Abstract
Mutations in the aryl hydrocarbon receptor-interacting protein-like 1 (AIPL1) gene have been found in patients with Leber congenital amaurosis (LCA), a severe, early-onset form of retinal degeneration. To determine the normal function of AIPL1 and to better understand how mutations in this gene cause disease, we performed a yeast two-hybrid screen to identify AIPL1-interacting proteins in the retina. One of the identified interacting proteins corresponds to NUB1 (NEDD8 Ultimate Buster 1), which is thought to control many biological events, especially cell cycle progression, by downregulating NEDD8 expression. The AIPL1–NUB1 interaction was verified by co-immunoprecipitation studies in Y79 retinoblastoma cells, demonstrating that this interaction occurs within cells that share a number of features with retinal progenitor cells. Furthermore, we examined the localization of the AIPL1 protein within developing and adult retinas, and found that AIPL1 is present in the developing photoreceptor layer of the human retina and within the photoreceptors of the adult retina. Similar to AIPL1, NUB1 is also expressed in the developing and adult retina. Therefore, it is possible that the early-onset form of retinal degeneration seen in LCA patients with AIPL1 mutations may be due to a defect in the regulation of cell cycle progression during photoreceptor maturation. These data raise the possibility that AIPL1 is important for appropriate photoreceptor formation during development and/or survival following differentiation.
INTRODUCTION
Leber congenital amaurosis (LCA) is a genetically heterogeneous, autosomal recessive retinal degenerative disease responsible for ~5% of all inherited retinopathies (1). LCA is often considered the most severe form of childhood retinopathy, and infants with this disease are usually blind at birth. To date, mutations in six genes have been found to cause LCA and two additional loci have been mapped (www.sph.uth.tmc.edu/RetNet/). Recently, we identified the fourth LCA-associated gene: aryl hydrocarbon receptor-interacting protein-like 1 (AIPL1) (2). Mutations in AIPL1 are estimated to account for ~7–9% of LCA cases worldwide, and have been found to cause autosomal dominant cone–rod dystrophy (3).
AIPL1 encodes a 384-amino-acid protein that contains three tetratricopeptide (TPR) motifs (2). These degenerate 34-amino-acid motifs have been found in proteins that participate in a variety of multiprotein complexes important for diverse biological processes (4). The human and primate forms of the AIPL1 protein also contain a C-terminal proline-rich region, which may serve to regulate the recruitment or exchange of proteins in multiprotein complexes (5,6). Furthermore, sequence analysis has shown that the AIPL1 protein shares considerable homology (49% identity and 69% similarity) to the aryl hydrocarbon receptor-interacting protein (AIP) (2), alternatively known as XAP2 (7) and ARA9 (8). AIP possesses a chaperone-like function that helps to mediate the stabilization and nuclear transport of the aryl hydrocarbon receptor (AhR) (7–15). It has also been found that binding of AIP to the AhR within the cytoplasm stabilizes and protects the AhR from ubiquitination (14). Collectively, these data suggest that AIPL1 may be important in protein trafficking and/or protein folding and stabilization.
Here we report the first identification of an AIPL1-interacting protein. Using a yeast two-hybrid approach, we find that bovine Aipl1 interacts with Nub1 (NEDD8 Ultimate Buster 1), a protein involved in regulation of cell cycle progression (16,17). The human AIPL1–NUB1 interaction was confirmed through co-immunoprecipitation experiments performed with mammalian COS cells and the Y79 retinoblastoma cell line. Previous studies have demonstrated that both AIPL1 and NUB1 are expressed within the photoreceptors of the retina (2,18). To further characterize the localization of AIPL1 during retinal development, we performed immunolocalization on human adult and developing retinas, and demonstrated that the AIPL1 protein is present in the developing photoreceptor layer and ultimately within mature photoreceptors of the adult retina. These data indicate the co-localization of AIPL1 with NUB1, which was previously shown to be localized within the cytoplasm and nuclei (16). Therefore, our data suggest that AIPL1 may function in the regulation of cell cycle progression through its interaction with NUB1, and that mutations in AIPL1 may lead to photoreceptor cell death during development by disrupting the normal regulation of the cell cycle.
RESULTS
Identification of AIPL1-interacting proteins
A GAL4 yeast two-hybrid system (ClonTech) was used to identify Aipl1-interacting proteins. Specifically, full-length bovine Aipl1 was used to screen a bovine retinal cDNA library (generously provided by Wolfgang Baehr) in yeast strain AH109, which contains Histidine-3 (HIS3), Adenine-2 (ADE2), LacZ and MEL1 gene reporters. Approximately 2 × 106 clones were screened for Aipl1-interacting proteins. A total of 57 clones grew on selection plates containing synthetic dropout medium lacking leucine (Leu), tryptophan (Trp) and histidine (His). All 57 colonies were then re-streaked onto selection plate, and of 57 colonies, 17 demonstrated activation of all three reporters (HIS3, MEL1 and ADE2), which indicates the presence of a strong protein–protein interaction. Bovine cDNA plasmids were isolated from these colonies, and re-tests were performed independently with each of the bovine cDNA plasmids along with the pGBKT7–Aipl1 bait plasmid or an empty pGBKT7 plasmid. These retests determined which clones were specific for and required the presence of Aipl1 to activate transcription of the reporter genes. False-positive clones, which were capable of activating the reporter genes without the presence of Aipl1, were eliminated from further studies.
Following elimination of false-positive clones, nine clones encoding putative AIPL1-interacting proteins remained. Upon sequencing and database analyses, we determined that two of the nine clones corresponded to bovine Nub1 (GenBank accession no. AF514279) (also known as NY-REN-18 and BS4). The bovine Nub1 clones are only capable of activating expression of the reporter genes in the presence of the pGBKT7–Aipl1 plasmid, thereby demonstrating that this interaction is specific for Aipl1 (Fig. 1). Furthermore, after sequencing the entire insert of the bovine Nub1 clones, we found that they only encode 221 amino acids, corresponding to the C-terminal portion of the human NUB1 protein, suggesting that it is this portion of the NUB1 protein that facilitates the interaction with AIPL1.
Figure 1.

Results of yeast two-hybrid retest with bovine Nub1. The pGAD10 plasmid containing bovine Nub1 was transformed into yeast strain AH109 with either the pGBKT7–bovine Aipl1 plasmid or the pGBKT7–empty plasmid. Transformants were grown on two different types of plates: (i) plates containing synthetic dropout media lacking Leu and Trp (−L/−W) to confirm transformation and (ii) plates containing dropout media with X-α-galactosidase (X-α-gal) and 3-AT (5 mM) but lacking Leu, Trp, and His (−L/−W/−H/+X-α-gal/+3AT) to assay for activation of the HIS3 and MEL1 reporters. (A) Transformation with pGBKT7–Aipl1 and pGAD10–Nub1 plasmids grown on −L/−W plates demonstrate that the transformation worked. (B) Transformation with pGBKT7–empty and pGAD10–Nub1 plasmids grown on −L/−W plates demonstrate that the transformation was successful. (C) Transformation with pGBKT7–Aipl1 and PGAD10–Nub1 plasmids. Formation of blue colonies on −L/−W/−H/+X-α-gal/+3AT plates indicates activation of MEL1 and HIS3 reporters and therefore the presence of a protein–protein interaction between AIPL1 and NUB1. (D) Transformation with pGBKT7–empty and pGAD10–bovine Nub1 plasmids. Lack of colony formation on −L/−W/−H/+X-α-gal/+3AT plates confirms that NUB1 requires the presence of AIPL1 to activate transcription of the HIS and MEL1 reporters.
Analysis of human protein interactions in yeast
We previously described a number of different mutations that were identified in patients with recessive Leber congenital amaurosis (2,3). To investigate the effect that these mutations may have on the interaction between AIPL1 and NUB1, nine LCA-associated mutations were introduced into the pGADT7–AIPL1 construct (Fig. 2). The ability of the mutant AIPL1 proteins to interact with the protein encoded by pGBK–NUB1 was determined through quantitative β-galactosidase assays (Fig. 2) and growth in liquid dropout media (−Leu, −Trp, −His), with the empty pGADT7 vector co-transformed with pGBKT7–NUB1 as a control. The majority of the mutations significantly reduced the interaction between these proteins. However, there was not a significant reduction of the interaction with the A197P mutation. Although the mutations 1206N and C239R appeared to cause reduced interaction with NUB1 by the quantitative β-galactosidase assay, the growth assay did not indicate a statistically significant deviation from wild-type control. In addition, to determine what regions of the AIPL1 protein were involved in the interaction with NUB1, pGADT7–AIPL1 constructs were designed to encode the sequence of (i) the N-terminal portion prior to the TPR motifs, (ii) the TPR motifs themselves or (iii) the C-terminal sequence following the TPR motifs (Fig. 2). While each construct was capable of interacting with NUB1, the construct encoding the C-terminal region of AIPL1 appeared to have the highest binding activity to NUB1. As a control, the empty pGADT7 vector was co-transformed with pGBKT7. As indicated in Figure 2, this control vector was unable to activate the reporter in either the quantitative β-galactosidase assay or the growth assay.
Figure 2.
Analysis of interaction in yeast. The pGADT7–AIPL1 deletion or mutant constructs shown on the left were co-transformed with the construct pGADT7–NUB1, and the activation of the HIS3 and LacZ reporter genes was determined (right) by a liquid growth assay (white bars) and a β-galactosidase assay (black bars), respectively.
Co-immunoprecipitation of the AIPL1–NUB1 complex from COS cells
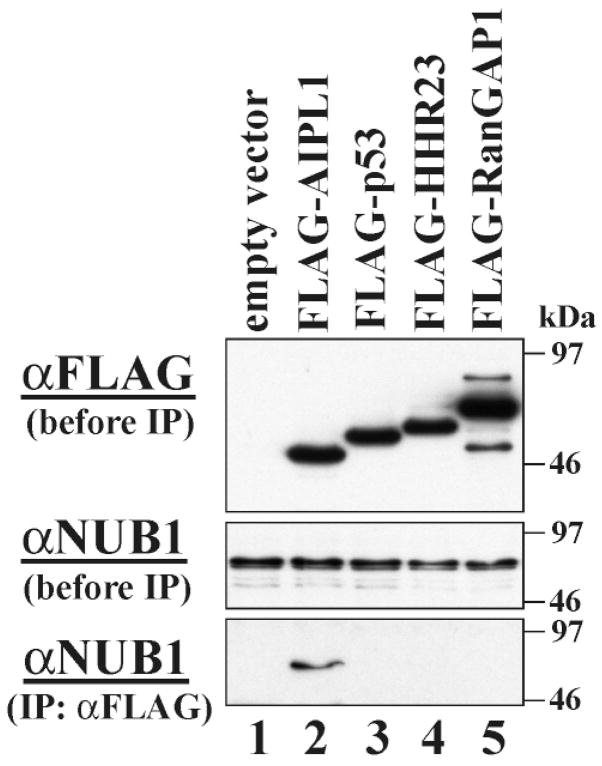
To initially confirm the AIPL1–NUB1 interaction within mammalian cells, we performed a series of immunoprecipitation experiments using a plasmid construct encoding FLAG-tagged human AIPL1. This construct was transiently transfected into COS-M6 cells, which endogenously express NUB1 (Fig. 3). Three other fusion FLAG-tagged proteins including p53, HHR23 and RanGAP1 and an empty vector were used as controls to ensure that NUB1 binding was specific to AIPL1. Prior to immunoprecipitation, immunoblotting was performed on cell lysates with both a mouse monoclonal anti-FLAG M5 antibody (Fig. 3: top panel) and a rabbit anti-NUB1 antibody (Fig. 3: middle panel) to detect transfected gene products and endogenous NUB1, respectively, within the cells (16,17). As expected, cell lysates probed with the anti-FLAG antibody indicated a band corresponding to FLAG-tagged AIPL1 protein at ~46 kDa (Fig. 3: top panel, lane 2), confirming translation of the AIPL1 protein in these cells, Endogenous NUB1 protein was detected in all transfected COS cells at an expected molecular weight of ~66 kDa (Fig. 3: middle panel, lanes 1–5). When AIPL1 was immunoprecipitated using the FLAG antibody, endogenous NUB1 was found to co-immunoprecipitate (Fig. 3: bottom panel). Only the FLAG-tagged AIPL1 protein could precipitate NUB1 (Fig. 3: bottom panel, lane 2). Co-immunoprecipitation of endogenous NUB1 and transfected mutant AIPL1 in this system was used to confirm the results of the mutation assays and to ensure that reduced binding was due to the mutation and not reduced protein stability. The mutants R53W, M79T, V96I, G262S, W278X and R302L failed to immunoprecipitate endogenous NUB1 (data not shown).
Figure 3.
Co-immunoprecipitation of endogenous NUB1 with FLAG–AIPL1. Empty vector (lane 1) or plasmid DNA encoding FLAG-tagged AIPL1 (lane 2), p53 (lane 3), HHR23 (lane 4) or RanGAP1 (lane 5) was overexpressed in COS cells. The total cell lysate was analyzed by antibody against FLAG (top panel) or NUB1 (middle panel). The total cell lysate was also incubated with anti-FLAG antibody for immunoprecipitation. Co-precipitated proteins were analyzed by immunoblotting using anti-NUB1 antibody (bottom panel). Only FLAG-tagged AIPL1 is capable of immunoprecipitating NUB1 (bottom panel, lane2), thereby demonstrating that these two proteins interact in vitro and in vivo. Molecular mass standards are expressed in kilodaltons.
Confirmation of AIPL1–NUB1 interaction in the retinoblastoma cell line Y79
Extensive molecular analysis has revealed that the Y79 retinoblastoma cell line resembles retina progenitor cells. Therefore, we also verified the AIPL1–NUB1 interaction within cells of retinal origin by using this cell line. Western blot analysis of total Y79 cell lysate with antibodies against NUB1 and AIPL1 demonstrates that both NUB1 and AIPL1 are expressed within this cell line (Fig. 4A and B: lanes 4). Immunoprecipitation was performed with preimmune serum, anti-AIPL1 serum or affinity-purified anti-AIPL1 antibody, followed by immunoblot analysis with either the NUB1 (Fig. 4A) or the AIPL1 antibody (Fig. 4B). The affinity-purified AIPL1 antibody co-precipitated bands of ~46 and ~50 kDa corresponding to the AIPL1 (Fig. 4B: lane 3) and NUB1 proteins (Fig. 4A: lane 3), respectively. The size of the NUB1 protein in retina is ~16 kDa smaller than that in other tissues. This result is consistent with identification of alternatively sliced NUB1 clones amplified from retinal cDNA libraries (ClonTech 7449-1, ATCC 77428, ATCC 87256 libraries; unpublished data). These data indicate that the AIPL1–NUB1 complex occurs within retinoblastoma cell line Y79, thereby providing further evidence of an interaction between these two proteins in vivo.
Figure 4.
Co-immunoprecipitation of NUB1 with AIPL1 in human Y79 retinoblastoma cells. Immunoprecipitates with preimmune serum (lane 1), anti-AIPL1 serum (lane 2) and affinity-purified anti-AIPL1 antibody (lane 3) were separated by SDS–PAGE and transferred for immunoblot analysis (IB) with either anti-NUB1 (A) or unpurified anti-AIPLl (B). Total Y79 cell lysate (lane 4) was also loaded on the gel.
Localization of AIPL1 and NUB1 within the retina
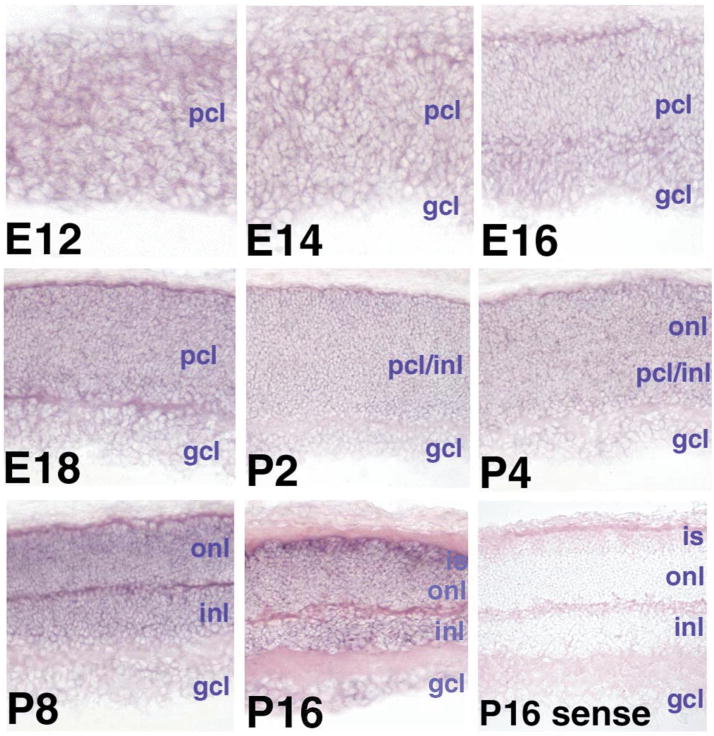
Previous studies have demonstrated through in situ hybridization with mouse retinal tissue sections that Aipl1 is expressed within the outer nuclear layer and photoreceptor inner segments of the retina as well as within pinealocytes of the pineal gland (2). To confirm photoreceptor specificity of AIPL1 within the human retina, we used full-length AIPL1 cDNA as a probe in a sectioned adult human retina, and found that the patterns of AIPL1 expression are the same in both mouse and human sections (data not shown). In situ hybridization in the developing mouse retina indicated low levels of Nub1 mRNA (mouse ortholog referred to as BS4) throughout the retinal progenitor and precursor cells, beginning at mouse embryonic day 12, roughly corresponding to human embryonic week 6–7 (Fig. 5) (18). This expression continues through retinal development, and is significantly greater in the photoreceptor inner segments beginning at mouse postnatal day 8. Furthermore, serial analysis of gene expression (SAGE) demonstrated that expression of both Aipl1 and Nub1 is reduced in Crx knockout (Crx−/−) retinas, which lack photoreceptors, compared with Crx wild type (Crx−/−) mice (18). Collectively, these data indicate that both AIPL1 and NUB1 transcripts are co-expressed in the developing and adult photoreceptors.
Figure 5.
Expression of Nub1 in the embryonic and postnatal mouse retina. Mouse Nub1 development in situ. Digoxygenin in situ hybridization of Nub1 mRNA is shown from embryonic day12 (E12) through to postnatal day 16 (P16). pcl, progenitor cell layer; gcl, ganglion cell layer; inl, inner nuclear layer; onl, outer nuclear layer; is, photoreceptor inner segments; P16 sense, sense control for background. Photographs were taken at 400×.
Because mutations in AIPL1 result in an early-onset form of retinal degeneration, we determined the AIPL1 localization pattern during retinal development by performing immunostaining on human retinas from several different developmental stages. At fetal week 10, there was faint AIPL1 protein in the developing inner plexiform layer (ipl) of the human retina. Four weeks later (fetal week 14), the AIPL1 protein was detected in a subset of cells just apical to the outer neuroblastic layer (onbl) in the region where newly postmitotic photoreceptors are beginning to differentiate (Fig. 6A). By fetal week 16, the outer nuclear layer had become morphologically distinct from the retinal progenitor cells in the onbl. At this stage, robust AIPL1 presence was found in most of the nuclei of the developing photoreceptors (Fig. 6B). While all the cells in this layer appeared to express some AIPL1 protein in their nuclei, the intensity of immunostaining varied dramatically from cell to cell (Fig. 6B). To demonstrate the specificity of the anti-AIPL1 antibody, specific (AIPL1) and non-specific (mouse cone arrestin) peptides were included in primary antibody reaction mixture (unpublished data). The AIPL1 peptide efficiently quenched antibody binding, while the non-specific peptide had no effect on the immunofluorescence shown in Figure 6.
Figure 6.
Expression of AIPL1 in the fetal, child and adult human retina. (A) Immunohistochemical analysis using a rabbit anti-AIPL1 antibody showed that human AIPL1 protein (green fluorescence) was expressed in a subset of the nuclei (red fluorescence) of the human retina at fetal week 14. Expression was only detected in the apical region of the retina, where the newly postmitotic photoreceptors are beginning to differentiate. Some nuclei expressed much higher levels of AIPL1 protein (arrow) than other nuclei (open arrowheads). (B) Approximately 2 weeks later at fetal week 16, the expression of AIPL1 was seen throughout the immature outer nuclear layer (ONL) containing the developing photoreceptors. While all the nuclei in this layer appear to express AIPL1, the level of AIPL1 appeared to be high in some cells and much lower in others as seen at fetal week 14. (C and D) Immunohistochemical analysis shows that the AIPL1 protein (dark brown stain) localizes to the ONL and inner segments (IS) of the photoreceptors within child (age 3) and adult retinas. The adult retinal image was counterstained (blue) to highlight the nuclei of the cells. onbl, outer neuroblastic layer; inbl, inner neuroblastic layer; GCL, ganglion cell layer, OS, outer segments, (A and B) scale bar = 10 μM; (C and D) scale bar = 20 μM.
Immunolabelling of AIPL1 was detected in the outer nuclear layer and photoreceptor inner segments of the child and adult retina (Fig. 6C and D, respectively). Figure 6C and D also demonstrate that AIPL1 is present within both the nucleus and cytoplasm of the photoreceptors unlike the nuclear specificity of AIPL1 in the developing retina. The NUB1 antibody used for the immunoprecipitation experiments is not useful for immunohistochemistry studies within retinal tissue (data not shown). However, subcellular location of the AIPL1 protein within the differentiated retina (Fig. 6C and D) is consistent with co-localization of AIPL1 and NUB1, since the NUB1 protein was previously localized to the nuclei and cytoplasm of cultured cells using this antibody (16).
DISCUSSION
Retinal photoreceptor degeneration comes in a variety of forms. In most cases, cell death begins in the totally differentiated retina and spreads throughout the photoreceptor layer, ultimately leading to loss of visual function. In other cases, the defect is developmental. For example, Crx-deficient mice fail to transcriptionally activate a subset of essential genes required for phototransduction and the photoreceptors die shortly after birth (19). Even earlier defects have been reported. Cyclin D1-deficient mice exhibit focal photoreceptor death beginning a few days after birth (20). This unique pattern of photoreceptor degeneration may be the result of a progenitor cell defect, because cyclin D1 is only expressed in retinal progenitor cells (20). Considering the early onset of vision loss in LCA patients and the expression of AIPL1 in developing photoreceptors, it is possible that AIPL1 function is critical during the period of photoreceptor development as with cyclin D1 or Crx.
Using a yeast-two hybrid approach with a retinal library, we determined that AIPL1 interacts with NUB1, a protein involved in regulating cell cycle progression and proliferation. Furthermore, we have shown with co-immunoprecipitation experiments using an AIPL1-specific antibody that AIPL1 specifically interacts with a 50 kDa NUB1 protein, which is 16 kDa smaller than that present in other tissues (16). This protein is likely encoded by an alternately spliced NUB1 RNA, since multiple alternate transcripts are present in retinal cDNA libraries. By immunohistochemistal studies, we demonstrated that the immunoreactive AIPL1 is present in both the developing and mature photoreceptors of the retina and that its subcellular localization overlaps with that of NUB1. The immunolocalization within the adult retina identified here is consistent with the recent results of van der Spuy et al. (21), who demonstrate that AIPL1 localizes to both the cytoplasm and the nucleus of the photoreceptor cells of the retina, and E. Deery et al. (manuscript in preparation), who also observe localization of the protein in the nucleus and cytoplasm of cultured mammalian cells. Moreover, the high perinuclear concentration of AIPL1 in the differentiated retina suggests that it may function similarly to AIP, in cytoplasmic stability and trafficking its target, such as NUB1, to the nucleus (7–15).
To determine if LCA-causing mutations in AIPL1 affect its ability to interact with NUB1, binding assays were performed with mutant AIPL1 and wild-type NUB1 proteins. Data presented here indicate that multiple mutations throughout the length of the AIPL1 protein result in reduced binding with NUB1. Co-immunoprecipitation assays with COS cells containing endogenous NUB1 and transfected mutant AIPL1 proteins confirm that the reduced interaction is not due to degradation of the AIPL1 protein. Furthermore, we determine that the most C-terminal 144 amino acids of AIPL1 have the highest ability to interact with NUB1. This is consistent with the finding that the sequence required for proper interaction of AIP with the aryl-hydrocarbon receptor is within the final 20 amino acids of AIP (22). Further studies will be necessary to determine if mutations in AIPL1 reduce binding to NUB1 as a result of improper AIPL1 protein folding or as a result of decreased binding affinities due to a change in a critical amino acid within the region directly involved in facilitating the interaction with NUB1. However, because the C-terminal portion of AIPL1 appears to facilitate the interaction with NUB1, it is likely that the N-terminal mutations result in a decrease in NUB1 binding due to improper AIPL1 folding rather than a decrease in the efficiency of the NUB1 binding site. Due to the similarity with AIP and the likelihood that AIPL1 is functioning as part of a multiprotein complex, it is also possible that the mutations not associated with reduced NUB1 interaction affect the interaction with another component of the AIPL1 complex or that these mutations do not interfere with protein binding, but result in mislocalization of the protein within the cell.
NUB1 functions in cell cycle progression through downregulating NEDD8 expression post-translationally by targeting NEDD8 and its conjugates for proteasomal degradation (16,17). NEDD8 is involved in the degradation of many proteins, including cyclin D1 and p27Kip1, two important proteins in the regulation of cell cycle progression in the developing retina (20,23–25). Furthermore, previous studies have demonstrated that overexpression of NUB1 can inhibit cell growth by up to 83%, providing further support for the importance of NUB1 in cell growth and proliferation (16). Given what is known about NUB1, it is also possible that NUB1 facilitates proteolytic degradation of AIPL1. Further studies will be needed to fully understand what role the AIPL1–NUB1 interaction plays in normal vision and/or retinal development and how mutations in AIPL1 affect the function of the AIPL1–NUB1 interaction. Experiments such as co-expression of AIPL1 and NUB1 within mammalian cells will be useful to determine if NUB1 targets AIPL1 to the proteosome, and to determine if AIPL1 expression leads to an increase in cytoplasmic stability of NUB1 or a change in the localization of NUB1 within the cell. Furthermore, it will be important to determine how LCA-causing mutations affect the normal localization patterns and stability of the individual AIPL1 and NUB1 proteins, as well as the AIPL1–NUB1 complex. In conclusion, based on the data presented here, we hypothesize that AIPL1 plays a role in cytosolic stability and/or nuclear transport of NUB1 during regulation of cell cycle progression during photoreceptor development. This function would be consistent with the severe, early-onset blindness observed in patients with LCA caused by mutations in AIPL1.
MATERIALS AND METHODS
Yeast two-hybrid screen
The full-length bovine Aipl1 (984 bp, 328 amino acids) was cloned into the GAL4 DNA-binding domain vector pGBKT7 (Clontech, Palo Alto, CA) and used as bait to screen a bovine retinal cDNA library in the pGAD10 vector (Clontech) (library kindly provided by Dr Wolfgang Baehr) (26,27). The bovine Aipl1 protein sequence is highly conserved when compared with human AIPL1 (88% identity and 94% similarity); however, unlike human AIPL1, it does not contain the C-terminal 56-amino-acid proline-rich region (5). The bovine Aipl1 coding sequence was amplified out of a bovine retinal cDNA library using bovine sequence-specific primers. Primers were designed such that EcoRI and BamHI restriction sites (underlined) were incorporated into the 5′and 3′ ends of the AIPL1 PCR products respectively (5′-ATTATGAATTCATGGATGCCACTCTGCTCCT-3′, 5′-ATTATGGATCCCTAGCCCAGCATGTTCCGG-3′). The PCR products were digested with EcoRI and BamHI and then ligated in-frame into the pGBKT7 vector, which was also digested with EcoRI and BamHI. Sequencing of the entire Aipl1 insert was performed using both vector-specific primers (ClonTech) and gene-specific primers using a BigDye Terminator Sequencing kit (Applied Biosystems, Foster City, CA) on an ABI 310 Prism automated sequencer to ensure that the insert was ligated inframe and would result in a GAL4 DNA-binding domain–Aipl1 fusion protein.
The yeast two-hybrid screen was performed in the Saccharomyces cerevisiae strain AH109 (Clontech) with a sequential transformation procedure using the YEASTMAKER lithium acetate method (Clontech) as described previously (26). AH109 contains four reporter genes, Histidine-3 (HIS3), Adenine-2 (ADE2), LacZ and MEL1, each under the control of a distinct GAL4-responsive promoter. These distinct promoter elements automatically eliminate three major categories of false positives: those caused by proteins that interact upstream of the reporter gene’s binding site, those that interact directly with the sequences flanking the GAL4 binding site, and those that interact with transcription factors bound to specific TATA boxes. Growth and α-galactosidase assays with synthetic dropout medium containing X-α-galactosidase (X-α-gal) but lacking tryptophan (Trp) and histidine (His) were performed with the pGBKT7–Aipl1 vector to ensure that Aipl1 could not autonomously activate the reporters prior to the screen. Once this was established, the YEASTMAKER lithium acetate method was used to transform AH109 cells already containing the pGBKT7–Aipl1 vector with 100 μg of the pGAD10 bovine retinal cDNA library. Yeast transformants were incubated at 30°C for 14 days on plates containing synthetic dropout medium lacking leucine (Leu), Trp, and His and plates containing medium lacking Leu, Trp, His and adenine (Ade). All plates contained 5 mM of 3-aminotriazole, an inhibitor of constitutive expression of the HIS protein within AH109 cells that suppresses background growth. Putative positive clones were selected and re-streaked onto plates with synthetic medium with X-α-gal but lacking Leu, Trp and His to confirm activation of the MEL1 and HIS3 reporters. Finally, yeast transformants that were positive for activation of the HIS3 and MEL1 reporters were streaked onto plates containing synthetic dropout medium with X-α-gal but lacking Leu, Trp, His and Ade to assay for the activation of a third reporter, ADE2.
pGAD10 plasmids with bovine retinal cDNA inserts that encode potential Aipl1-interacting proteins were then isolated from the yeast using a standard glass bead, phenol–chloroform extraction method as previously described (28). Once isolated from AH109, the prey plasmids were then amplified by transformation into Escherichia coli SURE electroporation competent cells (Stratagene).
Verification of positive clones in the yeast two-hybrid system
A yeast two-hybrid screen is capable of resulting in false positives; therefore, once pGAD10 plasmids containing putative Aipl1-interacting proteins were isolated, they were re-tested for their ability to activate the reporters both in the presence and in the absence of Aipl1 to eliminate false-positive clones. Yeast two-hybrid re-tests were performed by transforming the isolated pGAD10 bovine retinal cDNA plasmids into AH109 with either the pGBKT7–Aipl1 vector or the empty pGBKT7 vector (negative control). Yeast transformants were incubated for 4–6 days at 30°C on plates containing synthetic dropout medium lacking Leu and Trp to ensure that the transformations had worked and plates containing medium lacking Leu, Trp and His to assay for activation of the HIS3 reporter. Prey plasmids that produced colonies only in the presence of the pGBKT7–Aipl1 vector and not with the negative control pGBKT7 vectors were then streaked onto plates containing medium with X-α-gal but lacking Leu, Trp and His to check for activation of the MEL1 reporter. We then sequenced the pGAD10 bovine retinal plasmids that could activate the reporter genes only when Aipl1 was present within the cell. The entire cDNA inserts of these plasmids were sequenced using both vector- and sequence-specific primers. Finally, in silico analysis was performed to determine the identity of the Aipl1-interacting proteins.
Analysis of interactions between wild-type and mutant human AIPL1 and NUB1 in yeast
The human AIPL1 and NUB1 coding sequences were subcloned into the pGADT7 and pGBKT7 plasmids, respectively, and the resulting constructs were sequenced to confirm that the sequences were full-length and inframe. Mutations were introduced using the QuickChange site-directed mutagenesis kit (Stratagene), and deletion constructs including the pre-TPR motif AIPL1 sequence (PRE, encoding amino acids 1–117), the TPR motif region only (TPR, amino acids 118–297) or the post-TPR sequence (POST, amino acids 298–384) were developed. Interactions of mutant and wild-type AIPL1 with NUB1 were qualitatively analyzed by observation of growth in −L/−W/−H dropout medium plates over the course of 10 days. Interactions were quantitatively analyzed using liquid o-nitrophenyl-β-D-galactopyranoside (OPNG), according to published methods (29). Briefly, pGAD–AIPL1 constructs including the desired mutation or deletion construct were co-transformed into AH109 yeast competent cells (ClonTech) and plated on −L/−W plates. After three days of growth, single colonies were used to inoculate 5 ml of −L/−W/−H liquid media and were allowed to grow to mid-log phase. Cells were pelleted by centrifugation at 2500g for 10 min at 4°C and were resuspended in 5 ml of Z buffer, and the OD600 was measured. Assays using 50 μl and 100 μl of the cell culture in Z buffer were performed in parallel. Twenty-five microliters of 0.001% SDS and 50 μl of chloroform were added to permeabilize the yeast cell wall, and a solution of 4 mg/ml OPNG in potassium phosphate buffer, pH 7.0, was added. After 3.5 h of incubation at 30°C, the cell debris was pelleted by centrifugation, the OD420 of the supernatant was measured, and the units of β-galactosidase were calculated using the formula U = (1000 × OD420)/(t × v ×OD600), where v = volume of culture used in the assay in ml, and t = time of assay in min. All assays were performed in parallel using three colonies from each transformation and three liquid assays from each colony. The results were then averaged. For growth assays, the transformants were first plated on −L/−W agar plates and incubated at 30°C for 4 days, and then the colonies were pooled together by scraping into 3 ml of −L/−W/−H medium. The yeast cells were diluted and vortexed for 30 s to disperse the cell clusters. The cells were counted and seeded at about 1000 cells/ml into either −L/−W or −L/−W/−H media in a final volume of 2 ml. The samples were incubated with shaking for 2 days, and then the OD600 values were recorded. Data represent the mean±SD of a representative experiment done in triplicate and are represented as a percentage of the OD600 of yeast grown in selective medium supplemented with histidine.
Co-immunoprecipitation from COS-M6 cells
COS-M6 cells were transfected with empty pcDNA3 vector (Invitrogen, Carlsbad, CA) or plasmid DNA encoding FLAG-tagged AIPL1, p53, HHR23 or RanGAP1. The cells were harvested 20 h after transfection by scraping, and the total cell lysate was prepared in TBS buffer (20 mM Tris-HCl pH 7.5, 137 mM NaCl and 0.5% NP-40) containing Complete protease inhibitor cocktail (Roche, Indianapolis, IN). The lysate was passed through a 22G needle to shear the DNA and then centrifuged at 100 000 g at 4°C for 30 min. Prior to immunoprecipitation, some of the supernatant was analyzed by immunoblotting using mouse monoclonal anti-FLAG M5 antibody to detect transfected gene products and rabbit anti-NUB1 antibody (16,17) to detect endogenous NUB1. For immunoprecipitation, the rest of the supernatant was incubated with anti-FLAG M2 agarose beads (Sigma, St Louis, MO) at 4°C for 2 h. After incubation, the beads were washed three times with TBS buffer. Precipitated proteins were solubilized in 2% SDS treating solution at 45°C for 1 h and analyzed by immunoblotting using rabbit anti-NUB1 antibody to detect co-immunoprecipitated endogenous NUB1.
Production of AIPL1 antibody
Rabbit antisera against the peptide of AIPL1 (amino acids 371–384, ATEPPPSPGHSLQH) was produced by Alpha Diagnostic International, Inc. (San Antonio, TX). The peptide conjugate was injected into rabbits, and ELISA was performed on bleeds at week 7 and 9. As the best ELISA results were obtained from the bleed at 9 weeks, l5 ml of this serum was affinity-purified against the peptide using a Sepharose 4-B column. Ability of this antiserum to specifically bind to AIPL1 was determined by immunoblot analysis of the protein expressed in E. coli, in human Y79 retinoblastoma cells and in human retina protein extracts.
Co-immunoprecipitation of the AIPL1/NUB1 complex from retinoblastoma Y79 cells
Y79 retinoblastoma cells (American Type Tissue Culture, Rockville, MD) were grown as previously described (30). Y79 cells were harvested at 2 × 107 cells/tube by centrifugation, washed twice with PBS and frozen at −80°C. Immunoprecipitation was performed with the Protein A Agarose IP Kit (KPL, Gaithersburg, MD). Briefly, each cell pellet (2 × 107 cells) was solubilized in 1 ml of lysis buffer (150 mM NaCl, 1% Triton X-100 and 50 mM Tris, 1 × protease inhibitor cocktail) for 30 min on ice, sonicated for 1 s × 5 to shear DNA, and centrifuged at 14 000 g for 10 min at 4°C to remove cell debris. The supernatants were transferred to fresh microcentrifuge tubes and incubated with 50 μl of 50% protein A agarose in lysis buffer with no protease inhibitor for 1 h at 4°C with gentle mixing. The suspension was centrifuged at 14 000 g for 20 s, and the precleared supernatant was incubated with pre-immune serum (5 μl), rabbit anti-AIPL1 serum (5 μl) or affinity-purified rabbit anti-AIPL1 antibody ~1 μg) overnight at 4°C with gentle mixing. The samples were further incubated for 1 h at 4°C with mixing, following the addition of 50 μl of 50% protein A agarose in lysis buffer with no protease inhibitor to each tube. The immunoprecipitates were collected by centrifugation at 14 000 g for 20 s, washed with 1 ml of ice cold lysis buffer three times, solubilized in SDS–PAGE sample buffer and subjected to immunoblot analysis with anti-NUBI or anti-AIPL1.
Proteins were resolved on 10% SDS–polyacrylamide gel electrophoresis (SDS–PAGE) and were electrophoretically transferred to limmobilon-P membranes (Millipore, Bedford, MA) using previously published protocols (30). Two identical blots were made, incubated first with anti-NUB1 and anti-AIPL1 rabbit polyclonal antibodies, respectively, then with horseradish peroxidase-conjucated goat anti-rabbit IgG (Bio-Rad Laboratories), and detected using an Enhanced Chemiluminescence (ECL) Kit (Amersham, Arlington Heights, IL).
In situ hybridization and immunohistochemistry
In situ hybridization was performed as reported previously (18) using a cRNA digoxygenin probemade from a BMAP EST of mouse Nub1 (GenBank accession no. AI844804).
For immunohistochemistry, human adult eyes were obtained from the Alzheimer’s Disease Center at the Keck School of Medicine of the University of Southern California and the A. Ray Irvine, Jr Ophthalmic Pathology Laboratory, Doheny Eye Institute, the University of Southern California. Eyes with a postmortem time of 3–8 h were fixed in either 10% buffered formaldehyde or 4% paraformaldehyde and processed as described previously (27,31). The eyes were dehydrated overnight using an automated Shandon Citadel 2000 dehydrator, embedded and oriented sagittally for 6 μm sections through the optic nerve with a JUNGRM2035 microtome (Leica, Foster City, CA), and subjected to immunohistochemical staining.
Immunohistochemistry was performed as previously described with a few modifications (27). After deparaffination and hydration through graded alcohols, the sections were heated in 0.01% sodium citrate buffer, pH 6.0, for 20 min for antigen retrieval. After cooling to room temperature and rinsing with PBS, they were blocked with 3% H2O2 and 5% normal goat serum before incubation with anti-AIPL1 peptide antibody (1:50–200) overnight at 4°C. Following the washing steps, the sections were reacted for 30 min at room temperature with biotinylated anti-rabbit IgG and for 50 min at room temperature with VECTASTAIN Elite ABC Reagent (Vector Laboratories, Burlingame, CA). After washing, the sections were reacted with freshly prepared peroxidase substrate (diaminobenzidine tetrahydrochloride, DAB, Vector Laboratories) for 3 or 4 min, rinsed with water, counterstained in hematoxylin QS (Vector Laboratories), dehydrated through graded alcohols, and cleared with xylene. Examination and photography were performed using a Nikon light microscope and a Spot digital camera.
Human fetal retinal samples were obtained from Harvard Medical School or the Nelson Mandela Medical School in Durban, South Africa. Immunostaining of fetal retinal cryosections was carried out as previously described (32,33). The affinity-purified rabbit anti-AIPL1 antibody was used at a dilution of 1:100 and the biotin-conjugated goat anti-rabbit secondary antibody (Vector Laboratories Inc.) was used at a dilution of 1:500. The avidin–biotin–peroxidase complex (Vectastain ABC) was used according to the manufacturer’s instructions (Vector Laboratories, Inc.). Detection was achieved by using FITC-conjugated tyramide (NEN) compound, and staining of the nuclei was accomplished by using propidium iodide (0.1%). Confocal microscopy was performed using a Leica DM-RBE microscope equipped with a TCSNT true confocal scanner.
Acknowledgments
The authors thank Chelsea G. Castellano, Bruce Brown and Jodi Watson for technical assistance. This work was supported by the Kirchgessner Foundation, the Knights Templar Eye Foundation, the Foundation Fighting Blindness, Fight for Sight, Research Division of Prevent Blindness America, the RPB and NIH Grants EY00395 (C.M.C.) and EY03040 and the generous continued support of Mary D. Allen and William R. Acquavella. C.M.C. is the Mary D. Allen Professor for Vision Research, Doheny Eye Institute, and M.M.S. is the William R. Acquavella Scholar of Ophthalmic Research, Columbia University.
References
- 1.Kaplan J, Bonneau D, Frezal J, Munnich A, Dufier JL. Clinical and genetic heterogeneity in retinitis pigmentosa. Hum Genet. 1990;85:635–642. doi: 10.1007/BF00193589. [DOI] [PubMed] [Google Scholar]
- 2.Sohocki MM, Bowne SJ, Sullivan LS, Blackshaw S, Cepko CL, Payne AM, Bhattacharya SS, Khaliq S, Mehdi SQ, Birch DG, et al. Mutations in the novel photoreceptor–pineal gene on l7p cause Leber congenital amaurosis. Nat Genet. 2000;24:79–83. doi: 10.1038/71732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sohocki MM, Perrault I, Leroy HP, Payne AM, Dharmaraj S, Bhattacharya SS, Kaplan J, Maumenee IH, Koenekoop R, Meire FM, et al. Prevalence of AIPL1 mutations in inherited retinal degenerative disease. Mol Genet Metab. 2000;70:142–150. doi: 10.1006/mgme.2000.3001. [DOI] [PubMed] [Google Scholar]
- 4.Blatch GL, Lassle M. The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. BioEssays. 1999;21:932–939. doi: 10.1002/(SICI)1521-1878(199911)21:11<932::AID-BIES5>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 5.Sohocki MM, Sullivan LS, Tirpat DL, Daiger SP. Comparative analysis of aryl-hydrocarbon receptor interacting protein-like 1 (Aip11), a gene associated with inherited retinal disease in humans. Mamm Genome. 2001;12:566–568. doi: 10.1007/s003350020024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kay BK, Williamson MP, Sudol M. The importance of being proline: the interaction of proline-rich motifs in signaling proteins with their cognate domains. FASEB J. 2000;14:231–241. [PubMed] [Google Scholar]
- 7.Kuzhandaivelu N, Cong YS, Inouye C, Yang WM, Seto E. XAP2, a novel hepatitis B virus X-associated protein that inhibits X transactivation. Nucleic Acids Res. 1996;24:4741–4750. doi: 10.1093/nar/24.23.4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carver LA, Bradfield CA. Ligand-dependent interaction of the arylhydrocarbon receptor with a novel immunophilin homolog in vivo. J Biol Chem. 1997;272:11452–11456. doi: 10.1074/jbc.272.17.11452. [DOI] [PubMed] [Google Scholar]
- 9.Ma Q, Whitlock JP. A novel cytoplasmic protein that interacts with the Ah receptor, contains tetratricopeptide repeat motifs, and augments the transcriptional response to 2,3,7,8-tetrachlorodibenzo-p-dioxin. J Biol Chem. 1997;272:8878–8884. [PubMed] [Google Scholar]
- 10.Myer BK, Pray-Grant MG, Vanden Heuvel JP, Perdew GH. Hepatitis B virus X-associated protein 2 is a subunit of the unliganded aryl hydrocarbon receptor core complex and exhibits transcriptional enhancer activity. Mol Cell Biol. 1998;18:978–998. doi: 10.1128/mcb.18.2.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Myer BK, Perdew GH. Characterization of the AhR–hsp90–XAP2 core complex and the role of the immunophilin-related protein XAP2 in AhR stabilization. Biochemistry. 1999;38:8907–8917. doi: 10.1021/bi982223w. [DOI] [PubMed] [Google Scholar]
- 12.LaPres JJ, Glover E, Dunham EE, Bunger MK, Bradfield CA. ARA9 modifies agonist signaling through an increase in cytosolic aryl hydrocarbon receptor. J Biol Chem. 2000;275:6153–6159. doi: 10.1074/jbc.275.9.6153. [DOI] [PubMed] [Google Scholar]
- 13.Petrulis JR, Hord NG, Perdew GH. Subcellular localization of the aryl hydrocarbon receptor is modulated by the immunophilin homolog of hepatitis B virus X-assocaited protein 2. J Biol Chem. 2000;257:37448–37453. doi: 10.1074/jbc.M006873200. [DOI] [PubMed] [Google Scholar]
- 14.Kazlauskas A, Poellinger L, Pongratz I. The immunophilin-like protein XAP2 regulates ubiquitination and subcellular localization of the dioxin receptor. J Biol Chem. 2000;275:41317–41324. doi: 10.1074/jbc.M007765200. [DOI] [PubMed] [Google Scholar]
- 15.Meyer BK, Petrulis JR, Perdew GH. Arly hydrocarbon (Ah) receptor levels are selectively modulated by hsp90-associated immunophilin homolog XAP2. Cell Stress Chaperones. 2000;5:243–254. doi: 10.1379/1466-1268(2000)005<0243:aharla>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kito K, Yeh ETH, Kamitani T. NUB1, a NEDD8-interacting protein, is induced by interferon and down-regulates the NEDD8 expression. J Biol Chem. 2001;276:20603–20609. doi: 10.1074/jbc.M100920200. [DOI] [PubMed] [Google Scholar]
- 17.Kamitani T, Kito K, Fukuda-Kamitani T, Yeh ETH. Targeting of NEDD8 and its conjugates for proteasomal degradation of NUB1. J Biol Chem. 2001;276:46655–46660. doi: 10.1074/jbc.M108636200. [DOI] [PubMed] [Google Scholar]
- 18.Blackshaw S, Fraioli RE, Furukawa T, Cepko CL. Comprehensive analysis of photoreceptor gene expression and the identification of candidate retinal disease genes. Cell. 2001;107:579–589. doi: 10.1016/s0092-8674(01)00574-8. [DOI] [PubMed] [Google Scholar]
- 19.Furukawa T, Morrow EM, Li T, Davis FC, Cepko CL. Retinopathy and attenuated circadian entrainment in Crx-deficient mice. Nat Genet. 1999;23:466–470. doi: 10.1038/70591. [DOI] [PubMed] [Google Scholar]
- 20.Dyer MA, Cepko CL. Regulating proliferation during retinal development. Nat Rev Neurosci. 2001;2:333–342. doi: 10.1038/35072555. [DOI] [PubMed] [Google Scholar]
- 21.van der Spuy J, Chapple JP, Clark BJ, Luthert PJ, Sethi CS, Cheetham ME. The Leber congenital amaurosis gene product AIPL1 is localized exclusively in rod photoreceptors of the adult human retina. Hum Mol Genet. 2002;11:823–831. doi: 10.1093/hmg/11.7.823. [DOI] [PubMed] [Google Scholar]
- 22.Bell DR, Poland A. Binding of aryl hydrocarbon receptor (AhR) to AhR-interacting protein. The role of hsp90. J Biol Chem. 2000;275:36407–36414. doi: 10.1074/jbc.M004236200. [DOI] [PubMed] [Google Scholar]
- 23.Yu ZK, Gervais JLM, Zhang H. Human CUL-1 associates with the SKP1/SKP2 complex and regulates p21CIP1/WAF1 and cyclin D proteins. Proc Natl Acad Sci USA. 1998;95:11324–11329. doi: 10.1073/pnas.95.19.11324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maeda I, Ohta T, Koizumi H, Fukuda M. In vitro ubiquitination of cyclin D1 by ROC1–CUL1 and ROC1–CUL3. FEBS Lett. 2001;494:181–185. doi: 10.1016/s0014-5793(01)02343-2. [DOI] [PubMed] [Google Scholar]
- 25.Podust VN, Brownell JE, Gladysheva TB, Luo RS, Wang C, Coggins MB, Pierce JW, Lightcap ES, Chau V. A Nedd8 conjugation pathway is essential for proteolytic targeting of p27Kip1 by ubiquitination. Proc Natl Acad Sci USA. 2000;97:4579–4584. doi: 10.1073/pnas.090465597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu X, Craft CM. Interaction of phosducin and phosducin isoforms with a 26S proteasomal subunit, SUG1. Mol Vis. 1998;4:13–20. [PubMed] [Google Scholar]
- 27.Zhu X, Craft CM. Modulation of CRX transactivation activity by phosducin isoforms. Mol Cell Biol. 2000;20:5216–5226. doi: 10.1128/mcb.20.14.5216-5226.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoffman C, Winston F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene. 1987;57:267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- 29.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. Current Protocols in Molecular Biology. Vol. 2. Wiley; New York: 1994–2001. pp. 13.6.2–13.6.5. [Google Scholar]
- 30.Craft CM, Xu J, Slepak VZ, Zhan-Poe X, Zhu X, Brown B, Lolley RN. PhLPs and PhLOPs in the phosducin family of G βγ binding proteins. Biochemistry. 1998;37:15758–15772. doi: 10.1021/bi980921a. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Y, Li A, Zhu X, Wang C, Brown B, Craft CM. Proceedings of the Ninth International Symposium on Retinal Degeneration Durango CO. Kluwer Academic/Plenum Publishers; New York: 2001. Cone arrestin expression and induction in retinoblastoma cells; pp. 309–316. [Google Scholar]
- 32.Dyer MA, Cepko CL. p27Kip1 and p57Kip2 regulate proliferation in distinct retinal progenitor cell populations. J Neurosci. 2001;21:4259–4271. doi: 10.1523/JNEUROSCI.21-12-04259.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dyer MA, Cepko CL. p57(Kip2) regulates progenitor cell proliferation and amacrine interneuron development in the mouse retina. Development. 2000;127:3593–3605. doi: 10.1242/dev.127.16.3593. [DOI] [PubMed] [Google Scholar]