Abstract
Enzyme isoforms are found in many cellular reactions, and can differ in the kind of reaction they catalyze, in their substrate affinity, or in their reaction rates. The evolutionary significance of enzyme isoforms is only partially understood. We used mathematical modeling to investigate the hypothesis that isoforms may be favored by selection because they can increase the phenotypic robustness of the system. We modify a model for circadian clock gene expression in Drosophila to incorporate the presence of isoforms in the phosphorylation pathway of the period gene. We consider the case where different isoforms catalyze the same reaction but have different affinities for the substrate. Stability is increased if there is dynamic control of the expression of isoforms relative to each other. Thus, we show that controlling isoform proportion can be a powerful mechanism for reducing the effects of variations in the values of system parameters, increasing system robustness.
Keywords: circadian rhythm, gene duplications, isozymes, Drosophila, robustness, canalization
Isozymes are variants of an enzyme with the same function that are found in the same individual (Hunter and Market 1957). These enzymes may have different kinetic rates, different regulatory properties, or be expressed in a tissue-specific manner. They are produced in several different ways: They may be products of duplicated genes or alternative splicing, or alleles at the same locus. The source of an isozyme, however, does not provide us with information about its impact on fitness. It is often assumed that isozymes produced by duplicated genes or alternative splicing are adaptive, but in most cases it is unclear whether this is so.
Recently, genomic data has focused a great deal of attention on gene duplications, which generate enzyme isoforms. Ideas of how gene duplicates may arise (Ohno 1970) and the conditions under which they invade have been previously proposed (Clark 1994). A gene duplicate can have one of three ultimate fates: loss of function, neofunctionalization or subfunctionalization (Force et al. 1999). In the first case, one of the duplicate copies acquires a disabling mutation. In the second case one copy evolves a new function. The third case happens when both copies become necessary because each copy has accumulated degenerative mutations. At least in the short term, a fourth outcome is preservation of function because of selection for increased flux through the step catalyzed by a protein (Papp et al. 2004).
While most have assumed a guiding role for natural selection in arriving at these outcomes, recent theory suggests that the increased number of retained gene duplicates and increased gene number in multicellular eukaryotes may be a byproduct of low effective population size and not the result of natural selection (Lynch and Conery 2003). Should this be the case, then isozymes from gene duplication can be regarded as an incidental byproduct of population genetic processes rather than adaptations. The detailed, selective mechanisms by which gene duplicates might become neo- or sub-functionalized has received less attention than describing the rates and fates of gene duplicates. This debate is recapitulating an older one about whether allelic isozyme variants are neutral (Kimura 1983) or subject to natural selection (Gillespie 1991).
While the distinction between neo- and sub-functionalized seem clear at one level, more detailed consideration of selective mechanisms suggest that the boundaries between subfunctionalization and neofunctionalization may not be well defined. Once isoforms become coexpressed, for instance, they can provide an easy route towards an adaptive peak where robustness, the invariance of the phenotype in the face of changes in the values of system parameters, is increased. This would represent a case of neofunctionalization (increased robustness) through diversification of function (isozymes with different substrate affinities).
Much work relevant to the adaptive value of isozymes has been directed at interpreting data on dispensability of genes in yeast. A knockout of a functional gene can be compensated by the presence of another gene with equivalent function, or by redundancy of the genetic network as a whole (Gu 2003). The first hypothesis provides a potential selective mechanism for the maintenance of isozymes by gene duplication. While compensation does occur (Gu et al. 2003), dispensability seems to be primarily due to the second mechanism (Wagner 2000). A detailed analysis of network function suggested the alternative hypothesis that duplication might be favored in high flux pathways (Papp et al. 2004). All of these hypotheses apply only to isozymes originating from gene duplication.
Quantitative robustness to perturbations can arise due to the presence of isozymes from any cause. In this paper we use mathematical modeling to investigate the potential for co-expressed isoforms to increase robustness to quantitative perturbation. Perturbations can arise from environmental or genetic sources, although we make no distinction between them in this manuscript. Theoretical arguments (Wagner et al. 1997, Proulx and Phillips 2005) suggest that robustness to environmental noise evolves more readily than does robustness to genetic noise. We only consider isoforms originating from alternative splicing, but the general conclusions apply to isoforms from other sources as well. Our goal is not to provide a comprehensive framework for the origin and successive evolution of isoforms but rather to investigate their potential benefit in a biological system. We use the Drosophila circadian rhythm network as a model system in our investigation. The circadian rhythm is important to fitness (Kumar et al. 2005) and its phenotype (period) is reliable and can be measured in vitro and behaviorally (Panda et al. 2002). A previous model of the mammalian circadian clock has noted that this system seems robust to variation in parameter values, but they did not investigate the effects of isozymes (Forger and Peskin 2003).
In Drosophila the molecular generator of the rhythm is the negative feedback of the genes period and timeless on their own transcription. This mechanism has been described with various levels of detail by mathematical models (Goldbeter 1995; Leloup and Goldbeter 1998, 2003; Leloup et.al. 1999; Smolen et. al. 2001; Ueda et. al. 2001). The proteins of the gene period (PER) and timeless (TIM) undergo successive phosphorylation events, introducing delays in the feedback of the proteins on transcription. Phosphorylation is crucial for PER and TIM translocation into the nucleus, a requirement for the production of the rhythm. Casein kinase II (CKII) is the protein largely responsible for phosphorylation of PER and is found in at least three coexpressed functional variants (Jauch et al. 2006). CKII has been proposed as an evolutionary link between different circadian systems of animals, plants and fungi, and the circadian clock seems to be highly sensitive to its activity (Lin, 2002). CKII is a heterotetramer, with two α and two β subunits. The α subunits have catalytic functions. The β subunits protect the α-subunit against denaturing agents or conditions (Meggio et al. 1992), alter the substrate specificity of the α-subunits (Bidwai et al., 1993) and modulate the activity of the enzyme (Guerra et al. 1999). Alternative splicing variants of the β subunit are recombined to form the isozymes. We modify a mechanistic model for circadian oscillations in Drosophila to test if CKII isoforms can increase system robustness.
THE MODEL
We consider a minimal molecular model for circadian oscillations in constant darkness conditions in Drosophila (Goldbeter 1995), describing the time course of the gene period and its products. What the minimal model does not describe is the time course of other genes, such as timeless, that are entrained by light. We modified the model to incorporate the presence of CKII isoforms. These isoforms are also present, and with the same function, in the pathway of genes not represented in the model. We chose this model because it captures the essential features of circadian oscillations while minimizing the mathematical complexity. The model consists of differential equations for the concentrations of period mRNA (Mp), the unphosphorylated form of PER protein (P0), its single (P1) and double (P2) phosphorylated forms, and the nuclear PER concentration (PN). We do not explicitly model transcription and regulation of CKII but assume that its gene expression level can be regulated. Analysis of the enzyme structure suggests that CKII is in fact regulated by transcription (Guerra and Issinger 1999) and that different isoforms have different substrate affinities or functional properties (Guerra et al. 1999; Jauch et al. 2006).
One example of a system containing isoforms with different substrate affinities is the pancreatic β-cell in the mouse. This cell contains two isoforms for the sarco-endoplasmic reticulum calcium ATPase (SERCA pump), which pumps Ca2+ from the cytosol to the endoplasmic reticulum. One isoform, SERCA-2b, has high Ca2+ affinity and is important for maintaining basal Ca2+ levels in the cytosol. The other, SERCA-3, has a lower affinity and is important during stimulation by glucose (Arredouani et al. 2002).
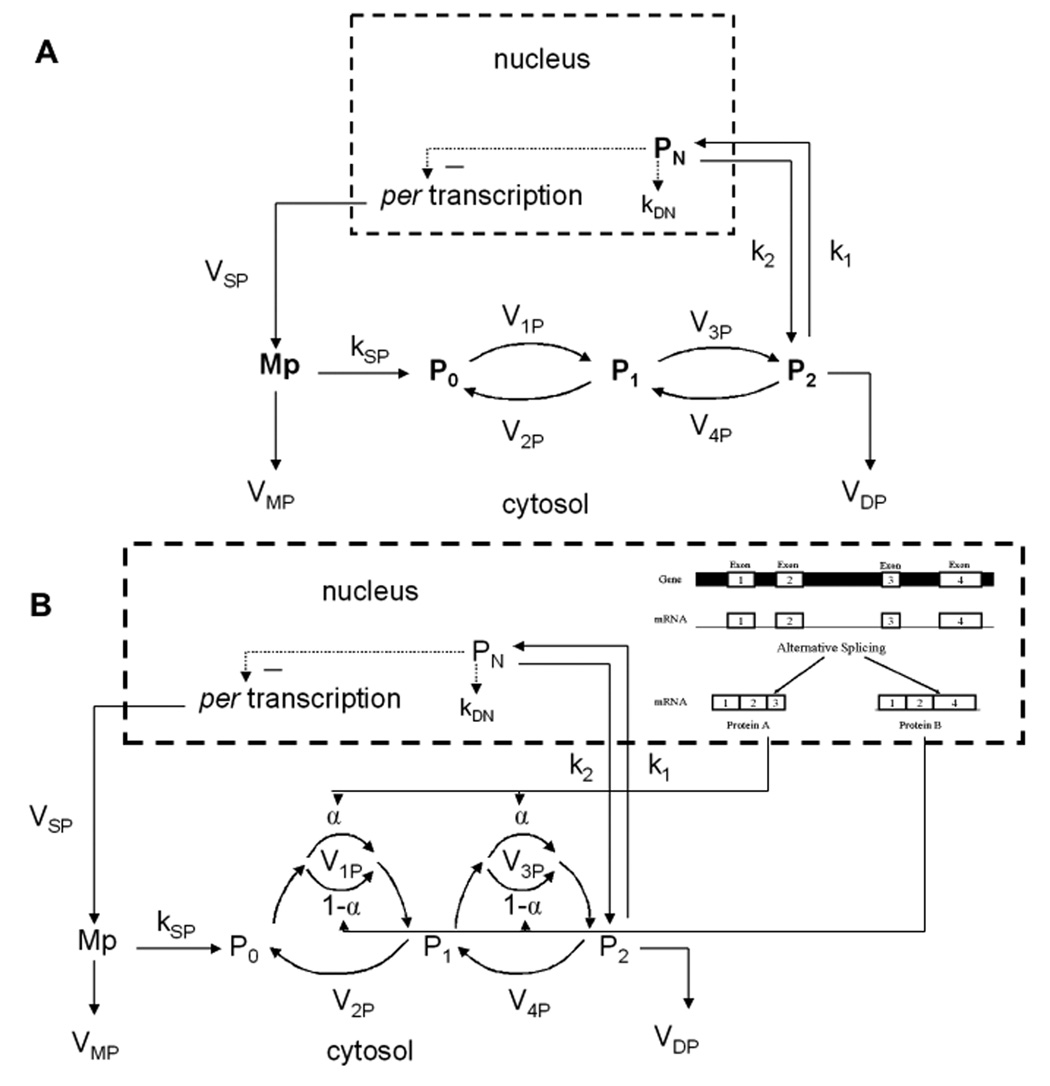
In our model we consider the presence of two CKII isoforms that differ in their affinity to the substrate, and separate the total amount of CKII into a fraction α of the high affinity isoform, and a fraction 1- α of the low affinity isoform. A schematic diagram illustrating the steps describing the circadian rhythm in Drosophila is shown in Fig. 1. The upper panel (1A) shows the system without isoforms while the lower panel (1B) shows the modified system with the presence of isoforms. The mRNA of period (Mp) is produced in the nucleus and transferred to the cytosol. Mp accumulates at maximum rate VSP, and is enzymatically degraded at maximum rate VMP, with Michaelis constant KmP. The synthesis of period protein (P0) is proportional to Mp and is characterized by a first-order rate constant kSP. Parameters ViP (i = 1, … 4) denote maximum rates of the kinases and phosphatases involved in the reversible phosphorylation of P0 to P1 and P1 to P2. Parameters K11P and K12P denote the Michaelis constants of the two kinase isoforms while KP denotes the Michaelis constant of the phosphatase. The fully phosphorylated form (P2) is enzymatically degraded at maximum rate VDP with Michaelis constant KdP, and is transported into the nucleus at rate k1. The reverse transport occurs with rate k2. Nuclear PER (PN) exerts negative feedback on period transcription, which is key to the generation of the rhythm. This feedback is described by a Hill equation with cooperativity degree n and repression constant KIP. The diagram also illustrates how the phosphorylation pathway of the gene period is connected with the production of CKII isoforms via alternative splicing (Fig. 1B).
1.
Model scheme for circadian oscillations in Drosophila. Upper panel (A) shows the system without isoforms, lower panel (B) shows the modified system with isoforms derived from alternative splicing.
The dynamics of the model variables are described by a system of five coupled ordinary differential equations (Goldbeter 1995). The first equation describes the mRNA transcription:
| (1) |
The first term describes mRNA synthesis and the negative feedback of the protein PER on its own transcription. The second term describes mRNA enzymatic degradation. The second equation describes the dynamics of the unphosphorylated protein P0:
| (2) |
The first term relates protein synthesis to the concentration of mRNA. The second term describes dephosphorylation of P1 to P0. The third and final terms describe P0 phosphorylation by the two isoforms of CKII. The mathematical system that describes the circadian rhythm without isoforms can be derived by either setting α to zero, or 1, and using only one Michaelis constant (K11P or K12P), or by setting α to 0.5 and using two identical Michaelis constants (K11P = K12P). The equation for the phosphorylated form P1 is similarly derived. P1 can be produced from its unphosphorylated form, P0, or from the dephosphorylation of P2. P1 can be phosphorylated into P2, or dephosphorylated to P0:
| (3) |
The fourth equation describes the dynamics of the double phosphorylated protein:
| (4) |
The first three terms represent phosphorylation and dephosphorylation. The fourth and fifth describe the transport of P2 into (k1) and out of (k2) the nucleus. The last term describes P2 enzymatic degradation.
The last equation shows the dynamics of the protein in the nucleus:
| (5) |
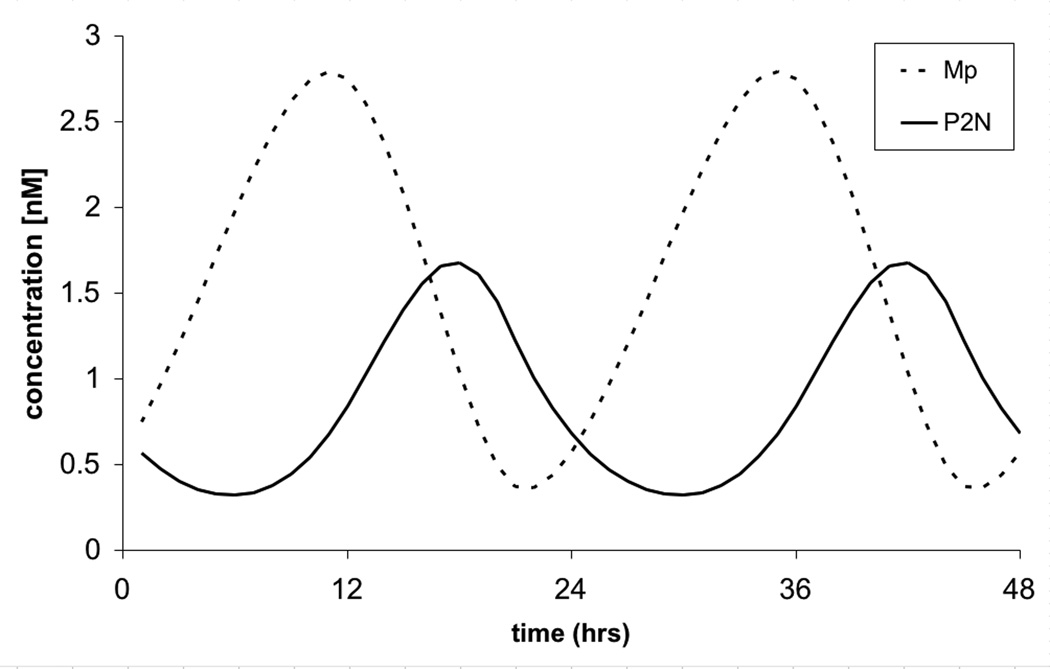
An example of the circadian oscillations produced by this model is shown in figure 2. The increase of messenger RNA (Mp), causes an increase in the amount of protein P0 (not shown), eventually leading to a delayed increase in nuclear protein PN. This feeds back negatively on period transcription, causing the messenger RNA to decrease. The delayed negative feedback of period protein on its own transcription is the mechanism for the rhythm. Importantly, it is the two phosphorylation steps that are primarily responsible for the delay.
2.
Circadian oscillations of Mp (dotted line) and PN (continuous line). The period of the oscillations is 24 hours. Parameter values are as in Table 1.
The differential equations were solved numerically with backward Euler method in the XPPAUT software package (Ermentrout 2002). The period was computed using Fast Fourier Transformations in the numerical package of Python.
RESULTS
CKII affinity
To determine if system robustness is increased by CKII isoforms we compare the dynamics of the model with isoforms to the dynamics of the model without. A system is considered to be robust if the period of the oscillation remains near 24 hours in spite of parameter variation. In this investigation we call a system “circadian” if it has a period of 24 ± 0.5 hours. While all the model variables oscillate with a circadian period, we use the concentration of mRNA to measure period length. For the purpose of this paper, we regard the total enzyme reaction rate (V1p), which is the product of the number of enzyme molecules and the reaction rate per molecule, as constant and characterize isoforms as having different affinities to the substrate.
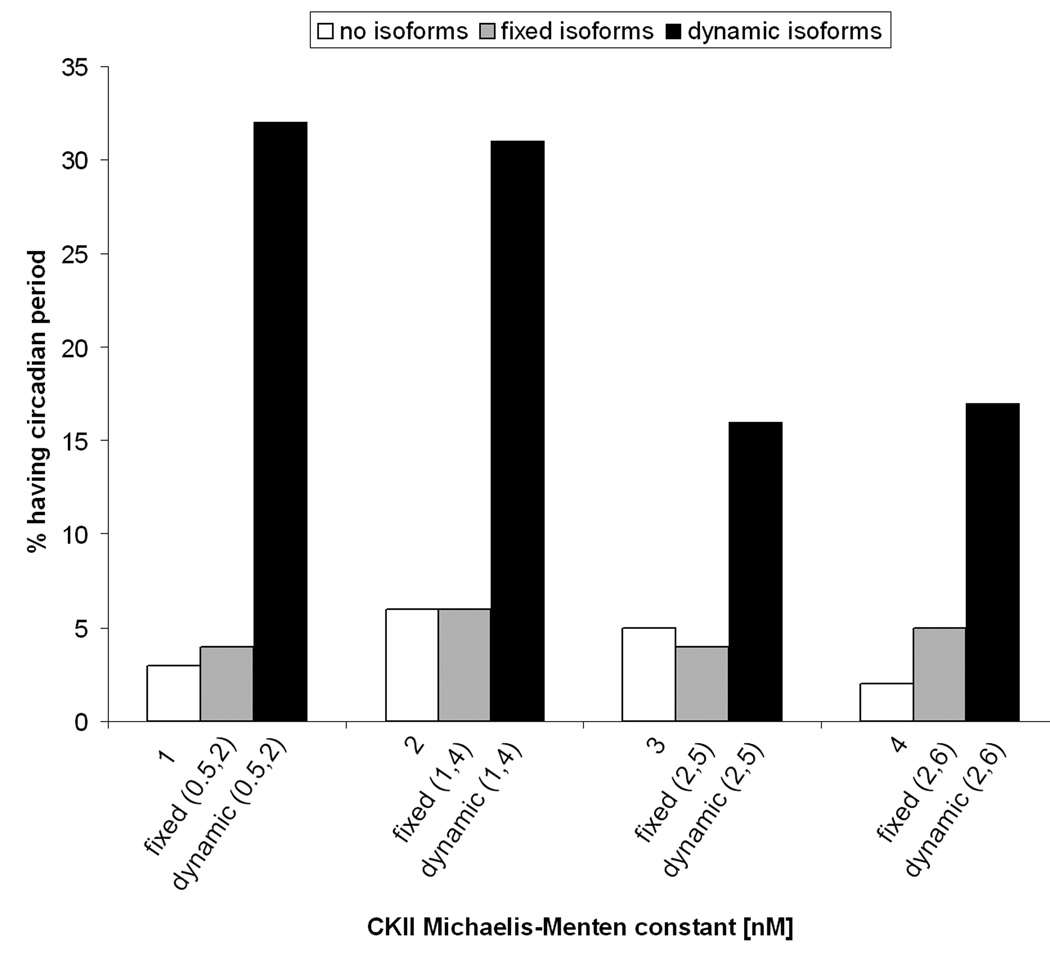
We begin by investigating the effect of enzyme affinity in a system without isoforms. We ask if the substrate affinity of CKII has any impact on the robustness of the system. For instance, is it better to have an enzyme with high affinity to the substrate or one with low affinity? We set α to 1 and select a baseline parameter set that has a 24 hour period (Leloup and Goldbeter 1998). We then generate 300 replicate systems that have different parameter values but the same CKII substrate affinity (i.e. the same K11P values). For each replicate a random set of values for all other parameters is generated. Each parameter value is drawn from a uniform distribution with the same mean as the baseline parameter set with a range of variation of plus or minus 50%. The values of the substrate affinities for CKII are shown as the first numbers under each column in Fig. 3. The vertical axis of Fig. 3 represents the percentage of replicates that produce a circadian rhythm.
3.
Fixed proportion of isoforms does not increase system robustness but dynamic regulation does. The x-axis shows systems with different combinations of enzyme affinities. The single number refers to the Michaelis constant of the enzyme in a system without isoforms, while the two numbers in the parenthesis refer to the Michaelis constants of the isoforms. The y-axis shows the percentage of the 300 replicates that exhibit a circadian rhythm. Replicates are generated using random uniform distributions of the values of system parameters.
In the absence of isoforms and with the Michaelis constant K11P = 2 nM only about 5% of the replicates produce an oscillation with a circadian period. Similar results are obtained with Michaelis constants 1, 3 and 4 nM (Fig.3, white bars). Thus, the affinity to the substrate alone does not affect the robustness of the system. Robustness could be increased if we allow enzyme production or maximum rate to be regulated according to the reaction needs (results not shown), but under the assumption of constant production, a high-affinity enzyme is neither better nor worse than a low-affinity enzyme.
Fixed proportion of isoforms
We now look at the effects of CKII isoforms that are produced at a constant rate and in constant relative proportion to each other, criteria that are satisfied by keeping the parameters V1p and α constant in the simulations. We then ask if this system with constant isoform expression is more robust then a system where isoforms are not present. Three hundred replicates of the system are obtained as described above. The values of the Michaelis constants are generated in the following way: given a Michaelis constant, say 2 nM, we obtain two values (K11P and K12P) that produce a saturation curve similar to a single enzyme with affinity 2 nM when the two isoforms are present in equal amounts (α=0.5). When isoforms are present, with Michaelis constants of K11P = 1 nM and K12P = 4 nM, shown in parenthesis in Fig. 3, the percentage of replicates yielding a circadian rhythm (gray bars) is almost the same as the system with no isoforms and Michaelis constant K11P = 2 nM. Similar results are obtained when different combinations of Michaelis constants are considered (Fig. 3, white vs gray bars).
In conclusion, replacing one enzyme with two isoforms chosen to give the same saturation curve has little effect on system robustness.
Dynamic regulation of isoform proportion
Now we investigate the effect of dynamic regulation of isoforms, asking if a system with the ability to regulate isoform proportion is more robust than a system without isoforms. We do this by first perturbing one parameter at a time to see how the system can compensate by an appropriate change in α. We then perturb multiple parameters in a random fashion, as was done earlier.
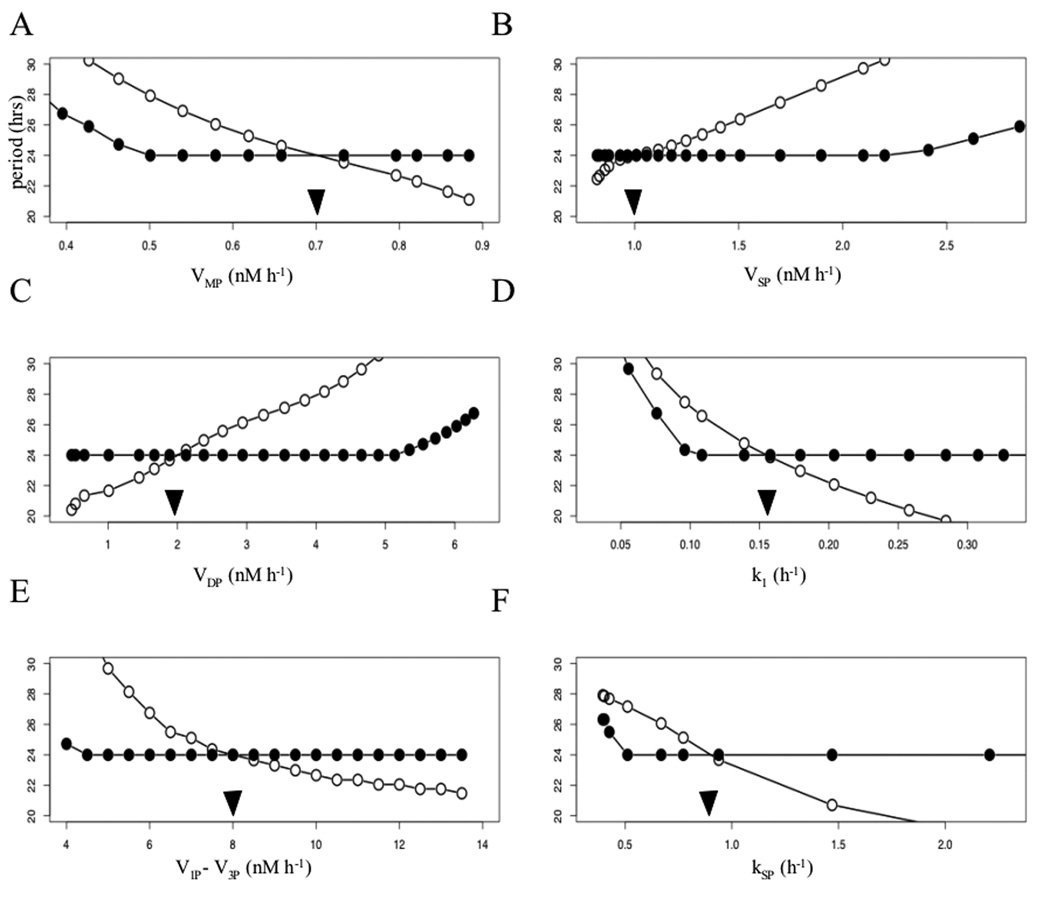
A convenient way of summarizing results of parameter variation is to plot the period of the oscillations as a bifurcation parameter is varied while keeping all the other parameters constant. In dynamical system theory this is referred to as a period bifurcation diagram (PBD). A robust system is identified by a flat PBD, corresponding to little change in the period over a range of parameter values. We contrast the PBD for the models with and without isoforms, varying one parameter at a time.
We begin by plotting the PBD of a system without isoforms (Fig. 4, unfilled circles). For each of the parameters considered the relation between the period and a change in parameter value can be increasing or decreasing. For instance, changing the enzymatic degradation rate of Mp (Fig.4, A) from 0.7 (arrow head) to 0.5 changes the period from 24 to about 28 hours, outside of the circadian range. We now consider a system with dynamic regulation of isoform proportions. For each of the parameter values considered above we ask if there is an α value that results in a circadian period. For instance, when VMP is changed to 0.5 a value of α = 0.93 provides a circadian period. Similar adaptations in α can be made so that the PBD is flat for a wide range of VMP values (Fig.4, A, filled circles). Thus, the system is robust to changes in VMP. Results are similar in the other panels.
4.
Period bifurcation diagrams of a system without isoforms (unfilled circles) and one with isoforms (filled circles). The relative proportion of different isoforms is regulated through α to achieve circadian periods. The presence of dynamically regulated isoforms provides an increase in stability to fluctuations of all parameters, changed one at the time. The arrow heads indicate the default values of the parameters.
We now look at how dynamic regulation can increase robustness by considering two parameters, k1 and kSP, as an illustrative example. A system should evolve towards the set of parameter combinations that provides a circadian period and makes it more robust. Isoform regulation, independently of parameter values, can increase canalization (Fig. 5). In panel A we show the contour of a fitness landscape, where fitness is modeled as a Gaussian function, of a system without isoforms. Regions with the same shading have the same fitness. Fitness is maximum when the period corresponding to the parameter combinations is 24 hours and decreases for deviations from the circadian period. For each parameter combination of k1 and kSP in panel A we compute first the period and then the fitness. In panel B we show a system with dynamically regulated isoforms, with Michaelis constants of K11P = 0.5 and K12P = 5. The number of parameter combinations that produce a circadian period, and thus maximum fitness (white region of plot), is greatly increased with dynamic regulation of isoforms.
5.
Dynamic regulation of isoforms can increase phenotypic robustness. Contour fitness landscape of systems without (A) and with (B) isoforms. Regions with same shading have equal fitness. Fitness has a maximum value of 1 (white regions) when the system has a period of 24 hours. Darker regions indicate parameter combinations that give periods that deviate from 24 hours. In panel B, K11P=0.5 and K12P=5 and for each combination of k1 and kSP α is adjusted to produce, if possible, a circadian period.
Dynamic regulation of isoforms also proves to be an effective way of compensating changes of multiple system parameter values (Fig. 3, white vs black columns). In this case we generate replicate systems with uniformly distributed parameter values as described earlier. When isoforms are present and can be regulated, the percentage of replicates yielding a circadian rhythm is greatly increased compared to the system with no isoforms, and the system with isoforms but no dynamic regulation. This is true for all combinations of Michaelis constants shown in Fig. 3.
Affinity difference between isoforms
Here we investigate the effects of affinity differences between isoforms that can be dynamically regulated. If isozymes are present they can be more or less similar in their amino acid sequence, or exon order, and will consequently have more or less similar affinities to the substrate. Increasing sequence divergence, or the number of possible exon combinations, can result in isozymes with increased difference in their substrate affinity.
We define the degree of affinity difference as the ratio between two isoform Michaelis constants, K12P/K11P. A ratio of one indicates two isoforms that are identical, and the more the ratio deviates from one the higher the degree of difference between two isoforms will be. We generate 300 system replicates with K11P fixed at 0.5 and change the value of K12P by increments of 0.5. We then randomly pick the other parameter values as described earlier. For each replicate system we compute the period. If the resulting period is not circadian we look for a value of α that reestablishes the circadian period. System robustness, indicated by the percentage of times the system oscillates with circadian period, increases with increasing difference between isoform affinities and saturates when the affinity ratio is 7 or greater (Fig. 6). Saturation occurs because as the Michaelis constant of the second isoform (K12P) reaches larger values its affinity becomes very low. In this case, the enzyme reaction curve has a small slope at typical substrate levels, so increasing K12P further will result in only a small reduction in substrate binding. The combination of different very low affinities enzymes with a high-affinity (k11P) enzyme will be very similar.
6.
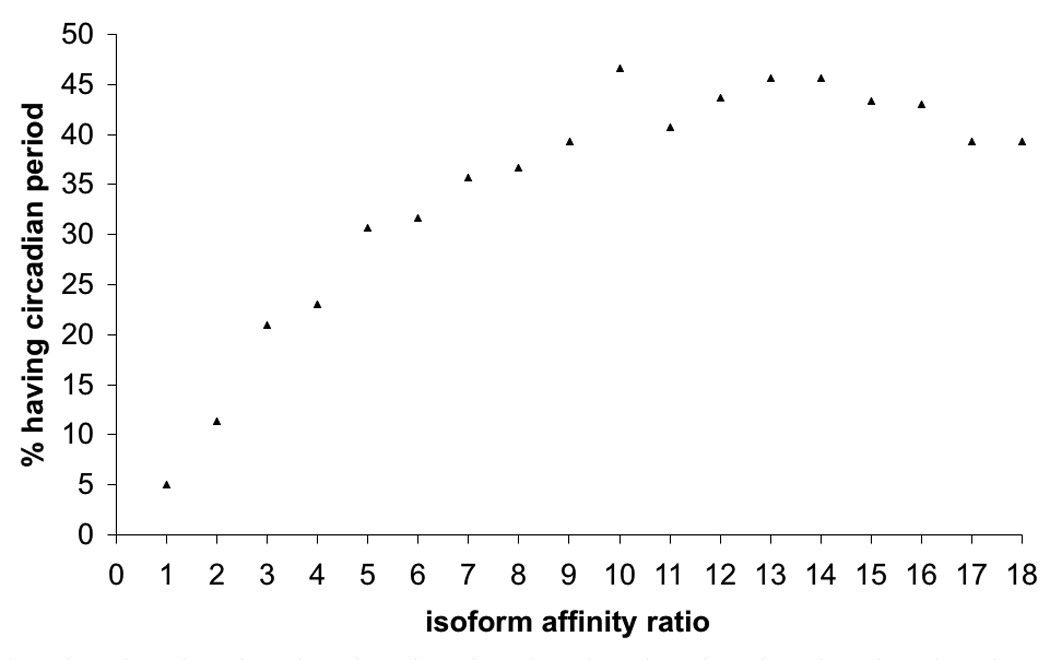
Isoforms with greater affinity differences provide more stability. The x-axis shows the ratio between isoform Michaelis constants (K11P=0.5 and K12P is varied). The y-axis shows the percentage of times a circadian period can be produced over 300 replicates with isoform dynamic regulation. The range of fluctuations that can be compensated (robustness) increases with increasing difference of isoform Michaelis constants, up to the point at which saturation occurs.
DISCUSSION
Enzyme isoforms are commonly found in organisms of every taxa but their evolutionary significance has not been thoroughly assessed. Here we used a model of the Drosophila circadian rhythm to investigate if known enzyme isoforms can increase the robustness of the system. We found that the presence of isoforms can increase the robustness only if dynamic regulation of isoform proportion is possible. Regulation at this level seems to be biologically plausible. First, feedback regulation of alternative splicing has been observed in Drosophila (Kumar and Lopez 2005). Second, external factors such as temperature can influence alternative splicing, causing a change in the ratio of alternative initiation codons (Colot et al. 2005). In addition, active regulation of CKIε levels in the Drosophila gene CLOCK pathway has been shown to occur (Kim and Edery 2006), providing evidence for dynamic regulation of a kinase in the circadian system.
We have shown that enzyme affinity per se, as well as the presence of isoforms without dynamic regulation, have no effect on robustness. Isoforms offer an increased dimension in the parameter space to match the target enzyme catalytic activity for a given set of parameter values. The isoform ratio may provide a flexible and parsimonious mechanism for adaptation to environmental fluctuations. The effectiveness of dynamic isoform regulation lies in its ability to change the pace of the reactions. If perturbations affect the rate at which some reactions occur, say there is average increase, then by increasing the proportion of the high affinity isoform, the pace of the entire cycle can be readjusted to stay within the circadian period.
One typical perturbation to the circadian oscillator is the presence of light, which enhances degradation of phosphorylated timeless protein (Myers et al. 1996). Although this occurs in the timeless protein pathway, which is excluded from our model, the possibility of dynamic isoform regulation is still a potential source of compensation for this environmental perturbation.
Alternative splicing, or mutations of duplicate gene copies, can generate enzyme isoforms that differ in substrate affinity, and there can be benefits to small or large differences. Producing two isoforms that are similar in affinity would be beneficial if flux perturbations are common because more enzymes result in more potential activity (Hurst and Randerson 2000, Bagheri and Wagner 2004). Similar isoforms, however, may not respond as well to other kinds of perturbations. Isoforms that differ greatly in their kinetic rates provide the potential for a more plastic system. Splicing events or mutations that result in the production of an enzyme that is very different from its original counterpart may foster selection for genetic variation. In this context, mutations of large effect that result in a viable phenotype (i.e. a working enzyme) will be better than small effect mutations. As the affinity difference among isoforms increases so does the flexibility in compensating larger changes in the values of other system parameters.
In this system, with multiple variables and parameters, the same period can be produced by different sets of parameter combinations. More generally in multidimensional systems it is often possible to produce the same phenotype from different underlying genotypes. Natural selection should then favor genotypes that are more canalized (Rice 1998). We have shown, however, that independently of the choice of parameter values (i.e. the genotypes), regulation of antagonistically expressed isoforms can always increase the robustness. If a system is already canalized, its degree of canalization could be further increased by regulatory elements that act on the relative expression of isoforms.
It has been shown, for instance, that antagonistic regulation of functionally redundant genes is not such a rare occurrence (for examples see Kafri et al. 2006). Antagonistic regulation is achieved by acting on the expression levels of paralog genes that alter their transcription levels to back each other up (Kafri et al. 2005). Our results suggest that such regulation can also occur by keeping expression levels constant while changing relative expression of isoforms. Gene dosage effects have also been proposed to play an important role in maintaining duplicate genes (Brown et al. 1998, Seoighe and Wolfe 1999, Kondrashov and Kondrashov 2006).
The existence of isoforms can thus provide the substrate for the emergence of a novel function. Isoform dynamic control drives the systems towards a larger adaptive peak and creates selection for their maintenance. This could represent another route towards subfunctionalization. In the traditional view of subfunctionalization the duplicate copies would be retained because degenerative mutations make it impossible for either copy to cover the role of the ancestral locus. This need not be the case. Degenerative mutations on only one copy, for instance, would produce a less effective enzyme. The copy carrying degenerative mutations could then be retained if in combination with the original copy it allows for increased robustness. The original copy would then accumulate mutations so that both copies become subfunctional. The difference between the two routes lies in that dynamically regulated isoforms would promote subfunctionalization through positive selection rather than by neutral population genetic processes.
Improving system robustness means that more of the underlying rates can vary without producing a phenotypic effect. This is true if variations are on a short time scale, such as fluctuations, or fixed such as different genetic backgrounds. The presence of dynamically regulated isoforms, therefore, could support more cryptic genetic variation at the population level. The robustness can improve by alternative means as well. Any of the rates, for instance, can be regulated to match the system’s current needs, provided the molecular machinery to do so is present. Robustness, for instance, can be increased by adjusting the CKII transcription rate, ViP. Transcription regulation, however, has its costs, such as energy and time. Even if we assume that energy expenditure is not the rate-limiting step, regulating cell processes only by means of transcription regulation takes time. After receiving a transcription signal, for instance, there can be a delay of anywhere from minutes to hours depending on cell turn over rates and gene size before the functional protein appears (Perez-Ortin et al. 2007). This delay includes the time needed for gene transcription and the time for post-transcriptional regulation, such as splicing processes. Bypassing transcription may thus be a faster route to achieve the same result. Alternative splicing is a candidate mechanism for such regulation. Splicing is used for both qualitative and quantitative regulation of gene expression (see Blencowe 2007 for review). In addition, splicing has been shown to be a fast mechanism for regulating gene expression in response to environmental stress (Pleiss et al. 2007). Regulating the kind of isozyme that results from alternative splicing does not require any change in gene transcription level, but only changes in post-transcriptional regulation, and may thus be an efficient way to control the rate at which a substrate is processed. Such regulation has no impact on the rate at which molecular machinery works and may thus be a potent and inexpensive way to increase system robustness.
The concept of dynamic isoform regulation as a means for increasing robustness could be tested in an empirical setting. It has long been known, for instance, that Drosophila circadian rhythm is robust with respect to temperature variation (Pittendrigh 1954). The nature of this compensation, however, is still debated (Roenneberg and Merrow 2003). Drosophila populations could be exposed to different temperature treatments and the corresponding protein isoform levels measured to see if the relative expression levels are different. This could indicate a dynamic regulation of isoforms to compensate for temperature differences.
Gene duplication and alternative splicing are recurrent events in the evolutionary history of most living organisms. Such events can provide the ingredients to modify, shape and improve biochemical networks as they are among the most basic levels of where selection operates. As isozymes first appear they may be neutral to selection, but this neutrality may be temporary. Any mechanism able to produce the raw material for evolution without compromising the current fitness of an organism may function as a highway to an adaptive peak.
Supplementary Material
Table 1.
Parameter values used to generate baseline parameter set with circadian oscillation of 24 hrs
| VSP | Accumulation rate of Mp in the cytosol | 1 nM h−1 |
| VMP | Maximum enzymatic degradation rate of Mp | 0.7 nM h−1 |
| kSP | Maximum rate of P0 synthesis | 0.9 h−1 |
| VDP | Maximum enzymatic degradation rate of P2 | 2 nM h−1 |
| k1 | Maximum transport rate of P2 in the nucleus | 0.155 h−1 |
| k2 | Maximum transport rate of PN in the cytosol | 0.2 h−1 |
| kIP | Threshold constant for transcription repression | 1 nM |
| n | Degree of cooperativity for Hill equation | 2 |
| k11P, k12P, | Michaelis constants for kinases | 2 nM |
| kP | Michaelis constants for phosphatases | 2 nM |
| V1P, V3P | Maximum rates for kinases | 8 nM h−1 |
| V2P, V4P | Maximum rates for phosphatases | 1 nM h−1 |
| kDN | Decay rate of PN | 0.01 h−1 |
| α | Proportion of isoforms | 0 |
ACKNOWLDGEMENTS
We thank Thomas F. Hansen, Alice Winn and the two anonymous reviewers for their insightful 3 comments on the manuscript. This work was partially supported by National Institute of Health 4 Grant DA-19356 to R.B. and M.E.F.
LITERATURE CITED
- Arredouani A, Guiot Y, Jonas JC, Liu LH, Nenquin M, Pertusa JA, Rahier J, Rolland JF, Shull GE, Stevens M, Wuytack F, Henquin JC, Gilon P. SERCA3 ablation does not impair insulin secretion but suggests distinct roles of different sarcoendoplasmic reticulum Ca2+ pumps for Ca2+ homeostasis in pancreatic β-cells. Diabetes. 2002;51:3245–3253. doi: 10.2337/diabetes.51.11.3245. [DOI] [PubMed] [Google Scholar]
- Bagheri HC, Wagner GP. Evolution of dominance in metabolic pathways. Genetics. 2004;168:1713–1735. doi: 10.1534/genetics.104.028696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blencowe BJ. Alternative splicing: new insights from global analyses. Cell. 2006;126:37–47. doi: 10.1016/j.cell.2006.06.023. [DOI] [PubMed] [Google Scholar]
- Brown CJ, Todd KM, Rosenzweig RF. Multiple duplications of yeast hexose transport genes in response to selection in a glucose-limited environment. Mol. Biol. Evol. 1998;15:931–942. doi: 10.1093/oxfordjournals.molbev.a026009. [DOI] [PubMed] [Google Scholar]
- Clark AG. Invasion and maintenance of a gene duplication. Proc. Natl. Acad. Sci. USA. 1994;91:2950–2954. doi: 10.1073/pnas.91.8.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colot HV, Loros JJ, Dunlap JC. Temperature-modulated alternative splicing and promoter use in the circadian clock gene frequency. Mol. Biol. Cell. 2005;16:5563–5571. doi: 10.1091/mbc.E05-08-0756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermentrout GB. Simulating, Analyzing, and Animating Dynamical Systems: A Guide to XPPAUT for Researchers and Students. Philadelphia, PA.: SIAM; 2002. [Google Scholar]
- Filhol O, Cochet C, Wedegaertner GN, Gill GN, Chambaz EM. Coexpression of both α and β subunits is required for assembly of regulated Casein Kinase II. Biochemistry. 1991;30:11133–11140. doi: 10.1021/bi00110a016. [DOI] [PubMed] [Google Scholar]
- Force A, Lynch M, Pickett FB, Amores A, Yan Y, Postlethwait J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forger DB, Peskin CS. A detailed predictive model of the mammalian circadian clock. Proc. Natl. Acad. Sci. U.S.A. 2003;100:14806–14811. doi: 10.1073/pnas.2036281100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie JH. The Causes of Molecular Evolution. New York: Oxford; 1991. [Google Scholar]
- Goldbeter A. A Model for Circadian Oscillations in the Drosophila Period Protein (PER) Proc. R. Soc. Lond. B. 1995;261:319–324. doi: 10.1098/rspb.1995.0153. [DOI] [PubMed] [Google Scholar]
- Gonze D, Halloy J, Goldbeter A. Robustness of circadian rhythms with respect to molecular noise. Proc. Natl. Acad. Sci. USA. 2002;99:673–678. doi: 10.1073/pnas.022628299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X. Evolution of duplicate genes versus genetic robustness against null mutations. Trends in Genetics. 2003;19:354–356. doi: 10.1016/S0168-9525(03)00139-2. [DOI] [PubMed] [Google Scholar]
- Gu Z, Steinmetz LM, Gu X, Scharfet C, Davis RW, Li W-H. Role of duplicate genes in genetic robustness against null mutations. Nature. 2003;421:63–66. doi: 10.1038/nature01198. [DOI] [PubMed] [Google Scholar]
- Guerra B, Issinger O-G. Protein kinase CK2 and its role in cellular proliferation, development and pathology. Electrophoresis. 1999;20:391–408. doi: 10.1002/(SICI)1522-2683(19990201)20:2<391::AID-ELPS391>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Guerra B, Boldyreff B, Sarno S, Cesaro L, Issinger O-G, Pinna LA. CK2: A Protein Kinase in Need of Control. Pharmacol. Ther. 1999;82:303–313. doi: 10.1016/s0163-7258(98)00064-3. [DOI] [PubMed] [Google Scholar]
- Hunter RL, Merkert CL. Histochemical demonstration of enzymes separated by zone electrophoresis in starch gels. Science. 1957;125:1294–1295. doi: 10.1126/science.125.3261.1294-a. [DOI] [PubMed] [Google Scholar]
- Hurst LD, Randerson JP. Dosage, deletions and dominance: simple models of the evolution of gene expression. J. Theor. Biol. 2000;205:641–647. doi: 10.1006/jtbi.2000.2095. [DOI] [PubMed] [Google Scholar]
- Jauch E, Wecklein H, Stark F, Jauch M, Raabe T. The Drosophila melanogaster DmCK2 β transcription unit encodes for functionally non-redundant protein isoforms. Gene. 2006;Vol. 374:142–152. doi: 10.1016/j.gene.2006.01.026. [DOI] [PubMed] [Google Scholar]
- Kafri R, Bar-Even A, Pilpel Y. Transcription control reprogramming in genetic backup circuits. Nat. Genet. 2005;37:295–299. doi: 10.1038/ng1523. [DOI] [PubMed] [Google Scholar]
- Kafri R, Levy M, Pilpel Y. The regulatory utilization of genetic redundancy through responsive backup circuits. Proc. Natl. Acad. Sci. 2006;103:11653–11658. doi: 10.1073/pnas.0604883103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim EY, Edery I. Balance between DBT/CKI0ε kinase and protein phosphatase activities regulate phosphorylation and stability of Drosophila CLOCK protein. Proc. Natl. Acad. Sci. USA. 2006;103:6178–6183. doi: 10.1073/pnas.0511215103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. The Neutral Theory of Molecular Evolution. Cambridge: Cambridge University Press; 1983. [Google Scholar]
- Kondrashov FA, Kondrashov AS. Role of selection in fixation of gene duplications. J. Theor. Biol. 2006;239:141–151. doi: 10.1016/j.jtbi.2005.08.033. [DOI] [PubMed] [Google Scholar]
- Kumar S, Mohan A, Sharma VK. Circadian dysfunction reduces lifespan in Drosophila melanogaster. Chronobiol. Int. 2005;22(4):641–653. doi: 10.1080/07420520500179423. [DOI] [PubMed] [Google Scholar]
- Kumar S, Lopez AJ. Negative feedback regulation among SR splicing factors encoded by Rbp1 and Rbp1-like in Drosophila. EMBO J. 2005;24:2646–2655. doi: 10.1038/sj.emboj.7600723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leloup JC, Goldbeter A. A model for circadian rhythms in Drosophila incorporating the formation of a complex between the PER and TIM proteins. J. Biol. Rhythms. 1998;13:70–87. doi: 10.1177/074873098128999934. [DOI] [PubMed] [Google Scholar]
- Leloup JC, Gonze D, Goldbeter A. Limit cycle models for circadian rhythms based on transcriptional regulation in Drosophila and Neurospora. J. Biol. Rhythms. 1999;14:433–448. doi: 10.1177/074873099129000948. [DOI] [PubMed] [Google Scholar]
- Leloup JC, Goldbeter A. Toward a detailed computational model for the mammalian circadian clock. Proc. Natl. Acad. Sci. U.S.A. 2003;100:7051–7056. doi: 10.1073/pnas.1132112100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JM, Kilman VL, Keegan K, Paddock B, Emery-Le M, Rosbash M, Allada R. A role for casein kinase 2α in the Drosophila circadian clock. Nature. 2002;420:816–820. doi: 10.1038/nature01235. [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery JS. The origins of genome complexity. Science. 2003;302:1401–1404. doi: 10.1126/science.1089370. [DOI] [PubMed] [Google Scholar]
- Myers MP, Wager-Smith K, Rothenfluh-Hilfiker A, Young MW. Light-induced degradation of TIMELESS and entrainment of the Drosophila circadian clock. Science. 1996;271:1736–1740. doi: 10.1126/science.271.5256.1736. [DOI] [PubMed] [Google Scholar]
- Ohno S. Evolution by gene duplication. New York, NY: Springer – Verlag; 1970. [Google Scholar]
- Panda S, Hogenesch JB, Kay SA. Circadian rhythms from flies to human. Nature. 2002;417:329–335. doi: 10.1038/417329a. [DOI] [PubMed] [Google Scholar]
- Perez-Ortin JE, Alepuz PM, Moreno J. Genomics and gene transcription kinetics in yeast. Trends in Genetics. 2007;23:250–257. doi: 10.1016/j.tig.2007.03.006. [DOI] [PubMed] [Google Scholar]
- Pittendrigh CS. On temperature independence in the clock-system controlling emergence time in Drosophila. Proc. Natl. Acad. Sci. 1954;40:1018–1029. doi: 10.1073/pnas.40.10.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleiss JA, Whitworth GB, Bergkessel M, Guthrie C. Rapid, Transcript-Specific Changes in Splicing in Response to Environmental Stress. Molecular cell. 2007;27:928–937. doi: 10.1016/j.molcel.2007.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proulx SR, Phillips PC. The opportunity for canalization and the evolution of genetic networks. Am. Nat. 2005;165:147–162. doi: 10.1086/426873. [DOI] [PubMed] [Google Scholar]
- Roenneberg T, Merrow M. The network of time: understanding the molecular circadian system. Curr. Biol. 2003;13:R198–R207. doi: 10.1016/s0960-9822(03)00124-6. [DOI] [PubMed] [Google Scholar]
- Rice SH. The evolution of canalization and the breaking of von Baer’s laws: modeling the evolution of developmental epistasis. Evolution. 1998;52(3):647–656. doi: 10.1111/j.1558-5646.1998.tb03690.x. [DOI] [PubMed] [Google Scholar]
- Seoighe C, Wolfe KH. Yeast genome evolution in the post-genome era. Curr. Opin. Microbiol. 1999;2:548–554. doi: 10.1016/s1369-5274(99)00015-6. [DOI] [PubMed] [Google Scholar]
- Smolen P, Baxter DA, Byrne JH. Modeling circadian oscillations with interlocking positive and negative feedback loops. J. Neurosci. 2001;21:6644–6656. doi: 10.1523/JNEUROSCI.21-17-06644.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda HR, Hagiwara M, Kitano H. Robust oscillations within the interlocked feedback model of Drosophila circadian rhythm. J. Theor. Biol. 2001;210:401–406. doi: 10.1006/jtbi.2000.2226. [DOI] [PubMed] [Google Scholar]
- Vilar JMG, Kueh HY, Barkai N, Leibler S. Mechanisms of noise-resistance in genetic oscillators. Proc. Natl. Acad. Sci. USA. 2002;99:5988–5992. doi: 10.1073/pnas.092133899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner GP, Booth G, Bagheri-Chaichian H. A population genetic theory of canalization. Evolution. 1997;51(2):329–347. doi: 10.1111/j.1558-5646.1997.tb02420.x. [DOI] [PubMed] [Google Scholar]
- Wagner A. Robustness against mutations in genetic networks of yeast. Nature Genetics. 2000;24:355–361. doi: 10.1038/74174. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.