Abstract
Heterochromatin formation at fission yeast centromeres is directed by RNA interference (RNAi). Noncoding transcripts derived from centromeric repeats are processed into small interfering RNAs (siRNAs) that direct the RNA-induced transcriptional silencing (RITS) effector complex to engage centromere transcripts, resulting in recruitment of the histone H3 lysine 9 methyltransferase Clr4, and hence silencing. We have found that defects in specific splicing factors, but not splicing itself, affect the generation of centromeric siRNAs and consequently centromeric heterochromatin integrity. Moreover, splicing factors physically associate with Cid12, a component of the RNAi machinery, and with centromeric chromatin, consistent with a direct role in RNAi. We propose that spliceosomal complexes provide a platform for siRNA generation and hence facilitate effective centromere repeat silencing.
RNA interference (RNAi) and related pathways regulate gene expression at both transcriptional and posttranscriptional levels. In fission yeast (Schizosaccharomyces pombe), RNAi directs the formation of heterochromatin (1, 2). Analogous to metazoan centromeres, fission yeast centromeres comprise a kinetochore domain flanked by outer repeat (otr) sequences that are assembled in heterochromatin. These otr regions are transcribed by RNA polymerase II (Pol II) and give rise to double-stranded RNA that is processed into small interfering RNAs (siRNAs) by Dicer (Dcr1). These siRNAs are loaded into Argonaute (Ago1), a component of the RNA-induced transcriptional silencing (RITS) effector complex (3). siRNAs target RITS to cognate nascent transcripts, resulting in recruitment of further factors including the RDRC complex (comprising Rdp1, Cid12, and Hrr1) (4), and ultimately Clr4, which methylates histone H3 on Lys9 (H3K9me2). H3K9me2 is bound by the HP1-related protein Swi6, which in turn recruits cohesin, critical for centromere function (5).
To further dissect the mechanism of RNAi-directed chromatin modification, we previously performed a screen that identified mutations at 12 loci termed csp (centromere: suppressor of position effect), which at 25°C alleviate silencing of marker genes inserted in the otr of centromere 1 (6). Several of the csp mutants are alleles of known RNAi components (7, 8). The csp4 and csp5 mutants are temperature-sensitive (ts) lethal alleles. Complementation and sequencing revealed that csp4 and csp5 are alleles of cwf10 and prp39, respectively, both of which encode splicing factors. csp4, now denoted cwf10-1, creates a missense mutation (C323Y) in the guanosine triphosphate-binding domain of Cwf10. Cwf10 is the homolog of the Saccharomyces cerevisiae U5 small nuclear ribonucleoprotein Snu114 (and of human EFTUD2) that is required for U4/U6 small nuclear RNA (snRNA) unwinding (9). csp5, now denoted prp39-1, makes a nonsense mutation in Prp39 (W550stop). S. cerevisiae Prp39 (homologous to human PRPF39) is associated with U1 snRNA and is required for commitment to splicing of pre-mRNA (10). Thus, mutations in two distinct essential splicing factors affect silencing at centromeres.
To further investigate possible links between splicing and centromere silencing, we surveyed several additional ts lethal splicing mutants for silencing defects at the permissive temperature (11-14). Only particular splicing mutants affected silencing. Silencing of a centromeric cen1:ade6+ marker gene (Fig. 1A) remained intact in the presence of prp1 (Prp6Sc/Hs), prp2 (U2AFHs), prp3 (Prp3Sc/PRPF3Hs), or prp4 (PRPF4BHs) mutations (where the superscripts Sc and Hs denotes S. cerevisiae and human, respectively). In contrast, mutations in prp5 (Prp46Sc/PLRG1Hs), prp8 (Prp2Sc/DHX16Hs), prp10 (Hsh155Sc/SF3B1Hs), and prp12 (Rse1Sc/SF3B2Hs), like cwf10 and prp39, alleviated cen1:ade6+ silencing (Fig. 1B) and increased cen1:ade6+ transcript accumulation (Fig. 1C, ade6). Moreover, mutants that alleviated cen1:ade6+ silencing also displayed increased levels of noncoding centromeric otr transcripts and concomitant reductions in centromeric siRNA accumulation, with prp10-1 showing the most severe silencing defects (Fig. 1C, cen-dh and cen-dg, and Fig. 1D). Thus, several specific splicing mutants affect processing of centromeric transcripts into siRNAs and impair centromere silencing.
Fig. 1.
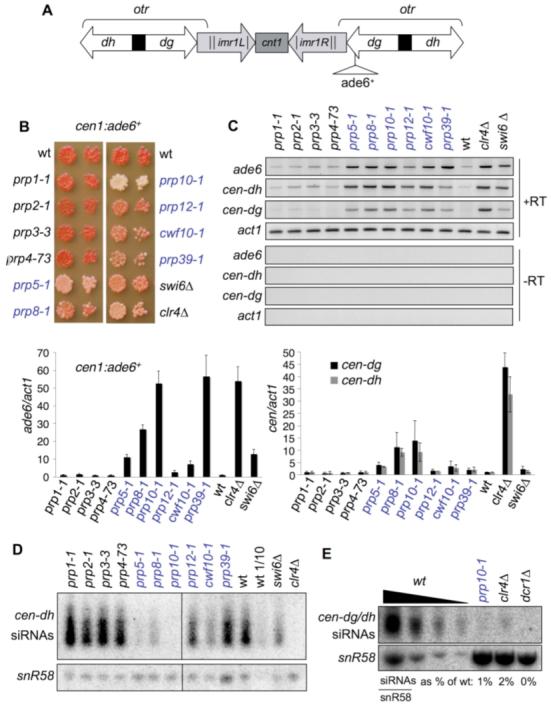
Specific splicing mutants affect RNAi-directed silencing at centromeres (A) Schematic of fission yeast centromere 1, indicating site of integration of cen1:ade6+ marker, outer repeat (otr) dg and dh elements, inner repeats (imr), and central core (cnt). (B) Silencing assay on strains bearing cen1:ade6+ (red, silent; pink/white, alleviated). (C) RT-PCR and quantitative RT-PCR (qRT-PCR) analysis of transcripts from ade6, cen-otr (dg and dh), and act1 control (-RT, no reverse transcriptase). Histograms show transcript levels relative to act1, normalized to the wild type. (D and E) Northern analysis of siRNAs corresponding to cen-dh or dg/dh. snoRNA58 (snR58) is a loading control.
A mundane explanation for these observations is that impaired splicing of mRNA encoding an RNAi component indirectly affects silencing. However, the silencing and splicing defects observed in these ts splicing mutants can be uncoupled. Reverse transcription polymerase chain reaction (RT-PCR) analysis of an mRNA (tbp1) that is highly sensitive to splicing defects (15) confirmed that these mutants accumulated increased levels of unspliced pre-mRNA at restrictive temperature (36°C). However, at the permissive temperature of 25°C (at which centromeric silencing was alleviated), splicing efficiency was similar to that in wild-type cells (Fig. 2A). Conversely, for mutants that did not alleviate silencing at 25°C, growth at semipermissive temperature impaired splicing of tbp1 but did not induce increased accumulation of cen1:ade6+ or otr (cen-dh) transcripts, as seen in prp10-1 (Fig. 2B). Even after prolonged incubation at 36°C, which strongly inhibits splicing (Fig. 2A), centromeric siRNAs were still readily detected in prp1-1, prp2-1, prp3-3, and prp4-73 (fig. S1). Thus, defective splicing does not inherently perturb the RNAi pathway.
Fig. 2.

Defective splicing does not cause defects in centromere silencing. (A and B) RT-PCR analysis of transcripts from tbp1, ade6, cen-dh, and act1. Spliced, mature (m), and unspliced, pre- (p) tbp1+ mRNA are indicated. Strains were grown at permissive (25°C) or semipermissive (30°C) temperatures or, for analysis at restrictive temperature, grown at 25°C and then shifted to 36°C for 6 hours. The histogram shows qRT-PCR analysis of tbp1+ mature transcript levels relative to act1, normalized to the wild type, at 25°C. (C) Silencing assay on strains bearing cen1:ade6+, with endogenous ago1+ or hrr1+ replaced by cDNAs.
The RNAi genes ago1+ and hrr1+ contain introns; if their splicing is particularly sensitive to defects, this could explain the observed phenotypes. We therefore constructed strains in which the endogenous ago1+ and hrr1+ genes were replaced by cDNAs. Even in these strains, the prp10-1 mutation alleviated silencing as in wild-type cells (Fig. 2C, white colonies). Thus, the silencing defects in prp10-1 do not result from inefficient splicing of these RNAi components. However, defective splicing of a gene encoding some other, unknown contributory factor cannot be excluded.
Because splicing is generally coupled to transcription (16), defective splicing might affect centromeric siRNA production by impairing otr transcription. However, in contrast to the rpb7-G150D mutation affecting RNA Pol II (7), splicing mutants did not affect the abundance or length of centromeric transcripts accumulating in a dcr1 background (fig. S2). Thus, in these splicing mutants the defect in RNAi-directed silencing lies downstream of otr transcription.
Strains lacking RNAi exhibit reduced H3K9 methylation and Swi6 association at centromeric otr chromatin (17). Chromatin immunoprecipitation (ChIP) revealed that splicing mutants cwf10-1 and prp10-1 (but not prp2-1) exhibited only a modest decrease in levels of H3K9me2 associated with both centromere repeats and cen1:ura4+ (Fig. 3A and fig. S3). Reductions in H3K9me2 in cwf10-1 and prp10-1 were greater on cen1:ura4+ than on otr repeats, and greater still on cen1:ade6+ (fig. S4); however, these mutants consistently retained more H3K9me2 than did dcr1 cells. Levels of centromeric Swi6 were also moderately reduced in cwf10-1 and prp10-1 cells (Fig. 3B). Thus, prp10-1 cells maintain relatively high levels of H3K9me2 and Swi6 at centromeres, even though siRNA generation is severely compromised. This is similar to cells lacking the polyA polymerase Cid14 (18).
Fig. 3.

Splicing mutants disrupt RNAi-dependent heterochromatin. (A and B) ChIP analysis of H3K9me2 and Swi6 at cen-dg or cen1:ura4+ relative to a euchromatic control locus (fbp1 or ura4 DS/E, respectively). Representative gels are shown. Relative enrichments were calculated as the ratio of product of interest to control product in immunoprecipitate (IP) relative to input (in). The histogram represents qPCR analysis of four independent experiments; relative enrichments (cen-dg/fbp1 and cen1:ura/fbp1) are shown as a percentage of the wild type. See also fig. S2. (C) ChIP analysis of H3K9me2 at cen-dg sequences on endogenous centromeres, or on plasmid pH-cc2 [see schematic and (20, 27)]. Plasmid was introduced by transformation and maintained under selection. Primers spanning the dg-plasmid backbone junction were used to specifically analyze H3K9me2 on the plasmid.
siRNAs are absolutely required for establishment, but not maintenance, of H3K9 methylation on centromeric repeats (19). To determine whether cwf10-1 and prp10-1 affect the establishment of H3K9me2 on naïve repeats, we transformed wild-type or mutant strains with a plasmid bearing part of a cen1 otr (dg) repeat (20). ChIP analysis using plasmid-specific primers confirmed that H3K9me2 was established on the plasmid in wild-type but not in dcr1 cells. In prp10-1 cells, H3K9me2 was still established on the plasmid (Fig. 3C, pH-cc2), albeit at a lower level than in wild-type cells, consistent with the lower level of H3K9me2 seen on endogenous centromeric repeats (Fig. 3, A and C, cen-dg). This result suggests that prp10-1 cells must retain a low level of siRNAs, and indeed we detected siRNAs at about 1% of wild-type levels in prp10-1 cells (Fig. 1E). Thus, very low levels of siRNAs are sufficient to establish H3K9me2 heterochromatin, but high siRNA levels are required to maintain robust heterochromatin, and this siRNA amplification requires splicing factors such as Prp10 and Cwf10.
Unlike at the centromeres, maintenance of silencing at the mating-type locus does not require RNAi components but does require Cid14 (18) and chromatin components such as Swi6 (21). Analysis of a ura4+ marker inserted within the mating-type locus revealed that, like dcr1, none of the tested splicing mutants alleviated mating-type silencing (Fig. 4A). We also tested the effect of the splicing mutants on a ura4+ locus silenced by tethering of the RITS component Tas3 to the ura4 transcript (22). As with RNAi mutants (dcr1), both prp10-1 and cwf10-1 mutants alleviated silencing, whereas prp2-1 did not (Fig. 4B). Together these observations confirm a specific role for Cwf10 and Prp10 in RNAi-mediated silencing that is downstream of RITS recruitment, consistent with a function related to amplification of the RNAi response.
Fig. 4.
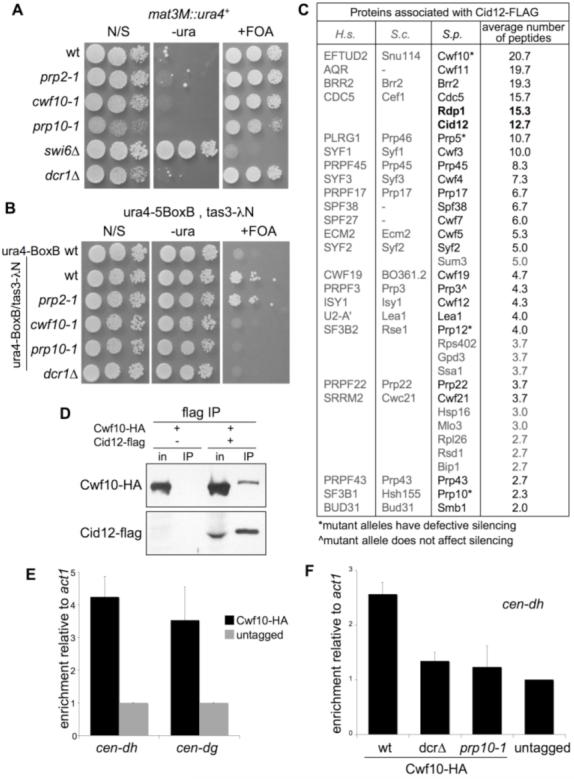
Splicing factors are specifically required for RNAi-dependent silencing and physically associate with the RNAi machinery and centromere repeats. (A and B) Assays for silencing of ura4+ at mating-type locus [mat3-M::ura4+ (21) (A)] or with tethered Tas3 [ura4-5BoxB/tas3-lN (22) (B)]. Plates are nonselective (N/S), lacking uracil (-ura), or supplemented with FOA (+FOA). (C) Proteins specifically associated with Cid12-FLAG, analyzed by LC-MS/MS, as described (27). For splicing factors, human (H.s.) and budding yeast (S.c.) homologs are shown. (D) Coimmunoprecipitation of Cwf10-HA with Cid12-FLAG. (E and F) ChIP analysis of Cwf10 enrichment at cen-dh or cen-dg relative to a euchromatic, unspliced control locus act1+, normalized to an untagged control strain.
The phenotypes of splicing mutants suggest that splicing factors may interact with the RNAi machinery. Indeed, in cells lacking Rdp1, spliceosome subunits have been reported to copurify with affinity-selected Cid12 (4). To examine this more closely, we analyzed immunoprecipitates of FLAG epitope-tagged Cid12 from wild-type cells by liquid chromatography-mass spectrometry (LC-MS)/MS. Many splicing factors were found to specifically associate with Cid12-FLAG. These included Cwf10, Prp10, Prp5, and Prp12, which were required for centromeric silencing, along with splicing factors such as Prp3 that did not affect silencing (Fig. 4C). Splicing factors were rarely identified in immunoprecipitates of numerous other FLAG-tagged proteins, indicating that this interaction is specific. The association of Cwf10 with Cid12 was also verified by coimmunoprecipitation (Fig. 4D). Thus, Cid12 may function in association with a large spliceosomal complex, with particular splicing factor mutants compromising its activity and there-by impairing RDRC-mediated siRNA amplification.
Because splicing factors interact with Cid12, they may (like other RNAi components) associate with centromeric repeats. To test this possibility, we performed ChIP with Cwf10, the splicing factor most strongly represented in Cid12 immunoprecipitates. Hemagglutinin epitope-tagged Cwf10 (Cwf10-HA) was found to be enriched on both dh and dg centromere repeats, relative to an unspliced control gene, act1 (Fig. 4E). Prp8-HA was also enriched on centromere repeats, and both Cwf10-HA and Prp8-HA were at least as strongly associated with cen-dh as was Cid12-HA (fig. S5). The association of Cwf10-HA with centromere repeats was essentially lost in dcr1 and prp10-1 cells, showing it to be linked to a functional RNAi pathway (Fig. 4F). We conclude that splicing factors associate with the RNAi machinery at the centromere to directly facilitate RNAi-mediated centromere silencing.
Processing of transcripts by both the splicing and RNAi machineries is thought to occur cotranscriptionally, implying that generation of siRNAs from centromere transcripts may occur in the context of larger RNA-processing “factories” (1, 16, 22). We propose that spliceosomal complexes provide a platform that promotes the processing of centromeric transcripts by RDRC, facilitating amplification of homologous siRNAs to high levels. The silencing and chromatin modification defects seen in particular splicing factor mutants are thus explained by disruption of this transcript-to-siRNA processing step, independently of splicing itself. Splicing factors have also been identified in screens for factors affecting RNAi in nematodes and plants (23, 24) and are required for processing of some microRNAs in nematodes, flies, and humans (25, 26). Thus, integration with other RNA processing events may be a conserved feature of RNAi-related pathways.
Supplementary Material
Acknowledgments
We thank J. Beggs, E. S. Choi, A. Pidoux, F. Simmer, I. Stancheva, and D. Tollervey for comments; N. Kaufer, D. Moazed, and T. Tani for materials; and the Allshire lab for support. Supported by Epigenome Network of Excellence (EC-FP6/Contract/LSHG-CT-2004-503433), of which R.C.A. and K.E. are members (E.B.); UK Medical Research Council grant G0301153/ID:69173 (A.K.); Wellcome Trust Prize Studentship 067844 (M.P.); Marie Curie Excellence Grant MEXT-CT-014171 from the European Commission (J.R.); and Wellcome Trust Principal Research Fellowship grant 065061/Z (R.C.A.).
References and Notes
- 1.Buhler M, Moazed D. Nat. Struct. Mol. Biol. 2007;14:1041. doi: 10.1038/nsmb1315. [DOI] [PubMed] [Google Scholar]
- 2.Grewal SIS, Jia ST. Nat. Rev. Genet. 2007;8:35. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- 3.Verdel A, et al. Science. 2004;303:672. doi: 10.1126/science.1093686. published online 2 January 2004 (10.1126/science.1093686). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Motamedi MR, et al. Cell. 2004;119:789. doi: 10.1016/j.cell.2004.11.034. [DOI] [PubMed] [Google Scholar]
- 5.Pidoux AL, Allshire RC. Philos. Trans. R. Soc. London Ser. B. 2005;360:569. doi: 10.1098/rstb.2004.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ekwall K, Cranston G, Allshire RC. Genetics. 1999;153:1153. doi: 10.1093/genetics/153.3.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Djupedal I, et al. Genes Dev. 2005;19:2301. doi: 10.1101/gad.344205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Volpe T, et al. Chromosome Res. 2003;11:137. doi: 10.1023/a:1022815931524. [DOI] [PubMed] [Google Scholar]
- 9.Bartels C, Klatt C, Luhrmann R, Fabrizio P. EMBO Rep. 2002;3:875. doi: 10.1093/embo-reports/kvf172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lockhart SR, Rymond BC. Mol. Cell. Biol. 1994;14:3623. doi: 10.1128/mcb.14.6.3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Potashkin J, Kim D, Fons M, Humphrey T, Frendewey D. Curr. Genet. 1998;34:153. doi: 10.1007/s002940050381. [DOI] [PubMed] [Google Scholar]
- 12.Urushiyama S, Tani T, Ohshima Y. Mol. Gen. Genet. 1996;253:118. doi: 10.1007/s004380050304. [DOI] [PubMed] [Google Scholar]
- 13.Rosenberg GH, Alahari SK, Kaufer NF. Mol. Gen. Genet. 1991;226:305. doi: 10.1007/BF00273617. [DOI] [PubMed] [Google Scholar]
- 14.Potashkin J, Li R, Frendewey D. EMBO J. 1989;8:551. doi: 10.1002/j.1460-2075.1989.tb03409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Habara Y, Urushiyama S, Shibuya T, Ohshima Y, Tani T. RNA. 2001;7:671. doi: 10.1017/s1355838201001200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bentley DL. Curr. Opin. Cell Biol. 2005;17:251. doi: 10.1016/j.ceb.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 17.Volpe TA, et al. Science. 2002;297:1833. doi: 10.1126/science.1074973. published online 22 August 2002 (10.1126/science.1074973) [DOI] [PubMed] [Google Scholar]
- 18.Buhler M, Haas W, Gygi SP, Moazed D. Cell. 2007;129:707. doi: 10.1016/j.cell.2007.03.038. [DOI] [PubMed] [Google Scholar]
- 19.Sadaie M, Iida T, Urano T, Nakayama J. EMBO J. 2004;23:3825. doi: 10.1038/sj.emboj.7600401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Folco HD, Pidoux AL, Urano T, Allshire RC. Science. 2008;319:94. doi: 10.1126/science.1150944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hall IM, et al. Science. 2002;297:2232. doi: 10.1126/science.1076466. published online 5 September 2002 (10.1126/science.1076466) [DOI] [PubMed] [Google Scholar]
- 22.Buhler M, Verdel A, Moazed D. Cell. 2006;125:873. doi: 10.1016/j.cell.2006.04.025. [DOI] [PubMed] [Google Scholar]
- 23.Kim JK, et al. Science. 2005;308:1164. published online 24 March 2005 (10.1126/science.1109267). [Google Scholar]
- 24.Herr AJ, Molnar A, Jones A, Baulcombe DC. Proc. Natl. Acad. Sci. U.S.A. 2006;103:14994. doi: 10.1073/pnas.0606536103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guil S, Caceres JF. Nat. Struct. Mol. Biol. 2007;14:591. doi: 10.1038/nsmb1250. [DOI] [PubMed] [Google Scholar]
- 26.Goymer P. Nat. Rev. Mol. Cell Biol. 2007;8:597. [Google Scholar]
- 27.See supporting material on Science Online.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


