC or SSEA-1 positive cells and the mean fluorescence intensity of the MHC-positive cells. Cell data were collected based on typical live ES cell and EB morphology, but without any further gating. The positive cell population was defined as cells with fluorescence intensity above the vast majority (8% or less) of the isotype control. The isotype control value (≤ 8%) was subtracted from this population of cells to give the total positive cell population.
Results
Differentiation of fES and pES cells
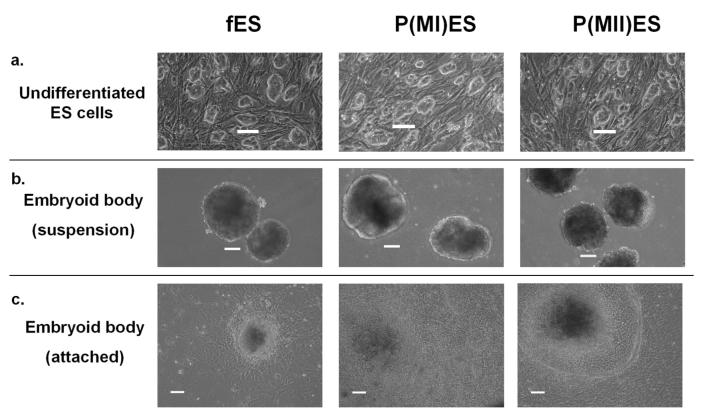
The fertilized and parthenogenetic embryonic stem cell lines underwent morphologically normal differentiation according to standard methods of embryoid body (EB) formation (in suspension) for 7 days, followed by attachment culture for an additional 7 days (day 14 time point). Differentiation was confirmed by morphological analysis of typical embryoid body morphology on day 6-7 and the observation of the presence of various differentiated cell types on day 13-14, including beating cardiomyocytes (Figure 1). All three cell lines showed decreasing expression of the stem cell antigen, SSEA-1, with differentiation (mean of approximately 83% SSEA-1 positive in day 0 undifferentiated ES cells, 26 % in day 7 EBs, and 11% in day 14 EBs), further confirming differentiation of the embryonic stem cells.
Figure 1.
Undifferentiated and differentiating embryonic stem cells derived from fertilized and parthenogenetic embryos. Fertilized ES cells (fES) and parthenogenetic ES cells [p(MI)ES and p(MII)ES] showed similar morphologies as undifferentiated cells (a), as embryoid bodies in suspension (b) and as attached embryoid bodies (c). Images were captured using a phase contrast microscope. Scale bar = 100 μm.
MHC class I and chaperone molecule mRNA levels in differentiating fES and pES cells
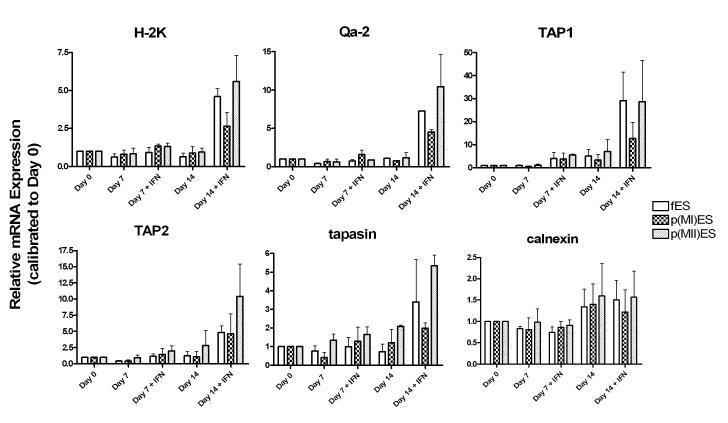
Quantitative real time RT-PCR analysis was used to quantify relative mRNA levels of an MHC class Ia gene (H-2K), an MHC class Ib gene (Qa-2) and antigen processing chaperone genes (TAP1, TAP2, tapasin and calnexin) in fES, p(MI)ES and p(MII)ES cells. We analyzed gene expression in day 0 undifferentiated ES cells, day 7 embryoid bodies and day 14 embryoid bodies. We also tested the effect of IFN-γ (10 ng/ml) treatment on mRNA expression in the embryoid bodies (both at day 7 and day 14). For graphical representation, the relative gene expression is shown for each gene, as calibrated to day 0 undifferentiated ES cells for each cell line (Figure 2).
Figure 2.
MHC class I and antigen processing chaperone mRNA expression in ES cells derived from fertilized (fES) and parthenogenetic [p(MI)ES and p(MII)ES] embryos. A quantitative analysis of relative mRNA expression was performed using Real Time PCR for six genes on undifferentiated (day 0) and differentiating (day7 and day 14) ES cells. On days 7 and 14, the effect of interferon-γ (IFN) treatment was also analyzed. Data are representative of triplicate samples from n=2 experiments. Note that the Y-axis scales differ between the different genes analyzed.
First, we analyzed the effects of differentiation on mRNA levels in the ES cells (no IFN-γ). Differentiation for 7 days in suspension did not result in any statistically significant changes in mRNA expression for any of the genes in the three cell lines, except for slight Qa-2 mRNA downregulation from day 0 to day 7 in fES cells only (P <0.05). Differentiation for 14 days resulted in significant upregulation of TAP1 chaperone mRNA in all three cell lines (P< 0.05 for all). All other genes remained relatively unchanged with differentiation to day 7 or day 14, with little change in relative expression and non-significant P values.
Second, we analyzed the effect of IFN-γ on mRNA levels in the ES cells. Table 2 presents the statistical analysis of the effect of IFN-γ treatment at day 7 and day 14. At day 7 of differentiation, only TAP1 and TAP2 showed significant upregulation after IFN-γ treatment in all three cell lines. The only other difference was a slight, but significant increase in tapasin mRNA expression in the p(MI)ES cells after treatment that was not significant in either the fES or p(MII)ES cells.
On day 14, five of the six genes analyzed, H-2K, Qa-2, TAP1, TAP2 and tapasin, were upregulated in fES cells with IFN-γ treatment. The p(MI)ES and p(MII)ES cells showed a similar pattern with a few exceptions: (1) The increase in H-2K mRNA expression was not significant in the p(MI)ES cells (P= 0.11); (2) the increase in tapasin mRNA was not significant in the p(MI)ES cells (P=0.17) or the p(MII)ES cells (P= 0.07).
Third, we analyzed mRNA levels of fES day 14 IFN-treated cells compared to p(MI) and p(MII)ES cells for each gene to determine if there were significant differences between the fES cells and the two parthenogenetic ES cell lines. The only significant change was found between TAP1 mRNA expression in fES and p(MI)ES cells, with the p(MI)ES cells exhibiting lower expression than the fertilized ES cells (p<0.05). Aside from this exception, the three cell lines showed remarkable similarity in their MHC class I and chaperone gene mRNA levels.
MHC class I and chaperone molecule mRNA levels in T cells
To put the level of MHC and chaperone molecule mRNA expression in undifferentiated (day 0) fES embryonic stem cells into perspective, we quantified expression of these same genes in ES cells relative to mouse T cells, which are known to express high mRNA levels for these MHC class I and antigen processing chaperone molecules. Table 3 shows the percentage of mRNA expression in day 0 fES cells compared to T cells for each gene. Only calnexin was expressed at similar levels in ES cells and T cells. All other genes were expressed at much lower levels in embryonic stem cells than in T cells, especially the non-classical MHC class Ib molecule Qa-2.
MHC class I protein expression in differentiating fES and pES cells
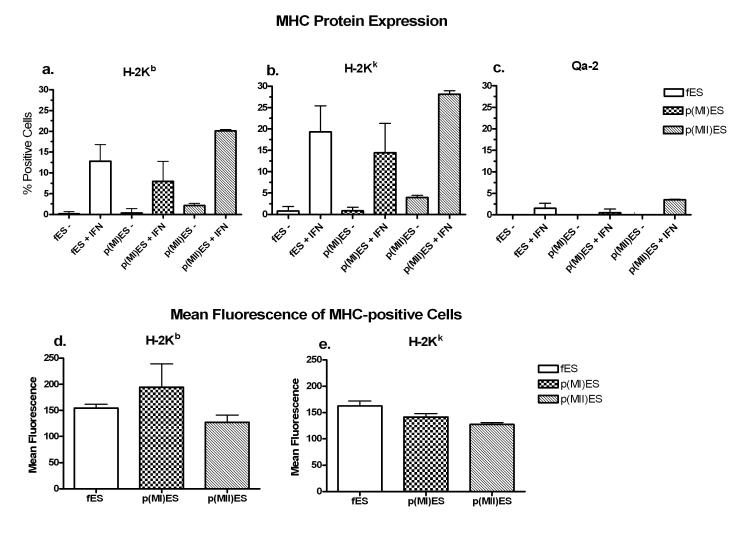
Since MHC class I protein surface expression is regulated both at the transcriptional level and at the post-translational level (by the MHC class I antigen processing pathway), we evaluated MHC class Ia (both H-2Kb and H-2Kk alleles) and class Ib (Qa-2) protein surface expression in embryonic stem cells derived from fertilized embryos (fES) and parthenogenetic embryos (pES) using flow cytometry. Based on the results shown in Figure 2, we decided to analyze protein expression of day 14 embryoid bodies, with or without IFN-γ. There was little to no MHC class I protein expression without IFN-γ in all three cell lines, despite 14 days of differentiation (Figure 3 a-c). However, a small population of cells expressed detectable levels of MHC class Ia proteins (H-2Kb and H-2Kk) with IFN-γ treatment in all three embryonic stem cell lines, without significant differences between the cell lines (Figure 3 a,b). The MHC class Ib protein Qa-2 remained low or absent in all three cell lines (≤ 4% positive or undetectable), even with IFN-γ treatment (Figure 3c). The MHC-positive cells (day 14 with IFN-γ), expressing H-2Kb or H-2Kk, also showed similar levels of mean fluorescence intensity by flow cytometry in all three cell lines (Figure 3 d,e). These data provide further evidence that the parthenogenetic ES cell lines have similar MHC expression patterns with differentiation and IFN-γ treatment compared to the traditional ES cells derived from fertilized embryos.
Figure 3.
MHC class I protein expression in day 14 embryoid bodies, with or without interferon-γ (IFN) treatment. Embryoid bodies were differentiated from embryonic stem cells derived from fertilized (fES) or parthenogenetic [p(MI)ES and p(MII)ES] embryos. Flow cytometry was used to evaluate the percentage of H-2Kb (panel a), H-2Kk (panel b) and Qa-2 (panel c) expression with or without IFN-γ in the three ES cell lines. The level of MHC expression was compared in the three cell lines treated with IFN-γ by analyzing the mean fluorescence intensity of the H-2Kb-positive (panel d) and H-2Kk-positive (panel e) cells. There were no significant differences between fES cells and either of the parthenogenetic ES cell lines. Data are representative of n=2 experiments.
Discussion
Embryonic stem cells posses unique immunological properties compared to typical transplantation grafts [15, 26, 27]. Therefore we decided to further characterize the expression of MHC class I proteins and antigen processing chaperones in both fertilized and novel MHC-matched parthenogenetic ES cell lines [9]. Our study is the first quantitative analysis of MHC class I and antigen processing chaperone mRNA expression during early differentiation of mouse embryonic stem cells and provides data in support of further analysis of parthenogenetic ES cells as source for transplantation therapy.
All genes analyzed, H-2K, Qa-2, TAP1, TAP2, tapasin and calnexin, were detected in undifferentiated ES cells using quantitative RT-PCR. Interestingly, mRNA expression levels of these molecules remained largely unchanged even with 14 days of differentiation (Figure 2). These mRNA levels correlated with flow cytometry data showing low to absent MHC class Ia (H-2Kb and H-2Kk alleles) and class Ib (Qa-2) protein expression on day 14 of differentiation (Figure 3a-c).
We also report quantitative data on the regulation of MHC class I and antigen processing mRNA levels in the presence of the IFN-γ, since this cytokine is produced by T cells in an immune response generated during transplantation. At the mRNA level, we showed that there was significant upregulation of both MHC class Ia (H-2K) and MHC class Ib (Qa-2) mRNA levels in day 14 EBs in the presence of IFN-γ. At the protein level, MHC class Ia protein levels (both H-2Kb and H-2Kk alleles) were also upregulated to detectable levels in day 14 EBs upon IFN-γ treatment (Figure 3a-c). Since these proteins are involved in graft rejection, their presence may contribute to an immune response following transplantation. In addition, we showed that treatment with IFN-γ resulted in only marginal expression of Qa-2 compared to the class Ia proteins (only 4 % or fewer cells were Qa-2 positive even with IFN-γ treatment). Qa-2 is an interesting molecule because it influences preimplantation embryo development even when expressed at levels that are too low to be detected by conventional techniques [28-30]. Therefore it is possible that Qa-2 may still exert influence on the immunological properties of ES cells even at low levels, possibly by acting as an inhibitory receptor for natural killer cells [31].
The MHC class I antigen processing and presentation chaperone genes TAP1, TAP2 and tapasin were upregulated in day 14 EBs with IFN-γ treatment (Figure 2 and Table 2), similar to reports in several other cell types [32-34] and human embryonic stem cells [35]. Cabrera et al. [35] found that undifferentiated human embryonic stem cells lacked TAP1 and TAP2 mRNA expression and had very low tapasin expression, but that expression of these genes was significantly upregulated with differentiation and with IFN-γ treatment. Therefore our data show that some differences in regulation of antigen processing and presentation genes exist between mouse and human embryonic stem cells, since we detected these chaperone molecules even in undifferentiated mouse embryonic stem cells (Figure 2).
We found that calnexin mRNA was not upregulated by IFN-γ treatment in embryonic stem cells (Figure 2 and Table 2). The three upregulated antigen processing chaperones, TAP1, TAP2, and tapasin, are involved in functions that are quite specific to MHC class I antigen processing. However, calnexin is a more generalized chaperone [reviewed in 36], which may account for its steady-state level in ES cells.
We also compared MHC class I and chaperone gene expression in undifferentiated ES cells to expression in T cells to provide some insight into the regulation of these genes and showed that, with the exception of calnexin, their mRNA levels were drastically lower in embryonic stem cells than in T cells (Table 3). Magliocca et al. [15] recently reported that MHC class I genes and some antigen processing chaperones (including TAP1 and TAP2) were easily detected in mouse ES cells and that the MHC class I genes were expressed at similar mRNA levels in lymphocytes. However, their data utilized a different fES cell line (D3) and appear to have been generated using qualitative rather than the quantitative methods that we used for analysis of mRNA levels. Overall, our data suggest that low expression of antigen processing machinery in undifferentiated and differentiating mouse embryonic stem cells may contribute to the low or absent levels of MHC class I protein expression. Both MHC class I genes (H-2K and Qa-2) and the antigen processing chaperones TAP1, TAP2 and tapasin appear to be transcriptionally regulated to low levels in embryonic stem cells, similar to findings in human embryonic stem cells [35].
In agreement with previous reports on conventional, fertilized ES cells, our data show low or absent MHC class I protein expression in undifferentiated and differentiating mouse fES cells as detected by flow cytometry [14-16]. These previous studies also showed that MHC class I proteins could be detected in differentiating embryoid bodies after IFN-γ treatment. Abdullah et al. provide evidence for the ability of ES cells to be recognized, but not lysed, in an MHC-specific manner by cytotoxic T lymphocytes (CTL), despite low or absent MHC class I protein levels as detected by flow cytometry or Western Blot analysis [14]. The study by Abdullah et al. [14], as well as other studies [26, 27, 37-39], point to unique and privileged immunological characteristics of embryonic stem cells that are just beginning to be explored. Our analysis of the expression of MHC protein and antigen processing machinery in mouse embryonic stem cells further adds to this emerging new field of research.
The major contribution of our study is the finding that mouse embryonic stem cells derived from fertilized embryos (fES cells) and parthenogenetic embryos (pES cells) express similar patterns of MHC class I and antigen processing chaperone expression (Figures 2 and 3). Although parthenogenetic embryonic stem cells have been shown to differentiate into cells from all three germ layers and to form teratomas in vivo, their potential for normal differentiation due to defects in genomic imprinting is still debated [5, 9, 40-42]. In particular, Allen et al. found that pES cells were severely limited in their potential to contribute to skeletal muscle tissue in vivo [5]. The extent to which pES cells are limited in potential and how this may affect their ability to produce high quality cells suitable for transplantation is not yet known [42]. Despite the unique immunological properties of embryonic stem cells and their derivatives, MHC protein expression is likely to play an important role in the success of ES cell-based transplantation therapies. Therefore, the similarities between the fES and pES cell lines in this regard provides important background data for the possible therapeutic use of parthenogenetic embryonic stem cells.
We used mouse MHC heterozygous parthenogenetic ES cells for our studies due to the exciting possibility that these cells could be a transplantation resource that is histocompatible to the oocyte donor and is also histocompatible to a limited population of potential transplant recipients [9]. As previously mentioned, MHC-heterozygosity may circumvent graft rejection associated with hybrid resistance. It seems likely that our study on MHC heterozygous pES cells will be applicable to MHC homozygous pES cells, although this remains to be tested. Parthenogenetic ES cell lines may be homozygous at the MHC if recombination does not include the MHC. The main advantage of homozygosity at the MHC is that it would provide a histocompatible cell source for a larger percentage of the population than heterozygosity at the MHC.
Recently, both HLA heterozygous and HLA homozygous parthenogenetic human embryonic stem cells have been derived, making the use of parthenogenetic ES cells for transplantation therapy one step closer to reality [43, 44]. The therapeutic relevance of homozygous and heterozygous pES relates to the idea of creating a bank of donor ES cell lines suitable for transplantation therapy. Taylor et al. [45] have estimated that with respect to HLA matching, 150 ES cell lines (derived from random donated heterozygous embryos) would be sufficient to maintain an adequate bank of ES cell lines for transplantation therapy. However, they calculated that only 10 highly selected HLA homozygous cell lines, such as from HLA homozygous pES cells, could maintain a similarly beneficial ES cell bank [45]. It remains to be determined whether MHC homozygous or MHC heterozygous pES cells would be better suited for transplantation therapies, although this may depend on the differentiated cell type to be used for the transplantation procedure. The research reported in this paper is an important step towards determining the extent to which embryonic stem cell therapies may include the use of parthenogenetic ES cells.
Acknowledgements
This research was supported by NIH grant HD39215 and by NSF grant EEC-9986821 through the Bernard M. Gordon Center for Subsurface Sensing and Imaging Systems at Northeastern University. PL was supported by the IGERT Nanomedicine Science and Technology program at Northeastern University, with funding by NCI and NSF grant 0504331. We sincerely thank Dr. George Q. Daley and Dr. Kitai Kim for the kind gift of MHC-matched parthenogenetic embryonic stem cell lines.
References
- 1.Thomson JA, Itskovitz-Eldor J, Shapiro SS, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–7. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 2.Wang Y, Yates F, Naveiras O, Ernst P, Daley GQ. Embryonic stem cell-derived hematopoietic stem cells. Proc Natl Acad Sci USA. 2005;102:19081–6. doi: 10.1073/pnas.0506127102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Keirstead HS, Nistor G, Bernal G, et al. Human embryonic stem cell-derived oligodendrocyte progenitor cell transplants remyelinate and restore locomotion after spinal cord injury. J Neurosci. 2005;25:4694–705. doi: 10.1523/JNEUROSCI.0311-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Passier R, van Laake LW, Mummery CL. Stem-cell-based therapy and lessons from the heart. Nature. 2008;453:322–9. doi: 10.1038/nature07040. [DOI] [PubMed] [Google Scholar]
- 5.Allen ND, Barton SC, Hilton K, Norris ML, Surani MA. A functional analysis of imprinting in parthenogenetic embryonic stem cells. Development. 1994;120:1473–82. doi: 10.1242/dev.120.6.1473. [DOI] [PubMed] [Google Scholar]
- 6.Strelchenko N, Verlinsky O, Kukharenko V, Verlinsky Y. Morula-derived human embryonic stem cells. Reprod Biomed Online. 2004;9:623–9. doi: 10.1016/s1472-6483(10)61772-5. [DOI] [PubMed] [Google Scholar]
- 7.Chung Y, Klimanskaya I, Becker S, et al. Embryonic and extraembryonic stem cell lines derived from single mouse blastomeres. Nature. 2006;439:216–9. doi: 10.1038/nature04277. [DOI] [PubMed] [Google Scholar]
- 8.Munsie MJ, Michalska AE, O’Brien CM, Trounson AO, Pera MF, Mountford PS. Isolation of pluripotent embryonic stem cells from reprogrammed adult mouse somatic cell nuclei. Curr Biol. 2000;10:989–92. doi: 10.1016/s0960-9822(00)00648-5. [DOI] [PubMed] [Google Scholar]
- 9.Kim K, Lerou P, Yabuuchi A, et al. Histocompatible embryonic stem cells by parthenogenesis. Science. 2007;315:482–6. doi: 10.1126/science.1133542. [DOI] [PubMed] [Google Scholar]
- 10.Kono T, Obata Y, Wu Q, et al. Birth of parthenogenetic mice that can develop to adulthood. Nature. 2004;428:860–4. doi: 10.1038/nature02402. [DOI] [PubMed] [Google Scholar]
- 11.Cudkowicz G, Bennett M. Peculiar immunobiology of bone marrow allografts. I. Graft rejection by irradiated responder mice. J Exp Med. 1971;134:83–102. doi: 10.1084/jem.134.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.George T, Yu YY, Liu J, et al. Allorecognition by murine natural killer cells: lysis of T-lymphoblasts and rejection of bone-marrow grafts. Immunol Rev. 1997;155:29–40. doi: 10.1111/j.1600-065x.1997.tb00937.x. [DOI] [PubMed] [Google Scholar]
- 13.Drukker M, Katz G, Urbach A, et al. Characterization of the expression of MHC proteins in human embryonic stem cells. Proc Natl Acad Sci USA. 2002;99:9864–9. doi: 10.1073/pnas.142298299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abdullah Z, Saric T, Kashkar H, et al. Serpin-6 expression protects embryonic stem cells from lysis by antigen-specific CTL. J Immunol. 2007;178:3390–9. doi: 10.4049/jimmunol.178.6.3390. [DOI] [PubMed] [Google Scholar]
- 15.Magliocca JF, Held IK, Odorico JS. Undifferentiated murine embryonic stem cells cannot induce portal tolerance but may possess immune privilege secondary to reduced major histocompatibility complex antigen expression. Stem Cells Dev. 2006;15:707–17. doi: 10.1089/scd.2006.15.707. [DOI] [PubMed] [Google Scholar]
- 16.Tian L, Catt JW, O’Neill C, King NJ. Expression of immunoglobulin superfamily cell adhesion molecules on murine embryonic stem cells. Biol Reprod. 1997;57:561–8. doi: 10.1095/biolreprod57.3.561. [DOI] [PubMed] [Google Scholar]
- 17.Drezen JM, Babinet C, Morello D. Transcriptional control of MHC class I and beta 2-microglobulin genes in vivo. J Immunol. 1993;150:2805–13. [PubMed] [Google Scholar]
- 18.Cresswell P, Ackerman AL, Giodini A, Peaper DR, Wearsch PA. Mechanisms of MHC class I-restricted antigen processing and cross-presentation. Immunol Rev. 2005;207:145–57. doi: 10.1111/j.0105-2896.2005.00316.x. [DOI] [PubMed] [Google Scholar]
- 19.Garbi N, Tan P, Diehl AD, et al. Impaired immune responses and altered peptide repertoire in tapasin-deficient mice. Nat Immunol. 2000;1:234–8. doi: 10.1038/79775. [DOI] [PubMed] [Google Scholar]
- 20.Van Kaer L, Ashton-Rickardt PG, Ploegh HL, Tonegawa S. TAP1 mutant mice are deficient in antigen presentation, surface class I molecules, and CD4-8+ T cells. Cell. 1992;71:1205–14. doi: 10.1016/s0092-8674(05)80068-6. [DOI] [PubMed] [Google Scholar]
- 21.Lampton PW, Goldstein CY, Warner CM. The role of tapasin in MHC class I protein trafficking in embryos and T cells. J Reprod Immunol. 2008;78:28–39. doi: 10.1016/j.jri.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ke X, Warner CM. Regulation of Ped gene expression by TAP protein. J Reprod Immunol. 2000;46:1–15. doi: 10.1016/s0165-0378(99)00053-4. [DOI] [PubMed] [Google Scholar]
- 23.Comiskey M, Domino KE, Warner CM. HLA-G is found in lipid rafts and can act as a signaling molecule. Hum Immunol. 2007;68:1–11. doi: 10.1016/j.humimm.2006.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muller PY, Janovjak H, Miserez AR, Dobbie Z.Processing of gene expression data generated by quantitative real-time RT-PCR Biotechniques 2002321372–4. 1376, 1378-9. [PubMed] [Google Scholar]
- 26.Bonde S, Zavazava N. Immunogenicity and engraftment of mouse embryonic stem cells in allogeneic recipients. Stem Cells. 2006;24:2192–201. doi: 10.1634/stemcells.2006-0022. [DOI] [PubMed] [Google Scholar]
- 27.Li L, Baroja ML, Majumdar A, et al. Human embryonic stem cells possess immune-privileged properties. Stem Cells. 2004;22:448–56. doi: 10.1634/stemcells.22-4-448. [DOI] [PubMed] [Google Scholar]
- 28.Warner CM, Gollnick SO, Flaherty L, Goldbard SB. Analysis of Qa-2 antigen expression by preimplantation mouse embryos: possible relationship to the preimplantation-embryo-development (Ped) gene product. Biol Reprod. 1987;36:611–6. doi: 10.1095/biolreprod36.3.611. [DOI] [PubMed] [Google Scholar]
- 29.McElhinny AS, Kadow N, Warner CM. The expression pattern of the Qa-2 antigen in mouse preimplantation embryos and its correlation with the Ped gene phenotype. Mol Hum Reprod. 1998;4:966–71. doi: 10.1093/molehr/4.10.966. [DOI] [PubMed] [Google Scholar]
- 30.Warner CM, Newmark JA, Comiskey M, et al. Genetics and imaging to assess oocyte and preimplantation embryo health. Reprod Fertil Dev. 2004;16:729–41. doi: 10.1071/rd04088. [DOI] [PubMed] [Google Scholar]
- 31.Chiang EY, Henson M, Stroynowski I. The nonclassical major histocompatibility complex molecule Qa-2 protects tumor cells from NK cell- and lymphokine-activated killer cell-mediated cytolysis. J Immunol. 2002;168:2200–11. doi: 10.4049/jimmunol.168.5.2200. [DOI] [PubMed] [Google Scholar]
- 32.Abarca-Heidemann K, Friederichs S, Klamp T, Boehm U, Guethlein LA, Ortmann B. Regulation of the expression of mouse TAP-associated glycoprotein (tapasin) by cytokines. Immunol Lett. 2002;83:197–207. doi: 10.1016/s0165-2478(02)00104-9. [DOI] [PubMed] [Google Scholar]
- 33.Bikoff EK, Jaffe L, Ribaudo RK, Otten GR, Germain RN, Robertson EJ. MHC class I surface expression in embryo-derived cell lines inducible with peptide or interferon. Nature. 1991;354:235–8. doi: 10.1038/354235a0. [DOI] [PubMed] [Google Scholar]
- 34.Seliger B, Wollscheid U, Momburg F, Blankenstein T, Huber C. Characterization of the major histocompatibility complex class I deficiencies in B16 melanoma cells. Cancer Res. 2001;61:1095–9. [PubMed] [Google Scholar]
- 35.Cabrera CM, Nieto A, Cortes JL, et al. The low rate of HLA class I molecules on the human embryonic stem cell line HS293 is associated with the APM components’ expression level. Cell Biol Int. 2007;31:1072–8. doi: 10.1016/j.cellbi.2007.03.015. [DOI] [PubMed] [Google Scholar]
- 36.Williams DB. Beyond lectins: the calnexin/calreticulin chaperone system of the endoplasmic reticulum. J Cell Sci. 2006;119:615–23. doi: 10.1242/jcs.02856. [DOI] [PubMed] [Google Scholar]
- 37.Drukker M, Katchman H, Katz G, et al. Human embryonic stem cells and their differentiated derivatives are less susceptible to immune rejection than adult cells. Stem Cells. 2006;24:221–9. doi: 10.1634/stemcells.2005-0188. [DOI] [PubMed] [Google Scholar]
- 38.Koch CA, Geraldes P, Platt JL. Immunosuppression by embryonic stem cells. Stem Cells. 2008;26:89–98. doi: 10.1634/stemcells.2007-0151. [DOI] [PubMed] [Google Scholar]
- 39.Wu D, Boyd AS, Wood KJ. Embryonic Stem Cells and their Differentiated Derivatives have a Fragile Immune Privilege, But Still Represent Novel Targets of Immune Attack. Stem Cells. 2008 doi: 10.1634/stemcells.2008-0078. [DOI] [PubMed] [Google Scholar]
- 40.Lengerke C, Kim K, Lerou P, Daley GQ. Differentiation potential of histocompatible parthenogenetic embryonic stem cells. Ann N Y Acad Sci. 2007;1106:209–18. doi: 10.1196/annals.1392.011. [DOI] [PubMed] [Google Scholar]
- 41.Jiang H, Sun B, Wang W, et al. Activation of paternally expressed imprinted genes in newly derived germline-competent mouse parthenogenetic embryonic stem cell lines. Cell Res. 2007;17:792–803. doi: 10.1038/cr.2007.70. [DOI] [PubMed] [Google Scholar]
- 42.Wilmut I. Embryo stem cells from parthenotes and embryos produced by nuclear transfer: the distinction between them and their potential value in cell therapy. Cloning Stem Cells. 2007;9:291–2. doi: 10.1089/clo.2007.00E1. [DOI] [PubMed] [Google Scholar]
- 43.Revazova ES, Turovets NA, Kochetkova OD, et al. HLA homozygous stem cell lines derived from human parthenogenetic blastocysts. Cloning Stem Cells. 2008;10:11–24. doi: 10.1089/clo.2007.0063. [DOI] [PubMed] [Google Scholar]
- 44.Revazova ES, Turovets NA, Kochetkova OD, et al. Patient-specific stem cell lines derived from human parthenogenetic blastocysts. Cloning Stem Cells. 2007;9:432–49. doi: 10.1089/clo.2007.0033. [DOI] [PubMed] [Google Scholar]
- 45.Taylor CJ, Bolton EM, Pocock S, Sharples LD, Pedersen RA, Bradley JA. Banking on human embryonic stem cells: estimating the number of donor cell lines needed for HLA matching. Lancet. 2005;366:2019–25. doi: 10.1016/S0140-6736(05)67813-0. [DOI] [PubMed] [Google Scholar]
- 46.Arcellana-Panlilio MY, Schultz GA. Temporal and spatial expression of major histocompatibility complex class I H-2K in the early mouse embryo. Biol Reprod. 1994;51:169–83. doi: 10.1095/biolreprod51.2.169. [DOI] [PubMed] [Google Scholar]
- 47.Wu L, Exley GE, Warner CM. Differential expression of Ped gene candidates in preimplantation mouse embryos. Biol Reprod. 1998;59:941–52. doi: 10.1095/biolreprod59.4.941. [DOI] [PubMed] [Google Scholar]
- 48.Pearce RB, Trigler L, Svaasand EK, Peterson CM. Polymorphism in the mouse Tap-1 gene. Association with abnormal CD8+ T cell development in the nonobese nondiabetic mouse. J Immunol. 1993;151:5338–47. [PubMed] [Google Scholar]
- 49.Wang X, Seed B. A PCR primer bank for quantitative gene expression analysis. Nucleic Acids Res. 2003;31:e154. doi: 10.1093/nar/gng154. [DOI] [PMC free article] [PubMed] [Google Scholar]