Abstract
In its simplest form, a DNA G-quadruplex is a four-stranded DNA structure that is composed of stacked guanine tetrads. G-quadruplex-forming sequences have been identified in eukaryotic telomeres, as well as in non-telomeric genomic regions, such as gene promoters, recombination sites, and DNA tandem repeats. Of particular interest are the G-quadruplex structures that form in gene promoter regions, which have emerged as potential targets for anticancer drug development. Evidence for the formation of G-quadruplex structures in living cells continues to grow. In this review, we examine recent studies on intramolecular G-quadruplex structures that form in the promoter regions of some human genes in living cells and discuss the biological implications of these structures. The identification of G-quadruplex structures in promoter regions provides us with new insights into the fundamental aspects of G-quadruplex topology and DNA sequence–structure relationships. Progress in G-quadruplex structural studies and the validation of the biological role of these structures in cells will further encourage the development of small molecules that target these structures to specifically modulate gene transcription.
1. Introduction
1.1. Beyond B-form duplex DNA
Since Watson and Crick proposed the double helix model as the basis for DNA replication [1], extensive studies have been carried out to determine a more complete picture of the DNA structure in cells. It is believed that the great majority of DNA is present as a B-form duplex helix, which is built up of Watson-Crick base pairs between two complementary DNA strands in cells [1]. Nevertheless, there are exceptions to this uniformity, because duplex DNA may transiently form alternative DNA secondary structures within certain sequences, fueled by dynamic molecular events. A number of non-B-form DNA secondary structures have been identified, which include left-handed DNA (Z-DNA), triplexes (H-DNA and sticky DNA), cruciforms, slipped hairpins, G-quadruplexes (G-tetraplexes), and i-motifs (i-tetraplexes) [2–6]. The dynamic polymorphism of DNA conformation is determined by DNA sequence, topology due to DNA supercoiling, ions, DNA binding proteins, and other modifications on DNA [7]. Publication of the complete draft of the human genome sequence provided a useful map with which to explore the occurrence and frequency of unusual DNA secondary structures [8–10]. One surprising conclusion from these studies is that only 1–2% of the human genome (~20,000–25,000 genes) codes for proteins. This raises the question of the function of the remaining 98% of non-coding genomic DNA [11, 12]. Bioinformatic studies on the human genome indicate that guanine-rich (G-rich) and cytosine-rich (C-rich) regions of chromosomes, including rDNA, single-copy genes (promoter and coding region), recombination sites, and repetitive sequences (satellite and telomeric DNA sequences), have the potential to form G-quadruplex structures [13–17].
1.2. DNA G-quadruplex structures
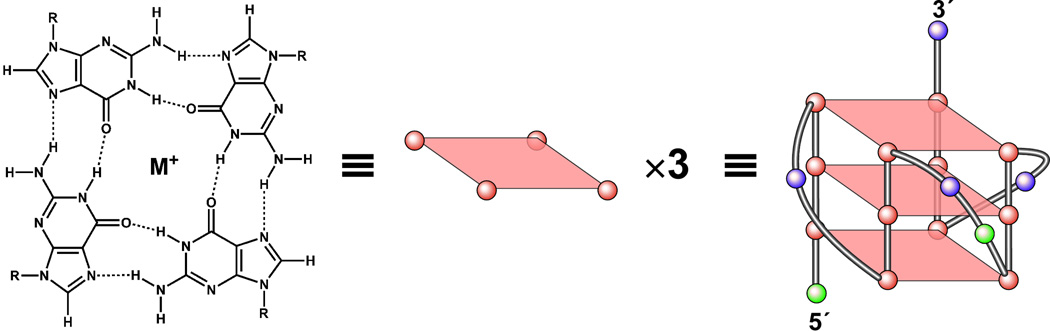
DNA G-quadruplex structures have received increased attention since the pioneering studies in the late 1980s from the Blackburn and Cech groups, showing that G-quadruplexes are capable of being formed from telomeric DNA and might be biologically relevant [18,19]. A G-quadruplex is a DNA secondary structure that consists of multiple vertically stacked guanine tetrads. Hoogsteen hydrogen bonds between the N1, N7, O6, and N2 guanine bases associate each of the four guanines to form a planar G-tetrad [20,21]. The involvement of N7 of guanine in the Hoogsteen base-pairing of a G-tetrad protects the site from chemical modification, such as methylation by dimethyl sulfate (DMS) [22]. This unique feature makes it possible to distinguish a G-quadruplex structure from single-stranded or B-form duplex DNA by DMS footprinting. G-quadruplexes can differ in their strand stoichiometry, strand orientation, and loop arrangement. The basic building block, the G-tetrad, and an example of a parallel G-quadruplex structure are shown in Figure 1. Numerous studies on G-quadruplex structure and function have been reported, including some excellent reviews [23–29].
Figure 1.
Structure of a G-tetrad and an example of the folding pattern of the known intramolecular G-quadruplex loop isomer (1:2:1) formed in the c-MYC promoter.
G-quadruplexes exhibit a remarkable dependency on alkali cations for their formation and stabilization. K+ and Na+ effectively bind to and stabilize many G-quadruplex structures [30]. In general, K+ is more potent than Na+ in binding to G-quadruplexes due to the better coordination of K+ with the eight carbonyl oxygen atoms present in the adjacent stacked tetrads [30–32]. However, the same DNA oligomer may form a different G-quadruplex species in the presence of K+ or Na+. Since K+ and Na+ are the prevalent alkali ions in cells, and the intracellular concentration of K+ is about 140 mM, while that of Na+ is about 5–15 mM [33,34], physiological conditions are favorable for the formation of a G-quadruplex.
Most of the published studies on G-quadruplexes have been carried out using synthetic oligomers, restriction fragments, or recombinant plasmids in a cell-free system. It has been particularly difficult to obtain convincing evidence of the existence of G-quadruplex structures in vitro by investigating these tiny segments of DNA chromosomes in the intact cell. However, studies have shown that G-quadruplexes do indeed have biological relevance and functional roles in living cells. Evidence for the formation of DNA/RNA G-quadruplexes in cells has been published in the past decade. One compelling piece of evidence comes from a study of the high-affinity antibody specific for the G-quadruplex formed by Stylonychia telomeric DNA [35]. The identification of proteins specific for G-quadruplex DNA, including MyoD, the tetrahymena G-quadruplex binding protein, and other telomeric proteins, provides further evidence for the existence of G-quadruplex DNA in cells [28,29,36–39]. The properties and biological roles of Gquadruplex-interactive proteins have been fully discussed in Michael Fry’s review [40]. Recent publications demonstrating the high density of putative G-quadruplex-forming regions in genomic regions adjacent to transcriptional start sites also support the biological significance of these structures [13–17]. Studies from our group have shown that a G-quadruplex can be formed within the promoter region of c-MYC and stabilized by a small molecule in living cells, which results in transcriptional repression of c-MYC [41–44]. It is now believed that certain GC-rich regions of gene promoters can transiently unwind and form single-stranded DNA conformations in superhelical conditions [45]. Notably, the guanine tracts of a G-rich strand can form G-quadruplex structures, and the cytosine tracts in the complementary strand may either form i-motif structures or remain as a single-stranded DNA loop.
In this review, we will summarize recently published contributions on the occurrence of G-quadruplex structures in the promoter regions of some important genes and their implications in the regulation of gene transcription.
2. Formation of intramolecular G-quadruplexes within promoter regions
G-quadruplex-forming motifs have been found in the promoter regions of a number of human genes, including the important protooncogenes c-MYC [41–44], VEGF [45], HIF-1α [46], Ret [47], KRAS [48], Bcl-2 [49,50], c-Kit [51–55], PDGF-A [56], and c-Myb [57], as well as a list of genes discussed in references 17, 41, and 58. Genes such as Rb [59–62], in which the G-quadruplex is in the 5′-coding region, have not been included. These polypurine/polypyrimidine motifs are located in the GC-rich regions of promoters and contain four or more runs of two or more contiguous guanines in the G-rich strand. The GC-rich region in the proximal region of these promoters is usually hypersensitive to nucleases and may form an altered structure with a single-stranded character, which is often a feature of transcriptionally active genes [45]. Compelling data show that intramolecular G-quadruplexes form within the promoter regions of some genes and play a critical role in transcriptional regulation (see later). G-quadruplexes that form in the promoter region of oncogenes have recently emerged as promising targets for the development of anticancer drugs [42]. It has been reported that the G-rich DNA sequences derived from the polypurine/polypyrimidine regions of the c-MYC, VEGF, HIF-1α, Ret, Bcl-2, c-Kit, and KRAS promoters form three-tetrad G-quadruplex structures in vitro [41–55], while PDGF-A and c-Myb form different types of G-quadruplex structures [56,57].
Here we will summarize some examples of intramolecular G-quadruplex structures that form in these promoter regions and, in some cases, their proposed biological roles in the control of gene transcription. Examples of intermolecular G-quadruplexes, most notably in the MyoD promoter [63], have also been documented and readers are referred to a recent review for a description of these structures and their biological significance [64].
3. Formation of three-tetrad G-quadruplexes in gene promoters
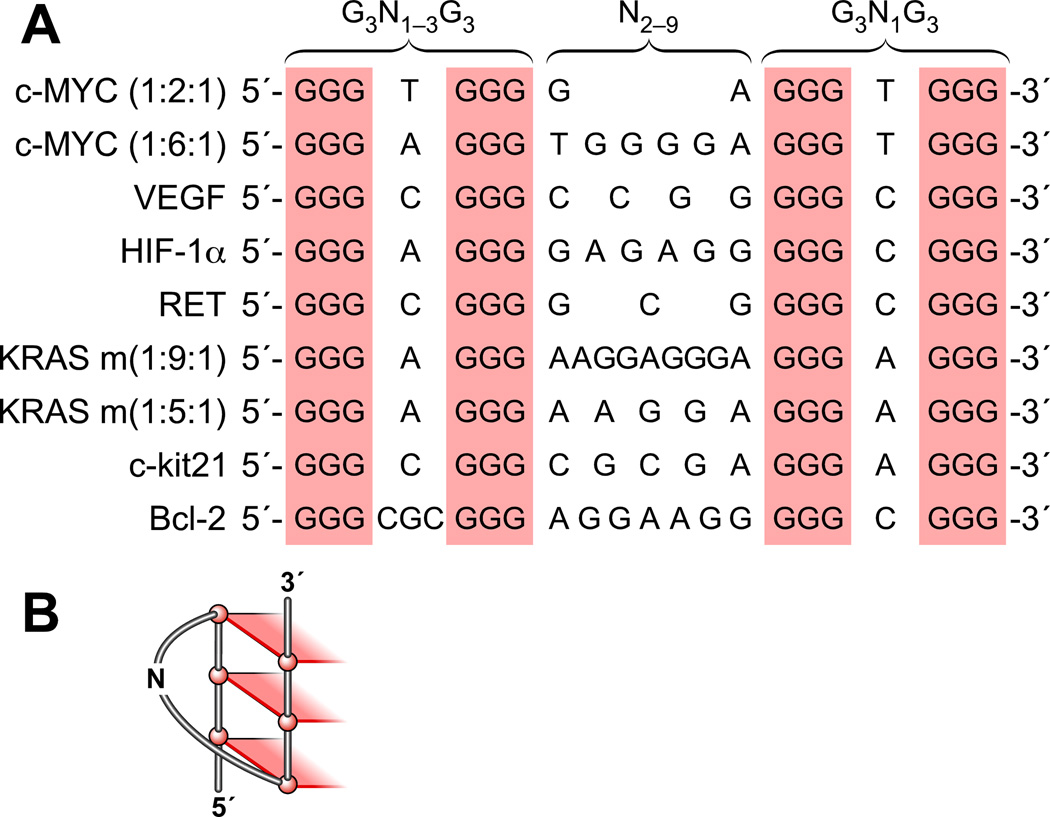
The formation of three-tetrad G-quadruplex structures in the polypurine/polypyrimidine regions of the c-MYC, VEGF, HIF-1α, Ret, c-Kit, KRAS, and Bcl-2 promoters has been observed in cell-free systems (Figure 2A). Notably, comparison of the G-quadruplex-forming motifs among these genes reveals an apparent sequence similarity. These unique DNA motifs provide a general transcriptional regulation mechanism involving interconversion between a G-quadruplex, unwound single-stranded DNA, and duplex DNA. One remarkable characteristic of these polypurine/polypyrimidine sequences is that the 3δ-end of each sequence is composed of an identical motif (G3N1G3), which is capable of forming a single-nucleotide double-chain-reversal loop (Figure 2B). It is proposed that this G3N1G3 motif is evolutionally selected to serve as a core-stabilizing G-quadruplex face around which to assemble the different intramolecular G-quadruplex structures. The middle loop, which bridges the second and third G-tracts (3′–5′), is a larger loop, containing between 2 and 9 nucleotides. The sequence of the loop at the 5′-end of the structure contains slightly more variation than the 3′-end loop, but in the majority of cases so far investigated, the 5′-end loop is composed of a single nucleotide. The size of this variant 5′-end loop plays a critical role in determining the loop folding and strand orientations of G-quadruplex structures in the different promoters. We thus propose that the general motif G3N1G3N2–9G3N1G3 (Bcl-2 being an exception) (where N represents the nucleotide(s) located in the G-quadruplex loop regions) is a common indicator for the potential of a G-rich DNA oligomer to form a three-tetrad intramolecular G-quadruplex with 3′- and 5′-end single-nucleotide double-chain reversal loops and a larger central loop. A second c-Kit G-quadruplex-forming region (c-Kit87up) also contains a 5′-end G3N1G3 motif and forms a intramolecular parallel G-quadruplex, but its folding pattern is more complex than that described here (see later) [53]. The importance of a single-nucleotide loop on the thermodynamic stability and topology of intramolecular G-quadruplexes has been noted previously by us [44,47,49], the Ebbinghaus group [46], and others, including the Fox group [65] and the Balasubramanian group [66]. In the latter study, UV melting and CD studies showed that G-quadruplexes containing two single-nucleotide loops are constrained to form a parallel double-chain reversal structure [66]. For a G-quadruplex-forming sequence with a combined loop length of five nucleotides, G-quadruplexes containing two single-nucleotide loops are more stable than G-quadruplexes with only one single-nucleotide loop [66].
Figure 2.
(A) Comparison of three-tetrad G-quadruplex-forming motifs (G3N1–3G3N2–9G3N1G3) within selected gene promoters. (B) One face of a three-tetrad G-quadruplex, showing a double-chain reversal loop containing one base.
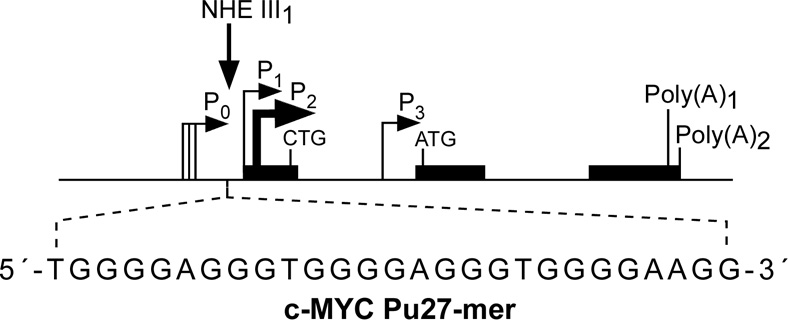
3.1. G-quadruplexes formed in the c-MYC promoter
c-MYC is an important oncoprotein and transcription factor and plays an essential role in cell proliferation and induction of apoptosis [67,68]. Overexpression of c-MYC is associated with a significant number of human malignancies, including breast, colon, cervix, and small-cell lung cancers, osteosarcomas, glioblastomas, and myeloid leukemias [69–73]. c-MYC transcription is under the complex control of multiple promoters. The nuclease hypersensitivity element III1 (NHE III1) in the proximal region of the c-MYC promoter (−142 to −115 base pairs) controls 80–90% of the total transcriptional activity of this gene [74–78]. The 27-base-pair core sequence of the wild-type c-Myc NHE III1 (c-MYC Pu27-mer) has an unusual strand asymmetry: the coding strand is a homopyrimidine tract and the noncoding strand is a homopurine tract. The sequence of the G-rich strand of c-MYC Pu27-mer is shown in Figure 3A. The sensitivity of NHE III1 to S1 nuclease, together with selective blocking of c-MYC transcription by a G-rich DNA oligomer, which can invade the NHE III1 duplex DNA, suggests that the NHE III1 is involved in a slow equilibrium between a typical duplex helix structure and an unwound non-B-form DNA structure [77–79]. Earlier studies on the atypical structure of the c-MYC NHE III1 led to a hypothetical model wherein the NHE III1 might adopt a tandem H-DNA structure that involves two intramolecular pyrimidine-purine-pyrimidine triplexes [80]. It was speculated that special transcription factors could activate c-MYC expression by recognizing and stabilizing the H-DNA conformation of the NHE III1. However, the formation of pyrimidine-purine-pyrimidine triplexes requires low pH, so this is unlikely to occur in living cells. There is no evidence to prove the existence or determine the biological function of H-DNA in the c-MYC promoter in cells. The observation that there are five consecutive G-tracts in the c-MYC Pu27-mer led to the realization that this strand can adopt an intramolecular G-quadruplex structure. Earlier studies from the Simonsson group showed that this G-rich strand can readily form a G-quadruplex structure under physiological conditions and that the formation of this structure is K+ dependent [81]. It was also postulated that the C-rich strand of the c-MYC NHE III1 might adopt an i-motif structure (see later) [82,83]. Although the formation of an i-motif structure requires acidic conditions, which makes their physiological relevance less compelling, we speculate that they may form in vitro under the driving force of negative superhelicity and/or G-quadruplex formation in the complementary strand, and there is some evidence for this in the VEGF promoter [45].
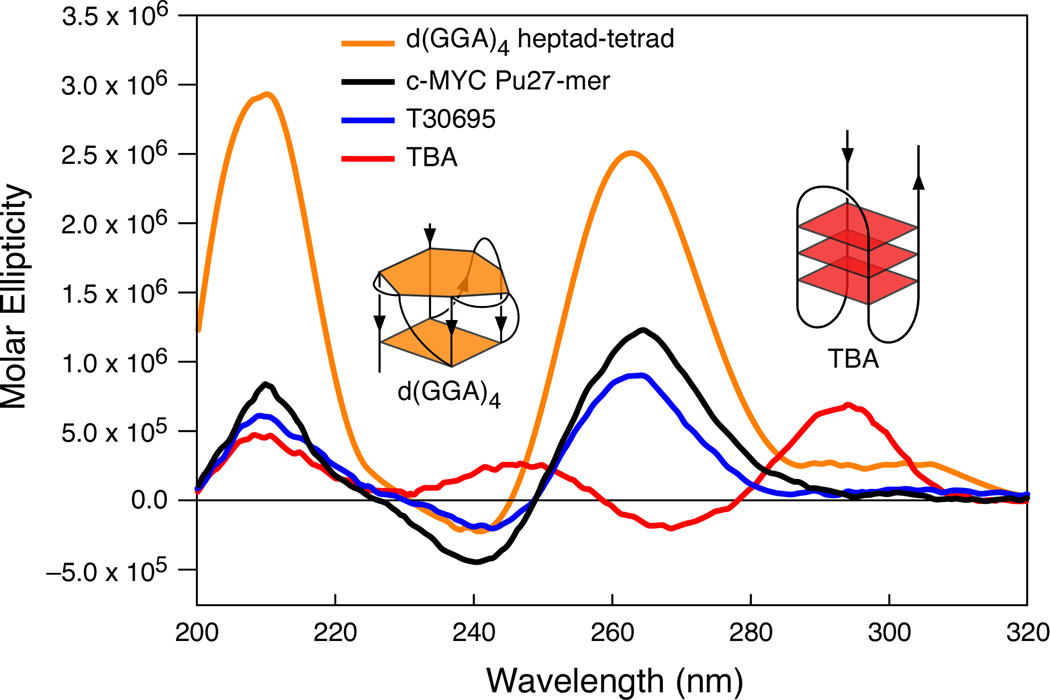
Figure 3.
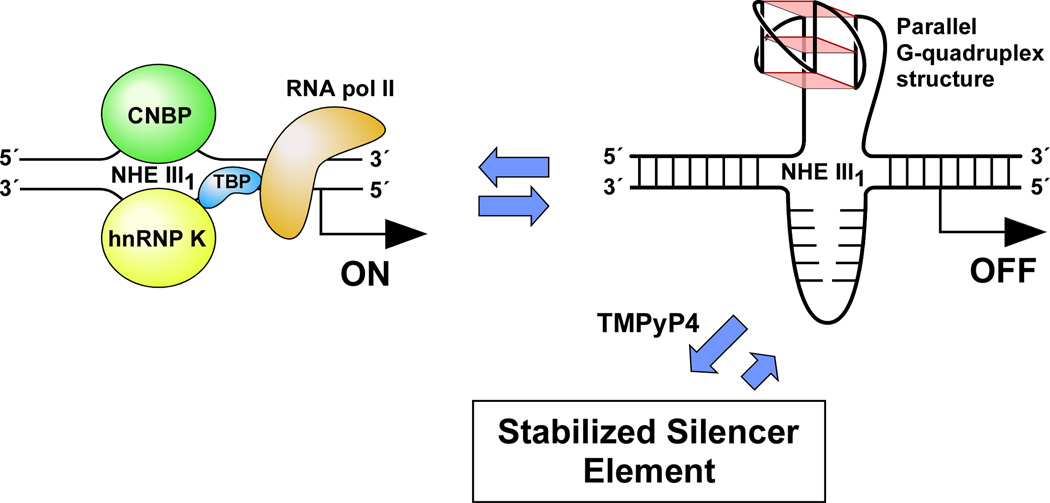
(A) Promoter structure of the c-MYC gene, showing the c-MYC Pu27-mer sequence of the guanine-rich strand upstream of the P1 promoter [41]. (B) CD spectra of the d(GGA)4 oligonucleotide, the c-MYC Pu27-mer, the T30695 oligonucleotide, and the thrombin binding aptamer (TBA). (C) Proposed structures of the four different c-MYC G-quadruplex loop isomers, in which dual G-to-T mutations at positions 11, 14, 20, and 23 result in defined loop isomers. The upper row shows the four proposed isomers and the lower row shows the results of dual G-to-T mutations. In this and subsequent figures, guanines = red, cytosines = yellow, thymines = blue, and adenines = green. (D) Schematic structures of c-MYC (1:2:1), c-MYC (1:2:1)-G14T/G23T, and c-MYC (1:6:1)-G11T–G14T determined by NMR in K+ solution. (E) Diagram of the rearrangements involved in the Ramos and CA46 Burkitt’s lymphoma cell lines (modified from ref. 72). Downward arrows indicate the breakage and rejoining points between chromosomes 14 and 8 for each translocation. RT-PCR for c-MYC and β-actin in Ramos (lanes 1–3) and CA46 (lanes 4–6) cell lines after no treatment (lanes 1 and 4) and treatment with 100 µM TMPyP2 (lanes 2 and 5) and TMPyP4 (lanes 3 and 6) for 48 hr. (F) Model for the activation and repression of c-MYC gene transcription involving the conversion of the paranemic secondary DNA structures (gene off) to purine and pyrimidine single-stranded DNA forms for transcriptional activation. hnRNP K and CNBP are single-stranded DNA binding proteins involved in transcriptional activation. Interaction of the parallel G-quadruplex structure with TMPyP4 stabilizes the gene-off form by conversion to the proposed double-loop G-quadruplex structure that stabilizes the silencer element and results in transcriptional repression [42].
Considerable efforts have been made to determine the biological role of the putative G-quadruplex structures in the c-MYC Pu27-mer in vitro. By extending our previous studies on the mechanistic basis of the effects of the G-quadruplex-interactive agent TMPyP4 on telomerase activity, we were fortuitously able to collect the first direct evidence of the formation of a G-quadruplex structure in the c-MYC Pu27-mer and its function as a silencer element to regulate c-MYC expression in cells [41]. We have previously shown that TMPyP4 can inhibit telomerase in both a cell-free system and living cells [84,85]. Subsequently, a cDNA microarray was performed to compare the gene expression profiles in HeLa cells after treatment with TMPyP4 and its positional isomer TMPyP2 [86]. (TMPyP2 lacks the ability to bind to the G-quadruplex structure because of the restricted rotation around the meso bonds in this molecule [87].) Most intriguingly, our data showed that TMPyP4, but not TMPyP2, can specifically downregulate c-MYC expression, as well as several genes downstream of c-MYC [86]. We also confirmed that TMPyP4 decreases c-MYC expression at the RNA and protein levels in MIA PaCa-2 and HeLa S3 cells [86]. This suggests that the unique effects of TMPyP4 on c-MYC expression may be mediated through interaction with a G-quadruplex structure formed in the c-MYC promoter. TMPyP4 has a greater affinity for the c-MYC G-quadruplex than duplex DNA by about 20 fold [88]. However, considering the overwhelming abundance of duplex versus G-quadruplex, it is surprising that we see such selectivity. In order to explain this unexpected selectivity, we propose the following. First, the apparent selectivity is affected by the dynamic equilibrium that exists between duplex, partially unwound single-stranded conformation, and secondary DNA structures such as G-quadruplex and i-motifs. This equilibrium is determined by the magnitude of the negative superhelicity resulting from transcriptional activation and the presence of proteins or ligands, which trap out the various forms (duplex, single-stranded, or G-quadruplex), together with proteins such as NM23-H2, which may sequentially trap out partially unfolded forms of the G-quadruplex in a step-wise manner. A high K+ concentration or drugs that stabilize the G-quadruplex will shift the equilibrium toward the G-quadruplex, which is of course the silenced form. In fact, enzymatic and chemical footprinting studies carried out in supercoiled plasmids show that the presence of G-quadruplex-stabilizing drugs or a high K+ concentration shifts the equilibrium toward the secondary DNA structures [45]. Once formed, the G-quadruplex is likely to act as a thermodynamic sink for TMPyP4, which binds tightly to the G-quadruplex with a relatively slow off-rate. When released, the TMPyP4 is trapped locally by the abundance of partially unwound or duplex DNA from which it has a relatively fast off-rate and can then rebind to the G-quadruplex. While the hemi-intercalated TMPyP4 bound into duplex DNA is unlikely to have a significant effect on enzymatic processes such as transcription or replication, binding to and stabilizing the c-MYC G-quadruplex by TMPyP4 is likely to have a much more profound effect on the equilibrium between a fully folded G-quadruplex and partially unfolded state required for conversion of the silenced form to the transcriptionally active form. Thus, we propose that it is the preferential trapping of the silenced G-quadruplex form by TMPyP4, which shifts this equilibrium between silenced and transcriptionally active form, that gives rise to an apparent higher selectivity of TMPyP4 than would otherwise be seen. Later, we showed that single G-to-A mutations in the c-MYC Pu27-mer, which destabilize the G-quadruplex-forming unit, result in a 3-fold increase in basal transcriptional activity of the c-MYC promoter [41]. This provided compelling evidence to confirm the intracellular formation of G-quadruplex structures and define their functions.
The critical role of G-quadruplex structures in the regulation of c-MYC expression and their potential as targets for small molecules to modulate c-MYC expression triggered considerable interest in studying the structural details of G-quadruplexes in the c-Myc promoter. Multiple biophysical and biochemical experimental methods have been applied to determine the biologically relevant G-quadruplex structure of the c-MYC Pu27-mer, including circular dichroism (CD), electrophoretic mobility shift assay (EMSA), DNA polymerase stop assay, DMS footprinting, mutational analysis, and nuclear magnetic resonance (NMR) (reviewed in reference 42). Comparative CD analysis of the c-MYC Pu27-mer, the HIV aptamer T30695, the thrombin binding aptamer (TBA), and the d(GGA)4 oligomer of the heptad-tetrad structure has been made to infer the topology of c-MYC G-quadruplex structures [44]. The chair-type antiparallel G-quadruplex structure of TBA (Figure 3B) has been proposed on the basis of NMR and X-ray crystallography [89,90], and a similar type G-quadruplex structure of T30695 was misassigned by NMR studies [91]. The CD spectrum of the c-MYC Pu27-mer is clearly different from TBA (Figure 3B). Moreover, the CD spectra of the c-MYC Pu27-mer and T30695 are coincident with the d(GGA)4 oligomer, which has been unequivocally shown by NMR to form a parallel-stranded heptad-tetrad structure [92,93] (Figure 3B). Thus, we were able to predict that the G-rich strands of the c-MYC Pu27-mer and T30695 form intramolecular parallel-stranded G-quadruplex structures. Indeed, NMR studies on the G-quadruplex formed by the DNA sequences derived from the c-MYC Pu27-mer [94] published back to back with our independent results [44] confirm that it is an intramolecular parallel-stranded G-quadruplex.
The five G-tracts in the c-MYC Pu27-mer have the potential to form multiple different G-quadruplex structures. In a previous study, we were able to separate two types of G-quadruplex structures in native gel by EMSA [41]. The DMS footprinting data identified the guanines involved in the formation of these two types of G-quadruplex structures. We proposed one type of G-quadruplex structure as G3N1–2G3N1–2G3N1–2G3, which is formed by the four consecutive 3′-end runs of guanines, while the other type of G-quadruplex structure, designated G3N1G3N6G3N1G3, is formed by the first, second, fourth, and fifth G-tracts of the c-MYC Pu27-mer (5′–3′) [see Figure 2A, labeled as c-MYC (1:2:1) and c-MYC (1:6:1)]. It has been shown that the G-quadruplex structures formed from the four consecutive 3′-end G-tracts of the c-MYC Pu27-mer in K+ solution are the predominant G-quadruplex structures isolated from the native gel [41]. Using a DNA polymerase stop assay with a series of single-guanine mutation sequences (G to A) derived from the c-MYC Pu27-mer, it was shown that the 5′-end run of guanines (G2–G5) is not required for formation of a stable G-quadruplex structure [44]. The degeneracy resulting from multiple G-tracts is somewhat analogous to different conformational forms of protein kinases, where only one form is catalytically active. Stabilization of the inactive conformational form, whether it is one conformation of a protein kinase or the minor G-quadruplex, may turn out to be an important concept for future drug targeting of G-quadruplex elements in promoters, since in each case the alternative conformations may have quite different biological functions.
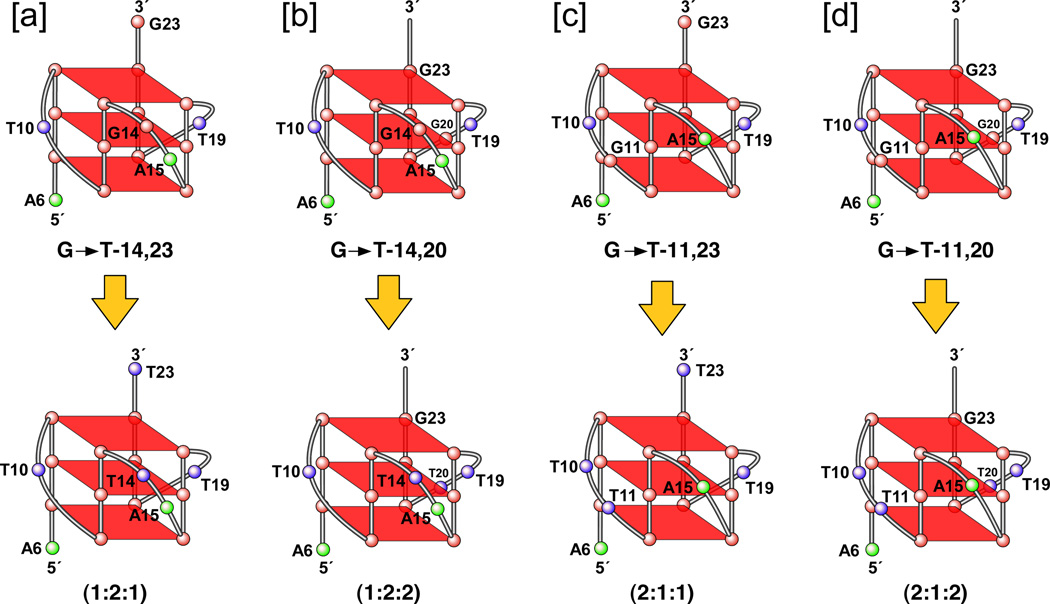
Since the two four-guanine G-tracts (G11–G14 and G20–G23) in the c-MYC Pu27-mer have more guanines than required for the formation of a stable three-tetrad G-quadruplex core, it is assumed that there may be a degeneracy in the use of these two guanine runs that results in guanine slippage and the formation of different loop isomers (G3N1–2G3N1–2G3N1–2G3). On the basis of DMS footprinting, mutational analysis, and a DNA polymerase stop assay, we were able to identify four loop isomers formed by the four consecutive 3′-end G-tracts of the c-MYC Pu27-mer (Figure 3C, upper panel) [44]. Dual-mutant studies and partial-protection patterns confirmed that guanine slippage could occur between G11 and G14 (and/or G20 and G23) in the c-MYC Pu27-mer, such that one guanine is involved in G-tetrad formation and the other guanine is located within a loop (Figure 3C, upper panel). Although all the guanines of the four 3′-end G-tracts in the c-MYC Pu27-mer are involved in the formation of G-tetrads, it is unlikely that all four isomers occur in equal proportion. The DMS footprinting and EMSA studies show that the 1:2:1 loop isomer ([a] isomer in Figure 3C) is the predominant isomer within the mixture. A luciferase reporter assay study proved that all four dual mutant loop isomers are biologically relevant to c-MYC transcriptional control and that each of these parallel G-quadruplex structures acts as a repressor element in the c-MYC NHE III1 [44].
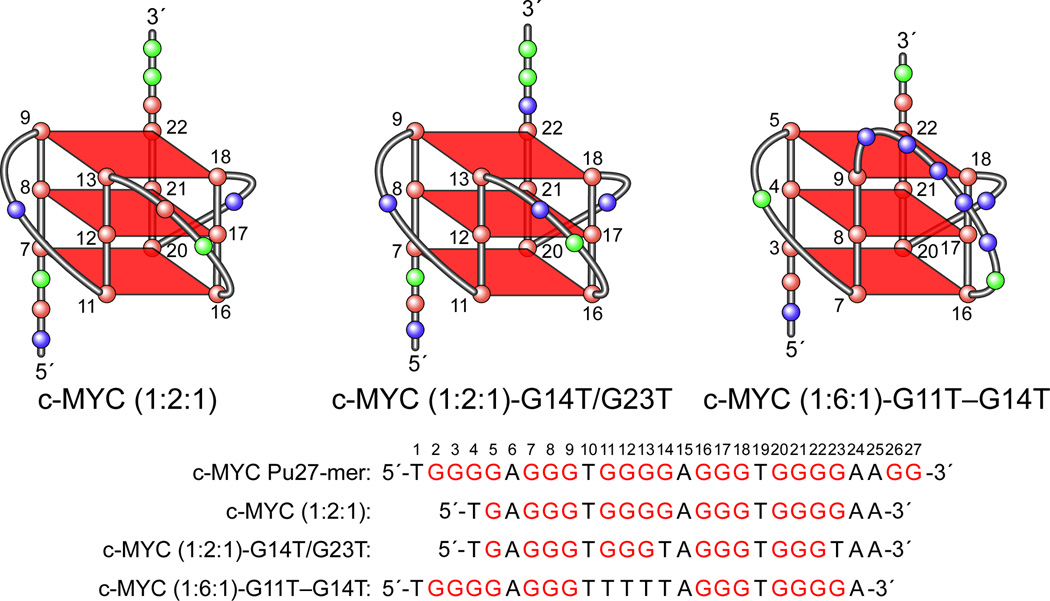
The formation of a parallel-stranded propeller-type G-quadruplex within the four biologically relevant 3′-end guanine runs in the c-MYC Pu27-mer has been confirmed by NMR spectroscopic studies from the Patel and Yang groups [94,95]. Using a 22-mer sequence [c-MYC (1:2:1), Figure 3D] that is derived from the four 3′-end guanine runs of the c-MYC Pu27-mer, Patel et al. have defined the folding pattern of an intramolecular parallel-stranded propeller-type G-quadruplex in K+-containing solution (Figure 3D). In similar experiments, Yang et al. used a sequence with dual G-to-T mutations at positions 14 and 23 [c-MYC (1:2:1)-G14T/G23T, Figure 3D], which represents the predominant loop isomer formed from the four 3′-end guanine runs of the c-MYC Pu27-mer, for their NMR structure determination. Both groups verified a parallel-stranded G-quadruplex with 1:2:1 side loops in K+-containing solution, and both NMR studies revealed the same predominant G-quadruplex structure for the four 3′-end guanine runs of the c-MYC Pu27-mer, which consists of three G-tetrads formed by four intramolecularly linked parallel DNA strands with all anti guanines and linked by three double-chain reversal side loops. Interestingly, Patel’s group also observed the formation of an intramolecular G-quadruplex by using a mutated sequence [c-MYC (1:6:1)-G11T–G14T, Figure 3D] derived from the c-MYC Pu27-mer [94]. This mutated sequence, in which the third G-tract in the c-MYC Pu27-mer is substituted with a T-tract (Figure 3D), directs the formation of a G3N1G3N6G3N1G3 G-quadruplex. The c-MYC (1:6:1)-G11T–G14T NMR study unequivocally confirmed that this sequence forms an intramolecular parallel-stranded G-quadruplex structure with a large six-nucleotide loop. The identification of two stable c-MYC G-quadruplex structures by NMR confirms that the c-MYC Pu27-mer can potentially form multiple G-quadruplex structures in cells.
While the G-rich strand of the c-MYC Pu27-mer can adopt several G-quadruplex structures, the complementary C-rich strand may form an i-motif structure. An i-motif structure is a tetraplex structure formed by intercalated cytosine+–cytosine base pairs between two parallel duplexes, which interact into each other in an antiparallel orientation. By employing CD and ultraviolet absorption spectroscopy in combination with EMSA, it has been shown that the C-rich strand of c-MYC NHE III1 can associate to form an intramolecular i-motif at moderately acidic or even neutral pH [83]. Normally, because of the requirement for acidic pH (i.e., ≤5.5), i-motif structures would not be considered to have biological relevance. However, in vitro, other factors, such as torsional stress, may play an equal or more important role in maintaining the i-motif structure at more physiological conditions. Preliminary data from our lab suggest that this i-motif may play a role in both silencing and remodeling events in c-MYC transcription in conjunction with a G-quadruplex structure (unpublished data).
3.2. Proof of concept that TMPyP4 targets G-quadruplexes to modulate c-MYC gene expression
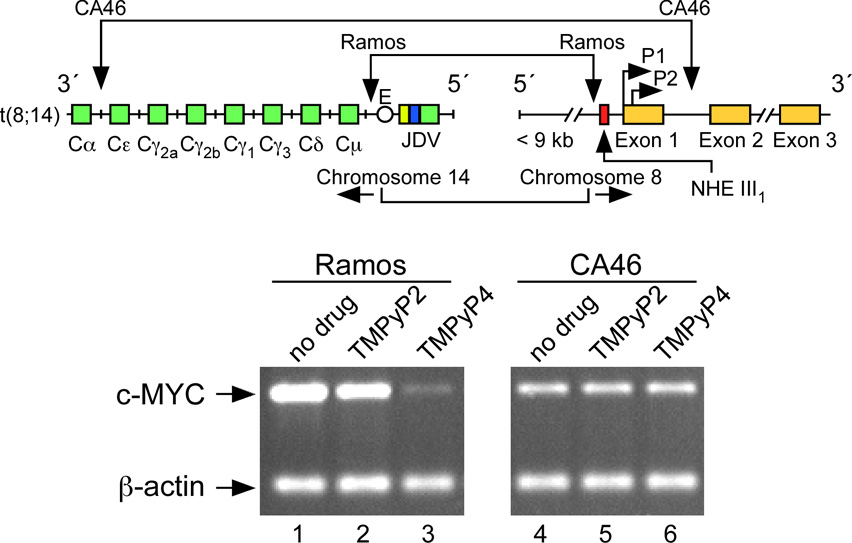
The biological significance of the intramolecular G-quadruplex structures formed in c-MYC NHE III1 has been evaluated for c-MYC transcriptional inhibition by TMPyP4 in Ramos and CA46, two Burkitt’s lymphoma cell lines [41]. The Ramos cell line retains the c-MYC NHE III1 during translocation, whereas the CA46 cell line loses this element, together with the P1 and P2 promoters [41,96]. As anticipated, in the CA46 cell line TMPyP4 had no effect on c-MYC transcriptional activation, whereas in the Ramos cell line, TMPyP4, but not TMPyP2, lowered c-MYC mRNA level [41] (Figure 3E). This result further confirms that TMPyP4 mediates its transcriptional inhibitory effect on c-MYC by interaction with the G-quadruplex formed in the c-MYC NHE III1. Recently, Ou et al. have confirmed the results of these experiments using a structurally distinct group of quindolines that bind to the c-MYC G-quadruplex and selectively inhibit c-MYC expression in the Ramos cell line [97]. We have proposed a model in which the transcriptionally active forms (duplex or single-stranded DNA conformations) are in equilibrium with the silenced form (G-quadruplex structure), and this can be maintained in the silenced form in the presence of TMPyP4, which shifts the equilibrium away from the transcriptionally active form [41,42] (Figure 3F). Our studies and those of Ou et al. on the G-quadruplex structures formed in the c-MYC promoter highlight the importance of secondary DNA structures in controlling gene expression and their utility as a receptor for external control of gene expression by small molecules.
The role of proteins in the control of gene expression through secondary DNA structures has been addressed by Fry [40]. In the case of c-MYC, at least four proteins are involved that bind to single-stranded DNA (hnRNP K and CNBP) [98,99], duplex DNA (Sp1) [99], and either single-stranded or duplex DNA (NM23-H2) [100,101]. Because the c-MYC G-quadruplex is a very stable species, it would seem necessary to identify proteins that unfold the G-quadruplex or more likely trap out transiently unwound DNA species in a step-wise manner. NM23-H2 may function in this regard [100,101]. The formation of the secondary DNA species from the duplex DNA may be mediated by negative superhelicity generated as a consequence of transcription [102]. In a very recent article from the Levens lab, the first direct evidence for the involvement of transcriptionally generated torsion superhelicity in unpairing DNA, which mediates the formation of non-B-DNA in the c-MYC promoter, has been elegantly demonstrated [102]. This experimental verification of the role of transcriptionally generated negative superhelicity, even in the presence of topoisomerases, provides a critical link in how secondary DNA structures can be generated from otherwise stable duplex DNA. This is an extremely important observation, which counters the commonly voiced concern that it is energetically very unfavorable for DNA secondary structures to emerge from stable duplex DNA. Additional support for this concept comes from the observation that single-stranded DNA binding proteins, such hnRNP K and CNBP, are important transcriptional factors involved in activation of c-MYC transcription.
3.3. G-quadruplexes formed in the VEGF promoter
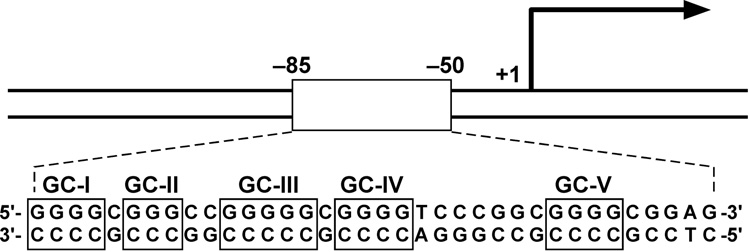
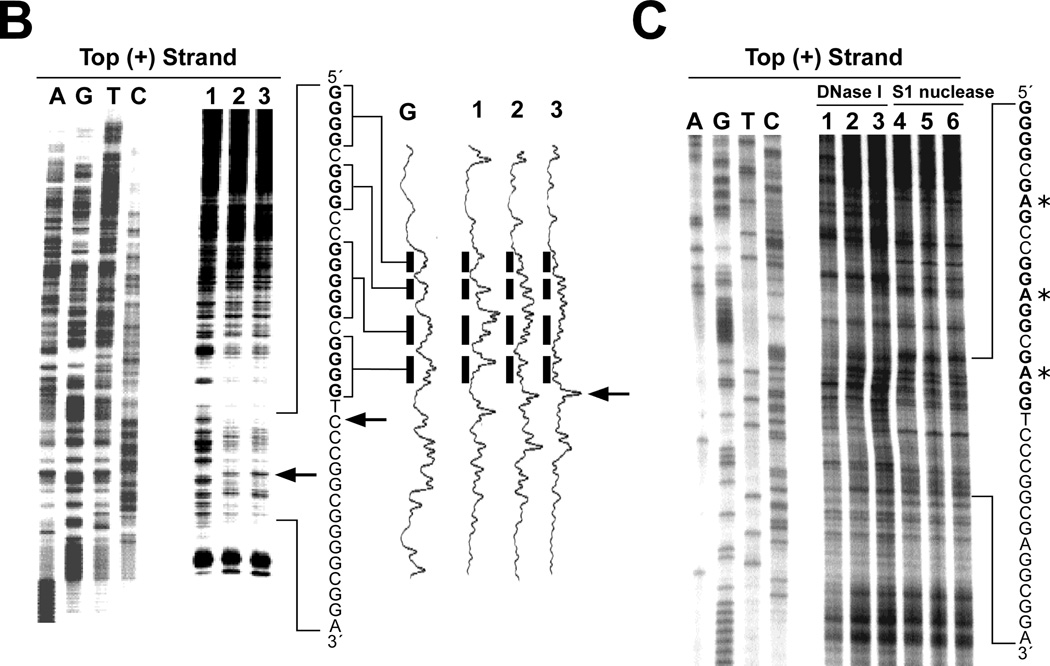
VEGF is an important angiogenic switch in starving cancer cells, and their survival often depends upon increased expression of this gene [103]. The polypurine/polypyrimidine tract in the proximal promoter region of the human vascular endothelial growth factor (VEGF) gene not only contains multiple binding sites for transcription factors, but also consists of five runs of at least three contiguous guanines (Figure 4A), corresponding to the general motif for the formation of an intramolecular G-quadruplex (Figure 2A) [45,103–105]. In duplex DNA oligomers we have observed the progressive unwinding of the duplex DNA containing the VEGF polypurine/polypyrimidine tract into single-stranded oligomers in the presence of K+, TMPyP4, or telomestatin. This suggests that the G-tracts of the VEGF promoter can transiently adopt G-quadruplex structures, which can be trapped out and stabilized by high concentrations of K+, TMPyP4, or telomestatin under physiological conditions [45]. Subsequent footprinting studies with DNase I and S1 nuclease on the supercoiled plasmid containing the human VEGF promoter region revealed a long protected region, including the G-quadruplex-forming region, in the presence of K+ and telomestatin [45]. The 3′-side residue of the G-quadruplex-forming region in the VEGF promoter has been shown to be hypersensitive to S1 nuclease (arrows in Figure 4B), suggesting the formation of a G-quadruplex structure within the VEGF promoter. In contrast, the mutant plasmid that contains the specific point mutations to abolish the G-quadruplex-forming ability in the VEGF proximal promoter does not present the same hypersensitivity toward DNase I or S1 nuclease in the polypurine/polypyrimindine region (Figure 4C). Collectively, these data provide first evidence that both supercoiled conformation and the wild-type G-quadruplex-forming sequence are required for the formation of nuclease hypersensitivity regions in the polypurine/polypyrimindine region. Subsequently, it was demonstrated that the CD spectrum of the G-rich strand in the VEGF promoter reveals a parallel-stranded G-quadruplex structure [45]. The formation of a G-quadruplex structure in this promoter has been further confirmed by a DNA polymerase stop assay, which shows a K+-dependent arrest of DNA polymerase extension through the template DNA containing the VEGF G-tract sequence (unpublished data). The presence of two arrest sites in the DNA polymerase stop assay suggests that two different G-quadruplex structures, made up of different runs of G-tracts, can be formed by the G-rich strand of the VEGF promoter. DMS footprinting was performed on the VEGF wild-type sequence containing the G-tracts, along with several site-mutation sequences, and the results indicate that the DNA secondary structures formed in the VEGF promoter are three-tetrad G-quadruplexes (unpublished data). Se2SAP has been shown to inhibit VEGF transcription in various human cell lines [106].
Figure 4.
(A) Promoter structure of the VEGF gene. The polypurine/polypyrimidine sequence is shown, together with the five GC boxes [45]. (B) Plasmid footprinting of the VEGF promoter region with S1 nuclease. The footprinting of the top strand of the supercoiled plasmid is shown in the left panel. The densitometric scanning of the S1 footprinting on top strand is shown in the right panel. Lane 1 = no salt, lane 2 = 100 mM KCl, lane 3 = 1 µM telomestatin. Arrows indicate the sites that are hypersensitive to S1 nuclease at the 3′-side of the G-quadruplex-forming region [45]. Guanines in bold are those associated with the quadruplex. (C) Plasmid footprinting of the top strand of the mutant VEGF promoter region with DNase I and S1 nuclease. The plasmid was incubated in the absence of salt (lanes 1 and 4), in the presence of 100 mM K+ (lanes 2 and 5), or with 1 µM K+ (lanes 3 and 6) at 37 °C for 1 hour before treating with nuclease [45]. Guanines in bold are those associated with the quadruplex. Asterisks indicate mutated guanines (G to A).
3.4. G-quadruplexes formed in the HIF-1α promoter
The formation of G-quadruplex structures has been observed in the promoter region of HIF-1α, which is an important transcription factor for regulating VEGF expression, and is overexpressed in many tumors [107–109]. The importance of the polypurine/polypyrimidine tract in the proximal promoter region of HIF-1α in gene transcriptional regulation has been confirmed by the observation that mutagenesis of this region results in lower basal HIF-1α expression [46]. To investigate G-quadruplex formation in the polypurine/polypyrimidine tract of the HIF-1α promoter, Ebbinghaus et al. analyzed G-rich DNA oligomers derived from this region using EMSA, CD, DNA polymerase stop assay, and DMS footprinting. The characteristics of the CD spectrum of the native G-tract sequence of the HIF-1α promoter suggest that this sequence forms an intramolecular parallel-stranded G-quadruplex structure in the presence of K+, and the DNA polymerase stop assay shows that TMPyP4 and telomestatin are capable of binding to and stabilizing the G-quadruplex [46]. In K+-containing buffer, DMS footprinting of the G-rich oligomer shows protection of all four runs of guanines, which is consistent with the formation of an intramolecular three-tetrad G-quadruplex with a central six-nucleotide loop [46] (Figure 5). The HIF-1α G-quadruplex structure, an intramolecular parallel-stranded three-tetrad G-quadruplex with a six-nucleotide central loop, is related to the general G-quadruplex-forming motif G3N1G3N2–9G3N1G3 (Figure 2A). This also suggests that the G-quadruplex structure formed in the HIF-1α promoter may play a role in regulating HIF-1α gene expression.
Figure 5.
DMS footprinting on the G-rich DNA oligomer derived from the HIF-1α promoter in the absence (−) or presence (+) of 140 mM K+. The sequence of the HIF-1α G-rich oligomer is shown adjacent to the gel. Open circles indicate the guanines that are fully protected, and the solid circles indicate the guanines that are cleaved [46].
3.5. G-quadruplexes formed in the RET promoter
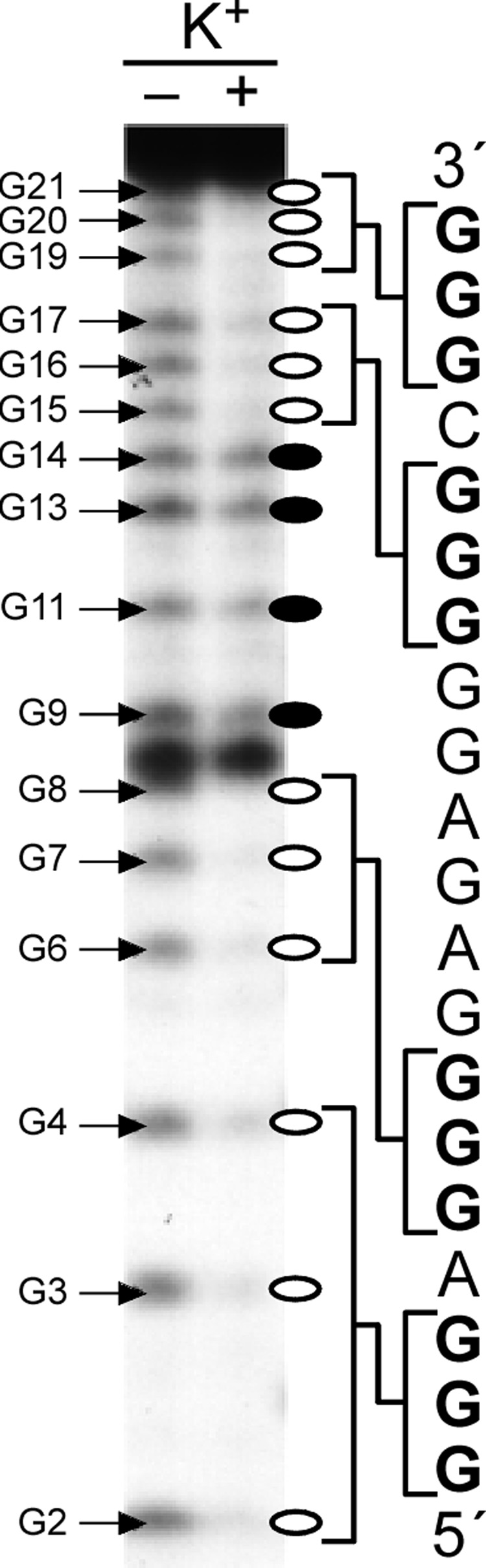
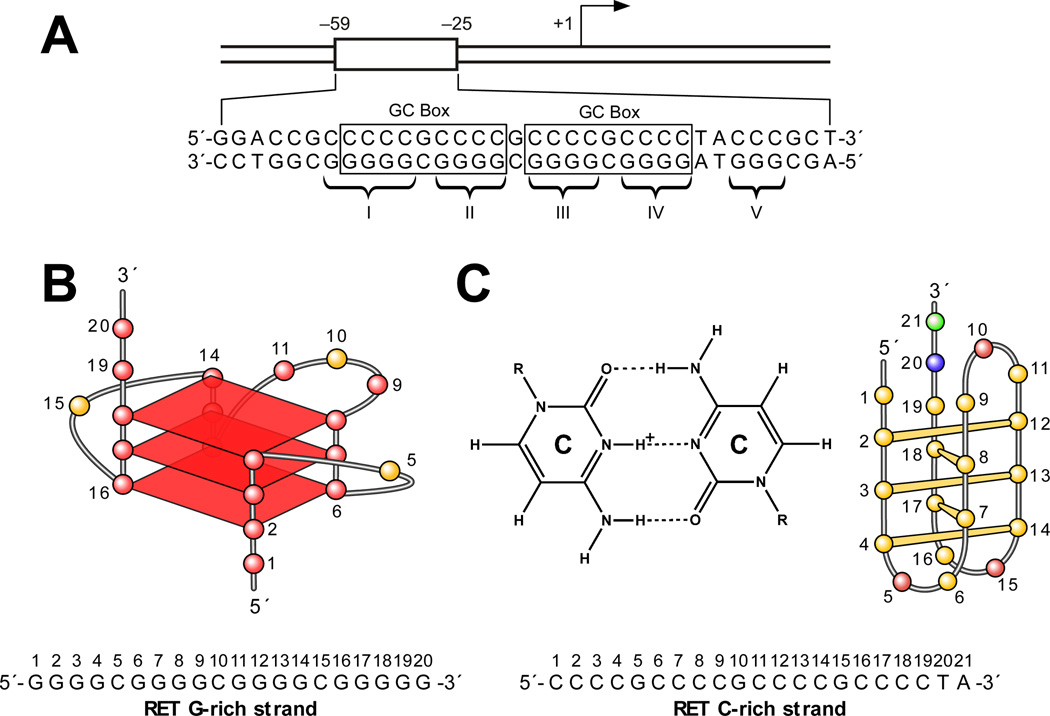
The RET proto-oncogene encodes a receptor-type tyrosine kinase that has been implicated in the development of several human cancers, especially thyroid cancer [110]. The two GC boxes in the proximal promoter region of RET are essential for its basal promoter activity (Figure 6A) [111,112]. The five polypurine/polypyrimidine tracts contained within this promoter element (Figure 6A) are very dynamic in nature and have the ability to adopt different non-B-form DNA secondary structures. We have shown that in this region the G-rich strand can fold into a G-quadruplex structure, while the C-rich strand can adopt an i-motif structure [47]. DNA polymerase stop assay studies demonstrate that, in the presence of 100 mM K+, the five G-tracts in the G-rich strand of RET have the ability to adopt two different intramolecular G-quadruplex structures [47]. One of these produces a Taq polymerase arrest site at G19 near the 3′-end of the core sequence (Figure 6B), whereas the other produces an arrest site at G14 immediately before the second 3′-end G-tract. The G-quadruplex that is responsible for the arrest site at G19 is the major arrest product formed in the DNA polymerase stop assay, suggesting that it is the major G-quadruplex structure formed in the RET proximal promoter element. Comparative CD and DMS footprinting studies have revealed that this structure is a parallel-stranded intramolecular structure containing three G-tetrads (Figure 6B). K+, TMPyP4, and telomestatin can further stabilize this G-quadruplex structure. By applying CD spectroscopy and Br2 footprinting, we have demonstrated that the complementary C-rich strand of the RET proximal promoter element forms an i-motif structure in vitro (Figure 6C) [47]. On the basis of these experimental studies, we constructed a molecular model for the potential DNA secondary structures formed in the duplex polypurine/polypyrimidine sequence of the RET promoter that comprises a stable intramolecular G-quadruplex and an i-motif structure having minimum symmetrical loop sizes of 1:3:1 and 2:3:2, respectively [47] (Figure 6D).
Figure 6.
(A) Promoter structure of the RET gene. Two GC boxes are highlighted. Five guanine tracts (I, II, III, IV, V) are indicated with braces [47]. (B) Proposed G-quadruplex structure for the G-rich DNA oligomer of the RET promoter. The sequence of this G-rich strand DNA is shown under the structure, together with the pol stop sites (arrows). (C) Cytosine+– cytosine base pair in the i-motif (left). Schematic structure of an i-motif formed in the C-rich DNA oligomer of the RET promoter (right). The sequence of this C-rich strand DNA is shown under the structure. (D) Molecular model of the RET promoter sequence (−66 to −19) with i-motif, G-quadruplex, and duplex DNA regions (adenine, green; guanine, red; thymine, blue; cytosine, yellow; potassium ions, white). The symmetrical arrangement of RET C-rich and G-rich sequences is shown below the model.
3.6. G-quadruplexes formed in the KRAS promoter
Activating mutants of KRAS are found in many human cancers [113]. The nuclease hypersensitive polypurine/polypyrimidine elements (NHPPE) in the mouse and human KRAS promoters are critical for gene transcription [114]. This is another compelling example of the parallel-stranded intramolecular G-quadruplex serving as a silencer element in the promoter region of an oncogene comes from the mouse KRAS promoter study by Cogoi and Xodo [48]. Consecutive runs of guanines in the mouse gene KRAS promoter, and possibly the human gene, suggest that it may assume an intramolecular G-quadruplex structure (Figure 2A). CD and EMSA studies on the G-rich strands of the mouse and human KRAS NHPPE show that the structural characteristics of a DNA secondary structure formed in NHPPE are consistent with those of an intramolecular parallel-stranded G-quadruplex structure [48]. The two arrest sites in the DNA polymerase stop assay of the template containing the G-quadruplex-forming motif of the mouse KRAS NHPPE indicate the formation of two different G-quadruplex structures from different runs of guanines. On the basis of DMS footprinting results on the G-rich DNA sequences of mouse KRAS NHPPE, along with CD, EMSA, and the DNA polymerase stop assay, two different parallel intramolecular three-tetrad G-quadruplex structures have been proposed. One of these structures consists of two one-base external loops and one nine-base external loop [KRAS m(1:9:1)], while the other consists of two one-base external loops and one five-base external loop [KRAS m(1:5:1)] (Figure 2A) [48]. These sequences closely resemble those found in the c-Myc G-quadruplexes. Thus, the folding topology of this structure is assumed to be similar to the c-MYC G-quadruplexes. DMS footprinting of the human KRAS G-quadruplex-forming motif did not reveal a clear picture. It has been shown that TMPyP4 can stack to the external G-tetrad of mouse KRAS G-quadruplexes and increase the Tm by ~20 °C. Transfection experiments show that the stabilization of the mouse KRAS G-quadruplexes with TMPyP4 results in the strong inhibition of mouse KRAS promoter activity, but TMPyP2 does not show any significant effect on KRAS transcription [48]. Cogoi and Xodo also describe the isolation of a nuclear protein that recognizes and binds to KRAS duplex DNA using the G-quadruplex-forming sequence of the mouse KRAS NHPPE. Furthermore, the transfection of KRAS G-quadruplex-forming oligomers in 293 cells induces inhibition of mouse KRAS promoter activity up to 40% of control [48]. All of these cellular assay data strongly suggest that a G-quadruplex structure forms within the mouse KRAS NHPPE and serves as a silencer in regulating KRAS transcription in living cells.
3.7. Typical and non-typical G-quadruplexes formed in the c-Kit promoter
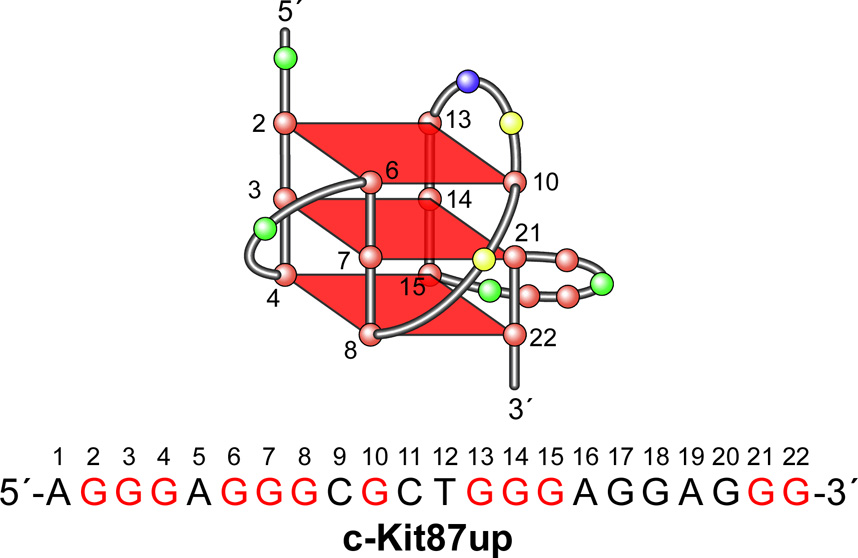
The receptor tyrosine kinase c-Kit is overexpressed in a number of cancers [115]. Recently two conserved G-quadruplex-forming sequences have been identified in the human c-Kit promoter [51,52]. They are both located within a nuclease hypersensitive region of the promoter, suggesting that they can form non-B-form DNA structures [116,117]. One G-quadruplex-forming unit (c-Kit21) (Figure 2A) is located within −140 to −160 base pairs upstream of the transcription initiation site, while the other (c-Kit87up) (Figure 7) is found between −87 and −109 [51,52].
Figure 7.
Schematic structure and folding topology of a c-kit87up G-quadruplex determined by NMR [53]. Sequence for c-kit87up is shown below the structure. Guanines in red are those involved in tetrad formation.
One-dimensional NMR studies and ultraviolet thermal melting analysis have confirmed that both c-Kit21 and c-Kit87up can form intramolecular G-quadruplexes in solution [51]. In the presence of 100 mM K+, the CD spectrum of c-Kit21 has a strong positive signal peak at 263 nm with a negative peak at 245 nm [52], which is characteristic of a parallel folded G-quadruplex structure. The c-Kit87up sequence in a 50 mM K+ solution showed a CD spectrum with a minimum absorbance at 240 nm and a maximum absorbance at 262 nm, which is also consistent with the spectrum of a parallel G-quadruplex [55]. On the basis of these studies, both c-Kit21 and c-Kit87up are expected to form parallel G-quadruplexes in the presence of K+.
Single-molecular fluorescence resonance energy transfer studies demonstrated that in the c-Kit21 fewer than 1% of the molecules in a duplex DNA molecule show real dynamic fluctuations and therefore interconversion between duplex and G-quadruplex is relatively rare [54]. We speculate that the negative superhelicity is probably required for efficient conversion. In a recent paper, the Balasubramanian group at Cambridge has shown that a group of 3,8,10-trisubstituted isoalloxazines show selective binding to the c-Kit21 G-quadruplex and inhibit c-Kit expression in MCF-7 and HGC-27 cells at 5 µM [118].
The c-Kit87up sequence contains four three-guanine tracts separated by linkers having one base and two sets of four bases (Figure 7). Since this motif, having only one (G3N1G3) sequence, would be expected to form a mixed parallel/antiparallel structure, it was surprising that the c-Kit87up sequence might form a conventional parallel-stranded three-tetrad G-quadruplex structure. Moreover, a mutational analysis of the predicted loop regions between the G-tracts showed that none of the mutated sequences could form a similar G-quadruplex structure under the physiological conditions that coincided with the native sequences [51]. This suggested that the c-Kit87up G-quadruplex may contain an unusual arrangement of guanines. An NMR-derived solution structure of the G-quadruplex structure formed by the c-Kit87up sequence in K+ solution has recently been reported (Figure 7) [53]. This study revealed a highly unusual G-quadruplex folding topology with several unique characteristics. This structure is composed of three G-tetrads, and the glycosidic conformations of all of the guanines are anti, both features that are consistent with the G-tetrad core containing all parallel G-tracts. One remarkable feature of the structure is the participation of an isolated single guanine (G10) in the formation of a G-tetrad, despite the presence of four three-guanine tracts in the sequence. There are four loops in the structure to bridge three consecutive G-tracts and a fourth G-tract made up of two consecutive guanines and one guanine from a different region of the sequence (i.e., G2–G4, G6–G8, and G21–G22, with G10 providing the additional guanine) (Figure 7). This unique arrangement of four loops leads to the formation of a snapback parallel-stranded G-quadruplex core. The NMR structure of c-Kit87up also revealed that three Watson-Crick base pairs (A1:T12, A16:G20, and G17:A19) contribute to the stabilization of this unique structure [53]. The involvement of most of the residues within the 22-nucleotide sequence in the formation of this unique stable G-quadruplex structure confirms the previous mutational analysis data, which showed that the loop regions play critical roles in c-Kit G-quadruplex formation [53]. This unique scaffold for the c-Kit87up G-quadruplex may provide a platform for designing G-quadruplex-interactive agents that specifically target the G-quadruplex in the c-Kit promoter [53]. A bioinformatic study also shows that the 22-nt c-Kit87up sequence is unique within the human genome [55]; furthermore, none of structurally closely related sequences are found immediately upstream of transcriptional start sites.
3.8. Mixed parallel/antiparallel G-quadruplexes formed in the Bcl-2 promoter
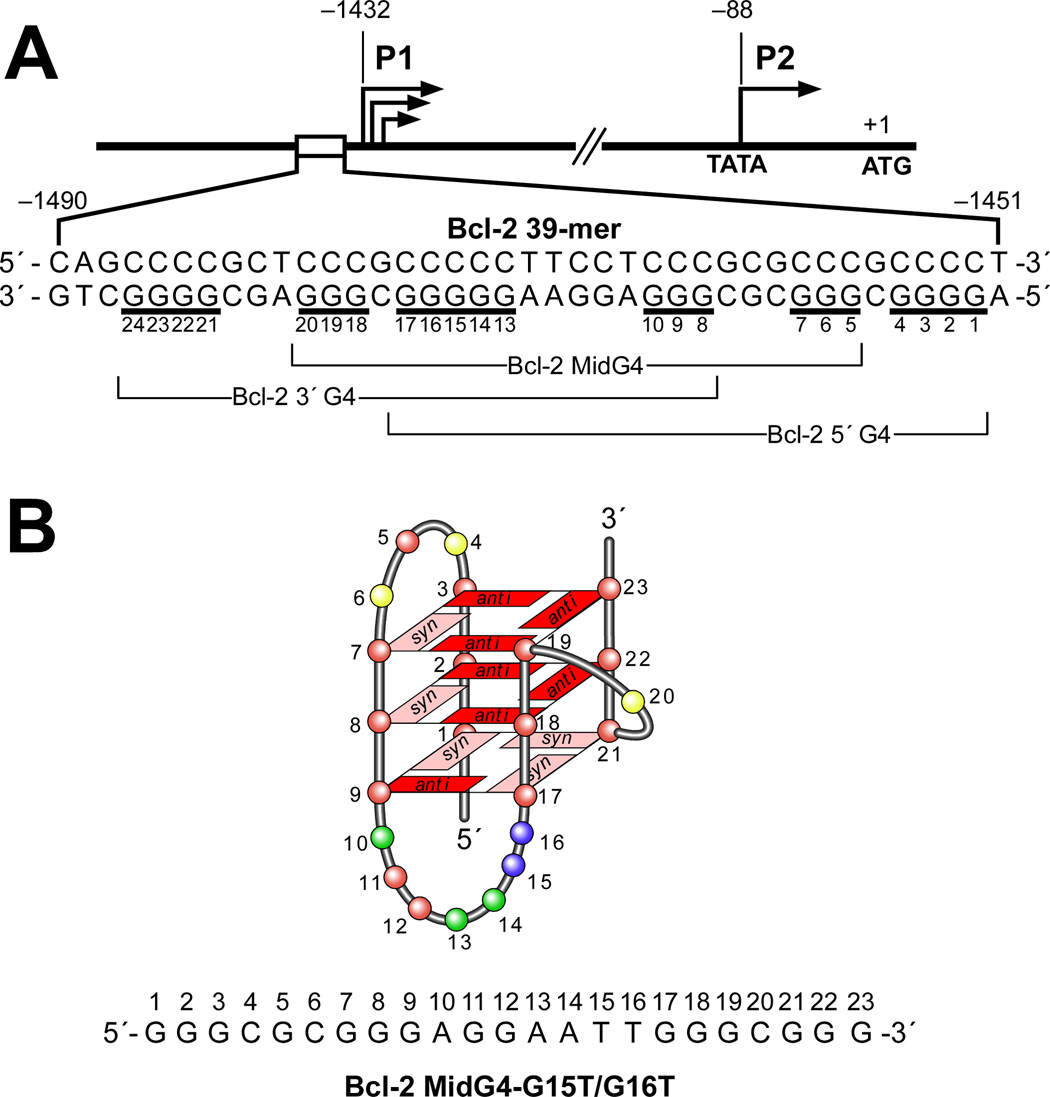
The bcl-2 (B-cell CLL/lymphoma 2) gene product is a mitochondrial membrane protein that exists in delicate balance with other related proteins and is involved in the control of programmed cell death, functioning as an apoptosis inhibitor [119]. In the Bcl-2 promoter, one region of particular interest is the GC-rich region located in the nuclease hypersensitive region − 1490 to −1451 upstream of the P1 promoter [120]. This polypurine/polypyrimidine tract has been shown to be critically involved in the regulation of Bcl-2 gene expression. The core sequence of the polypurine/polypyrimidine tract in the Bcl-2 P1 promoter (Bcl-2 Pu39WT) contains seven runs of at least two contiguous guanines separated by one or more bases (Figure 8A), which is different to the proposed general G-quadruplex-forming motif G3N1G3N2–9G3N1G3. A DNA polymerase stop assay was used to investigate the ability of the G-rich strand in Bcl-2 Pu39WT to form intramolecular G-quadruplex structures [49]. Three primer extension arrest sites, which increased in a K+-dependent manner, were observed, which indicates that Bcl-2 Pu39WT has the capability to form three separate but overlapping intramolecular G-quadruplex structures. Subsequent studies using this assay show that the G-quadruplex-interactive agents TMPyP4, Se2SAP, and telomestatin can stabilize the Bcl-2 G-quadruplex structures, with individual selectivity toward one or more of the three G-quadruplex structures [49]. The formation of multiple intramolecular G-quadruplex structures in Bcl-2 Pu39WT was also confirmed by EMSA and DMS footprinting [49].
Figure 8.
(A) Diagram of the promoter region of the Bcl-2 gene. Shown in the inset is the core sequence of the G-quadruplex-forming region in the Bcl-2 promoter. Guanine tracts are underlined. Different groups of five contiguous G-tracts (Bcl-2 3′G4, Bcl-2 MidG4, Bcl-2 5′G4,) within the G-rich strand are indicated with braces [49]. (B) G-quadruplex folding pattern of the Bcl-2 MidG4-G15T/G16T determined by NMR [50].
To identify the guanine tracts involved in the formation of each of the three different Bcl-2 G-quadruplex structures, a mutational analysis of the seven G-tracts in Bcl-2 Pu39WT was carried out. These studies confirm that three different G-quadruplex structures (Bcl-2 3′G4, Bcl-2 MidG4, Bcl-2 3′G4) could form by using three different sets of five consecutive G-tracts within Bcl-2 Pu39WT (Figure 8A). As shown in Figure 8A, the Bcl-2 3′G4 contains five consecutive G-tracts (G8–G10, G11/G12, G13–G17, G18–G20, and G21–G24) at the 3′-end of the G-quadruplex-forming region; the Bcl-2 5′G4 utilizes five consecutive G-tracts at the 5′-end (G1–G4, G5–G7, G8–G10, G11/G12, and G13–G17); and the Bcl-2 MidG4 is constructed using the five internal consecutive G-tracts (G5–G7, G8–G10, G11/G12, G13–G17, and G18–G20) of the Bcl-2 Pu39WT. Based upon the relative amount of K+ required to produce a primer extension arrest, the Bcl-2 MidG4 is the most stable G-quadruplex structure and subsequent DMS footprinting confirmed this [49]. Like Bcl-2 3′G4 and Bcl-2 5′G4, Bcl-2 MidG4 has the ability to form different loop isomers because one of its G-tracts contains five guanines. This was confirmed by studies using a series of dual G-to-T mutation sequences within the five-guanine G-tract in Bcl-2 MidG4. The mutant sequence, which contains G-to-T mutations in the two 5′-end guanines in the five-guanine tract (MidG4-G15T/G16T, sequence shown in Figure 8B), generates the most stable G-quadruplex structure, based on the minimal amount of K+ required to produce a DNA polymerase arrest [49]. Although the Bcl-2 Pu39WT closely resembles the general three-tetrad G-quadruplex-forming motif G3N1G3N2–9G3N1G3, which has been found in c-MYC, Ret, VEGF, HIF1-α, RET, c-Kit21, and KRAS (Figure 2), the CD spectrum of Bcl-2 Pu39WT resembles most closely that of the Tetrahymena telomeric sequence d(T2G4)4 by displaying an absorption maximum at 264 nm and a broad shoulder at 295 nm, which is different from the typical spectra of parallel or antiparallel intramolecular G-quadruplexes represented by c-MYC and TBA sequences respectively (see Figure 3B and reference 53). The mixed parallel/antiparallel G-quadruplex structure of the Tetrahymena telomeric sequence in Na+-containing buffer has been well defined by NMR studies [121]. This suggests that the Bcl-2 Pu39WT forms a mixed parallel/antiparallel G-quadruplex structure.
On the basis of mutational analysis, we proposed that the dual mutant Bcl-2 MidG4-G15T/G16T (Figure 8B) is the ideal NMR candidate to determine the structure of the major G-quadruplex formed in the Bcl-2 promoter. The unambiguous results from the NMR studies demonstrate a novel folding of Bcl-2 MidG4-G15T/G16T into an intramolecular G-quadruplex structure with mixed parallel/antiparallel G-strands [50]. This unique G-quadruplex structure contains three G-tetrads connected with a single-nucleotide double-chain reversal side loop and two lateral loops, and four grooves of different widths (Figure 8B). The loop conformations of the NMR structure fully agree with CD and DMS footprinting data The complementary C-rich strand of the Bcl-2 MidG4 has been examined primarily by molecular absorption and CD and found to form two intramolecular i-motif structures formed maximally at pH 4 and 6 [122]. Although the precise biological function of the G-quadruplexes and i-motifs in the Bcl-2 promoter still need to be explored, we suspect that the presence of these structures within the Bcl-2 promoter play a critical role in regulating gene transcription.
4. Formation of four-tetrad G-quadruplexes in gene promoters
4.1. G-quadruplexes formed in the PDGF-A promoter
Overexpression of PDGF activity has been implicated in the pathogenesis of a number of serious diseases, including cancer, as well as other disorders characterized by excessive cell growth, such as atherosclerosis and various fibrotic conditions [123]. The proximal 5′-flanking region of the human platelet-derived growth factor A (PDGF-A) promoter contains one nuclease hypersensitive element (NHE) (−120 to −33) that accounts for 80% of basal promoter activity [124–126]. This GC-rich region is structurally dynamic and capable of adopting non-B-DNA conformations. The importance of the unwound paranemic structure of the NHE in PDGF-A promoter activity was confirmed by demonstration that a single-stranded oligomer, derived from the G-rich strand (−74 to −51), inhibits the promoter activity of PDGF-A [127]. This G-rich strand contains five runs of contiguous guanines, each run separated by one base (Figure 9A), and this motif has some similarity to that of the G-quadruplex-forming region in the c-MYC promoter. By analogy, the G-rich strand of the PDGF-A NHE has the potential to form a G-quadruplex under physiological conditions. We propose that the equilibrium between the B-form DNA, the single-stranded DNA, and the intramolecular G-quadruplex is instrumental in the determination of the binding of transcription factors to the PDGF-A NHE [56].
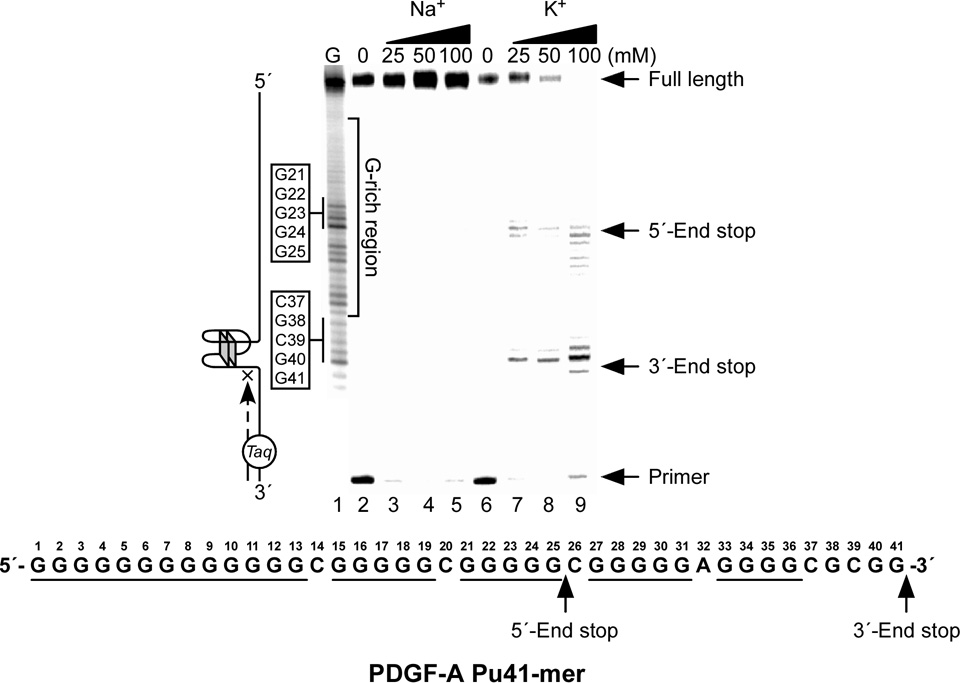
Figure 9.
Effect of K+ and Na+ on the formation of the PDGF-A NHE G-quadruplex in a Taq polymerase stop assay. Two stop products are designated as the 5′-end product and 3′-end product. The corresponding arrest sites are indicated on the core PDGF-A Pu41-mer sequence [56].
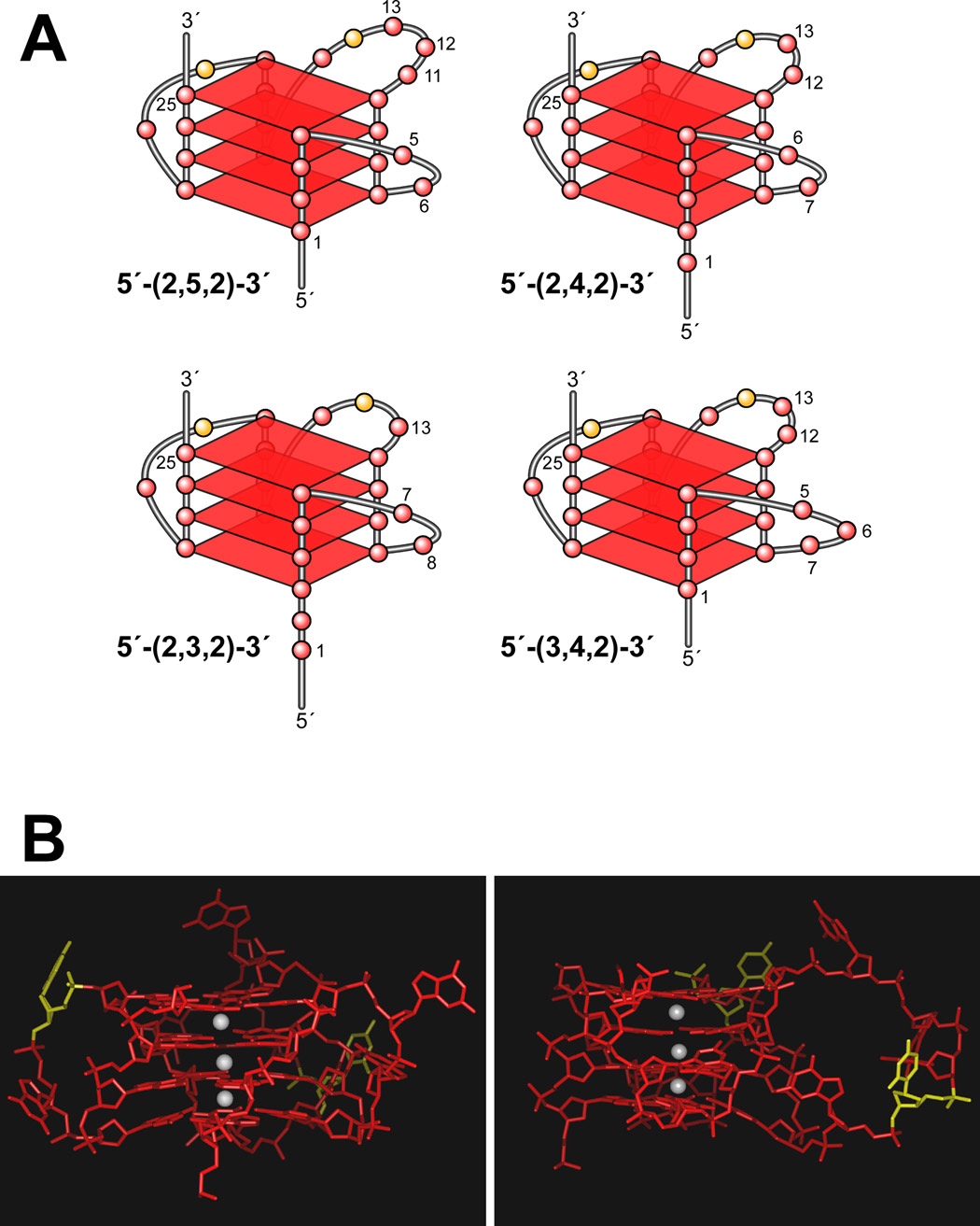
The DNA oligomer PDGF-A Pu41-mer (sequence shown in Figure 9A) represents the whole core sequence of the G-rich strand of the PDGF NHE region and exhibits a CD spectrum characterized by a maximum positive ellipticity at 266 nm, a negative band at 240 nm, and a minor positive band at 212 nm, which is similar to that of the c-MYC G-quadruplex [56]. This suggests that the G-quadruplexes formed by the G-rich strand of the PDGF-A Pu41-mer are parallel-stranded. A DNA polymerase stop assay and EMSA and DMS footprinting studies have shown that the PDGF-A Pu41-mer can form two major intramolecular four-tetrad G-quadruplex structures that are in dynamic equilibrium [56]. As shown in Figure 9, K+ can facilitate the formation of two G-quadruplex structures in the PDGF-A Pu41-mer. One G-quadruplex forms at the 3′-end of the PDGF-A Pu41-mer, while the other forms at the 5′-end. The four-tetrad intramolecular G-quadruplex formed by the three consecutive G-tracts at the 5′-end of the PDGF-A Pu41-mer appears to be the most stable G-quadruplex structure, based on the minimal amount of K+ required to produce DNA polymerase arrest, suggesting that it may be the predominant G-quadruplex structure formed in the PDGF-A promoter (Figure 9) [56].
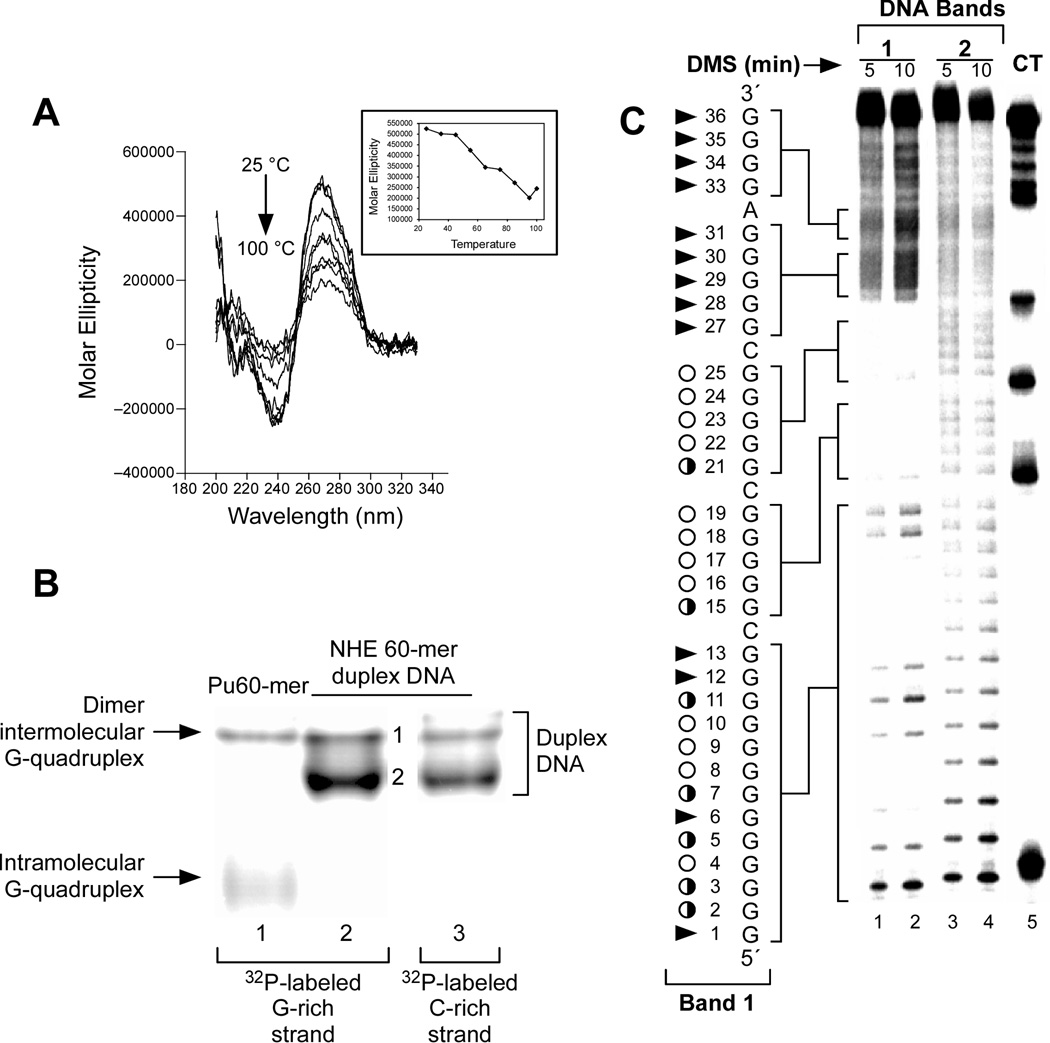
Surprisingly, CD spectroscopy has shown that a stable G-quadruplex can exist within the duplex DNA of PDGF-A NHE, even up to 100 °C, in K+ buffer (Figure 10A). The major G-quadruplex- forming region in the double-stranded DNA of the NHE has been identified by EMSA and DMS footprinting (Figure 10, B and C). This G-quadruplex structure corresponds to that at the 5′-end found in the single-stranded DNA form of the PDGF-A. This is the first example of a G-quadruplex structure that exists in duplex DNA. This unique structure is likely to represent the dominant biologically relevant G-quadruplex structure formed in the duplex region of the PDGF-A promoter, while the complementary C-rich strand forms a single-stranded DNA loop, which may be in equilibrium with an i-motif structure [56]. On the basis of the previous data, the overall folding pattern of the parallel G-quadruplex structure in the PDGF-A NHE duplex DNA has been defined as a G-quadruplex with four tetrads and a two-base internal loop, which is the minimum required to bridge two runs of four guanines. In our molecular models for the PDGF-A G-quadruplex, the 3′-face of this structure has two runs of four guanines, which are separated by a two-base (CG) internal loop; and the opposite 5′-face of this structure is constructed from a run of thirteen contiguous guanines, in which the DMS footprinting pattern predicts a mixture of at least four different double-chain reversal isomers. Each of these loop isomers (Figure 11A) has a different arrangement of nucleotides in the 5′-end double-chain reversal loop and in the large intervening loop. A molecular model of one of the PDGF-A G-quadruplex isomers (loop isomer [a] in Figure 11A) is shown in Figure 11B. To provide further insight into the sequence requirement for the stability of PDGF-A G-quadruplexes, a full mutational analysis of the three consecutive 5′-runs of guanines has been performed. Surprisingly, all the studied oligomer having these guanine mutations, which were originally designed to trap out different stable loop isomers, showed significant destabilization and dramatic changes in the CD spectra, implying the formation of different G-quadruplex structures [56]. Therefore, we suggest that the equilibrium between the different loop isomers in the G-quadruplexes plays a critical role in the overall stabilization of these structures [56].
Figure 10.
(A) Effect of temperature (25–100 °C) on the CD spectra of 90-bp duplex DNA containing the PDGF-A Pu41-mer. At 100 °C, the 90-bp duplex DNA still generates a strong G-quadruplex CD signal. (B) Nondenatured gel analysis of 60-mer G-rich single-stranded DNA containing the PDGF-A Pu41-mer (lane 1) and 60-bp double-stranded DNA, also containing the PDGF-A Pu41-mer (lanes 2 and 3). In lanes 1 and 2, the G-rich strand was 5′-end-radiolabeled with 32P, and the C-rich strand was 5′-end-radiolabeled with 32P in lane 3. (C) DMS footprinting of G-rich strands of 60-bp duplex DNA band 1 (lanes 1 and 2) and band 2 (lanes 3 and 4). Lane 5 shows the CT sequencing on the G-rich strand of the 60-bp duplex DNA. Open circles indicate the guanines that are fully protected, partially open circles indicate the guanines that are partially protected, and arrowheads indicate the guanines that are cleaved [56].
Figure 11.
(A) Proposed folding patterns of the four different loop isomers formed in the core sequence of PDGF-A. (B) Model of the biologically relevant PDGF-A NHE G-quadruplex (loop isomer 5′-(2,5,2)-3′), which contains two 2-base double-chain reversal loops and one 5-base intervening loop (K+ ions = white). For clarity, hydrogen atoms have not been shown. In the left panel, the two 2-base double-chain reversal loops are shown on each side of the model, and in the right panel, the model has been rotated to show the 5-base intervening loop on the right side of the model [56].
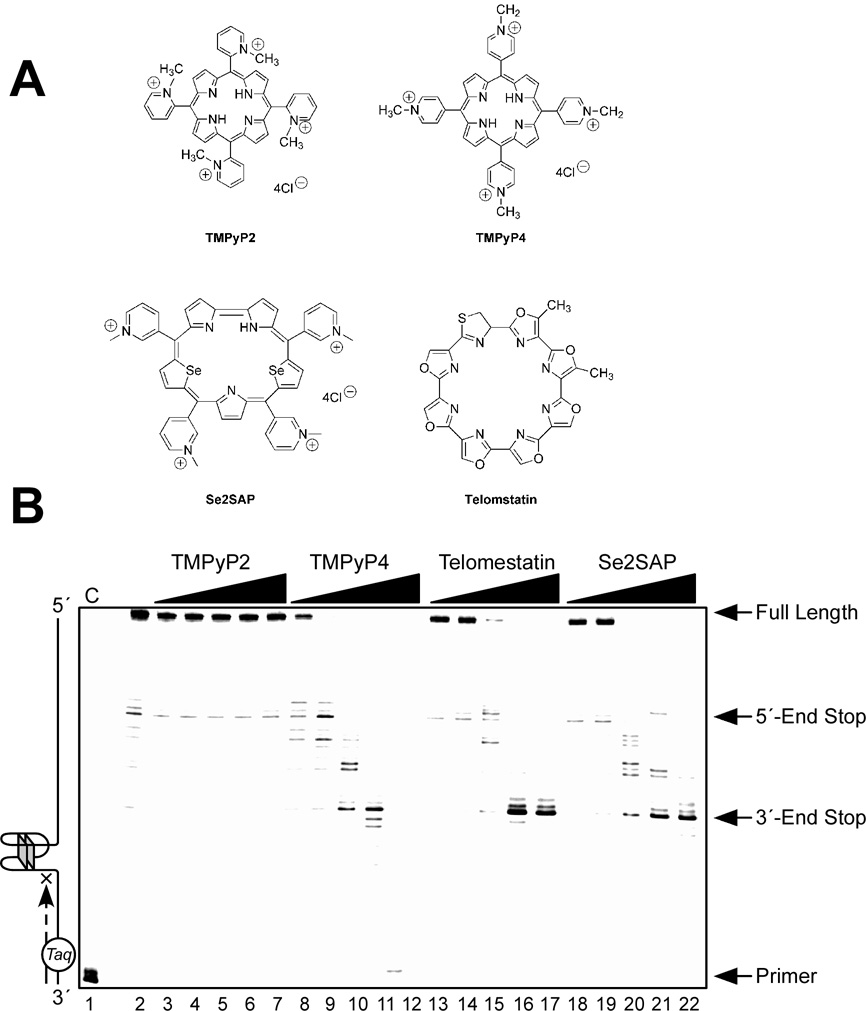
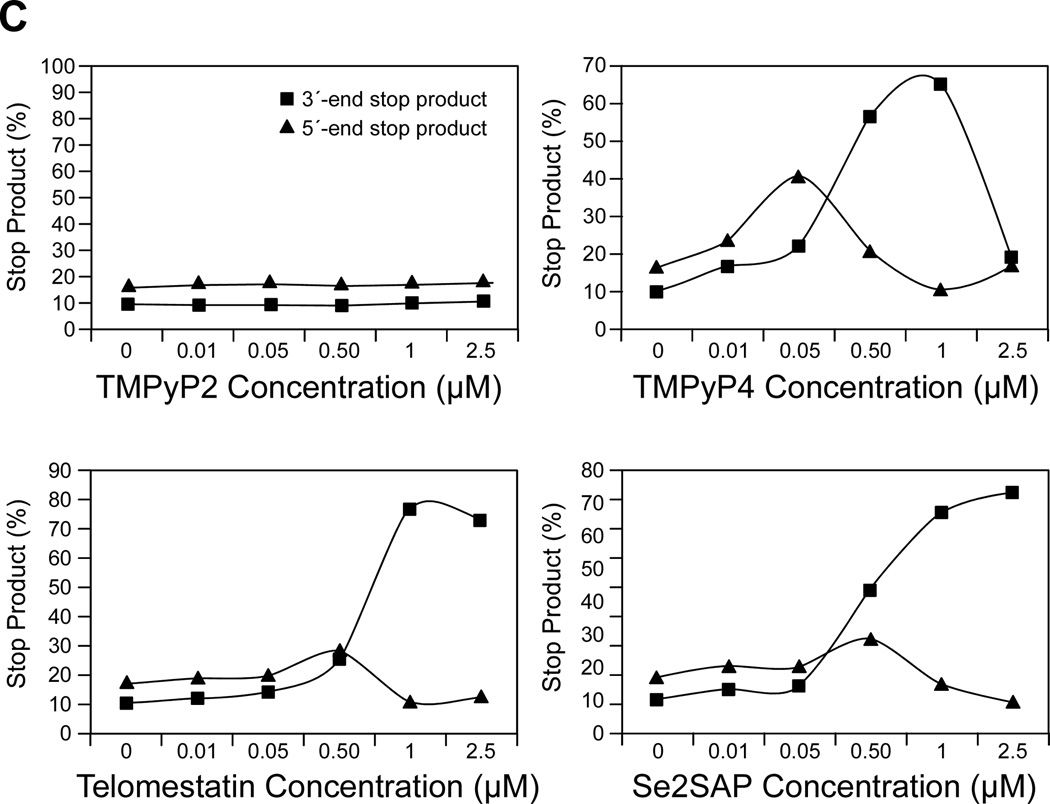
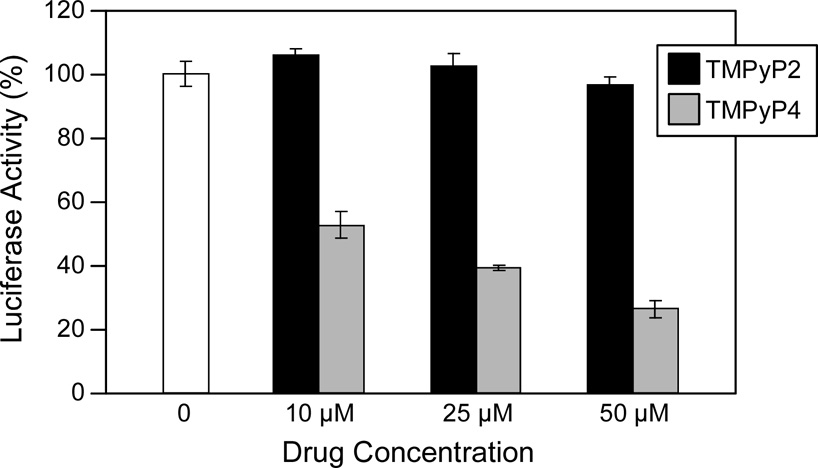
We have examined the affinities of TMPyP4, Se2SAP, and telomestatin for the PDGF-A G-quadruplex (Figure 12A). The results from DNA polymerase stop assays reveal that these three drugs preferentially interact with and stabilize the 5′-end PDFG-A G-quadruplex (Figure 12B). TMPyP4 exhibited stronger selectivity than either Se2SAP or telomestatin for stabilization of the 5′-end stop product (Figure 12C). Thus, TMPyP4 appears to be an ideal candidate for modulating PDGF-A transcription. In transfection experiments, 10 µM TMPyP4 reduced the activity of the basal promoter of PDGF-A by about 40%, relative to the control, while TMPyP2 did not have a significant effect on the PDGF-A promoter activity, indicating that G-quadruplex structures can form in the PDGF-A NHE in living cells (Figure 12D). On the basis of our studies, we established that ligand-mediated stabilization of G-quadruplex structures within the PDGF-A NHE can silence PDGF-A expression [56].
Figure 12.
(A) Structures of the G-quadruplex-interactive compounds TMPyP4, telomestatin, and Se2SAP, and the control compound TMPyP2. (B) The Taq polymerase stop assay was used to compare the stabilization of the PDGF-A G-quadruplexes by TMPyP2 (lanes 3–7), TMPyP4 (lanes 8–12), telomestatin (lanes 13–17), and Se2SAP (lanes 18–22) by using increasing concentrations of drugs (0.01, 0.05, 0.5, 1, and 2.5 µM) at 60 °C. Lane 1 is control and lane 2 is without drug. (C) The ratios of the major arrest products of each sample to the total product were plotted against drug concentrations. (D) Dual luciferase assay to determine the effect of TMPyP4 and TMPyP2 on the transcriptional activity of PDGF-A basal promoter containing the NHE. The comparative firefly luciferase expressions (firefly/renilla) of TMPyP2 and TMPyP4 are shown in the histograms [56].
5. Formation of two-tetrad G-quadruplexes downstream of gene promoters
5.1. G-quadruplexes formed in the c-Myb promoter
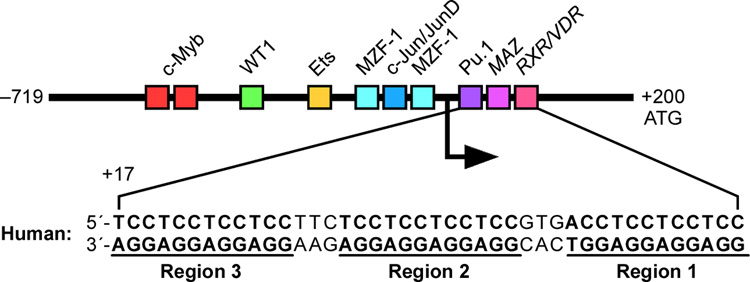
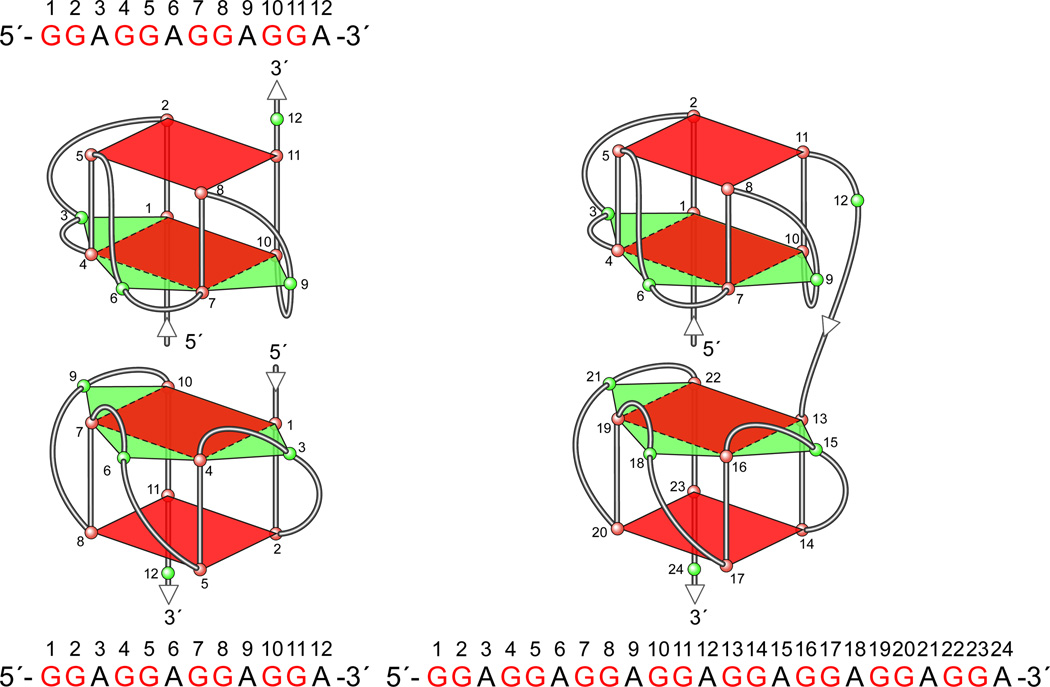
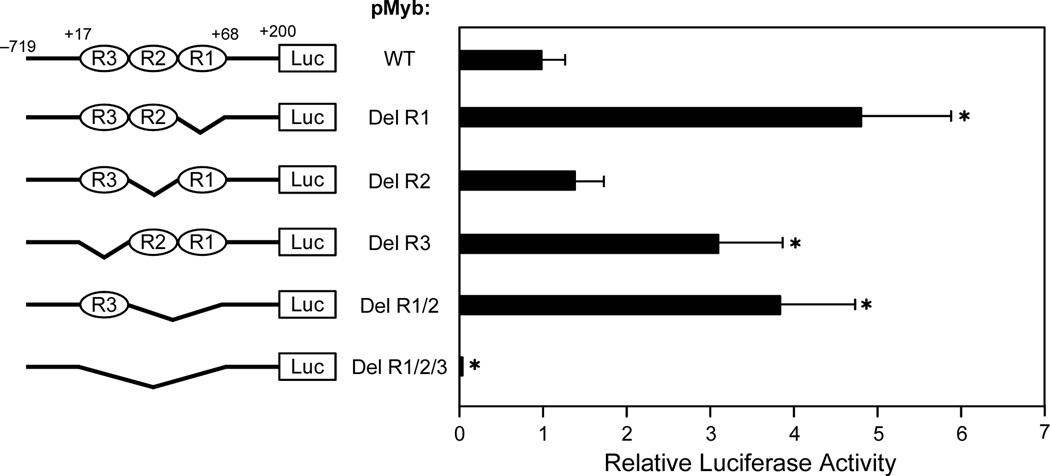
The proto-oncoprotein c-Myb is a critical transcriptional factor for proliferation, differentiation, and survival of hematopoietic progenitor cells [128]. The overexpression of c-Myb has been observed in many leukemias and some solid tumors [129–131]. c-Myb transcription is under the strict control of several transcription factors and cis-elements in the gene promoter. The human c-Myb promoter contains a GC-rich region with three imperfect copies of (GGA)4 located 17 base pairs downstream of the transcription initiation site (Figure 13A) [57]. This region plays a crucial role in regulating c-Myb transcription. A DNA secondary structure of d(GGAGGAGGAGGA), containing four tandem repeats of a GGA triplet sequence, has been determined under physiological K+ conditions by NMR [92,93]. This sequence folds into an intramolecular DNA secondary structure composed of a guanine tetrad (T) and a G(:A):G(:A):G(:A):G heptad (H), with four parallel G–G segments [92] (Figure 13B, top left structure). Two d(GGA)4 sequences can form an intermolecular G-quadruplex consisting of a dimer tetrad:heptad:heptad:tetrad (T:H:H:T) stabilized through a stacking interaction between the heptads (Figure 13B, left), and a d(GGA)8 forms an intramolecular T:H:H:T structure (Figure 13B, right) [93]. By sequence analogy, the G-rich strand of the GGA repeat sequence in the c-Myb promoter has the potential to form several different T:H:H:T G-quadruplex structures. DMS footprinting and CD spectroscopy analysis confirm that the intramolecular T:H:H:T G-quadruplex structure is formed within the GGA repeat region of the c-Myb promoter, and any two of the three GGAGGAGGAGG repeats can form this dimer structure (T:H:H:T) [57]. Mutational studies on the (GGA)4 repeats in combination with a DNA polymerase stop assay show that there are three stable intramolecular T:H:H:T G-quadruplexes formed by the whole core (GGA)4 repeat sequence of the c-Myb promoter [57]. This most-preferred intramolecular T:H:H:T G-quadruplex structure is formed with two consecutive 5′-end (GGA)4 repeats. On the basis of luciferase reporter studies, deletion of one or two (GGA)4 repeats increases c-Myb promoter activity (Figure 13C) [57]. However, deletion of all three (GGA)4 repeats results in complete abrogation of c-Myb promoter activity, indicating the dual roles of the c-Myb (GGA)4 repeats as both a transcriptional repressor and an activator [57]. It is speculated that equilibrium between transcription factor binding and T:H:H:T G-quadruplex formation plays a critical role in the regulation of c-Myb transcription. An analysis of the c-Myb (GGA)4 repeats reveals several consensus binding sites for the Myc-associated zinc finger protein (MAZ). EMSA and DNase I footprinting studies confirm that MAZ can bind to both double-stranded DNA and the intramolecular T:H:H:T G-quadruplexes in the c-Myb promoter [57]. The transfection of MAZ expression plasmid and forced overexpression of MAZ in cells result in a decrease of c-Myb promoter activity, indicating that MAZ is a repressor of the c-Myb promoter. However, it is still not clear if MAZ functions as a transcription factor by binding to duplex DNA or to the G-quadruplex structures [57].
Figure 13.
(A) Promoter structure of the c-Myb gene and the location of key transcription factors; shown in the inset is the sequence of three (GGA)4 repeats downstream of the transcriptional initiation site of the c-Myb promoter. (B) The intermolecular and intramolecular T:H:H:T G-quadruplexes formed by d(GGA)4 and d(GGA)8. (C) Luciferase activity driven by the wild-type and GGA-deleted c-Myb promoter constructs in CCRF-CEM cells. Deletion of R1, R2, R3, or both R1 and R2 from the c-myb promoter increases luciferase activity in CCRT-CEM cells. The R1, R2, and R3 deletion mutant pMybDelR1/2/3 markedly reduces luciferase activity (* P value < 0.01) [57].
Conclusions
In this review we have described the diversity of G-quadruplex structures formed in the promoter regions of a number of important oncogenes. These intramolecular G-quadruplexes consist of two, three, or four tetrads, but in the most part form parallel G-quadruplexes in which at least one of the faces is made up of three tetrads with a one-base double-chain reversal loop that provides considerable thermal stability, which is a hallmark of promoter G-quadruplexes. In all the cases described here, the region that forms the G-quadruplexes has been shown to have importance in the control of gene expression. In one case (VEGF), a requirement for superhelicity and the precise sequence for formation of a stable G-quadruplex has been shown to be necessary for G-quadruplex formation in plasmid DNA. In a second case (PDGF-A), the G-quadruplex can coexist within a duplex region. Both of these examples are important in validating the role of G-quadruplex in the control of gene expression. Finally, direct evidence that small molecules which target these G-quadruplexes in the case of c-Myc can repress gene expression is critical for proof of concept that this is a valid approach to modulation of gene expression.
Bioinformatic studies showing a concentration of putative G-quadruplex-forming elements either upstream or downstream of the transcription start site provide additional support for the biological relevance of these structures in transcriptional control. Our observation that these sequences conform in large part to conserved sequences (Figure 2A) further supports this premise. A recent paper [132], which “challenges” the idea that G-quadruplex-forming sequences upstream of the transcription start site are likely to be involved in transcriptional control based on a predominance of methylation sites and Sp1 binding sites in this region, appears to us largely to disregard the evidence presented in papers published by us and groups in Europe and Asia that are discussed in this review.
Most often these G-quadruplexes exist as a mixture of loop isomers that can generally be trapped out by appropriate substitution of loop guanines with other bases. The region of most diversity is in the central loop (see Figure 2A), where both loop size (2–9 bases) and composition are different in the examples so far examined. Adenines and guanines predominate within this loop, perhaps reflecting the need for cytosines to form i-motif structures on the opposite strand. c-Myb is an example of a G-quadruplex structure where two tetrads suffice for stability. The heptad structure and its dimer formation explain the stability of Myb in the double-chain reversal. One of the c-Kit G-quadruplexes is the most surprising, containing an isolated guanine together with a two-guanine tract to form one side of the tetrad. Last, for PDGF-A, where a four-tetrad G-quadruplex replaces the three-tetrad G-quadruplex, a two-base double-chain reversal loop is required.
Those G-quadruplexes so far examined appear to act as silencer elements in the promoter regions, and therefore stabilization should lead to inhibition of oncogene expression, which has been most clearly shown for c-Myc. In the case of RET, the folding pattern of the accompanying i-motif has been inferred from footprinting and base substitution studies.
The publication of further examples will help define the requirements for stability of these G-quadruplexes in promoter regions, and subsequent examples of ligands that bind to these structures and inhibit gene expression will further validate the concept that drug modulation of gene expression can be achieved by targeting these structures. In a recent international meeting on G-quadruplexes held in Louisville, Kentucky (April 21–24, 2007), scientists from Cylene Pharmaceuticals reported that a first-in-class G-quadruplex-interactive compound, aptly named Quarfloxin, is in phase II clinical trials [133]. This compound, a fluoroquinolone derived from QQ58 [134], is proposed to bind to G-quadruplexes in rDNA, which displaces nucleolin from its binding sites on the non-template strand.
The biggest challenge to be faced by the G-quadruplex community involved in drug targeting will be specificity. However, in this regard we are at the same point as in the early development of kinase inhibitors, where similar reservations were expressed. We believe that this challenge will be successfully met and G-quadruplex-interactive compounds will assume their place in molecular therapeutics. The specificity challenge is really at two levels, the first being discrimination between G-quadruplexes and other forms of DNA, most notably duplex DNA. Compounds that interact with G-quadruplexes generally do so through binding to external faces of the quadruplexes in pockets created by adjacent bases, which may provide capping structures. This is loosely analogous to intercalation between bases pairs in duplex, but both the electronic and steric constraints are quite different. For example, intercalation between base pairs is expected to be much more constrained by rigid features of a dinucleotide base pair than by flexible capping structures that are likely to exist in the G-quadruplex structures. Moreover, the positively charged center of a tetrad and its size and shape are quite different to adjacent base pairs. The second level of selectivity is between different G-quadruplexes, which itself has two levels of discrimination, between different folding patterns, e.g., c-MYC, Bcl-2, and c-kit87up, and between G-quadruplexes having the same folding pattern, e.g., c-MYC, VEGF, Hif-1α, RET, and KRAS. For the former, the different juxtaposition of the loops to the external tetrads should provide opportunities for discrimination, whereas in the latter the heterogeneity of the central loop (see Figure 2) is a potential opportunity. Consequently, the design strategy to develop selective agents should be in two steps: first, consideration of the “intercalation” binding pocket and its steric and electronic characteristics, and second, placement of side chains to achieve selectivity through interactions with the loops and different sized grooves in the stacked tetrads.
Acknowledgments
This research has been supported by grants from the National Institutes of Health (CA94166, CA95060) and the Arizona Biomedical Research Commission (9006). We thank our coworkers, who provided the bulk of the examples described here. Finally, we are grateful to David Bishop for preparing, proofreading, and editing the final versions of the text and figures.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Watson JD, Crick FHC. Molecular structure of nucleic acids: a structure for deoxyribose nucleic acid. Nature. 1953;171:737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- 2.Wells RD. Unusual DNA structures. J Biol Chem. 1988;263:1095–1098. [PubMed] [Google Scholar]
- 3.Wells RD. Non-B DNA conformations, mutagenesis and disease. Trends Biochem Sci. 2007;32:271–278. doi: 10.1016/j.tibs.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 4.Raghavan SC, Lieber MR. DNA structure and human diseases. Front Biosci. 2007;12:4402–4408. doi: 10.2741/2397. [DOI] [PubMed] [Google Scholar]
- 5.Bacolla A, Wells RD. Non-B DNA conformations, genomic rearrangements, and human disease. J Biol Chem. 2004;279:47411–47414. doi: 10.1074/jbc.R400028200. [DOI] [PubMed] [Google Scholar]
- 6.Majumdar A, Patel DJ. Identifying hydrogen bond alignments in multistranded DNA architectures by NMR. Acc Chem Res. 2002;35:1–11. doi: 10.1021/ar010097+. [DOI] [PubMed] [Google Scholar]
- 7.Wells RD, Blakesley RW, Hardies SC, Horn GT, Larson JE, Selsing E, Burd JF, Chan HW, Dodgson JB, Jensen KF, Nes IF, Wartell RM. The role of DNA structure in genetic regulation. CRC Crit Rev Biochem. 1977;4:305–340. doi: 10.3109/10409237709102561. [DOI] [PubMed] [Google Scholar]
- 8.Olivier M, Aggarwal A, Allen J, Almendras AA, Bajorek ES, Beasley EM, Brady SD, Bushard JM, Bustos VI, Chu A, Chung TR, De Witte A, Denys ME, Dominguez R, Fang NY, Foster BD, Freudenberg RW, Hadley D, Hamilton LR, Jeffrey TJ, Kelly L, Lazzeroni L, Levy MR, Lewis SC, Liu X, Lopez FJ, Louie B, Marquis JP, Martinez RA, Matsuura MK, Misherghi NS, Norton JA, Olshen A, Perkins SM, Perou AJ, Piercy C, Piercy M, Qin F, Reif T, Sheppard K, Shokoohi V, Smick GA, Sun WL, Stewart EA, Fernando J, Tejeda, Tran NM, Trejo T, Vo NT, Yan SC, Zierten DL, Zhao S, Sachidanandam R, Trask BJ, Myers RM, Cox DR. A high-resolution radiation hybrid map of the human genome draft sequence. Science. 2001;291:161298–161302. doi: 10.1126/science.1057437. [DOI] [PubMed] [Google Scholar]
- 9.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann N, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M, Gibbs RA, Muzny DM, Scherer SE, Bouck JB, Sodergren EJ, Worley KC, Rives CM, Gorrell JH, Metzker ML, Naylor SL, Kucherlapati RS, Nelson DL, Weinstock GM, Sakaki Y, Fujiyama A, Hattori M, Yada T, Toyoda A, Itoh T, Kawagoe C, Watanabe H, Totoki Y, Taylor T, Weissenbach J, Heilig R, Saurin W, Artiguenave F, Brottier P, Bruls T, Pelletier E, Robert C, Wincker P, Smith DR, Doucette-Stamm L, Rubenfield M, Weinstock K, Lee HM, Dubois J, Rosenthal A, Platzer M, Nyakatura G, Taudien S, Rump A, Yang H, Yu J, Wang J, Huang G, Gu J, Hood L, Rowen L, Madan A, Qin S, Davis RW, Federspiel NA, Abola AP, Proctor MJ, Myers RM, Schmutz J, Dickson M, Grimwood J, Cox DR, Olson MV, Kaul R, Raymond C, Shimizu N, Kawasaki K, Minoshima S, Evans GA, Athanasiou M, Schultz R, Roe BA, Chen F, Pan H, Ramser J, Lehrach H, Reinhardt R, McCombie WR, de la Bastide M, Dedhia N, Blöcker H, Hornischer K, Nordsiek G, Agarwala R, Aravind L, Bailey JA, Bateman A, Batzoglou S, Birney E, Bork P, Brown DG, Burge CB, Cerutti L, Chen HC, Church D, Clamp M, Copley RR, Doerks T, Eddy SR, Eichler EE, Furey TS, Galagan J, Gilbert JG, Harmon C, Hayashizaki Y, Haussler D, Hermjakob H, Hokamp K, Jang W, Johnson LS, Jones TA, Kasif S, Kaspryzk A, Kennedy S, Kent WJ, Kitts P, Koonin EV, Korf I, Kulp D, Lancet D, Lowe TM, McLysaght A, Mikkelsen T, Moran JV, Mulder N, Pollara VJ, Ponting CP, Schuler G, Schultz J, Slater G, Smit AF, Stupka E, Szustakowski J, Thierry-Mieg D, Thierry-Mieg J, Wagner L, Wallis J, Wheeler R, Williams A, Wolf YI, Wolfe KH, Yang SP, Yeh RF, Collins F, Guyer MS, Peterson J, Felsenfeld A, Wetterstrand KA, Patrinos A, Morgan MJ, de Jong P, Catanese JJ, Osoegawa K, Shizuya H, Choi S, Chen YJ. International Human Genome Sequencing Consortium, Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 10.McPherson JD, Marra M, Hillier L, Waterston RH, Chinwalla A, Wallis J, Sekhon M, Wylie K, Mardis ER, Wilson RK, Fulton R, Kucaba TA, Wagner-McPherson C, Barbazuk WB, Gregory SG, Humphray SJ, French L, Evans RS, Bethel G, Whittaker A, Holden JL, McCann OT, Dunham A, Soderlund C, Scott CE, Bentley DR, Schuler G, Chen HC, Jang W, Green ED, Idol JR, Maduro VV, Montgomery KT, Lee E, Miller A, Emerling S, Kucherlapati , Gibbs R, Scherer S, Gorrell JH, Sodergren E, Clerc-Blankenburg K, Tabor P, Naylor S, Garcia D, de Jong PJ, Catanese JJ, Nowak N, Osoegawa K, Qin S, Rowen L, Madan A, Dors M, Hood L, Trask B, Friedman C, Massa H, Cheung VG, Kirsch IR, Reid T, Yonescu R, Weissenbach J, Bruls T, Heilig R, Branscomb E, Olsen A, Doggett N, Cheng JF, Hawkins T, Myers RM, Shang J, Ramirez L, Schmutz J, Velasquez O, Dixon K, Stone NE, Cox DR, Haussler D, Kent WJ, Furey T, Rogic S, Kennedy S, Jones S, Rosenthal A, Wen G, Schilhabel M, Gloeckner G, Nyakatura G, Siebert R, Schlegelberger B, Korenberg J, Chen XN, Fujiyama A, Hattori M, Toyoda A, Yada T, Park HS, Sakaki Y, Shimizu N, Asakawa S, Kawasaki K, Sasaki T, Shintani A, Shimizu A, Shibuya K, Kudoh J, Minoshima S, Ramser J, Seranski P, Hoff C, Poustka A, Reinhardt R, Lehrach H. International Human Genome Mapping Consortium. Nature. 2001;409:934–941. doi: 10.1038/35057157. [DOI] [PubMed] [Google Scholar]
- 11.Lander ES, Linton LM, Birren B, Nusbaum C. International Human Genome Sequencing Consortium, Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 12.McConkey EH, Varki A. Genomics, thoughts on the future of great ape research. Science. 2005;309:1499–1501. doi: 10.1126/science.1113863. [DOI] [PubMed] [Google Scholar]
- 13.Eddy J, Maizels N. Gene function correlates with potential for G4 DNA formation in the human genome. Nucleic Acids Res. 2006;34:3887–3896. doi: 10.1093/nar/gkl529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huppert JL, S. Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huppert JL, Balasubramanian S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007;35:406–413. doi: 10.1093/nar/gkl1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Todd AK, Johnston M, Neidle S. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005;33:2901–2907. doi: 10.1093/nar/gki553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang R, Lin Y, Zhang CT. Greglist: a database listing potential G-quadruplex regulated genes. Nucleic Acids Res. 2008;36(Database issue):D372–D376. doi: 10.1093/nar/gkm787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henderson E, Hardin CC, Walk SK, Tinoco I, Jr, Blackburn EH. Telomeric DNA oligonucleotides form novel intramolecular structures containing guanine-guanine base pairs. Cell. 1987;51:899–908. doi: 10.1016/0092-8674(87)90577-0. [DOI] [PubMed] [Google Scholar]
- 19.Williamson JR, Raghuraman MK, Cech TR. Monovalent cation-induced structure of telomeric DNA: the G-quartet model. Cell. 1989;59:871–880. doi: 10.1016/0092-8674(89)90610-7. [DOI] [PubMed] [Google Scholar]
- 20.Gellert M, Lipsett MN, Davies DR. Helix formation by guanylic acid. Proc Natl Acad Sci USA. 1962;48:2013–2018. doi: 10.1073/pnas.48.12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guschlbauer W, Chantot JF, Thiele D. Four-stranded nucleic acid structures 25 years later: from guanosine gels to telomer DNA. J Biomol Struct Dyn. 1990;8:491–511. doi: 10.1080/07391102.1990.10507825. [DOI] [PubMed] [Google Scholar]
- 22.Vialas C, Pratviel G, Meunier B. Oxidative damage generated by an oxo-metalloporphyrin onto the human telomeric sequence. Biochemistry. 2000;39:9514–9522. doi: 10.1021/bi000743x. [DOI] [PubMed] [Google Scholar]
- 23.Simonsson T. G-quadruplex DNA structures—variations on a theme. Biol Chem. 2001;382:621–628. doi: 10.1515/BC.2001.073. [DOI] [PubMed] [Google Scholar]
- 24.Gilbert DE, Feigon J. Multistranded DNA structures. Curr Opin Struct Biol. 1999;9:305–314. doi: 10.1016/S0959-440X(99)80041-4. [DOI] [PubMed] [Google Scholar]
- 25.Keniry MA. Quadruplex structures in nucleic acids. Biopolymers. 2000–2001;56:123–146. doi: 10.1002/1097-0282(2000/2001)56:3<123::AID-BIP10010>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 26.Burge S, Parkinson GN, Hazel P, Todd AK, Neidle S. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 2006;34:5402–5415. doi: 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davis JT. G-quartets 40 years later: from 5′-GMP to molecular biology and supramolecular chemistry. Angew Chem Int Ed Engl. 2004;43:668–698. doi: 10.1002/anie.200300589. [DOI] [PubMed] [Google Scholar]
- 28.Arthanari H, Bolton PH. Functional and dysfunctional roles of quadruplex DNA in cells. Chem Biol. 2001;8:221–230. doi: 10.1016/s1074-5521(01)00007-2. [DOI] [PubMed] [Google Scholar]
- 29.Shafer RH, Smirnov I. Biological aspects of DNA/RNA quadruplexes. Biopolymers. 2000–2001;56:209–227. doi: 10.1002/1097-0282(2000/2001)56:3<209::AID-BIP10018>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 30.Sen D, Gilbert W. A sodium-potassium switch in the formation of four-stranded G4-DNA. Nature. 1990;344:410–414. doi: 10.1038/344410a0. [DOI] [PubMed] [Google Scholar]
- 31.Hud NV, Smith FW, Anet FA, Feigon J. The selectivity for K+ versus Na in DNA quadruplexes is dominated by relative free energies of hydration: a thermodynamic analysis by 1H NMR. Biochemistry. 1996;35:15383–15390. doi: 10.1021/bi9620565. [DOI] [PubMed] [Google Scholar]
- 32.Xu Q, Deng H, Braunlin WH. Selective localization and rotational immobilization of univalent cations on quadruplex DNA. Biochemistry. 1993;32:13130–13137. doi: 10.1021/bi00211a023. [DOI] [PubMed] [Google Scholar]
- 33.Alberts B. Molecular Biological of the Cell. 4th ed. Garland, New York: 2002. [Google Scholar]
- 34.Sten-Knudsen O. Biological membranes: theory of transport, potentials and electric impulses. Cambridge University Press; 2002. pp. 396–397. [Google Scholar]
- 35.Schaffitzel C, Berger I, Postberg J, Hanes J, Lipps HJ, Plückthun A. In vitro generated antibodies specific for telomeric guanine-quadruplex DNA react with Stylonychia lemnae macronuclei. Proc Natl Acad Sci USA. 2001;98:8572–8577. doi: 10.1073/pnas.141229498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Macaya RF, Schultze P, Smith FW, Roe JA, Feigon J. Thrombin-binding DNA aptamer forms a unimolecular quadruplex structure in solution. Proc Natl Acad Sci USA. 1993;90:3745–3749. doi: 10.1073/pnas.90.8.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walsh K, Gualberto A. MyoD binds to the guanine tetrad nucleic acid structure. J Biol Chem. 1992;267:13714–13718. [PubMed] [Google Scholar]
- 38.Schierer T, Henderson E. A protein from Tetrahymena thermophila that specifically binds parallel-stranded G4-DNA. Biochemistry. 1994;33:2240–2246. doi: 10.1021/bi00174a034. [DOI] [PubMed] [Google Scholar]
- 39.Williamson JR. G-quartet structures in telomeric DNA. Annu Rev Biophys Biomol Struct. 1994;23:703–730. doi: 10.1146/annurev.bb.23.060194.003415. [DOI] [PubMed] [Google Scholar]
- 40.Fry M. Tetraplex DNA and its interacting proteins. Front Biosci. 2007;12:4336–4351. doi: 10.2741/2391. [DOI] [PubMed] [Google Scholar]
- 41.Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc Natl Acad Sci USA. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hurley LH, Von Hoff DD, Siddiqui-Jain A, Yang D. Drug targeting of the c-MYC promoter to repress gene expression via a G-quadruplex silencer element. Semin Oncol. 2006;33:498–512. doi: 10.1053/j.seminoncol.2006.04.012. [DOI] [PubMed] [Google Scholar]
- 43.Yang D, Hurley LH. Structure of the biologically relevant G-quadruplex in the c-MYC promoter. Nucleosides Nucleotides Nucleic Acids. 2006;25:951–968. doi: 10.1080/15257770600809913. [DOI] [PubMed] [Google Scholar]
- 44.Seenisamy J, Rezler EM, Powell TJ, Tye D, Gokhale V, Joshi CS, Siddiqui-Jain A, Hurley LH. The dynamic character of the G-quadruplex element in the c-MYC promoter and modification by TMPyP4. J Am Chem Soc. 2004;126:8702–8709. doi: 10.1021/ja040022b. [DOI] [PubMed] [Google Scholar]
- 45.Sun D, Guo K, Rusche JJ, Hurley LH. Facilitation of a structural transition in the polypurine/polypyrimidine tract within the proximal promoter region of the human VEGF gene by the presence of potassium and G-quadruplex-interactive agents. Nucleic Acids Res. 2005;33:6070–6080. doi: 10.1093/nar/gki917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.De Armond R, Wood S, Sun D, Hurley LH, Ebbinghaus SW. Evidence for the presence of a guanine quadruplex forming region within a polypurine tract of the hypoxia inducible factor 1alpha promoter. Biochemistry. 2005;44:16341–16350. doi: 10.1021/bi051618u. [DOI] [PubMed] [Google Scholar]
- 47.Guo K, Pourpak A, Beetz-Rogers K, Gokhale V, Sun D, Hurley LH. Formation of pseudosymmetrical G-quadruplex and i-motif structures in the proximal promoter region of the RET oncogene. J Am Chem Soc. 2007;129:10220–10228. doi: 10.1021/ja072185g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cogoi S, Xodo LE. G-quadruplex formation within the promoter of the KRAS proto-oncogene and its effect on transcription. Nucleic Acids Res. 2006;34:2536–2549. doi: 10.1093/nar/gkl286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dexheimer TS, Sun D, Hurley LH. Deconvoluting the structural and drug-recognition complexity of the G-quadruplex-forming region upstream of the bcl-2 P1 promoter. J Am Chem Soc. 2006;128:5404–5415. doi: 10.1021/ja0563861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dai J, Chen D, Jones RA, Hurley LH, Yang D. NMR solution structure of the major G-quadruplex structure formed in the human BCL2 promoter region. Nucleic Acids Res. 2006;34:5133–5144. doi: 10.1093/nar/gkl610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rankin S, Reszka AP, Huppert J, Zloh M, Parkinson GN, Todd AK, Ladame S, Balasubramanian S, Neidle S. Putative DNA quadruplex formation within the human c-kit oncogene. J Am Chem Soc. 2005;127:10584–10589. doi: 10.1021/ja050823u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fernando H, Reszka AP, Huppert J, Ladame S, Rankin S, Venkitaraman AR, Neidle S, Balasubramanian S. A conserved quadruplex motif located in a transcription activation site of the human c-kit oncogene. Biochemistry. 2006;45:7854–7860. doi: 10.1021/bi0601510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Phan AT, Kuryavyi V, Burge S, Neidle S, Patel DJ. Structure of an unprecedented G-quadruplex scaffold in the human c-kit promoter. J Am Chem Soc. 2007;129:4386–4392. doi: 10.1021/ja068739h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shirude PS, Okumus B, Ying L, Ha T, Balasubramanian S. Single-molecule conformational analysis of G-quadruplex formation in the promoter DNA duplex of the proto-oncogene c-kit. J Am Chem Soc. 2007;129:7484–7485. doi: 10.1021/ja070497d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Todd AK, Haider SM, Parkinson GN, Neidle S. Sequence occurrence and structural uniqueness of a G-quadruplex in the human c-kit promoter. Nucleic Acids Res. 2007;35:5799–5808. doi: 10.1093/nar/gkm609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Qin Y, Rezler EM, Gokhale V, Sun D, Hurley LH. Characterization of the G-quadruplexes in the duplex nuclease hypersensitive element of the PDGF-A promoter and modulation of PDGF-A promoter activity by TMPyP4. Nucleic Acids Res. 2007;25:7698–7713. doi: 10.1093/nar/gkm538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee Palumbo S, Memmott RM, Uribe DJ, Krotova-Khan Y, Hurley LH, Ebbinghaus SW. A novel G-quadruplex forming GGA repeat region in the c-myb promoter is a critical regulator of promoter activity. Nucleic Acids Res. 2008 doi: 10.1093/nar/gkm1069. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Simonsson T, Pribylova M, Vorlickova M. A nuclease hypersensitive element in the human c-myc promoter adopts several distinct i-tetraplex structures. Biochem Biophys Res Commun. 2000;278:158–166. doi: 10.1006/bbrc.2000.3783. [DOI] [PubMed] [Google Scholar]
- 59.Murchie AI, Lilley DM. Retinoblastoma susceptibility genes contain 5′ sequences with a high propensity to form guanine-tetrad structures. Nucleic Acids Res. 1992;20:49–53. doi: 10.1093/nar/20.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xu Y, Sugiyama H. Highly efficient photochemical 2′-deoxyribonolactone formation at the diagonal loop of a 5-iodouracil-containing antiparallel G-quartet. J Am Chem Soc. 2004;126:6274–6279. doi: 10.1021/ja031942h. [DOI] [PubMed] [Google Scholar]
- 61.Xu Y, Sugiyama H. Structural and functional characterizations of the G-quartet and i-motif elements in retinoblastoma susceptibility genes (Rb) Nucleic Acids Symp Ser. 2005;49:177–178. doi: 10.1093/nass/49.1.177. [DOI] [PubMed] [Google Scholar]
- 62.Xu Y, Sugiyama H. Formation of the G-quadruplex and i-motif structures in retinoblastoma susceptibility genes (Rb) Nucleic Acids Res. 2006;34:949–954. doi: 10.1093/nar/gkj485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shklover J, Etzioni S, Weisman-Shomer P, Yafe A, Bengal E, Fry M. MyoD uses overlapping but distinct elements to bind E-box and tetraplex structures of regulatory sequences of muscle-specific genes. Nucleic Acids Res. 2007;35:7087–7095. doi: 10.1093/nar/gkm746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dexheimer TS, Sun D, Fry M, Hurley LH. DNA quadruplexes and gene regulation. In: Neidle S, editor. Quadruplex Nucleic Acids. Cambridge: Royal Society of Chemistry, RSCPublishing; 2006. pp. 180–207. [Google Scholar]
- 65.Risitano A, Fox KR. Influence of loop size on the stability of intramolecular DNA quadruplexes. Nucleic Acids Res. 2004;32:2598–2606. doi: 10.1093/nar/gkh598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bugaut A, Balasubramanian S. A sequence-independent study of the influence of short loop lengths on the stability and topology of intramolecular DNA G-quadruplexes. Biochemistry. 2008;47:689–697. doi: 10.1021/bi701873c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Marcu KB, Bossone SA, Patel AJ. Myc function and regulation. Annu Rev Biochem. 1992;61:809–860. doi: 10.1146/annurev.bi.61.070192.004113. [DOI] [PubMed] [Google Scholar]
- 68.Pelengaris S, Rudolph B, Littlewood T. Action of Myc in vivo—proliferation and apoptosis. Curr Opin Genet Dev. 2000;10:100–105. doi: 10.1016/s0959-437x(99)00046-5. [DOI] [PubMed] [Google Scholar]
- 69.Pelengaris S, Khan M. The many faces of c-MYC. Arch Biochem Biophys. 2003;416:129–136. doi: 10.1016/s0003-9861(03)00294-7. [DOI] [PubMed] [Google Scholar]
- 70.Pelengaris S, Khan M. The c-MYC oncoprotein as a treatment target in cancer and other disorders of cell growth. Expert Opin Ther Targets. 2003;7:623–642. doi: 10.1517/14728222.7.5.623. [DOI] [PubMed] [Google Scholar]
- 71.Cole MD, McMahon SB. The Myc oncoprotein: a critical evaluation of transactivation and target gene regulation. Oncogene. 1999;18:2916–2924. doi: 10.1038/sj.onc.1202748. [DOI] [PubMed] [Google Scholar]
- 72.Spencer CA, Groudine M. Control of c-myc regulation in normal and neoplastic cells. Adv Cancer Res. 1991;56:1–48. doi: 10.1016/s0065-230x(08)60476-5. [DOI] [PubMed] [Google Scholar]
- 73.Facchini LM, Penn LZ. The molecular role of Myc in growth and transformation: recent discoveries lead to new insights. FASEB J. 1998;12:633–651. [PubMed] [Google Scholar]
- 74.Sakatsume O, Tsutsui H, Wang Y, Gao H, Tang X, Yamauchi T, Murata T, Itakura K, Yokoyama KK. Binding of THZif-1, a MAZ-like zinc finger protein to the nuclease-hypersensitive element in the promoter region of the c-MYC protooncogene. J Biol Chem. 1996;271:31322–31333. doi: 10.1074/jbc.271.49.31322. [DOI] [PubMed] [Google Scholar]
- 75.Postel EH, Mango SE, Flint SJ. A nuclease-hypersensitive element of the human c-myc promoter interacts with a transcription initiation factor. Mol Cell Biol. 1989;9:5123–5133. doi: 10.1128/mcb.9.11.5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Siebenlist U, Hennighausen L, Battey J, Leder P. Chromatin structure and protein binding in the putative regulatory region of the c-myc gene in Burkitt lymphoma. Cell. 1984;37:381–391. doi: 10.1016/0092-8674(84)90368-4. [DOI] [PubMed] [Google Scholar]
- 77.Boles TC, Hogan ME. DNA structure equilibria in the human c-myc gene. Biochemistry. 1987;26:367–376. doi: 10.1021/bi00376a006. [DOI] [PubMed] [Google Scholar]
- 78.Tomonaga T, Levens D. Activating transcription from single stranded DNA. Proc Natl Acad Sci USA. 1996;93:5830–5835. doi: 10.1073/pnas.93.12.5830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cooney M, Czernuszewicz G, Postel EH, Flint SJ, Hogan ME. Site-specific oligonucleotide binding represses transcription of the human c-myc gene in vitro. Science. 1988;241:456–459. doi: 10.1126/science.3293213. [DOI] [PubMed] [Google Scholar]
- 80.Kinniburgh AJ, Firulli AB, Kolluri R. DNA triplexes and regulation of the c-myc gene. Gene. 1994;149:93–100. doi: 10.1016/0378-1119(94)90416-2. [DOI] [PubMed] [Google Scholar]
- 81.Simonsson T, Pecinka P, Kubista M. DNA tetraplex formation in the control region of c-myc. Nucleic Acids Res. 1998;26:1167–1172. doi: 10.1093/nar/26.5.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Simonsson T, Pribylova M, Vorlickova M. A nuclease hypersensitive element in the human c-myc promoter adopts several distinct i-tetraplex structures. Biochem Biophys Res Commun. 2000;278:158–166. doi: 10.1006/bbrc.2000.3783. [DOI] [PubMed] [Google Scholar]
- 83.Mathur V, Verma A, Maiti S, Chowdhury S. Thermodynamics of i-tetraplex formation in the nuclease hypersensitive element of human c-myc promoter. Biochem Biophys Res Commun. 2004;320:1220–1227. doi: 10.1016/j.bbrc.2004.06.074. [DOI] [PubMed] [Google Scholar]
- 84.Izbicka E, Wheelhouse RT, Raymond E, Davidson KK, Lawrence RA, Sun D, Windle BE, Hurley LH, Von Hoff DD. Effects of cationic porphyrins as G-quadruplex interactive agents in human tumor cells. Cancer Res. 1999;59:639–644. [PubMed] [Google Scholar]
- 85.Shammas MA, Shmookler Reis RJ, Akiyama M, Koley H, Chauhan D, Hideshima T, Goyal RK, Hurley LH, Anderson KC, Munshi NC. Telomerase inhibition and cell growth arrest by G-quadruplex interactive agent in multiple myeloma. Mol Cancer Ther. 2003;2:825–833. [PubMed] [Google Scholar]
- 86.Grand CL, Han H, Muñoz RM, Weitman S, Von Hoff DD, Hurley LH, Bearss DJ. The cationic porphyrin TMPyP4 down-regulates c-MYC and human telomerase reverse transcriptase expression and inhibits tumor growth in vivo. Mol Cancer Ther. 2002;1:565–573. [PubMed] [Google Scholar]
- 87.Han H, Langley DR, Rangan A, Hurley LH. Selective interactions of cationic porphyrins with G-quadruplex structures. J Am Chem Soc. 2001;123:8902–8913. doi: 10.1021/ja002179j. [DOI] [PubMed] [Google Scholar]
- 88.Seenisamy J, Bashyam S, Gokhale V, Vankayalapati H, Sun D, Siddiqui-Jain A, Streiner N, Shin-Ya K, White E, Wilson WD, Hurley LH. Design and synthesis of an expanded porphyrin that has selectivity for the c-MYC G-quadruplex structure. J Am Chem Soc. 2005;127:2944–2959. doi: 10.1021/ja0444482. [DOI] [PubMed] [Google Scholar]
- 89.Schultze P, Macaya RFJ, Feigon Three-dimensional solution structure of the thrombin-binding DNA aptamer d(GGTTGGTGTGGTTGG) J Mol Biol. 1994;235:1532–1547. doi: 10.1006/jmbi.1994.1105. [DOI] [PubMed] [Google Scholar]
- 90.Padmanabhan K, Padmanabhan KP, Ferrara JD, Sadler JE, Tulinsky A. The structure of alpha-thrombin inhibited by a 15-mer single-stranded DNA aptamer. J Biol Chem. 1993;268:17651–17654. doi: 10.2210/pdb1hut/pdb. [DOI] [PubMed] [Google Scholar]
- 91.Jing N, Hogan ME. Structure-activity of tetrad-forming oligonucleotides as a potent anti-HIV therapeutic drug. J Biol Chem. 1998;273:34992–34999. doi: 10.1074/jbc.273.52.34992. [DOI] [PubMed] [Google Scholar]
- 92.Matsugami A, Ouhashi K, Kanagawa M, Liu H, Kanagawa S, Uesugi S, Katahira M. An intramolecular quadruplex of (GGA)(4) triplet repeat DNA with a G:G:G:G tetrad and a G(:A):G(:A):G(:A):G heptad, and its dimeric interaction. J Mol Biol. 2001;313:255–269. doi: 10.1006/jmbi.2001.5047. [DOI] [PubMed] [Google Scholar]
- 93.Matsugami A, Okuizumi T, Uesugi S, Katahira M. Intramolecular higher order packing of parallel quadruplexes comprising a G:G:G:G tetrad and a G(:A):G(:A):G(:A):G heptad of GGA triplet repeat DNA. J Biol Chem. 2003;278:28147–28153. doi: 10.1074/jbc.M303694200. [DOI] [PubMed] [Google Scholar]
- 94.Phan AT, Modi YS, Patel DJ. Propeller-type parallel-stranded G-quadruplexes in the human c-myc promoter. J Am Chem Soc. 2004;126:8710–8716. doi: 10.1021/ja048805k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ambrus A, Chen D, Dai J, Jones RA, Yang D. Solution structure of the biologically relevant G-quadruplex element in the human c-MYC promoter Implications for G-quadruplex stabilization. Biochemistry. 2005;44:2048–2058. doi: 10.1021/bi048242p. [DOI] [PubMed] [Google Scholar]
- 96.Simonsson T, Henriksson M. c-myc Suppression in Burkitt's lymphoma cells. Biochem Biophys Res Commun. 2002;290:11–15. doi: 10.1006/bbrc.2001.6096. [DOI] [PubMed] [Google Scholar]
- 97.Ou TM, Lu YJ, Zhang C, Huang ZS, Wang XD, Tan JH, Chen Y, Ma DL, Wong KY, Tang JC, Chan AS, Gu LQ. Stabilization of G-quadruplex DNA and down-regulation of oncogene c-myc by quindoline derivatives. J Med Chem. 2007;50:1465–1474. doi: 10.1021/jm0610088. [DOI] [PubMed] [Google Scholar]
- 98.Bomsztyk K, Van Seuningen I, Suzuki H, Denisenko O, Ostrowski J. Diverse molecular interactions of the hnRNP K protein. FEBS Lett. 1997;403:113–115. doi: 10.1016/s0014-5793(97)00041-0. [DOI] [PubMed] [Google Scholar]
- 99.Michelotti EF, Tomonaga T, Krutzsch H, Levens D. Cellular nucleic acid binding protein regulates the CT element of the human c-myc protooncogene. J Biol Chem. 1995;270:9494–9499. doi: 10.1074/jbc.270.16.9494. [DOI] [PubMed] [Google Scholar]
- 100.Berberich SJ, Postel EH. PuF/NM23-H2/NDPK-B transactivates a human c-myc promoter-CAT gene via a functional nuclease hypersensitive element. Oncogene. 1995;10:2343–2347. [PubMed] [Google Scholar]
- 101.Agou F, Raveh S, Mesnildrey S, Véron M. Single strand DNA specificity analysis of human nucleoside diphosphate kinase B. J Biol Chem. 1999;274:19630–19638. doi: 10.1074/jbc.274.28.19630. [DOI] [PubMed] [Google Scholar]
- 102.Kouzine F, Sanford S, Elisha-Feil Z, Levens D. The functional response of upstream DNA to dynamic supercoiling in vivo. Nat Struct Mol Biol. 2008;15:146–154. doi: 10.1038/nsmb.1372. [DOI] [PubMed] [Google Scholar]
- 103.Goodsell DS. The molecular perspective: VEGF and angiogenesis. Stem Cells. 2003;21:118–119. doi: 10.1634/stemcells.21-1-118. [DOI] [PubMed] [Google Scholar]
- 104.Finkenzeller G, Sparacio A, Technau A, Marmé D, Siemeister G. Sp1 recognition sites in the proximal promoter of the human vascular endothelial growth factor gene are essential for platelet-derived growth factor-induced gene expression. Oncogene. 1997;15:669–676. doi: 10.1038/sj.onc.1201219. [DOI] [PubMed] [Google Scholar]
- 105.Shi Q, Le X, Abbruzzese JL, Peng Z, Qian CN, Tang H, Xiong Q, Wang B, Li XC, Xie K. Constitutive Sp1 activity is essential for differential constitutive expression of vascular endothelial growth factor in human pancreatic adenocarcinoma. Cancer Res. 2001;61:4143–4154. [PubMed] [Google Scholar]
- 106.Sun D, Liu W-J, Guo K, Rusche JJ, Ebbinghaus S, Gokhale V, Hurley LH. Proximal promoter region of the human vascular endothelial growth factor gene has a G-quadruplex structure which can be targeted by G-quadruplex-interactive agents. Mol Cancer Ther. 2008 doi: 10.1158/1535-7163.MCT-07-2119. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Semenza GL, Agani F, Booth G, Forsythe J, Iyer N, Jiang BH, Leung S, Roe R, Wiener C, Yu A. Structural and functional analysis of hypoxia-inducible factor 1. Kidney Int. 1997;51:553–555. doi: 10.1038/ki.1997.77. [DOI] [PubMed] [Google Scholar]
- 108.Ferrara N. Vascular endothelial growth factor: basic science and clinical progress. Endocr Rev. 2004;25:581–611. doi: 10.1210/er.2003-0027. [DOI] [PubMed] [Google Scholar]
- 109.Ebbinghaus SW, Gordon MS. Renal cell carcinoma: rationale and development of therapeutic inhibitors of angiogenesis. Hematol Oncol Clin North Am. 2004 Oct;18:1143–1159. doi: 10.1016/j.hoc.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 110.Kouvaraki MA, Shapiro SE, Perrier ND, Cote GJ, Gagel RF, Hoff AO, Sherman SI, Lee JE, Evans DB. RET proto-oncogene: a review and update of genotype-phenotype correlations in hereditary medullary thyroid cancer and associated endocrine tumors. Thyroid. 2005;15:531–544. doi: 10.1089/thy.2005.15.531. [DOI] [PubMed] [Google Scholar]
- 111.Andrew SD, Delhanty PJ, Mulligan LM, Robinson BG. Sp1 and Sp3 transactivate the RET proto-oncogene promoter. Gene. 2000;256:283–291. doi: 10.1016/s0378-1119(00)00302-4. [DOI] [PubMed] [Google Scholar]
- 112.Munnes M, Patrone G, Schmitz B, Romeo G, Doerfler W. A 5′-CG-3′-rich region in the promoter of the transcriptionally frequently silenced RET protooncogene lacks methylated cytidine residues. Oncogene. 1998;17:2573–2583. doi: 10.1038/sj.onc.1202165. [DOI] [PubMed] [Google Scholar]
- 113.Kranenberg O. The KRAS oncogene. Past, present, and future. Biochim Biophys Acta. 2005;176:81–82. doi: 10.1016/j.bbcan.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 114.Hoffman EK, Trusko SP, Murphy M, George DL. An S1 nuclease-sensitive homopurine/homopyrimidine domain in the c-Ki-ras promoter interacts with a nuclear factor. Proc Natl Acad Sci USA. 1990;87:2705–2709. doi: 10.1073/pnas.87.7.2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sakurai S, Fukasawa T, Chong JM, Tanaka A, Fukayama M. C-kit gene abnormalities in gastrointestinal stromal tumors (tumors of interstitial cells of Cajal) Jpn J Cancer Res. 1999;90:1321–1328. doi: 10.1111/j.1349-7006.1999.tb00715.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yamamoto K, Tojo A, Aoki N, Shibuya M. Characterization of the promoter region of the human c-kit proto-oncogene. Jpn J Cancer Res. 1993;84:1136–1144. doi: 10.1111/j.1349-7006.1993.tb02813.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Park GH, Plummer HK, 3rd, Krystal GW. Selective Sp1 binding is critical for maximal activity of the human c-kit promoter. Blood. 1998;92:4138–4149. [PubMed] [Google Scholar]
- 118.Bejugam M, Sewitz S, Shirude PS, Rodriguez R, Shahid R, Balasubramanian S. Trisubstituted isoalloxazines as a new class of G-quadruplex binding ligands: small molecule regulation of c-kit oncogene expression. J Am Chem Soc. 2007;129:12926–12927. doi: 10.1021/ja075881p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Chao DT, Korsmeyer SJ. BCL-2 FAMILY: regulators of cell death. Annu Rev Immunol. 1998;16:395–419. doi: 10.1146/annurev.immunol.16.1.395. [DOI] [PubMed] [Google Scholar]
- 120.Young RL, Korsmeyer SJ. A negative regulatory element in the bcl-2 5′-untranslated region inhibits expression from an upstream promoter. Mol Cell Biol. 1993;13:3686–3697. doi: 10.1128/mcb.13.6.3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wang Y, Patel DJ. Solution structure of the Tetrahymena telomeric repeat d(T2G4)4 G-tetraplex. Structure. 1994;2:1141–1156. doi: 10.1016/s0969-2126(94)00117-0. [DOI] [PubMed] [Google Scholar]
- 122.Khan N, Aviñó A, Tauler R, González C, Eritja R, Gargallo R. Solution equilibria of the i-motif-forming region upstream of the B-cell lymphoma-2 P1 promoter. Biochimie. 2007;89:1562–1572. doi: 10.1016/j.biochi.2007.07.026. [DOI] [PubMed] [Google Scholar]
- 123.Yu J, Ustach C, Kim HR. Platelet-derived growth factor signaling and human cancer. J. Biochem Mol Biol. 2003;36:49–59. doi: 10.5483/bmbrep.2003.36.1.049. [DOI] [PubMed] [Google Scholar]
- 124.Takimoto Y, Wang ZY, Kobler K, Deuel TF. Promoter region of the human platelet-derived growth factor A-chain gene. Proc Natl Acad Sci USA. 1991;88:1686–1690. doi: 10.1073/pnas.88.5.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lin X, Wang Z, Gu L, Deuel TF. Functional analysis of the human platelet-derived growth factor A-chain promoter region. J Biol Chem. 1992;267:25614–25619. [PubMed] [Google Scholar]
- 126.Kaetzel DM, Maul RS, Jr, Liu B, Bonthron D, Fenstermaker RA, Coyne DW. Platelet-derived growth factor A-chain gene transcription is mediated by positive and negative regulatory regions in the promoter. Biochem. J. 1994;301:321–327. doi: 10.1042/bj3010321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wang ZY, Lin XH, Nobuyoshi M, Qiu QQ, Deuel TF. Binding of single-stranded oligonucleotides to a non-B-form DNA structure results in loss of promoter activity of the platelet-derived growth factor-A chain gene. J Biol Chem. 1992;267:13669–13674. [PubMed] [Google Scholar]
- 128.Oh IH, Reddy EP. The myb gene family in cell growth, differentiation and apoptosis. Oncogene. 1999;18:3017–3033. doi: 10.1038/sj.onc.1202839. [DOI] [PubMed] [Google Scholar]
- 129.Ramsay RG, Thompson MA, Hayman JA, Reid G, Gonda TJ, Whitehead RH. Myb expression is higher in malignant human colonic carcinoma and premalignant adenomatous polyps than in normal mucosa. Cell Growth Differ. 1992;3:723–730. [PubMed] [Google Scholar]
- 130.Griffin CA, Baylin SB. Expression of the c-myb oncogene in human small cell lung carcinoma. Cancer Res. 1985;45:272–275. [PubMed] [Google Scholar]
- 131.Slamon DJ, Boone TC, Murdock DC, Keith DE, Press MF, Larson RA, Souza LM. Studies of the human c-myb gene and its product in human acute leukemias. Science. 1986;233:347–351. doi: 10.1126/science.3014652. [DOI] [PubMed] [Google Scholar]
- 132.Eddy J, Maizels N. Conserved elements with potential to form polymorphic G-quadruplex structures in the first intron of human genes. Nucleic Acids Res. doi: 10.1093/nar/gkm1138. Advance access published 2008 Jan 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Bates P, Mergny J-L, Yang D. Quartets in G-major. The first international meeting on quadruplex DNA. EMBO Reports. 2007;8:1003–1010. doi: 10.1038/sj.embor.7401073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Duan W, Rangan A, Vankayalapati H, Kim M-Y, Zeng Q, Sun D, Han H, Yu Fedoroff O, Nishioka D, Rha SY, Izbicka E, Von Hoff DD, Hurley LH. Design and synthesis of fluoroquinophenoxazines that interact with human telomeric G-quadruplexes and their biological effects. Mol Cancer Ther. 2001;1:103–120. [PubMed] [Google Scholar]