Summary
How genes are regulated to produce the correct assortment of proteins for every cell type is a fundamental question in biology. For many genes, regulation begins at the DNA level using promoter sequences to control transcription. Regulation can also occur after transcription using sequences in the 3’ untranslated region (UTR) of the mRNA to affect mRNA stability and/or translation. Using a transgenic assay, we have compared the contribution of promoters and 3’UTRs to gene regulation in the C. elegans germline. We find that for most genes tested, 3’ UTRs were sufficient for regulation. With the exception of promoters activated during spermatogenesis, promoters were permissive for expression in all germ cell types (from undifferentiated progenitors to mature oocytes and sperm) and showed no cell-type specificity. In progenitors, 3’ UTRs inhibit the production of meiotic and oocyte proteins by post-transcriptional mechanisms involving two classes of RNA-binding proteins (PUF and KH domain proteins). Our findings indicate that many genes rely primarily on 3’UTRs, not promoters, for regulation during germline development.
RESULTS AND DISCUSSION
3’ UTRS are sufficient to specify most expression patterns in the germline
In the C. elegans adult hermaphrodite gonad, germ cells are arranged in two U-shaped tubes in order of differentiation, with progenitors at one end (distal), cells that have initiated meiotic prophase in the middle (“pachytene region”), and gametes at the other end (proximal) [1] (Fig. 1). Proteins expressed during specific stages of germline development localize to specific regions of the gonad. Both transcriptional and post-transcriptional mechanisms have been implicated in the generation of these patterns [2–5], but direct in vivo evidence for the involvement of specific cis-regulatory sequences has been obtained for only 4 genes so far (among genes with known protein distributions). In each case, 3’ UTR sequences were found to be necessary (gld-1, cep-1) or sufficient (glp-1, pal-1) for regulation [6–9].
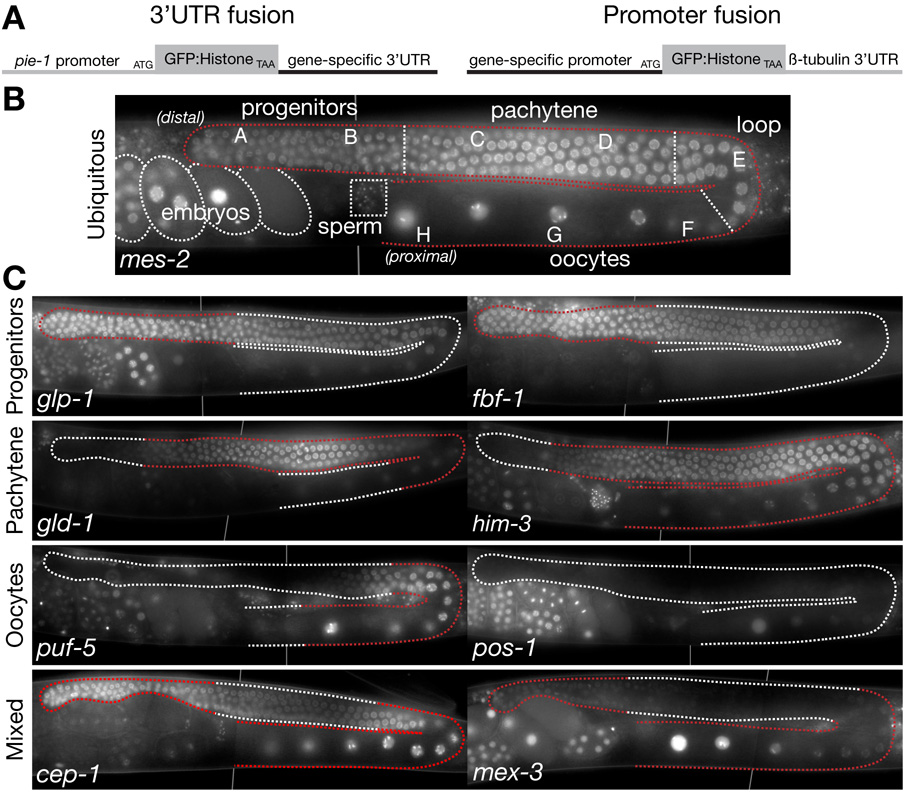
Figure 1. 3’UTR fusions in adult hermaphrodites.
A. Schematics showing the design of 3’UTR and promoter fusions. Both contain green fluorescent protein (GFP) fused to histone H2B fusion as the protein reporter. H2B associates dynamically with chromatin [31]. Chromatin localization facilitates detection and turnover is fast enough to visualize changes in gene expression even in non-dividing cells ([10]; this study). The pie-1 promoter and β-tubulin 3’ UTR (tbb-2) are permissive for expression in all germ cells.
B. Photomicrograph of a single adult gonadal arm (outlined with stippled line) expressing the mes-2 3’UTR fusion (ubiquitous expression). By this stage, hermaphrodites are producing oocytes. Sperm (produced during the L4 larval stage) are stored in the spermatheca. Gonad is divided into different zones used for systematic scoring of expression patterns as in Fig. 2: A is first half of mitotic zone, containing germline stem cells, B is second half of mitotic zone containing cells that have initiated meiotic S phase, C and D are pachytene regions, E is loop region where cells transition from pachytene to diakinesis and initiate oocyte formation, F–H is region of oocyte growth, H denotes last oocyte [1].
C. Photomicrographs as in B showing examples of different 3’ UTR fusions, arranged based on the primary site of protein expression as reported in the literature. For each gene, red stippling outlines the region of the gonad where high levels of endogenous protein have been detected (Sup. Table 1, for references). See Fig. 3 and Sup. Fig.1 for photomicrographs of all other 3’ UTR fusions examined in this study and Fig. 2 for a schematic summary. In this and all subsequent figures, vertical lines are used to indicate where two photomicrographs of the same animal have been merged.
To investigate the extent to which 3’ UTRs contribute to gene regulation in the germline, we selected 30 genes representative of every germline expression patterns reported in the literature, including 25 of the 35 genes known to encode proteins expressed in specific germ cell types (Methods). We cloned the 3’UTR of each gene downstream of green fluorescent protein (GFP) fused to Histone H2B, and used the pie-1 promoter to drive all 30 fusions (Fig. 1A). The pie-1 gene encodes a maternal protein expressed specifically in oocytes and embryos, but the pie-1 promoter permits expression in all germ cell types (reference [10] and see below). Each fusion was randomly integrated in the genome by microparticle bombardment [11]. 30/30 fusions yielded lines that expressed GFP:H2B in the germline, and for 24/30 fusions, the GFP:H2B expression pattern matched that reported for the corresponding endogenous protein (compare nuclear GFP fluorescence to red stippling in Fig. 1, Fig. 3A and Sup. Fig. 1). For example, the glp-1, gld-1 and puf-5 3’ UTR fusions (Fig. 1C) express GFP:H2B preferentially in the distal, pachytene, and proximal regions, respectively, as has been reported for the GLP-1, GLD-1 and PUF-5 proteins (Sup Table S1). Although all 24 patterns generally matched endogenous patterns, in many cases pattern “boundaries” (areas where GFP:H2B levels rise/decline) appeared expanded compared to those reported in the literature (pattern “expansion” is denoted by hatching in Fig. 2). For example, p53 (CEP-1) levels are high in progenitors, low in pachytene, and high in oocytes. The cep-1 3’UTR fusion approximates this pattern, except that GFP:H2B steadily declines through the first half of the pachytene region and reaches minimal levels only in the second half (Fig. 1C). Replacement of H2B with the native ORF led to a more accurate pattern for cep-1 and several other fusions (Sup. Fig. 2), suggesting that the expanded boundaries are a GFP:H2B-specific artifact, perhaps due to slow turnover of the fusion. In total, among the 30 3’ UTRs surveyed, we could identify 17 distinct expression patterns (Fig. 2), spanning all stages of germ cell development (with one exception, see below). We conclude that 3’UTRs are sufficient to specify a wide range of expression patterns in the germline.
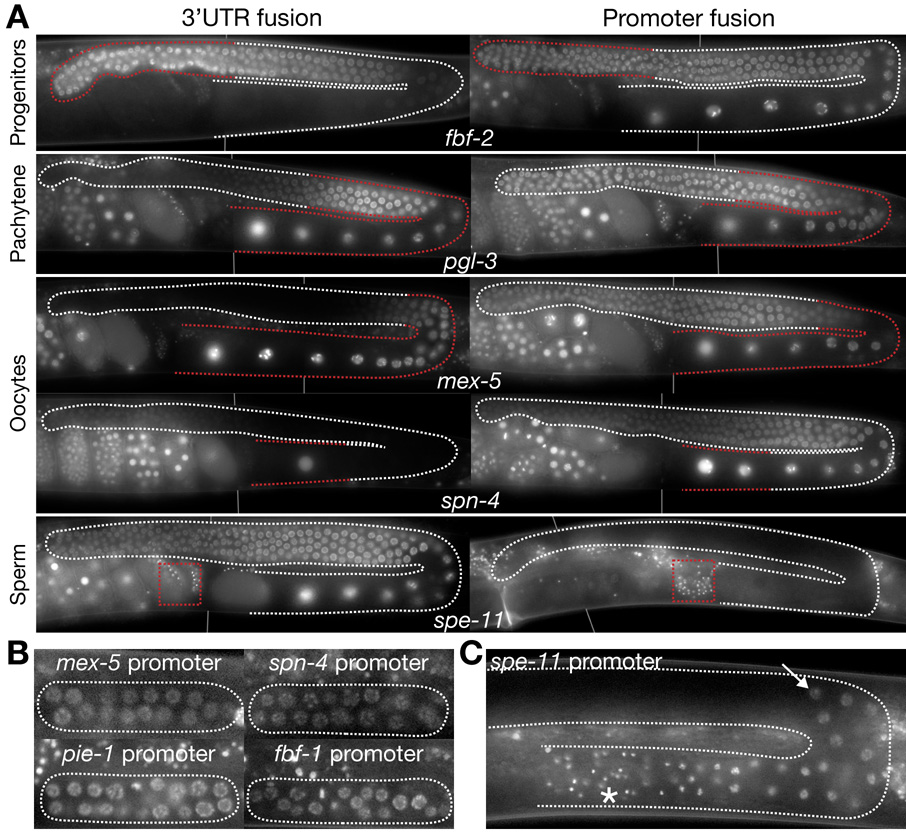
Fig. 3. Comparison of 3’ UTR and promoter fusions.
A. Photomicrographs of adult gonads expressing 3’UTR or promoter fusions for the genes indicated. Additional promoter fusions are shown in Sup. Fig. 3. For fbf-2, pgl-3, mex-5 and spn-4, the 3’UTR fusions show the expected cell-type specificity (highest GFP fluorescence matches red stippling), but the promoter fusions do not. In contrast for spe-11, the promoter fusion shows the expected cell type specificity (sperm, see box), but the 3’UTR fusion does not.
B and C. Photomicrographs of larval gonadal arms expressing the indicated promoter fusions. All fusions contain the tubulin 3’UTR.
B.The mex-5, spn-4, pie-1 and fbf-1 promoter fusions are active in all germline progenitors in L2 gonads.
C. In contrast, the spe-11 promoter fusion does not become active until the L4 stage when spermatogenesis begins. GFP:H2B is first detected in late pachytene cells (arrow) and is maintained through mature sperm (asterisk).
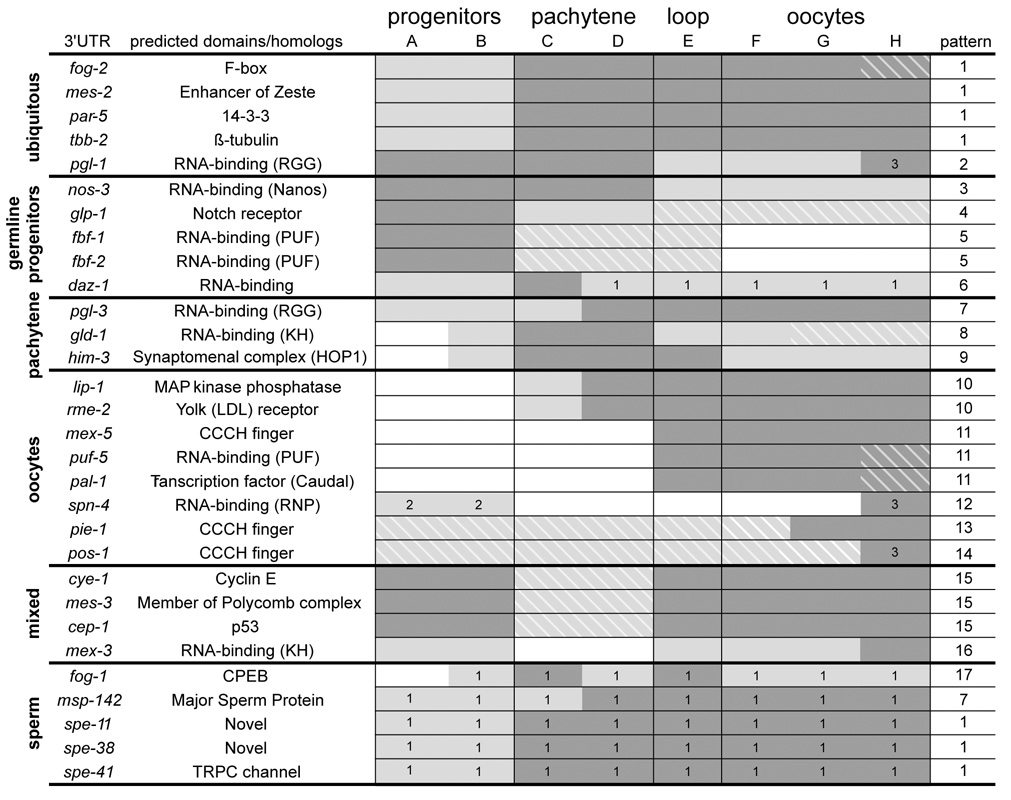
Figure 2. Summary of all 3’UTR fusions examined in this study.
Genes are arranged based on the primary site of protein expression as reported in the literature. GFP:H2B expression was scored for each gonadal region (A–H) as defined in Fig. 1B. Dark grey indicates strongest domain(s) of GFP:H2B expression, light grey: weaker domain(s) of GFP expression, and white: no GFP expression. (Evaluation of strong and weak expression was made by comparing GFP intensities directly within individual gonads; at least 20 hermaphrodites were examined for each fusion). Hatching denotes regions where GFP:H2B levels are low/declining and where the endogenous protein has not been reported to be expressed. This type of discrepancy is likely due to perdurance of the H2B fusion or to undetected low levels of endogenous protein. For fog-2, daz-1, gld-1, pal-1, pie-1, and cep-1, replacing H2B with the native ORF gave a pattern more closely matching the endogenous pattern (Sup. Fig. 2).
1. GFP:H2B levels are steady in this region, but endogenous protein has not been reported here. This type of discrepancy suggests that the 3’UTR is insufficient for proper regulation. 5 of 6 3’UTRs in this category come from genes coding for proteins expressed specifically during spermatogenesis. Note that the fog-1 3’UTR is sufficient to confer proper regulation (repression) in germline progenitors, but is insufficient to inhibit expression in germlines undergoing oogenesis. The only other gene in this category is daz-1. DAZ-1 protein has been reported to be expressed in the distal germline (Sup. Table 1), but the daz-1 3’UTR fusion is also expressed in oocytes. The DAZ-1ORF: 3’UTR fusion shows reduced expression in oocytes (Sup. Fig. 2).
2. Whether SPN-4 is expressed or not in this region has not been reported.
3. GFP:H2B expression was most prominent in oocytes undergoing maturation.
3’ UTRS are not sufficient to specify sperm-specific expression
The only cell-type specificity not reproduced in our survey was sperm-specific expression. Five genes in our survey encode proteins expressed only during spermatogenesis. FOG-1 is expressed in pachytene cells during spermatogenesis and MSP-142, SPE-11, SPE-38 and SPE-41 are expressed in differentiating spermatocytes and mature sperm (Sup. Table 1 for references). The msp-142, spe-11, spe-38 and spe-41 3’UTR fusions caused GFP:H2B to be expressed in all germ cells, including progenitors, oocytes and sperm (Fig. 2, Fig. 3A and Sup. Fig. 1). Replacement of H2B with the native ORF (for msp-142, spe-11, and spe-38) did not correct the ubiquitous expression (Sup. Fig. 2). The fog-1 3’UTR fusion showed the expected pachytene enrichment, but remained ON in pachytene cells during oogenesis (Sup. Fig. 1). Downregulation of fog-1 in oogenic germlines depends on the transcription factor tra-1 [12], and four consensus tra-1 binding sites are present in the fog-1 promoter [2]. We conclude that 3’ UTRs are not sufficient to direct sperm-specific expression and that promoters may be required.
Promoters are permissive for expression in all germ cells, with the exception of promoters activated during spermatogenesis
To examine the role of promoters directly, we constructed promoter fusions for 12 genes encoding proteins with restricted patterns, including fbf-1, fbf-2, daz-1 (distal germline), gld-1, lip-1, him-3 (meiotic prophase), pgl-3, mex-5, pie-1, spn-4 (oocytes) and spe-11 and msp-56 (sperm) (Fig. 3A and Sup. Fig. 3). The promoters were fused to GFP:H2B and a 3’UTR permissive for expression in all germ cells (3’ UTR of tbb-2, coding for beta-tubulin) (Fig. 1A). Of the 12 promoters tested, only spe-11 and msp-56 drove GFP:H2B in the predicted pattern (Fig. 3A and Sup. Fig. 3), confirming that sperm-specific expression depends on promoter sequences (also see [5]). Surprisingly, all other promoter fusions caused GFP:H2B to be expressed in all germ cells. GFP:H2B levels often appeared highest in the pachytene region (most obvious for spn-4, Fig. 3A). The pachytene region is the most transcriptionally active domain in the adult gonad as determined by rates of tritiated uridine incorporation [13]. Furthermore, functional genomic studies have suggested that genes coding for oocyte proteins, such as spn-4, are transcriptionally upregulated during pachytene in adult hermaphrodites [14]. Our results are consistent with those findings, but suggest that these genes are also transcribed (at least at basal levels) in other cells, including germline progenitors.
To determine whether promoters coding for oocyte proteins are active in progenitors before the onset of gametogenesis, we examined the expression of 5 promoter fusions in larval gonads containing only germline progenitors (L2 stage). We examined promoter fusions from three genes encoding oocyte proteins (spn-4, mex-5, pie-1), one gene encoding a sperm protein (spe-11), and one gene encoding a protein enriched in progenitors (fbf-1) (Fig. 3B). We detected GFP:H2B expression from the spn-4, mex-5, pie-1 and fbf-1 fusions, but not from the spe-11 fusion. In the case of pie-1, we detected GFP:H2B in gonads containing as few as 8–12 germ cells (but not earlier), indicating that this promoter becomes active after the primordial germ cells begin proliferation in the first larval stage (data not shown). In contrast, expression of the spe-11 fusion was first detected in the L4 stage, in cells at the end of the pachytene region and continuing through mature sperm (Fig. 3C). In adult hermaphrodites (which have switched to oogenesis), spe-11:GFP:H2B was no longer detected in pachytene cells but was still present in mature sperm stored in the spermatheca (Fig. 3A). In males, which produce sperm continuously, spe-11:GFP:H2B remain expressed in late pachytene cells and later stages of spermatogenesis through adulthood (Sup. Fig. 3). In contrast, pie-1:GFP:H2B was detected in all cells in both hermaphrodite and male germlines (Sup. Fig. 3). Therefore, with the exception of sperm-specific genes, the promoters in our survey are not sufficient to specify cell-type specific expression when paired with a permissive 3’UTR such as tbb-2. We conclude that 3’UTRs are the primary source of gene regulation in the germline, with the exception of genes expressed during spermatogenesis. We note, however, that since all promoters were tested in transgenes, we cannot exclude that some promoters do in fact exhibit specificity when in their endogenous chromatin context.
3’ UTRs function post-transcriptionally and depend on two classes of RNA binding proteins to inhibit expression in progenitors
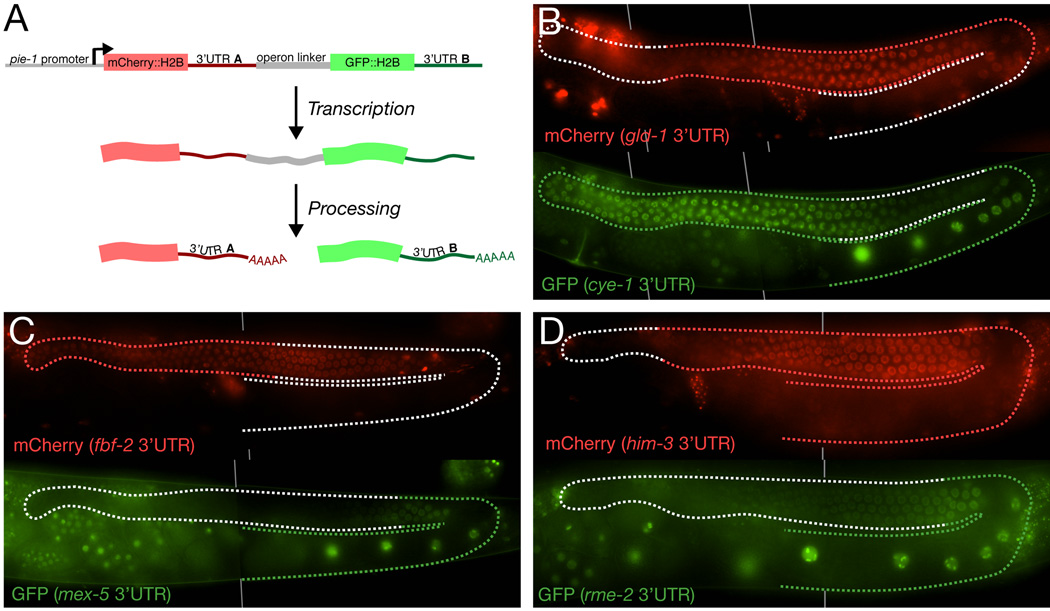
3’UTR regulatory elements are expected to function after transcription at the mRNA level, but in principle could also function at the DNA level as distal enhancers to modulate transcription. To distinguish between these possibilities, we tested six 3’UTRs in the context of an “operon” (Fig. 4A). C. elegans operons are transcribed into polycistronic primary transcripts, which are processed by transplicing into monocistronic mRNAs, each with its own 3’UTR and poly(A) tail. We constructed three operon transgenes containing an mCherry fusion linked to a first 3’ UTR (gld-1, fbf-2 or him-3), and a GFP fusion linked to a second 3’UTR (cye-1, mex-5 or rme-2). If the 3’UTRs regulate transcription, we would expect the two fusions to be expressed in the same pattern. Instead we found that each reporter behaved independently and was expressed in the pattern expected for its 3’ UTR (Fig. 4B–D). We conclude that the 3’ UTRs tested function post-transcriptionally to regulate protein expression.
Figure 4. 3’UTRs function post-transcriptionally.
A. Schematic showing the design of operon transgenes. The operon linker is the intercistronic region from the gpd-2/gpd-3 operon. This 138bp region promotes transplicing and has no promoter activity [32].
B–D. Photomicrographs of adult hermaphrodite gonads expressing the indicated operon transgenes. Top and bottom figures show expression of mCherry and GFP, respectively, in the same animal. Red and green stippling indicated regions of endogenous expression.
To investigate the mechanisms by which 3’UTRs confer cell-type specificity, we first examined the ability of a “somatic” 3’ UTR to support germline expression. The unc-54 gene encodes myosin and is only expressed in muscle. The unc-54 3’ UTR is the standard 3’ UTR used in transgenes in C. elegans and is permissive for expression in all somatic cells [15]. We found that a 3’UTR fusion containing the pie-1 promoter and the unc-54 3’UTR fusion was expressed in all germ cells (Sup. Fig. 1). This result suggests that the “default” state in germ cells is ON, and that 3’ UTRs confer specificity primarily by blocking expression in specific cell types (although activating mechanisms may also be needed in the context of germline 3’ UTRs [16]).
Several post-transcriptional repressors have been implicated in gene regulation in the germline [17]. The best characterized are two classes of RNA-binding proteins that function in the distal gonad: the PUF domain proteins FBF-1 and FBF-2, and the KH domain proteins MEX-3 and GLD-1. FBF-1 and FBF-2 are nearly identical and are required redundantly for the post-transcriptional repression of several genes in the mitotic zone, including fog-1, gld-1 and lip-1 in our survey [18]. MEX-3 and GLD-1 are two unrelated KH domain proteins expressed in complementary patterns in the mitotic (MEX-3) and pachytene (GLD-1) regions. MEX-3 and GLD-1 have been implicated in the negative regulation of several mRNAs, including rme-2, pal-1, mex-5 and puf-5 in our survey [7, 19, 20]. Consistent with regulation by these proteins, the fog-1, gld-1, lip-1, rme-2, pal-1, mex-5 and puf-5 3’ UTR fusions all showed low or no GFP:H2B expression in the distal region of the gonad (Fig. 2). To test directly whether these fusions are regulated by FBF-1/2 or MEX-3/GLD-1, we co-depleted FBF-1and FBF-2 or MEX-3 and GLD-1 by RNAi. Complete loss of these proteins cause dramatic cell transformations in the adult stage, including loss of all progenitors (fbf-1; fbf-2 double mutant [21]) and transdifferentiation of cells in the pachytene region to somatic fates (gld-1;mex-3 mutant [22]). To minimize secondary effects due to cell fate transformations, we used RNA-mediated interference to partially inactivate each gene in young adults (Methods). We found that depletion of FBF-1/2 strongly de-repressed the gld-1 and fog-1 3’ UTR fusions, but had no effect on the other fusions (Fig. 5A, Sup. Table 2). Depletion of MEX-3 and GLD-1 caused all fusions to expand their expression distally. Most fusions maintained a small domain of no or low expression at the distal-most tip, except for rme-2 which was de-repressed throughout the distal region (Fig. 5A, and Sup. Table 2). To confirm that expression in the distal-most region is due to derepression in germline progenitors and not secondary to cell fate transformations, we repeated the RNAi depletions in L2 larvae, which contain only proliferating germ cells (Fig. 5B and Sup. Table 3). These experiments confirmed that fog-1 and gld-1 require FBF-1/2 for repression in progenitors, whereas rme-2 requires GLD-1/MEX-3. None of the RNAi treatments affected the expression of the spe-11 promoter fusion in adults or in L2 larvae (Sup. Tables 2 and 3). We conclude that 3’ UTR-dependent regulation in progenitors involves inhibitory mechanisms mediated by FBF-1/2 and GLD-1/MEX-3.
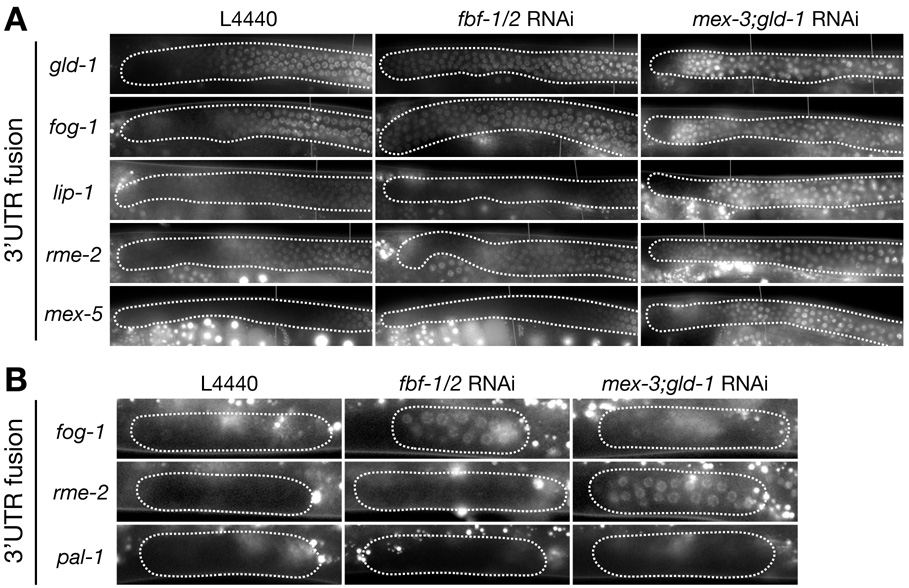
Figure 5. Regulation of 3’UTR fusions in the distal gonad by FBF-1/2, MEX-3 and GLD-1.
A. Fluorescence photomicrographs of distal arms (A, B and C regions in Fig. 1b) from adult hermaphrodites expressing the indicated 3’ UTR fusions, and fed either control bacteria (L4440) or bacteria expressing double-stranded RNA against fbf-1 and fbf-2 or mex-3 and gld-1. See Sup. Table 2 for numbers and additional 3’ UTR fusions examined. Note that lip-1 has low, but non-zero expression throughout the distal region.
B. Photomicrographs of L2 gonadal arms expressing the indicated fusions, and fed either control bacteria (L4440) or bacteria expressing double-stranded RNA against fbf-1 and fbf-2 or mex-3 and gld-1. The RNAi conditions used in these experiments allow the small pool of L2 germline progenitors (~30 cells per arm) to proliferate as in wild-type (~1000 cells per arm by adult stage), indicating that the cells retain mitotic potential and have not yet been transformed. See Sup. Table 3 for numbers and additional 3’UTR fusions examined. Bright foci outside of the gonad are autofluorescent gut granules.
FBF-1 and FBF-2 bind in vitro to a sequence motif (termed FBF-binding element or FBE) present in gld-1, fog-1 and lip-1 [18, 23]. Consistent with the RNAi results, we found that mutating the FBEs derepressed the gld-1 3’ UTR fusion, but did not affect the lip-1 fusion (Sup. Fig. 4). Unlike gld-1, which is repressed in a narrow distal domain contained within the mitotic zone, lip-1 is repressed in a broader region, perhaps reflecting more complex regulation (also see [24]). We note that FBEs were common in 3’ UTRs with no expression in the distal region (7/8), but were also present in a significant number of 3’ UTRs in our survey (17/30), including 3’ UTRs with strong expression in the distal region (5/8) (Sup. Table 4). Our results provide new evidence that, as suggested previously [21] [19], FBF-1/2 directly regulate the meiosis-promoting protein GLD-1, and that FBF-1/2 and MEX-3/GLD-1 regulate the expression of several genes via 3’ UTRs, although whether this regulation is direct remains to be tested.
Conclusion
In summary, our findings demonstrate that post-transcriptional mechanisms acting on 3’ UTRs are the main source of regulation throughout germ cell development, with the exception of spermatogenesis. FBF-1, FBF-2, GLD-1 and MEX-3 mediate 3’UTR-dependent repression in germline progenitors and are themselves dependent on 3’UTR sequences for proper expression (Fig. 2), suggesting the existence of complex tiered networks of RNA-binding proteins and RNA target sites (see also [18]). Germ cells contain unique RNA-rich organelles (germ granules), which may represent a specialization for post-transcriptional regulation [25]. Whether similar RNA regulatory networks function in somatic lineages and how these interact with transcriptional networks [26] will be interesting to investigate. Changes in steady-state mRNA levels as detected by microarray analyses are often presumed to be due to transcriptional regulation but may also be due to post-transcriptional effects on mRNA stability [27]. Our survey demonstrates that 3’ UTRs are a rich source of regulatory diversity that can bypass the need for most promoter specificity. Biochemical and computational methods have identified many potential functional sites in 3’UTRs, including binding sites for RNA-binding proteins and microRNAs [28–30]. Many such sites are present in the 3’UTRs in our survey (Sup. Table 4), although none correlate perfectly with a specific expression pattern. Systematic analysis of 3’UTR fusions, as initiated here, will be necessary to test the function of these sequences in vivo and elucidate the regulatory logic of 3’UTRs.
Supplementary Material
Acknowledgments
We thank Jane Hubbard (NYU), Tim Schedl (Washington U.) and Judith Kimble (U. of Wisconsin) and the anonymous reviewers for comments on the manuscript and Chris Gallo for pCG150. This work was supported by NIH grant R01HD37047 to G. S., and NICHD training grant 2T32HD007276 to C. M. G. Seydoux is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Hubbard EJ, Greenstein D. Introduction to the germ line. WormBook. 2005:1–4. doi: 10.1895/wormbook.1.18.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ellis R, Schedl T. Sex determination in the germ line. WormBook. 2007:1–13. doi: 10.1895/wormbook.1.82.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kimble J, Crittenden SL. Germline proliferation and its control. WormBook. 2005:1–14. doi: 10.1895/wormbook.1.13.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reinke V. Germline genomics. WormBook. 2006:1–10. doi: 10.1895/wormbook.1.74.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shim YH. elt-1, a gene encoding a Caenorhabditis elegans GATA transcription factor, is highly expressed in the germ lines with msp genes as the potential targets. Mol. Cells. 1999;9:535–541. [PubMed] [Google Scholar]
- 6.Jones AR, Francis R, Schedl T. GLD-1, a cytoplasmic protein essential for oocyte differentiation, shows stage- and sex-specific expression during Caenorhabditis elegans germline development. Dev. Biol. 1996;180:165–183. doi: 10.1006/dbio.1996.0293. [DOI] [PubMed] [Google Scholar]
- 7.Mootz D, Ho DM, Hunter CP. The STAR/Maxi-KH domain protein GLD-1 mediates a developmental switch in the translational control of C. elegans PAL-1. Development. 2004;131:3263–3272. doi: 10.1242/dev.01196. [DOI] [PubMed] [Google Scholar]
- 8.Marin VA, Evans TC. Translational repression of a C. elegans Notch mRNA by the STAR/KH domain protein GLD-1. Development. 2003;130:2623–2632. doi: 10.1242/dev.00486. [DOI] [PubMed] [Google Scholar]
- 9.Schumacher B, Hanazawa M, Lee MH, Nayak S, Volkmann K, Hofmann ER, Hengartner M, Schedl T, Gartner A. Translational repression of C. elegans p53 by GLD-1 regulates DNA damage-induced apoptosis. Cell. 2005;120:357–368. doi: 10.1016/j.cell.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 10.D'Agostino I, Merritt C, Chen PL, Seydoux G, Subramaniam K. Translational repression restricts expression of the C. elegans Nanos homolog NOS-2 to the embryonic germline. Dev. Biol. 2006;292:244–252. doi: 10.1016/j.ydbio.2005.11.046. [DOI] [PubMed] [Google Scholar]
- 11.Praitis V, Casey E, Collar D, Austin J. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics. 2001;157:1217–1226. doi: 10.1093/genetics/157.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lamont LB, Kimble J. Developmental expression of FOG-1/CPEB protein and its control in the Caenorhabditis elegans hermaphrodite germ line. Dev. Dyn. 2007;236:871–879. doi: 10.1002/dvdy.21081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Starck J. Radioautographic study of RNA synthesis in Caenorhabditis elegans (Bergerac variety) oogenesis. Biologie Cellulaire. 1977;30:181–182. [Google Scholar]
- 14.Chi W, Reinke V. Promotion of oogenesis and embryogenesis in the C. elegans gonad by EFL-1/DPL-1 (E2F) does not require LIN-35 (pRB) Development. 2006;133:3147–3157. doi: 10.1242/dev.02490. [DOI] [PubMed] [Google Scholar]
- 15.Hunt-Newbury R, Viveiros R, Johnsen R, Mah A, Anastas D, Fang L, Halfnight E, Lee D, Lin J, Lorch A, et al. High-throughput in vivo analysis of gene expression in Caenorhabditis elegans. PLoS biology. 2007;5:e237. doi: 10.1371/journal.pbio.0050237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suh N, Jedamzik B, Eckmann CR, Wickens M, Kimble J. The GLD-2 poly(A) polymerase activates gld-1 mRNA in the Caenorhabditis elegans germ line. Proc Natl Acad Sci U S A. 2006;103:15108–15112. doi: 10.1073/pnas.0607050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee MH, Schedl T. RNA-binding proteins. WormBook. 2006:1–13. doi: 10.1895/wormbook.1.79.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kimble J, Crittenden SL. Controls of germline stem cells, entry into meiosis, and the sperm/oocyte decision in Caenorhabditis elegans. Annu Rev Cell Dev Biol. 2007;23:405–433. doi: 10.1146/annurev.cellbio.23.090506.123326. [DOI] [PubMed] [Google Scholar]
- 19.Lee MH, Schedl T. Identification of in vivo mRNA targets of GLD-1, a maxi-KH motif containing protein required for C. elegans germ cell development. Genes Dev. 2001;15:2408–2420. doi: 10.1101/gad.915901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ciosk R, DePalma M, Priess JR. ATX-2, the C. elegans ortholog of ataxin 2, functions in translational regulation in the germline. Development. 2004;131:4831–4841. doi: 10.1242/dev.01352. [DOI] [PubMed] [Google Scholar]
- 21.Crittenden SL, Bernstein DS, Bachorik JL, Thompson BE, Gallegos M, Petcherski AG, Moulder G, Barstead R, Wickens M, Kimble J. A conserved RNA-binding protein controls germline stem cells in Caenorhabditis elegans. Nature. 2002;417:660–663. doi: 10.1038/nature754. [DOI] [PubMed] [Google Scholar]
- 22.Ciosk R, DePalma M, Priess JR. Translational regulators maintain totipotency in the Caenorhabditis elegans germline. Science. 2006;311:851–853. doi: 10.1126/science.1122491. [DOI] [PubMed] [Google Scholar]
- 23.Bernstein D, Hook B, Hajarnavis A, Opperman L, Wickens M. Binding specificity and mRNA targets of a C. elegans PUF protein, FBF-1. RNA. 2005;11:447–458. doi: 10.1261/rna.7255805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee MH, Hook B, Lamont LB, Wickens M, Kimble J. LIP-1 phosphatase controls the extent of germline proliferation in Caenorhabditis elegans. EMBO J. 2006;25:88–96. doi: 10.1038/sj.emboj.7600901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seydoux G, Braun RE. Pathway to totipotency: lessons from germ cells. Cell. 2006;127:891–904. doi: 10.1016/j.cell.2006.11.016. [DOI] [PubMed] [Google Scholar]
- 26.Levine M, Davidson EH. Gene regulatory networks for development. Proc Natl Acad Sci U S A. 2005;102:4936–4942. doi: 10.1073/pnas.0408031102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Foat BC, Houshmandi SS, Olivas WM, Bussemaker HJ. Profiling condition-specific, genome-wide regulation of mRNA stability in yeast. Proc Natl Acad Sci U S A. 2005;102:17675–17680. doi: 10.1073/pnas.0503803102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chan CS, Elemento O, Tavazoie S. Revealing posttranscriptional regulatory elements through network-level conservation. PLoS computational biology. 2005;1:e69. doi: 10.1371/journal.pcbi.0010069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, Lander ES, Kellis M. Systematic discovery of regulatory motifs in human promoters and 3' UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Elemento O, Slonim N, Tavazoie S. A universal framework for regulatory element discovery across all genomes and data types. Mol Cell. 2007;28:337–350. doi: 10.1016/j.molcel.2007.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kimura H. Histone dynamics in living cells revealed by photobleaching. DNA repair. 2005;4:939–950. doi: 10.1016/j.dnarep.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 32.Huang T, Kuersten S, Deshpande AM, Spieth J, MacMorris M, Blumenthal T. Intercistronic region required for polycistronic pre-mRNA processing in Caenorhabditis elegans. Mol. Cell. Biol. 2001;21:1111–1120. doi: 10.1128/MCB.21.4.1111-1120.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.