Abstract
The presence of Fe(II) α-ketoglutarate hydroxylases in rat and human pancreatic islets and INS-1 832/13 cells was demonstrated with RT-PCR (PHD1, 2 and 3, lysyl hydroxylases 1, 2 and 3 and phytanoyl-CoA hydroxylase were seen) and/or immunoblotting (High levels of proline hydroxylase P4Hα1, PHD2 and PHD4 and low levels of PHD2 and PHD3 in human islets and high levels of PHD2 in rat islets and INS-1 cells were seen). Prolyl hydroxylase enzyme activity in INS-1 832/13 cells was purified with polyproline affinity chromatography. Inhibitors of α-ketoglutarate hydroxylases lowered glucose-induced and leucine-plus-glutamine-induced insulin release in rat pancreatic islets, suggesting that there may be acute unknown effects of α-ketoglutarate hydroxylases in insulin secretion. It is possible that an increase in mitochondrially-generated α-ketoglutarate derived from insulin secretagogue carbon and translocated to the cytosol may be part of the signal for insulin secretion.
Keywords: Inhibitors of α-ketoglutarate hydroxylases, human and rat pancreatic islets, INS-1 832/13 cells, mitochondrially generated α-ketoglutarate
Introduction
Fuel secretagogues stimulate insulin secretion by their metabolism in beta cell mitochondria. Not only does this produce ATP which powers cellular processes, but also mitochondria synthesize citric acid cycle intermediates (anaplerosis) which are exported from mitochondria to the cytosol, where they are converted to metabolites that have signaling and supporting roles in insulin exocytosis (1). Roles have been proposed for several citric acid cycle intermediates, including malate and citrate which may participate in shuttles of equivalents of reduced and/or oxidized pyridine nucleotides across the inner mitochondrial membrane (2–8), or the export of short chain acyl groups for synthesis of short chain acyl-CoAs in the cytosol (9–13) or regulate cellular metabolic oscillations (14). α-Ketoglutarate formed in the mitochondria can be transported across the inner mitochondrial membrane to the cytosol in exchange for malate (1). However, except for the participation of α-ketoglutarate in the malate aspartate shuttle (2), no specific extramitochondrial role for α-ketoglutarate in insulin secretion has been previously mentioned. In the current study, we investigated whether the α-ketoglutarate hydroxylase family of enzymes might have a role in insulin secretion. Fe(II) α-ketoglutarate dependent hydroxylases catalyze a diverse array of reactions in non-islet tissues (15). Primary substrates include prolyl, lysyl and aspartyl residues in proteins, as well as lipids. Oxidative decomposition of α-ketoglutarate forms CO2 plus succinate and leads to the generation of an oxoiron radical or other activated oxygen species that hydroxylate the primary substrate (15). The current study shows that inhibitors of α-ketoglutarate hydroxylases markedly decreased insulin release from pancreatic islets possibly indicating that α-ketoglutarate translocated from mitochondria is necessary for insulin secretion by serving as a substrate for these cytosolic hydroxylases. RT-PCR experiments indicated that transcripts for prolyl and lysyl hydroxylases and phytanoyl-CoA hydroxylases are present in human and rat pancreatic islets and INS-1 cells. In addition, we detected α-ketoglutarate hydroxylase activity with endogenous protein substrates in INS-1 cell cytosol. Prolyl hydroxylase enzyme activity was identified in INS-1 cells by purifying the enzyme activity with polyproline affinity chromatography.
The rapid inhibition of insulin release by inhibitors of α-ketoglutarate hydroxylases indicates that these hydroxylases have an acute influence on insulin secretion. Well-known actions of α-ketoglutarate hydroxylases are in slower processes, including collagen formation and transcription factor activation (15). Thus, the results may indicate that there are new and, as yet, unidentified substrates for (a) α-ketoglutarate hydroxylase(s) in the pancreatic beta cell.
EXPERIMENTAL PROCEDURES
Materials
(Pro-pro-gly)10 was from Peptides International Inc., Louisville, Kentucky. (Ile-lys-gly)3 was synthesized by the University of Wisconsin Biotechnology Center. All other chemicals were from Sigma Chemical Co. in the highest purity available. Human pancreatic islets were from the Islet Isolation Core at Washington University School of Medicine, St. Louis. INS-1 832/13 cells were from Chris Newgard (16).
General methods
Pancreatic islet isolation, insulin release studies, glucose oxidation studies and subcellular fractionation of islets and INS-1 cells were done as previously described (3, 17, 18, 19).
Reverse transcription – PCR analysis
Total RNA from pancreatic islets and INS-1 cells was isolated with the Qiagen RNeasy kit. RNA (1–2 μg) was reverse transcribed with the Ambion RETROscript kit in a volume of 20 μl. Reverse transcription reaction mixture (1 μl) was then added to 10 μl of PCR reaction mixture and amplification was performed with 1 unit of Sigma REDtaq genomic DNA polymerase and 200 μM dNTP with sets of forward and reverse primers designed from DNA sequences identified from the following GenBank accession numbers. For the rat hydroxylases: prolyl hydroxylases 1, 2 and 3, AY229997, AY228140 and NM_019371; lysl hydroxylases 1, 2, and 3, NM_053827, AJ430861 and NM_178101; phytanoyl-CoA hydroxylase, NM_053674 and mouse aspartyl hydroxylase, NM_133723. For the human hydroxylases: prolyl hydroxylases 1, 2, 3 and 4, NM_053046.2, NM_022051, NM_022073.2 and NM_177938; lysyl hydroxylases 1, 2 and 3, NM_000302.2, NM_182943.1, NM_001084.3; phytanoyl-CoA hydroxylase, IVM_006214.2, and aspartyl hydroxylases NM_004318, NM_032468.1 and NM_032466.1. Primer sequences are available upon request.
SDS-Polyacrylamide gel electrophoresis and immunoblot analyses
Total cellular protein or cytosol was boiled in Laemmli’s sample buffer and added in amounts equivalent to 50 pancreatic islets, or more islets as designated in individual legends, to 1.5 mm thick 10% SDS-polyacrylamide gels and electrophoresed in a Mini-Protean II gel apparatus (BioRad). Samples were transferred to nitrocellulose membranes in a Mini Trans-Blot apparatus (BioRad), probed with antibodies and ECL reagents and exposed to Kodak BioMax MR film for 1 or 5 min. Antibodies were used at a 1:500 dilution and were raised against proteins with the following GenBank protein ID numbers: Prolyl hydroxylase P4Hα1 (ICN Biomedicals, Inc.) (NP_000908 (human) and NP_742059 (rat); HIF prolyl hydroxylases (Novus Biologicals, Inc.) (HIF prolyl hydroxylase 1 (PHD1, Egln2) (NP_444274 (human) and AA046039 (rat); HIF PHD 2 (Egln1) (NP_071334 (human) and AA034711 (rat)); HIF PHD 3 (Egln3) (NP_071356 (human) and NP_062244 (rat)), and HIF PHD 4 (NP_80887 (human)).
Affinity chromatography of prolyl hydroxylase
Polyproline was coupled to agarose as previously described (20). Poly-L-proline (Mr 30,000) (36 mg) (Sigma Chemical Co. catalog number p3886) was dissolved in 2 ml of 0.15 M NaCl, 0.1 M NaHCO3 buffer, pH 9.3, and added to a suspension of 2.5 ml of CNBr-activated Sepharose 4B (250 mg dry weight) (Sigma). The mixture was gently rotated for 20 h at 4° and then the gel was washed with 10 volumes of 0.1 M NaCl, 0.1 M glycine, 10 μM dithiothreitol and 10 mM Tris-HCl buffer, pH 7.8, for two hours. The gel was kept in the same buffer for 20 h before it was washed with column buffer and used for affinity chromatography.
About 0.5 ml of packed 832/13 INS-1 cells that were cultivated as monolayers in INS-1 medium (21) (RPMI 1640 tissue culture medium (the glucose concentration in this medium is 11.1 mM) supplemented with 10% fetal bovine serum, 1 mM pyruvate, 50 μM β-mercaptoethanol and 10 mM Hepes buffer (12, 13)) and penicillin (100 units/ml) and streptomycin (100 μg/ml) were washed three times in PBS and homogenized in 1.2 ml of 0.1 M NaCl, 0.1 M glycine and 0.01 M Tris-HCl buffer, pH 7.8 (column buffer), containing 0.2% Triton-X-100. The homogenate was centrifuged at 16,000 x g × 10 min and the supernatant fraction was saved. The resulting pellet was rehomogenized in 0.6 ml of the same buffer solution, centrifuged and the supernatant fractions combined. The supernatant fraction volume (2 ml) was increased to 3.4 ml with buffer and applied to a (0.14 cm2 × 3.5 cm) (bed volume 0.5 ml) poly-L-proline Sepharose-4B column at a flow rate of 0.1 ml/minute. The column flow-through was collected and applied to the column a second time. The second column flow-through was collected and saved. The column was then washed with 2 ml of buffer and 1 ml of 5 mg/ml poly-L-proline (Mr 8,900) was applied to the column at a flow rate of 0.05 ml/min to elute the prolyl hydroxylase. Column fractions were concentrated 3–4 fold by centrifuging at 14,000 × g for 40–60 min in a Microcon YM-10 Centrifugal Filter Device (10,000 molecular weight cutoff) (Millipore Corp.).
Enzyme assays
α-Ketoglutarate hydroxylase activity was estimated in a final volume of 50 μl reaction mixture containing 18–25 μl of enzyme sample and 200 μM Tris(2-carboxyethyl) phosphine hydrochloride, 100 μM FeS04, 2 mM ascorbic acid, 0.2 mg/ml catalase, 2 mg/ml bovine serum albumin, 50 μM [1-14C] α-ketoglutarate (specific radioactivity 55 mCi/mmol), with or without an added prolyl-containing peptide substrate (100 μg/ml) (or a lysyl containing peptide as a control) in 100 mM Tris-chloride buffer, pH 7.8, at 37° (22). The reaction was carried out in a microfuge test tube inside a rubber-stoppered 20 ml scintillation vial and started by the addition of enzyme sample and stopped after 40 min by adding 100 μl of 5% trichloracetic acid. Tissue solubilizer (500 μl, Solvable, Perkin Elmer Corp.) was added to the scintillation vial outside the microfuge test tube and the CO2 evolved was collected for 3 h. After the microfuge test tube was removed, 14CO2 trapped in the solubilizer was estimated by liquid scintillation spectrometry as previously described (18, 19). Activities of α-ketoglutarate dehydrogenase, glutamate dehydrogenase and NAD- and NADP-dependent isocitrate dehydrogenase were estimated by standard assays as previously described (2, 13, 23, 24, 25).
RESULTS
Insulin release
Table 1 shows that inhibitors of α-ketoglutarate hydroxylases (26) were potent inhibitors of glucose- and leucine-plus-glutamine-induced insulin release from rat pancreatic islets. Ethyl 3,4 dihydroxybenzoate was the most potent inhibitor followed by 2-methyl α-ketoglutarate and 3-methyl α-ketoglutarate.
Table 1. Inhibition of insulin release from rat pancreatic islets by inhibitors of α-ketoglutarate hydroxylases.
Islets were incubated in the presence of the secretagogues for 1 h. Inhibitors were added 10 min before the secretagogues. The concentration of glucose was 16.7 mM and the concentrations of leucine and glutamine were 10 mM. Results are the mean ± SE (n = 6 to 12 replicate incubations for each condition)
| Secretagogue | Inhibitor | Insulin Release(μU Insulin/5 islets/ 60 min.) |
|---|---|---|
| None | None | 20 ± 2 |
| Glucose | None | 841 ± 63 |
| Glucose | 1 mM Ethyl-3,4-dihydroxybenzoate | 48 ± 5a |
| Glucose | 10 mM 2-Methyl-α-ketoglutarate | 54 ± 7a |
| Glucose | 5 mM 2-Methyl-α-ketoglutarate | 291 ± 21a |
| Glucose | 10 mM 3-Methyl-α-ketoglutarate | 315 ± 29a |
| Leucine + glutamine | None | 1094 ± 105 |
| Leucine + glutamine | 1 mM Ethyl-3,4-dihydroxybenzoate | 72 ± 24a |
p < 0.001 vs same secretagogue condition without inhibitor
Hydroxylase expression
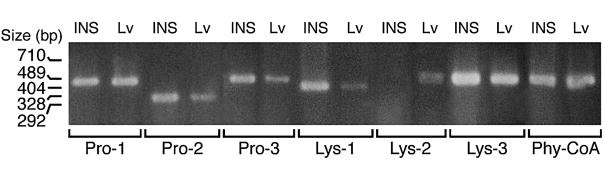
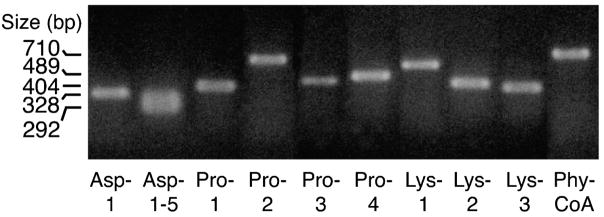
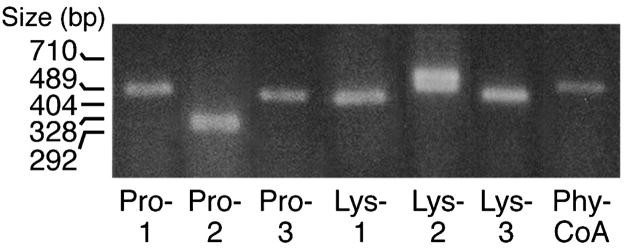
Figures 1–3 show that mRNA transcripts for four known prolyl hydroxylases and two of three lysyl hydroxylases, as well as phytanoyl-CoA hydroxylase, are present in human and rat pancreatic islets and INS-1 832/13 cells as judged from RT-PCR analysis. Aspartyl hydroxylases were seen in human pancreatic islets, but not in rat pancreatic islets or INS-1 832/13 cells. It is likely that the presence of aspartyl hydroxylase transcripts was due to non-islet cellular contamination of the human islet preparations because aspartyl hydroxylase protein was previously found not to be present in human islets, as judged from immunohistochemistry of human pancreas (J. Dinchuk, personal communication). The relative expressions of the hydroxylase genes in beta cells and liver as a positive control are summarized in Table 2.
Figure 1. RT-PCR products of various α-ketoglutarate hydroxylases in INS-1 cells and liver.
Liver (LV) is shown as a positive control. The band of the lysine hydroxylase-2 PCR product is a doublet.
Figure 3. RT-PCR products of various α-ketoglutarate hydroxylase genes in human pancreatic islets.
The primers used for aspartyl hydroxylase (Asp 1–5) amplify up to 5 different aspartyl hydroxylase transcripts
Table 2. Relative expression of hydroxylase genes in human and rat pancreatic islets, INS-1 cells and rat liver.
Expression is summarized from RT-PCR analyses shown in Figures 1–3 and other experiments. Relative levels are described on a scale of 0 to 3 with 3 representing the highest level and 0 indicating undetectable. ND, not done
| Relative Level of Expression by PCR(Scale of 0–3) | ||||
|---|---|---|---|---|
| Tissue | ||||
| RT-PCR Gene Product | Human Pancreatic Islets | Rat Pancreatic Islets | INS-1 | Rat Liver |
| Prolyl hydroxylase -1 | 2.5 | 2 | 3 | 3 |
| Prolyl hydroxylase -2 | 1 | 2.5 | 2 | 2 |
| Prolyl hydroxylase -3 | 1 | 2.5 | 2 | 1 |
| Prolyl hydroxylase -4 | 1.5 | ND | ND | ND |
| Lysyl hydroxylase -1 | 2 | 2 | 1.5 | 2 |
| Lysyl hydroxylase -2 | 1 | 0 | 0 | 0 |
| Lysyl hydroxylase -2 variant | ND | 2 | 1.5 | 2 |
| Lysyl hydroxylase -3 | 2.5 | 2.5 | 3 | 2.5 |
| Phytanoyl-CoA hydroxylase | 1 | 2.5 | 2.5 | 3 |
| Aspartyl hydroxylase -1 | 1 | ND | 0 | ND |
| Aspartyl hydroxylases 1–5 | 2.5 | ND | ND | ND |
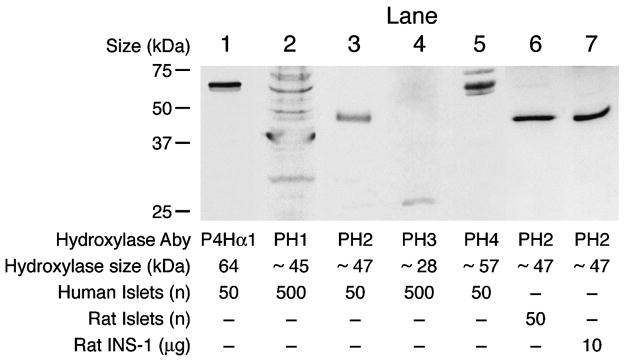
Immunoblot analysis showed that prolyl hydroxylase proteins P4 Hα1, PHD2 and PHD4 are present in human pancreatic islets and PHD2 is present in rat pancreatic islets and INS-1 832/13 cells. Prolyl hydroxylases PHD1 and PHD3 were present at very low levels in human islets and these two proteins plus PHD4 could not be detected in rat pancreatic islets (Figure 4).
Figure 4. Immunoblots of prolyl hydroxylases in human and rat pancreatic islets and INS-1 cells.
The figure shows lanes from immunoblots probed with antibodies against the various prolyl hydroxylases. Abbreviations: Prolyl hydroxylase (P4Hα1), PHD prolyl hydroxylase 1 (PH 1), PHD prolyl hydroxylase 2 (PH 2), PHD prolyl hydroxylase 3 (PH 3), PHD prolyl hydroxylase 4 (PH 4). The known sizes of the various hydroxylases are shown at the bottom of the figure. To show the absence or a low level of a hydroxylase, 500 islets instead of 50 islets were used.
α-Ketoglutarate hydroxylase activity with endogenous substrates
Table 3 shows that INS-1 cell cytosol has the ability to catalyze decarboxylation of α-ketoglutarate and that inhibitors of α-ketoglutarate hydroxylases lower the decarboxylase activity. This indicates that α-ketoglutarate hydroxylase enzyme activity as well as endogenous substrate(s) for these hydroxylases, are present in INS-1 cells. It is unlikely that the decarboxylase activity is due to other enzymes that use α-ketoglutarate, such as α-ketoglutarate dehydrogenase or glutamate dehydrogenase because these are mitochondrial enzymes and cytosol was used in these experiments. In addition, the inhibitors of the α-ketoglutarate hydroxylases did not inhibit glutamate dehydrogenase. It is also unlikely that activities of these enzymes would be detected in the enzyme reaction mixture because the endogenous levels of substrates and cofactors for these two enzymes would have been diluted > 1000 fold in the hydroxylase enzyme reaction mixture. Although α-ketoglutarate is a substrate for the reverse reaction of NADP-isocitrate dehydrogenase and this enzyme is present in the cytosol of islets and clonal beta cells, this enzyme was not inhibited by high levels of these inhibitors (see below). In addition, α-ketoglutarate is not decarboxylated in its conversion to isocitrate.
Table 3. Summary of effects of inhibitors of α-ketoglutarate hydroxylases on enzyme activity in the presence of endogenous substrates in INS-1 cell cytosol.
Results are typical of six separate experiments
| Incubation Condition | Relative enzyme activity(Expressed as a % of control = 100%) |
| Cytosol | 100% |
| No cytosol | 10–18% |
| Cytosol, heated | 5% |
| Cytosol + 60 mM ZnCl2 | 15–34% |
| Cytosol + 5 mM 2-methyl-α-ketoglutarate | 25% |
| Cytosol + 5 mM 3-methyl-α-ketoglutarate | 48% |
| Cytosol + 2.5 mM ethyl-3,4-dihydroxybenzoate | 49% |
Prolyl hydroxylase activity
To ascertain whether some of the α-ketoglutarate hydroxylase activity was due to prolyl hydroxylase, INS-1 cell cytosol was fractionated with ammonium sulfate and/or by affinity chromatography on a poly-L-proline column and then tested with inhibitors of α-ketoglutarate hydroxylases. The fraction precipitated by 30% to 60% ammonium sulfate contained prolyl hydroxylase activity, while the zero to 30% fraction and the 60% supernatant fraction contained little or no prolyl hydroxylase activity (Table 4). A supernatant fraction of INS-1 cells was also applied to a poly-L-proline Sepharose-4B column. The column was washed with buffer and poly-L-proline was added to the column to elute prolyl hydroxylases adhering to the column. Table 5 shows that there was prolyl hydroxylase enzyme activity in the fraction eluted with poly-L-proline (Mr 8,900) and that concentrating the eluate increased the enzyme activity in direct proportion to the extent of concentration of the eluate. This fraction contained no activity when (Ile-lys-gly)3 was the substrate (Data not shown). In addition, adding (pro-pro-gly)10 (which is a better substrate for prolyl hydroxylase than the longer chain proline polymer, which was present in the eluate because it was used for elution of the enzyme from the column) to the enzyme reaction mixture increased the prolyl hydroxylase activity of both the unconcentrated and concentrated fractions even more. In contrast, concentrating the flow-through fraction obtained while adding the cytosol sample to the column, or also the column wash fraction obtained while washing the column with buffer after adding the sample and before eluating with poly-proline, did not result in an increase in enzyme activity. Also, adding (pro-pro-gly)3 to either of the latter two unconcentrated or concentrated fractions did not increase enzyme activity. Enzyme activity was not present in column fractions collected subsequent to the eluate obtained in the presence of poly-L-proline. Adding ethyl-3,4-dihydroxybenzoate or 2-methyl-α-ketoglutarate to the concentrated polyproline eluate column fraction inhibited the enzyme activity (Table 5) to extents proportional to their effects on insulin release (Table 1).
Table 4. α-Ketoglutarate hydroxylase activity in various ammonium sulfate fractions of INS-1 cell cytosol.
INS-1 cell cytosol was fractionated with ammonium sulfate (AS) and dialyzed into 100 mM Tris-Cl buffer, pH 7.8, 20 % glycerol, 1 mM dithiothreitol and α-ketoglutarate hydroxylase activity was estimated in the presence and absence of (pro-gly-pro)10 (PGP) or EDTA which chelates iron and inhibits the enzyme. Results are the mean of ± SE of triplicate estimates on each fraction
| Ammonium Sulfate Fraction | α-Ketoglutarate Hydroxylase Activity(pmol [1-14C] α-ketoglutarate decarboxylated /mg protein |
|---|---|
| Buffer only | 0 |
| 0–30% | 0.70 ± 0.06 |
| 0–30% + PGP | 0.85 ± 0.06 |
| 30–60% | 4.9 ± 0.25 |
| 30–60% + PGP | 7.3 ± 0.06 |
| 60% Supernatant | 0.75 ± 0.09 |
| 60% Supernatant + PGP | 0.72 ± 0.09 |
| Heated 30–60% + PGP | 0.12 ± 0.01 |
| 30–60% + PGP + 5 mM EDTA | 0.84 ± 0.10 |
Table 5. Prolyl hydroxylase enzyme activity in various fractions eluted from INS-1 cell cytosol on a polyproline agarase column.
An INS-1 832/13 cell supernatant fraction was applied to a poly-L-proline Sepharose 4B. The column was washed with buffer solution and prolyl hydroxylase was eluted with poly-L-proline (Mr 8,900) as described under Experimental Procedures. The fractions were collected from the column; part of each fraction was concentrated and part of each fraction was assayed for hydroxylase activity in the absence or presence of (pro-gly-pro)10 (PGP). Note that prolyl hydroxylases are known to possess enzyme activity in the presence of long polyproline polymers as substrates, but less than with (pro-gly-pro)10 as a substrate. The hydroxylation of poly-L-proline, plus hydroxylation of any endogenous proteins, explains enzyme activity in the absence of PGP. ND indicates not done
| α-Ketoglutarate Hydroxylase Activity | |||
|---|---|---|---|
| Column Fraction | (pmol [1-14C] α-ketoglutarate decarboxylated/ 25 μl of fraction) | ||
| Fraction alone | Fraction plus PGP | Specific enzyme activity in presence of PGP(nmol [1-14C] α-ketoglutarate decarboxylated/mg protein) | |
| Flow through unconcentrated | 4.0 | 3.0 | 0.05 |
| Flow through concentrated | 3.5 | 3.4 | 0.01 |
| Wash unconcentrated | 4.4 | 4.2 | 0.13 |
| Wash concentrated | 2.7 | 1.1 | 0.01 |
| Polyproline eluate unconcentrated | 4.3 | 14.9 | 14.9 |
| Polyproline eluate concentrated | 18.3 | 38.6 | 12.9 |
| Polyproline eluate concentrated plus 1 mM ethyl-3, 4- dihydroxybenzoate | ND | 6.1 | 2.0 |
| Polyproline eluate concentrated plus 10 mM 2-methyl α-ketoglutarate | ND | 22.0 | 7.3 |
Little or no inhibition by hydroxylase inhibitors on various enzymes that utilize α-ketoglutarate
The inhibitors of α-ketoglutarate hydroxylases were tested for inhibition of other enzymes that use α-ketoglutarate. α-Ketoglutarate dehydrogenase was not inhibited by 2-dimethyl-α-ketoglutarate or 3-methyl-α-ketoglutarate at concentrations of up to 10 mM and when assayed at α-ketoglutarate concentrations of 0.5 mM or 5 mM. Only when 10 mM ethyl 3, 4-dihydroxybenzoic acid (which is 10 fold the highest concentration used in the insulin release study) was added to the enzyme reaction mixture before 5 mM of the substrate α-ketoglutarate, or 1 mM ethyl 3, 4-dihydroxybenzoic acid was added before 0.5 mM α-ketoglutarate, was inhibition seen and then it was only 30%. The three inhibitors did not inhibit pure glutamate dehydrogenase from bovine liver or pure NADP-isocitrate dehydrogenase from porcine heart or NAD-isocitrate dehydrogenase in an INS-1 cell mitochondrial supernatant fraction (data not shown).
Lack of effect of inhibitors of hydroxylases on glucose metabolism - 2
Methyl-α-ketoglutarate at 1 mM or 10 mM, which inhibited insulin release up to 67% (Table 1), did not inhibit glucose oxidation. Ethyl-3,4-dihydroxybenzoate (0.25 mM and 0.5 mM) did not inhibit glucose oxidation by pancreatic islets and 1 mM ethyl-3,4-dihydroxybenzoate inhibited glucose oxidation by about 37% (Table 6). Inhibition of insulin release by 1 mM ethyl-3,4-dihydroxybenzoate (94–97%, Table 1) was proportionately much larger than its inhibition of glucose metabolism.
Table 6. Little or slight inhibition of glucose oxidation in pancreatic islets by ethyl 3, 4-dihydroxybenzoic acid or 2-methylα-ketoglutarate.
Islets were incubated with 16.7 mM [U-14C]glucose (0.4 mCi/mmol) with or without either inhibitor for 90 minutes and glucose oxidation to CO2 was measured. Abbreviations: DKG (2-dimethyl-α-ketoglutarate) and EDB (ethyl-3,4-dihydroxybenzoate). Results are the mean ± SE with the number of replicate incubations in parentheses
| Condition | Glucose Oxidation (nmol CO2 formed/100 islets/90 minutes) |
|---|---|
| Glucose alone | 3.2 ± 0.2 (3) |
| Glucose + 1 mM DKG | 3.7 ± 0.3 (4) |
| Glucose + 10 mM DKG | 3.0 ± 0.1 (4) |
| Glucose + 0.25 mM EDB | 3.4 ± 0.2 (3) |
| Glucose + 0.5 mM EDB | 2.9 ± 0.4 (4) |
| Glucose + 1 mM EDB | 2.0 ± 0.3 (4) |
DISCUSSION
RT-PCR analyses of human and rat pancreatic islets and INS-1 832/13 cells indicated that all three tissues possess mRNA transcripts for prolyl hydroxylases PHD1, 2 and 3 and lysyl hydroxylases 1, 2 and 3 as well as phytanoyl-CoA hydroxylase. The expression of the lysyl-hydroxylase 2 was very low in human islets and undetectable in rat islets and INS-1 cells. RT-PCR is very sensitive and does not accurately estimate the rate of translation of a protein and its level in a tissue. Therefore, the RT-PCR analysis was corroborated by estimating the levels of proteins by immunoblotting. Antibodies for lysyl hydroxylases and phytanoyl-CoA hydroxylase were not available for use in immunoblotting. However, immunoblot analyses of human islets indicated that they possess high levels of proline hydroxylase P4Hα1, PHD2 and PHD4 and low levels of PHD1 and PHD3. Rat islets and INS-1 cells possessed only PHD2 as judged from immunoblot analyses. The presence of prolyl hydroxylase enzyme activity in INS-1 832/13 cells was demonstrated by purifying the enzyme with polyproline affinity chromatography indicating the enzyme is present in beta cells.
Inhibitors of α-ketoglutarate hydroxylases lowered glucose-induced and leucine-plus-glutamine-induced insulin release in rat pancreatic islets and inhibited hydroxylase enzyme activity with endogenous protein substrates in homogenates of INS-1 832/13 cells and also inhibited the affinity-purified INS-1 cell prolyl hydroxylase. Since various metabolic enzymes besides α-ketoglutarate hydroxylase utilize α-ketoglutarate as a substrate, the hydroxylase inhibitors were tested for effects on the other enzymes and on glucose oxidation by pancreatic islets. The inhibitors either did not inhibit the other enzymes and glucose oxidation or else inhibition of insulin release was proportionately much greater than inhibition of glucose metabolism and the metabolic enzymes. 2-Methyl-α-ketoglutarate and 3-methyl-α-ketoglutarate, which inhibited α-ketoglutarate hydroxylase activity by about 50% and insulin release 60–90%, did not inhibit α-ketoglutarate dehydrogenase, NADP- or NAD-dependent isocitrate dehydrogenase enzyme activity or glucose metabolism. Ethyl-3,4-dihydroxybenzoate inhibited prolyl hydroxylase enzyme activity with endogenous substrates by 50% and insulin release by > 90%, but inhibited α-ketoglutarate dehydrogenase by 30% and glucose metabolism by 37%. Although these data support the idea that there are novel actions of α-ketoglutarate hydroxylases in supporting insulin release in the beta cell, there are reports of inhibition by 3,4-dihydroxybenzoate (not the ethyl ester) of other enzymes, such as hepatic P450 enzymes. However, the inhibition with very high doses was only 20–30% (27). 3,4-Dihydroxybenzoate has also been reported to have a moderate effect on protein kinase C (28) and to have an apoptotic effect in gastric carcinoma cells (29).
Since the known actions of α-ketoglutarate hydroxylases are in collagen synthesis (P4Hα1) or on activation of gene transcription (HIF prolyl hydroxylases), which are non-acute mechanisms, but the inhibition of insulin release was immediate, the results suggest that there may be acute unknown effects of α-ketoglutarate hydroxylases in insulin secretion. Insulin secretagogues increase α-ketoglutarate in pancreatic islets and INS-1 cells (12). Thus it is possible that an increase in mitochondrially-generated α-ketoglutarate derived from insulin secretagogue carbon and translocated to the cytosol may be part of the signal for insulin secretion. We are attempting to identify targets of hydroxylase enzymes in insulin-producing cells. However, this is proving difficult because there is no assay for selecting peptides not previously known to undergo hydroxylation.
Figure 2. RT-PCR products of various α-ketoglutarate hydroxylases in rat pancreatic islets.
The band of the lysine hydroxylase-2 PCR product is a doublet.
Acknowledgments
This work was supported by NIH grant DK28348 and the Oscar C. Rennebohm Foundation.
The authors thank Scott Stoker and David Julian Buss for technical assistance and Barbara Olach for providing the human islets.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.MacDonald MJ, Fahien LA, Brown LJ, Hasan NM, Buss JD, Kendrick MA. Perspective: Emerging evidence for signaling roles of mitochondrial anaplerotic products in insulin secretion. Am J Physiol Endocrinol Metab. 2005;288:E1–E15. doi: 10.1152/ajpendo.00218.2004. [DOI] [PubMed] [Google Scholar]
- 2.MacDonald MJ. Evidence for the malate aspartate shuttle in pancreatic islets. Arch Biochem Biophys. 1982;213:643–649. doi: 10.1016/0003-9861(82)90594-x. [DOI] [PubMed] [Google Scholar]
- 3.MacDonald MJ. Feasibility of a mitochondrial pyruvate malate shuttle in pancreatic islets: further implication of cytosolic NADPH in insulin secretion. J Biol Chem. 1995;270:20051–20058. [PubMed] [Google Scholar]
- 4.Schuit F, De Vos A, Farfari S, Moens K, Pipeleers D, Brun T, Prenki M. Metabolic fate of glucose in purified islet cells. Glucose-regulated anaplerosis in beta cells. J Biol Chem. 1997;272:18572–18579. doi: 10.1074/jbc.272.30.18572. [DOI] [PubMed] [Google Scholar]
- 5.Farfari S, Schulz V, Corkey B, Prentki M. Glucose-regulated anaplerosis and cataplerosis in pancreatic beta-cells: possible implication of a pyruvate/citrate shuttle in insulin secretion. Diabetes. 2000;49:718–726. doi: 10.2337/diabetes.49.5.718. [DOI] [PubMed] [Google Scholar]
- 6.Flamez D, Berger V, Kruhoffer M, Orntoft T, Pipeleers D, Schuit FC. Critical role for cataplerosis via citrate in glucose-regulated insulin release. Diabetes. 2002;51:2018–24. doi: 10.2337/diabetes.51.7.2018. [DOI] [PubMed] [Google Scholar]
- 7.Lu D, Mulder H, Zhao P, Burgess SC, Jensen MV, Kamzolova S, Newgard CB, Sherry AD. 13C NMR isotopomer analysis reveals a connection between pyruvate cycling and glucose-stimulated insulin secretion (GSIS) Proc Natl Acad Sci. 2002;99:2708–2713. doi: 10.1073/pnas.052005699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cline GW, Lepine RL, Papas KK, Kibbey RG, Shulman GI. 13C NMR isotopomer analysis of anaplerotic pathways in INS-1 cells. J Biol Chem. 2004;279:44370–44375. doi: 10.1074/jbc.M311842200. [DOI] [PubMed] [Google Scholar]
- 9.Corkey BE, Glennon MC, Chen KS, Deeney JT, Matschinsky FM, Prentki M. A role for malonyl-CoA in glucose-stimulated insulin secretion from clonal pancreatic β-cells. J Biol Chem. 1989;264:21608–21612. [PubMed] [Google Scholar]
- 10.Liang L, Matschinsky FM. Content of CoA-esters in perifused rat islets stimulated by glucose and other fuels. Diabetes. 1991;40:327–333. doi: 10.2337/diab.40.3.327. [DOI] [PubMed] [Google Scholar]
- 11.Brun T, Roche E, Assimacopoulos-Jeannet F, Corkey BE, Kim KH, Prentki M. Evidence for an anaplerotic malonyl-CoA pathway in pancreatic beta-cell nutrient signaling. Diabetes. 1996;45:190–198. doi: 10.2337/diab.45.2.190. [DOI] [PubMed] [Google Scholar]
- 12.MacDonald MJ. Synergistic potent insulin release by combinations of weak secretagogues in pancreatic islets and INS-1 cells. J Biol Chem. 2007;282:6043–6052. doi: 10.1074/jbc.M606652200. [DOI] [PubMed] [Google Scholar]
- 13.MacDonald MJ, Smith AD, Hasan NM, Sabat G, Fahein LA. Feasibility of pathways for transfer of acyl groups from mitochondria to the cytosol to form short chain acyl-CoAs in the pancreatic beta cell. J Biol Chem. 2007;282:30596–30606. doi: 10.1074/jbc.M702732200. [DOI] [PubMed] [Google Scholar]
- 14.MacDonald MJ, Fahien LA, Buss JD, Hasan NM, Fallon MJ, Kendrick MA. Citrate oscillates in liver and pancreatic beta cell mitochondria and in INS-1 insulinoma cells. J Biol Chem. 2003;278:51894–51900. doi: 10.1074/jbc.M309038200. [DOI] [PubMed] [Google Scholar]
- 15.Hausinger RP. FeII/alpha-ketoglutarate-dependent hydroxylases and related enzymes. Crit Rev Biochem Mol Biol. 2004;39:21–68. doi: 10.1080/10409230490440541. [DOI] [PubMed] [Google Scholar]
- 16.Hohmeier HE, Mulder H, Chen G, Henkel-Rieger R, Prentki M, Newgard CB. Isolation of INS-1-derived cell lines with robust ATP- sensitive K+ channel-dependent and –independent glucose-stimulated insulin secretion. Diabetes. 2000;49:424–30. doi: 10.2337/diabetes.49.3.424. [DOI] [PubMed] [Google Scholar]
- 17.MacDonald MJ, Fahien LA, McKenzie DI, Moran SM. Novel effects of insulin secretagogues on capacitation of insulin release and survival of cultured pancreatic islets. Am J Physiol. 1990;259:E548–554. doi: 10.1152/ajpendo.1990.259.4.E548. [DOI] [PubMed] [Google Scholar]
- 18.MacDonald MJ. Metabolism of the insulin secretagogue methyl succinate by pancreatic islets. Arch Biochem Biophys. 1993;300:201–205. doi: 10.1006/abbi.1993.1028. [DOI] [PubMed] [Google Scholar]
- 19.MacDonald MJ. Estimates of glycolysis, pyruvate (de)carboxylation, pentose phosphate pathway and methyl succinate metabolism in incapacitated pancreatic islets. Arch Biochem Biophys. 1993;305:205–214. doi: 10.1006/abbi.1993.1413. [DOI] [PubMed] [Google Scholar]
- 20.Tuderman L, Kuutti E-R, Kivirikko K. An Affinity-Column Procedure Using Poly(L-proline) for the Purification of the Prolyl Hydroxylase. Eur J Biochem. 1975;52:9–16. doi: 10.1111/j.1432-1033.1975.tb03967.x. [DOI] [PubMed] [Google Scholar]
- 21.Asfari M, Janjic D, Meda P, Li G, Halban PA, Wollheim CB. Establishment of 2-mercaptoethanol-dependent differentiated insulin-secreting cell lines. Endocrinology. 1992;130:167–178. doi: 10.1210/endo.130.1.1370150. [DOI] [PubMed] [Google Scholar]
- 22.Gronke RS, Welsch DJ, VanDusen WJ, Garsky VM, Sardana MK, Stern AM, Friedman PA. Partial purification and characterization of bovine liver aspartyl beta-hydroxylase. J Biol Chem. 1990;265:8558–8565. [PubMed] [Google Scholar]
- 23.MacDonald MJ, Marshall LK. Survey of normal appearing mouse strain which lacks malic enzyme and NAD+-linked glycerol phosphate dehydrogenase: normal pancreatic beta cell function, but abnormal metabolite pattern in skeletal muscle. Mol. Cell. Biochem. 2001;220:117–125. doi: 10.1023/a:1010821821921. [DOI] [PubMed] [Google Scholar]
- 24.MacDonald MJ. Flavin content of intracellular compartments of pancreatic islets compared with acinar tissue and liver. Endocrinology. 1981;108:1899–1902. doi: 10.1210/endo-108-5-1899. [DOI] [PubMed] [Google Scholar]
- 25.MacDonald MJ, McKenzie DI, Kaysen JH, Walker TM, Moran SM, Fahein LA, Towle HC. Glucose regulates leucine-induced insulin release and the expression of the branched chain ketoacid dehydrogenase E1 alpha subunit gene in pancreatic islets. J Biol Chem. 1991;266:1335–1340. [PubMed] [Google Scholar]
- 26.Sasaki T, Majamaa K, Uitto J. Reduction of collagen production in keloid fibroblast cultures by ethyl-3,4-dihydroxybenzoate. Inhibition of prolyl hydroxylase activity as a mechanism of action. J Biol Chem. 1987;262:9397–9403. [PubMed] [Google Scholar]
- 27.Baer-Dubowska W, Szaefer H, Krajka-Kuzniak V. Inhibition of murine hepatic cytochrome P450 activities by natural and synthetic phenolic compounds. Xenobiotica. 1998;28:735–743. doi: 10.1080/004982598239155. [DOI] [PubMed] [Google Scholar]
- 28.Szaefer H, Kaczmarek J, Rybczynska M, Baer-Dubowska W. The effect of plant phenols on the expression and activity of phorbol ester-induced PKC in mouse epidermis. Toxicology. 2007;230:1–10. doi: 10.1016/j.tox.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 29.Lin HH, Chen JH, Huang CC, Wang CJ. Apoptotic effect of 3,4-dihydroxybenzoic acid on human gastric carcinoma cells involving JNK/p38 MAPK signaling activation. Int J Cancer. 2007;120:2306–2316. doi: 10.1002/ijc.22571. [DOI] [PubMed] [Google Scholar]