Abstract
Outer membrane (OM) cytochromes OmcA (SO1779) and MtrC (SO1778) are the integral components of electron transfer used by Shewanella oneidensis for anaerobic respiration of metal (hydr)oxides. Here the OmcA–MtrC interaction was identified in vivo using a novel hydrophobic chemical cross-linker (MRN) combined with immunoprecipitation techniques. In addition, identification of other OM proteins from the cross-linked complexes allows first visualization of the OmcA–MtrC interaction network. Further experiments on omcA and mtrC mutant cells showed OmcA plays a central role in the network interaction. For comparison, two commercial cross-linkers were also used in parallel, and both resulted in fewer OM protein identifications, indicating the superior properties of MRN for identification of membrane protein interactions. Finally, comparison experiments of in vivo cross-linking and cell lysate cross-linking resulted in significantly different protein interaction data, demonstrating the importance of in vivo cross-linking for study of protein–protein interactions in cells.
Keywords: protein interactions, Protein Interaction Reporter, PIR, electron transport, metal ion reduction, membrane proteins, FTICR-MS, outer membrane cytochrome C, OmcA, OmcB, MtrC, SO-0404
Introduction
Shewanella oneidensis MR-1, a facultatively anaerobic bacterium, is able to use a wide range of terminal electron acceptors during anaerobic respiration. The metal reducing activity of S. oneidensis provides an alternative strategy for remediation of the sites contaminated with toxic metals such as uranium, technetium, and chromium.1–3 A greater understanding of the underlying mechanisms of this metal reducing activity will enable utilization of S. oneidensis in bioremediation, bioenergy production, or other areas of biotechnology that can benefit from electron transfer.
Because of their cell surface location,4 outer membrane (OM) decaheme c-type cytochromes OmcA (SO1779) and MtrC (SO1778) play critical roles in the reduction of extracellular electron acceptors. Deletion of the genes impairs the ability of Shewanella to reduce Fe(III) and Mn(VI) oxides.4–12 Purified OmcA and MtrC are functional metal reductases with the ability to bind and reduce solid metal oxide such as hematite.8,13 In addition, OmcA and MtrC are directly involved in extracellular reduction of uranyl carbonate complexes to uraninite.14 A previous study showed that MtrC was copurified with recombinant OmcA, and purified OmcA and MtrC formed a stable complex in vitro.13 Using “in vivo” cross-linking by formalde-hyde and Western blot, the OmcA–MtrC interaction has also been reported.15 Despite recent advances made in understanding the roles of OmcA and MtrC in reduction of metals, the other components of the OmcA/MtrC-mediated electron transfer pathway have yet to be identified.
Mapping protein–protein interaction networks in vivo is crucial for understanding the nature of biological processes at the systems-level. However, determination of protein interactions in native living systems is largely an unmet challenge for today's technology. The difficulties of this type of analysis stem from the fact that the primary physical property that must be detected is the close proximity of interacting partners. Often times, this is difficult to detect even for specific isolated interactions. For large-scale determinations, this becomes improbable, if not impossible. When specific antibodies are available, immunoprecipitation of the target protein along with its noncovalent binding protein partners, i.e., coimmunoprecipitation (co-IP), has been a commonly used technique for identifying potential interacting proteins surrounding the target proteins.16–21 Co-IP methods have generated large-scale protein interaction data in yeast, mammalian, and many other organisma, and the validation of many of these results with orthogonal methods confirms the utility of these methods. However, prior to analysis, co-IP-related methods require cell lysis during which the native environment is disrupted, and nonspecific binding to the antigen may occur resulting in false identification of protein–protein interactions. In fact, nonspecific interactions are one of the most challenging impediments to nearly every protein interaction determination.
Chemical cross-linking coupled with immunoprecipitation provides an alternative strategy for in vivo identification of protein–protein interactions,22–36 which has been extensively reviewed.37–39 Cross-linking reactions can be carried out with intact cells and chemically “freeze” protein–protein interactions with stable covalent bonds that allow subsequent purification steps to be carried out under much harsher or more stringent conditions. Consequently, nonspecific binding can be reduced considerably. In addition, immunoprecipitation in conjunction with cross-linking is well suited for investigating the interactions of membrane proteins. Isolation and purification of membrane proteins usually requires use of detergents that can sometimes disrupt interactions among membrane proteins. Thus, stabilization of the complexes with cross-linkers prior to immunoprecipitation of membrane proteins significantly increases the chances of identification of the proteins bound to the antigens.
In this study, we report the development and application of a novel type of cross-linker coupled with immunoprecipitation techniques to specifically identify OM protein–protein interactions. OmcA and MtrC interactions were targeted in this study due to the critical importance of these proteins in the electron transport pathway of S. oneidensis. For comparison, two commercial cross-linkers, DSS and DST, were also applied in parallel. We previously reported a new family of cross-linkers, from a class of reagents that we call protein interaction reporter (PIR) molecules, with the incorporation of tunable spacer chain chemistry, mass spectrometry-cleavable bonds, affinity tag, and hydrophobic groups.40,41 We further demonstrated that the hydrophobic PIR cross-linkers preferentially label membrane proteins.42 However protein–protein interactions identified in vivo with PIR cross-linkers have not yet been reported. This research illustrates that novel hydrophobic PIR cross-linkers allow identification of more OM protein interactions as compared to DSS and DST, indicating hydrophobicity of the PIR cross-linker is advantageous for mapping OM protein–protein interactions. Collectively, these cross-linking studies not only showed that OmcA and MtrC closely interact with each other in cells but also revealed the identities of many other OM proteins that are likely components of a larger OmcA–MtrC extracellular electron transfer complex or interaction network.
Experimental Procedures
Chemical Cross-Linkers
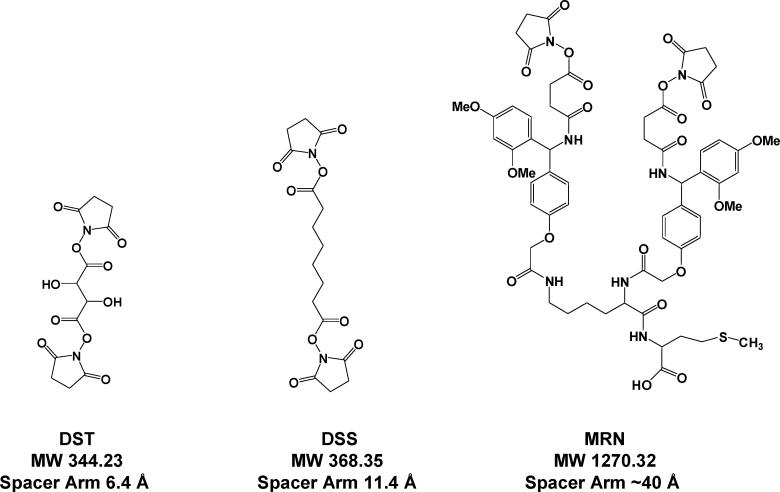
The novel hydrophobic PIR cross-linker, MRN (Figure 1), was synthesized using solid phase peptide synthesis methodology. First, methionine was coupled to HMPB-MBHA (4-hydroxymethyl-3-methoxyphenoxybutyric acid) resin. Then, the lysine in the form of Fmoc-Lys-ε-Fmoc was coupled to methionine and formed a branch point for the cross-linker. Finally, the Rink groups, succinic anhydride (SA), and N-hydroxysuccinimides (NHS) were coupled as described previously.41 The crude product was cleaved using either 0.5 or 1.0% TFA in chloroform and then neutralized with pyridine. The chloroform and TFA pyridine salts were removed under a vacuum. The crude product was purified using reversed-phase HPLC, and the final product had a purity of approximately 90%.
Figure 1.
Structures of the novel hydrophobic PIR cross-linker, MRN, and two commercial cross-linkers, DSS and DST.
Two commercial cross-linkers (Pierce, Rockford, IL), disuccinimidyl suberate (DSS) and disuccinimidyl tartarate (DST), were used to label S. oneidensis cells without further purification (Figure 1).
All three chemical cross-linkers contain two N-hydroxysuccinamide (NHS) esters, which react with primary amine groups in proteins and form stable covalent bonds. DSS, DST, and MRN have a maximum spacer arm length of 11.4, 6.4, and 40 Å, respectively. DST is soluble both in water and in organic solvents. DSS and MRN can only be dissolved in organic solvents such as DMSO or DMF. Thus, DST is less hydrophobic than DSS and MRN. HPLC was used to further characterize the hydrophobicity of DSS and MRN. The retention time of MRN is 50% higher than that of DSS under the same LC conditions, which indicates significantly higher hydrophobicity of MRN as compared to DSS (data not shown).
Cell Culture
S. oneidensis MR-1 cells, ATCC 700550, were obtained from American Type Culture Collection (Manassas, VA) and maintained in Luria–Bertani (LB) medium. A single colony was transferred with an inoculating loop to 40 mL LB broth and was incubated overnight at room temperature with rotary shaking (100 rpm). An amount of 4 mL of the overnight culture was transferred to 200 mL fresh LB in a 1 L flask and incubated with a rotary shaker at 100 rpm at room temperature for 8 h. Cells were harvested by centrifuging at 3000g for 20 min. Cell pellets were washed 4 times with 40 mL of ice-chilled PBS buffer (150 mM sodium phosphate, 100 mM NaCl, pH 7.5).
OmcA and MtrC mutant cells of S. oneidensis MR-1 were generated as described previously14,43 and were cultured under the same conditions as wild type cells.
Cross-Linking Labeling and Cell Lysis
After the last wash, about 1 × 107 cells were suspended in 1 mL of PBS buffer, and the cross-linkers were added to the suspended cell pellets to produce a final concentration of 1 mM. The cross-linking reaction was carried out at 4 °C for 1 h and quenched by 1 M ammonium bicarbonate. Extensive washing steps with PBS after cross-linking reactions were used to eliminate most nonspecific contamination as reported previously.42 Then, cells were lysed in 0.1% NP-40 PBS solution by sonication for 2 min. The cell lysates were centrifuged at 15 000 g at 4 °C for 45 min. The pellets were discarded, and supernatants were collected in a clean tube. The protein concentration of the supernatants was determined by a Bradford Assay (Bio-Rad, Hercules, CA). Cell-lysate cross-linking experiments were performed under the same experimental conditions as in vivo cross-linking except that the cross-linkers were added after cells were lysed.
Immunoprecipitation (IP), SDS-PAGE, In-Gel Digestion, and Western Blot Analysis
Recombinant MtrC or OmcA were individually purified from S. oneidensis cells,13 and respective polyclonal antibodies toward each protein were commercially generated and purified by affinity chromatography (BioSyn-thesis, Inc., Lewisville, TX). Antibody specificities were validated by immunoblot analysis of S. oneidensis cell lysate (Supplementary Figure 5).
An amount of 10 μg of OmcA or MtrC antibody was added to 400 μL of cross-linker-labeled cell lysate, and the reaction was incubated at 4 °C overnight. An amount of 50 μL of immobilized protein G gel slurry (Pierce, Rockford, IL) was added to antigen–antibody complex solution, and the reaction was incubated for 2 h with gentle mixing at room temperature. Then, the protein G beads were washed twice with 500 μL of IP buffer (25 mM Tris, 150 mM NaCl, pH 7.2) and once with 25 mM Tris buffer (pH 7.2). After the last wash, protein G beads were incubated with 50 μL of SDS-PAGE sample loading buffer (Bio-Rad, Hercules, CA) for 5 min at 95 °C. The supernatant was collected after centrifugation (2500g) and then analyzed by 8% SDS-PAGE. Protein bands were visualized with Coo-massie Blue R250 (Bio-Rad, Hercules, CA). In-gel digestion was performed as described previously.42
For Western blot analysis, the eluents from Protein G beads were separated by 8% SDS-PAGE gel and then transferred to a nitrocellulose membrane (Whatman, Sanford, ME) with a Trans-Blot semidry transfer cell (BioRad, Hercules, CA). After transferring, the membranes were blocked with 5% nonfat milk in TBS overnight at 4 °C followed by incubation with the primary antibody anti-OmcA/anti-MtrC at 1:5000 dilution for 1 h at room temperature. Finally, the membranes were probed by HRP-conjugated secondary antibody and chemiluminescence peroxidase substrate (Sigma, St. Louis, MO).
LC/MS/MS and Protein Identification
LC/MS/MS analysis of in-gel digest was performed using an ion trap mass spectrometer (Esquire HCT, Bruker Daltonics, Billerica, MA) equipped with a nano ESI source and nano HPLC systems (Ultimate, Dionex, Sunnyvale, CA). Data analysis was performed with Bruker Daltonics DataAnalysis software (version 3.1). Protein identification was carried out by searching against the complete genome database of S. oneidensis MR-1 (4,854 ORFs) downloaded from The Institute for Genomic Research (TIGR) (http://www.tigr.org/), using Mascot44 (version 2.1.0, MatrixScience Ltd., London) licensed in house. Database search parameters were used as previously reported;42 briefly: enzyme, trypsin; allowed missed cleavages up to 3; fixed modifications, carbamidomethyl (C); variable modifications, oxidation (M); peptide tolerance, 1.6 Da; and MS/MS tolerance, 0.8 Da. The auto hits option was selected to allow reporting of all the protein hits with the probability-based Mowse scores that exceeded their thresholds (p < 0.05), indicating significance above the 95% confidence level. The search output results were further filtered using more stringent MudPIT scoring and an ion score cutoff of 0.05, which removed all the peptides with expected values (E) > 0.1. All proteins identified with more than 2 unique peptides which had a minimal total peptide–ion score of 45 were accepted as true identifications without additional manual spectral inspection. There is one exception, which is that the major outer membrane lipoprotein (SO1295) which has a mass of 9 kDa was identified by a single peptide with high score (>78) in all experiments. To evaluate false positive identification, searches were also performed against all mammalia of MSDB, which contains 280 595 sequences. In most searches, only trypsin and various forms of keratins (common contamination during in-gel digestion) were identified with significance; other searches resulted in no significant protein hits. Protein interaction visualization was carried out using Cytoscape (version 2.4.0) downloaded from http://www.cytoscape.org/.45
Results
Anti-OmcA and Anti-MtrC Immunoprecipitation
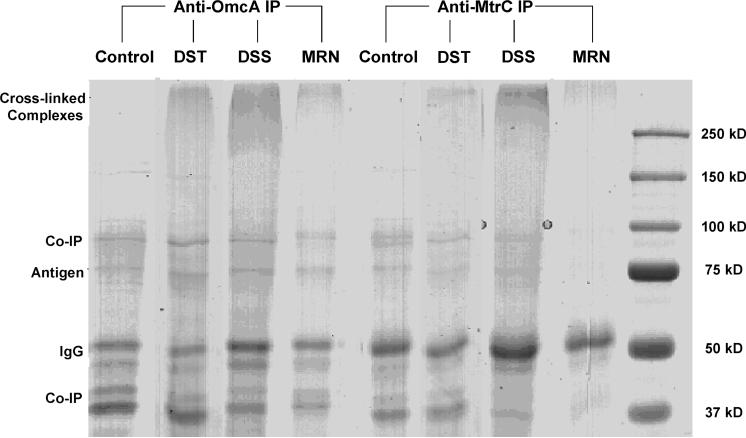
The structures of three cross-linkers, MRN, DSS, and DST, are shown in Figure 1. Cells are harvested, washed, and then reacted with each cross-linker separately. After IP from cross-linked cell lysates, five types of proteins were expected to be eluted from protein G beads, i.e., (i) antigen (either OmcA or MtrC), (ii) proteins that bind with antigen in strong noncovalent bonds (co-IP products), (iii) cross-linked protein complexes with a component of antigen, (iv) nonspecific binding proteins, and (v) IgGs. As shown in Figure 2, the antigen (either OmcA or MtrC) band appeared at the expected molecular weight (MW) of ∼75 kD in every sample. These bands were excised, and the presence of antigens was confirmed by LC/MS/MS following in-gel digestion (Supplementary Table 1). The bands at ∼50 kD were identified to be IgGs. Other bands which appeared in both control and labeled samples were the results of co-IP and/or nonspecific binding. Some of these bands were also selected for protein identification (Supplementary Figure 1 and Supplementary Table 1). It is worth noting that in all labeled samples there were bands above a MW of 250 kD which were not observed in the control samples. These high MW bands are likely to be cross-linked protein complexes with either OmcA or MtrC. To further verify this, we investigated the protein mass distribution of the entire genome database in S. oneidensis cells and plotted protein counts vs MW (Supplementary Figure 2). This histogram shows most proteins have a MW between 10 and 100 kD, and only four proteins have a MW of about 250 kD (SO0189, SO1602, SO4149, and SO4317). This further suggests that the bands above 250 kD can only be from cross-linked proteins.
Figure 2.
SDS-PAGE analysis of the proteins immunoprecipitated with the OmcA or MtrC antibody from in vivo cross-linked cells by DST, DSS, or MRN. IP of uncross-linked cells served as a control.
Identification of Cross-Linked Complexes
The stained regions that appeared above 250 kD for labeled cells and the corresponding blank regions for unlabeled cells (Figure 2) were excised and subjected to in-gel digestion and LC/MS/MS. No proteins were identified with significant Mascot scores for unlabeled cells. The Mascot scores of identified proteins for labeled cells are reported in Table 1. In most cases, both OmcA and MtrC were identified with the highest Mascot scores, especially those cross-linked by MRN and DSS. This provides strong evidence of close interaction between OmcA and MtrC in S. oneidensis cells and is consistent with the results that were obtained previously from in vitro or in vivo measurements.13,15 As for DST-labeled samples, MtrC was identified in the anti-OmcA IP; however, OmcA was not identified in the anti-MtrC IP. In addition to OmcA and MtrC, a few other OM proteins, as well as some periplasmic, inner membrane, and cytoplasmic proteins, were identified and are listed in Table 1.
Table 1.
Identified Cross-Linked Proteins from > 250 kD Gel Bands in Anti-OmcA and Anti-MtrC IP of MRN, DSS, and DST in Vivo Labelinga
| Anti-OmcA IP |
Anti-MtrC IP |
MW |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| gene | MRN | DSS | DST | MRN | DSS | DST | annotation | categoryb | (kDa) |
| SO1779 | 789 | 432 | 523 | 811 | 132 | decaheme cytochrome c, OmcA | OM | 79 | |
| SO1778 | 378 | 49 | 118 | 343 | 307 | 136 | decaheme cytochrome c, MtrC | OM | 72 |
| SOa0110 | 253 | 322 | lipoprotein, putative | OM | 145 | ||||
| SO3545 | 228 | 45 | 376 | 123 | 189 | OmpA family protein | OM | 40 | |
| SO3896 | 149 | 67 | 90 | 97 | outer membrane porin, putative | OM | 39 | ||
| SO1295 | 126 | 78 | 202 | 153 | 96 | 153 | major outer membrane lipoprotein | OM | 9 |
| SO0404 | 112 | hypothetical protein | OM | 129 | |||||
| SO1429 | 58 | anaerobic dimethyl sulfoxide reductase | OM | 91 | |||||
| SO3099 | 57 | long chain fatty acid transport, putative | OM | 47 | |||||
| SO2001 | 46 | 5-nucleotidase | OM | 61 | |||||
| SO1824 | 134 | 51 | conserved hypothetical | P | 25 | ||||
| SO1825 | 174 | 103 | 87 | MotA/TolQ/ExbB proton channel protein | IM | 48 | |||
| SO3286 | 105 | 76 | 126 | cytochrome c ubiquinol oxidase | IM | 58 | |||
| SO4747 | 100 | ATP synthase F1, beta subunit | C | 49 | |||||
| SO0217 | 79 | 94 | 231 | 91 | 146 | 120 | translation elongation factor Tu | C | 43 |
| SO4749 | 54 | ATP synthase F1, alpha subunit | C | 55 | |||||
| SO0237 | 141 | 125 | ribosomal protein S3 | C | 25 | ||||
| SO1930 | 72 | 2-oxoglutarate dehydrogenase, sucA | C | 105 | |||||
The numbers in the table are the Mascot scores.
OM denotes outer membrane; P denotes periplasmic; IM denotes inner membrane; and C denotes cytoplasmic.
Western Blot Analysis to Confirm OmcA–MtrC Interaction
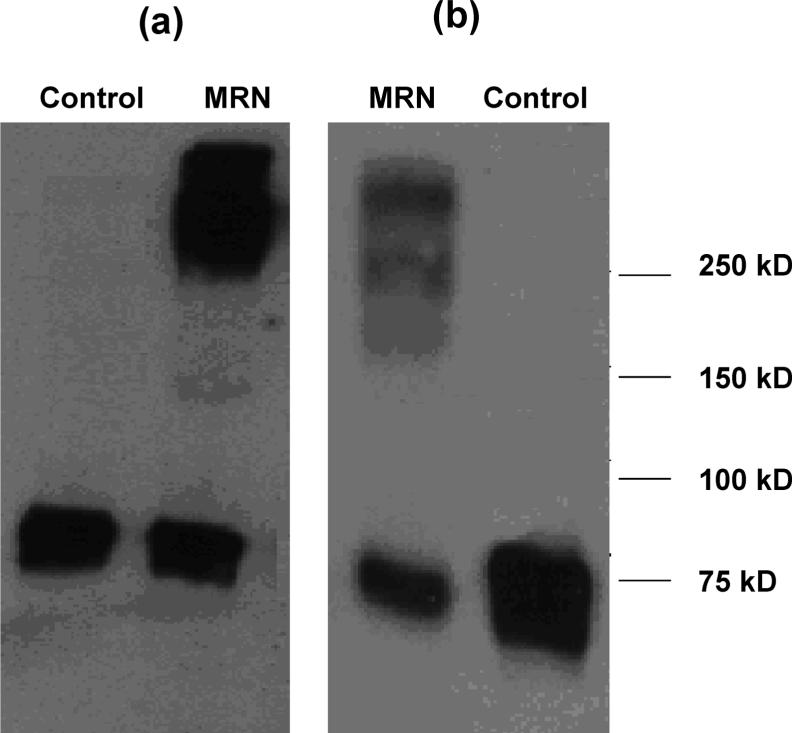
To further verify OmcA and MtrC interaction, anti-OmcA and anti-MtrC Western blot analyses were done with MtrC and OmcA IP eluents, respectively, which showed a strong contrast between labeled and unlabeled samples (Figure 3). Both Western blot images show their corresponding antigen bands, either OmcA or MtrC, that appear near 75 kDa MW, resulting from copurification from MtrC or OmcA IP. More importantly, the cross-linked MtrC/OmcA complexes >250 kDa only appeared in MRN labeled cells, which confirms the OmcA–MtrC interaction.
Figure 3.
(a) Anti-OmcA Western blot analysis after anti-MtrC immunoprecipitation. (b) Anti-MtrC Western blot analysis after anti-OmcA immunoprecipitation.
In Vivo Cross-Linking on omcA and mtrC Mutant Cells
To further investigate protein–protein interactions among OmcA, MtrC, and other OM proteins, in vivo cross-linking as well as anti-OmcA and anti-MtrC IP experiments were carried out with mtrC and omcA mutant cells, respectively. SDS-PAGE images from MRN and DSS labeled mutant cell samples appeared similar to that of wild type cells except that the observed intensity of the cross-linking complex band above 250 kDa from omcA mutant cells was lower (Supplementary Figure 3). All cross-link-specific bands were excised and subjected to protein identification (Table 2). OmcA and MtrC were not identified in corresponding immunoprecipitated cross-linked complexes from either mutant cells, as one might expect since their genes are deleted. As for the mtrC mutant, four OM proteins (SO0404, SO1429, SO3099, and SO2001), one inner membrane protein (SO1825), and two cytosolic proteins (SO1930, SO4749) were missing as compared to the results from experiments with wild type cells. In strong contrast, however, all the proteins except for the major outer membrane lipoprotein (SO1295) and translation elongation factor Tu (SO0217) were not observed in cross-linking results from anti-MtrC IP experiments with omcA deletion mutant cells. In fact, this striking change was observed in both MRN and DSS experiments.
Table 2.
Identified Cross-Linked Proteins from > 250 kD Gel Bands in Anti-OmcA and Anti-MtrC IP of MRN and DSS in Vivo Labeling on MtrC and OmcA Mutant Cells, Respectivelya
| anti-OmcA |
anti-MtrC |
||||||
|---|---|---|---|---|---|---|---|
| ΔMtrC |
ΔOmcA |
MW |
|||||
| gene | MRN | DSS | MRN | DSS | annotation | categoryb | (kDa) |
| SO1779 | 971 | 476 | decaheme cytochrome c, OmcA | OM | 79 | ||
| SO1778 | 333 | 312 | decaheme cytochrome c, MtrC | OM | 72 | ||
| SOa0110 | 308 | lipoprotein, putative | OM | 145 | |||
| SO3545 | 472 | 149 | OmpA family protein | OM | 40 | ||
| SO3896 | 100 | 125 | outer membrane porin, putative | OM | 39 | ||
| SO1295 | 183 | 153 | 179 | 106 | major outer membrane lipoprotein | OM | 9 |
| SO1824 | 69 | conserved hypothetical | P | 25 | |||
| SO3286 | 68 | cytochrome c ubiquinol oxidase | IM | 58 | |||
| SO1490 | 306 | alcohol dehydrogenase II, AdhB | C | 40 | |||
| SO4747 | 108 | ATP synthase F1, beta subunit | C | 49 | |||
| SO0217 | 82 | 179 | translation elongation factor Tu | C | 43 | ||
| SO0237 | 105 | ribosomal protein S3 | C | 25 | |||
| SO0314 | 57 | ornithine decarboxylase | C | 82 | |||
The numbers in the table are the Mascot scores.
OM denotes outer membrane; P denotes periplasmic; IM denotes inner membrane; and C denotes cytoplasmic. ΔMtrC denotes mtrC mutant, and ΔOmcA denotes omcA mutant.
Identification of Cross-Linked Protein Complexes from Cell-Lysate Cross-Linking
Protein–protein interactions can occur transiently, and sometimes only a few amino acids of proximal proteins are required for the interactions. The local environments close to the interaction sites play a critical role for the stability of noncovalent protein interactions. When the environment is changed, such as during cell lysis, the original interactions may be disturbed and new interactions may occur, some of which may hold no physiological relevance. However, to the best of our knowledge, no comparison has been made to determine the degrees of difference between the protein–protein interactions measured from intact cells cross-linked prior to cell lysis with those observed when cross-linking was performed after cell lysis. To investigate this issue, we performed similar anti-OmcA IP experiments with cell lysates that were cross-linked with MRN and DSS immediately after lysis.
As with in vivo cross-linking experiments, we observed high mass gel bands (MW > 250 kD) in these cell-lysate cross-linked samples that were not present in control samples (Supplementary Figure 4). These high MW bands were excised for protein identification, and the results are summarized in Table 3. In contrast to the results obtained with in vivo cross-linking, IP experiments with DSS resulted in identification of many more cross-linked proteins from cell lysate application as compared with MRN. Furthermore, a large number of cytosolic proteins were identified with significantly high Mascot scores, indicating their high abundance and/or high reactivity with cross-linkers.
Table 3.
Identified Cross-Linked Proteins from >250 kD Gel Bands in Anti-OmcA IP of MRN and DSS Cell-Lysate Labelinga
| gene | DSS | MRN | annotation | categoryb | MW (kDa) |
|---|---|---|---|---|---|
| SO0225 | 1928 | DNA-directed RNA polymerase, rpoC | C | 156 | |
| SO0224 | 1431 | 74 | DNA-directed RNA polymerase, rpoB | C | 150 |
| SO1490 | 987 | alcohol dehydrogenase II, AdhB | C | 40 | |
| SO0229 | 928 | 359 | translation elongation factor Tu, TufA | C | 43 |
| SO0217 | 888c | 367c | translation elongation factor Tu, TufB | C | 43 |
| SO4749 | 467 | ATP synthase F1, alpha subunit | C | 55 | |
| SO1142 | 321 | 88 | carbamoyl-phosphate synthase, CarB | C | 119 |
| SO4747 | 238 | 45c | ATP synthase F1, beta subunit | C | 50 |
| SO2638 | 170 | leucine dehydrogenase, Ldh | C | 37 | |
| SO0314 | 161 | 130 | ornithine decarboxylase, SpeF | C | 82 |
| SO3032 | 160 | NTP-dependent alcaligin synthetase, AlcC | C | 69 | |
| SO1679 | 120 | branched-chain acyl-CoA dehydrogenase | C | 42 | |
| SOA0112 | 117 | lipoprotein, putative | OM | 146 | |
| SO1521 | 100 | conserved hypothetical protein | C | 103 | |
| SO3532 | 91 | isoleucyl-tRNA synthetase, IleS | C | 106 | |
| SO0021 | 86 | fatty oxidation complex, alpha subunit | C | 77 | |
| SO0256 | 81 | DNA-directed RNA polymerase, RpoA | C | 36 | |
| SO4030 | 73 | excinuclease ABC, A subunit, UvrA | C | 106 | |
| SO1295 | 68c | 138c | major outer membrane lipoprotein | OM | 9 |
| SO3440 | 56 | enolase, Eno | C | 42 | |
| SO1779 | 41c | 140c | OmcA | OM | 80 |
| SO3542 | 55 | conserved hypothetical protein | C | 90 |
The numbers in the table are the Mascot scores.
C denotes cytoplasmic, and OM denotes outer membrane.
Indicates that those proteins were also identified from on-cell labeling.
Discussion
To identify OmcA and MtrC interactions and their interaction networks, we have developed an in vivo cross-linking strategy that includes novel hydrophobic compounds which facilitate cross-linking proteins within cell membranes. Analysis of the results presented in Table 1 shows that for both anti-OmcA and anti-MtrC IP cross-linking with MRN resulted in more identified proteins than that with DSS or DST. More importantly, the identified OM proteins resultant from DSS and DST experiments are a subset of those identified using MRN, indicating superior cross-linking properties of MRN for membrane protein interaction identification as compared with either DSS or DST. All three cross-linkers are cell membrane permeable. However, MRN is larger in size and much more hydrophobic than DSS and DST. Thus, MRN may penetrate cell membranes at a different rate than DSS or DST and may remain closer to hydrophobic membranes after cell penetration. The different penetration rate of MRN may also allow more reaction time with membrane proteins.
As shown in Figure 1, MRN has much longer spacer chain than DSS and DST. However, hydrophobicity of MRN seems to play a more important role than spacer length for enhanced labeling efficiency of OM proteins. This observation is supported by comparison of the results of in vivo cross-linking and cell lysate cross-linking experiments. Anti-OmcA IP using DSS resulted in identification of more proteins from cell lysate cross-linking (21 proteins) as opposed to those identified from in vivo cross-linking (9 proteins). On the other hand, anti-OmcA IP using MRN resulted in fewer cross-linked protein identifications in cell lysates (9 proteins) than was observed with intact cells (16 proteins). This further reflects the selectivity of the hydrophobic property of the cross-linkers for labeling OM proteins in cells. If MRN length were the dominant difference, one would expect proportionately more cross-linked proteins from cell lysate experiments, rather than fewer proteins. DSS is less hydrophobic than MRN and more soluble in cell lysates; thus, DSS exhibits good cross-linking activity with soluble proteins in cell lysates and likely results in higher cross-linked protein concentration than MRN (Supplementary Figure 4), whereas MRN works better under the hydrophobic environment of cell membranes leading to detection of more cross-linked membrane proteins in cells. It should also be noted that other PIR compounds we have synthesized with similar overall size as compared to MRN, but with decreased hydrophobic properties, do not provide the same performance for OM protein interaction identification as we observe with MRN (data not shown). Therefore, the novel PIR cross-linker, MRN, with increased hydrophobic properties allows greater capability of OM protein–protein interaction identification. In contrast, in vivo cross-linking experiments with DST failed to result in identification of the OmcA–MtrC interaction in the anti-MtrC IP experiment, most likely due to its weak hydrophobicity (Table 1). In all experiments, a few cytosolic proteins were identified which could result from cross-linker labeling after cell membrane penetration. However, we do not eliminate the possibility of nonspecific contamination of highly abundant proteins. For example, the translation elongation factor Tu has previously been observed as a contaminant in other proteomic studies.13,42
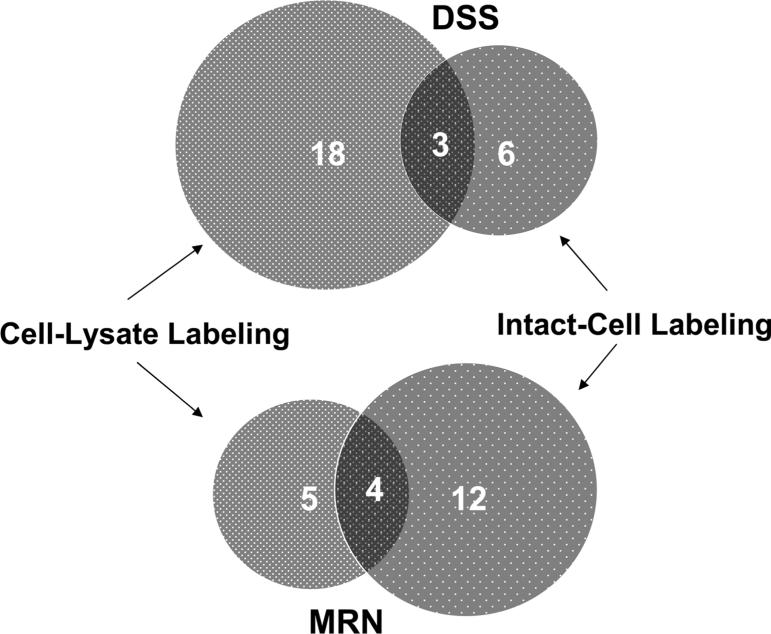
After cell lysis, proteins are extracted into the buffer solution. In contrast to the native environment of intact cells, protein conformation and its interacting partners may change dramatically in the cell lysate solution. Accordingly, cross-linked protein identities from cell-lysate cross-linking appear significantly different from those observed with in vivo cross-linking, as is indicated in the results shown in Table 1 and Table 3. For example, cell-lysate cross-linking experiments resulted in identification of mostly cytosolic proteins (18 out of 21 using DSS, and 7 out of 9 using MRN), while in vivo cross-linking experiments identified more membrane proteins (7 out of 9 using DSS, and 13 out of 16 using MRN). Interestingly, MtrC was not identified from either DSS or MRN cell-lysate labeling experiments, indicating either OmcA and MtrC interaction was interrupted or the cross-linking became less efficient after the cells were lysed. This comparison is further illustrated in the Venn diagram (Figure 4). Few proteins (3 using DSS and 4 using MRN) were identified both from cell-lysate and in vivo cross-linking including the antigen protein itself, OmcA. Copurified proteins identified from unlabeled cell lysates can be considered as traditional IP results (Supplementary Figure 1 and Supplementary Table 1). Both OmcA and MtrC were identified from unlabeled control samples at band B2 in Supplementary Figure 1 further supporting the identified OmcA and MtrC interaction from this work and that of others.13,15 However the IP of unlabeled samples also resulted in many other proteins that were different from those cross-linked proteins identified from in vivo cross-linking. Traditional co-IP experiments for identifying protein–protein interactions can only be applied after cells are lysed. Our results demonstrate that the interruption of living cells can change protein–protein interactions dramatically, especially for membrane proteins. Therefore, caution must be exercised when interpreting results from co-IP experiments or cell-lysate cross-linking experiments. Furthermore, this study also demonstrated the advantages of cross-linking methods which can provide snap-shots of the native protein–protein interactions in vivo.
Figure 4.
Venn-diagram analysis on cross-linked protein numbers of cell-lysate labeling and intact-cell labeling with anti-OmcA IP using DSS and MRN. The numbers in the overlaid area are the numbers of proteins that were identified in both cell-lysate labeling and intact-cell labeling.
OmcA and MtrC have been shown to be exposed on the outer membranes of S. oneidensis cells to allow direct electron transfer to extracellular electron acceptors during anaerobic respiration. Four identified proteins in Table 1 may be involved in either OmcA or MtrC transport across the OM or are components of a large protein complex that facilitates extracellular electron transport. Based on the identified proteins in Table 1, an OmcA–MtrC interaction network map was constructed with Cytoscape (Figure 5a). In addition to OmcA and MtrC, three OM proteins including the OmpA family protein (SO3545), the outer membrane porin (SO3896), and the major outer membrane lipoprotein (SO1295) were identified by cross-linking and immunoprecipitation of OmcA or MtrC with all three chemical cross-linkers. The first two OM proteins are predicted to be transmembrane β-barrel proteins,46,47 and the major outer membrane lipoprotein (SO1295) is known as one of the most abundant OM proteins in another Gram-negative bacterium, E. coli,48 that, like S. oneidensis, is a member of the γ-proteobacteria. The other five OM proteins, putative lipoprotein (SOa0110), hypothetical protein (SO0404), anaerobic DMSO reductase (SO1429), putative long chain fatty acid transport (SO3099), and 5-nucleotidase (SO2001), were only identified from MRN experiments presumably due to their more hydrophobic localization environment. Among all the identified OM proteins, the putative lipoprotein (SOa0110) as well as three other proteins, SO1295, SO3896, and SO3545, were identified in both anti-OmcA and anti-MtrC IP experiments, and the other four proteins (SO0404, SO1429, SO3099, and SO2001) were only identified in anti-OmcA IP experiments. One periplasmic protein (SO1824), two inner membrane proteins (SO1825, SO3286), and five cytosolic proteins (SO4749, SO1930, SO4747, SO0217, and SO0237) were identified as shown in Table 1. Identified cytosolic proteins could be the result of cross-linker penetration, nonspecific binding, and indirect binding through formation of larger complexes as discussed previously.42 Furthermore, results from in vivo cross-linking with omcA and mtrC deletion mutant cells strongly suggest many of the identified protein interactions, except for the major outer membrane lipoprotein (SO1295), are mediated by OmcA. Figure 5b and 5c illustrate the Cytoscape visualization of the subset of interactions observed from the mtrC and omcA deletion mutant cell experiments. In both cases, four OM proteins (SO0404, SO1429, SO3099, and SO2001) were missing as compared to the results from experiments with wild type cells and appear in gray in the figures, which suggests that both OmcA and MtrC are required to maintain these interactions. However, OmcA and MtrC could possibly play different roles in maintaining these interactions. Several cross-linked OM proteins (SOa0110, SO1295, SO3896, and SO3545) that were identified with high scores for wild type cells were also identified with the use of mtrC deletion mutant cells (except for MtrC). In addition, one inner membrane protein (SO1825) and two cytosolic proteins (SO1930, SO4749) were also missing for mtrC mutant cells (Figure 5b). In strong contrast, however, all proteins except for the major outer membrane lipoprotein (SO1295) and the translation elongation factor Tu (SO0217) were not observed in cross-linking results from anti-MtrC IP experiments with omcA deletion mutant cells, as illustrated in Figure 5c. It should be noted that translation elongation factor Tu (SO0217) has been reported as a contamination in other proteomic studies.13,42 These results strongly suggest that deletion of OmcA from cells has a strong influence on the interaction network of MtrC. A possible explanation for this observation is that the binding stoichiometry for the OmcA–MtrC interaction in cell membranes may involve more than one OmcA molecule per MtrC molecule, as observed in vitro by Shi et al.13 If this were the case, then one might expect that multiple OmcA molecules might occupy most interaction sites on MtrC. Therefore, MtrC might form cross-linking complexes with other OM proteins through the mediation of OmcA. The comprehensive protein complex network involved in electron transfer in S. oneidensis cells is still far from being fully determined, and most interacting protein components remain unknown. Nonetheless, our initial characterization of the OmcA–MtrC network provides a critical first step to help unveil the interaction map and further understand electron transfer mechanisms within S. oneidensis cells.
Figure 5.
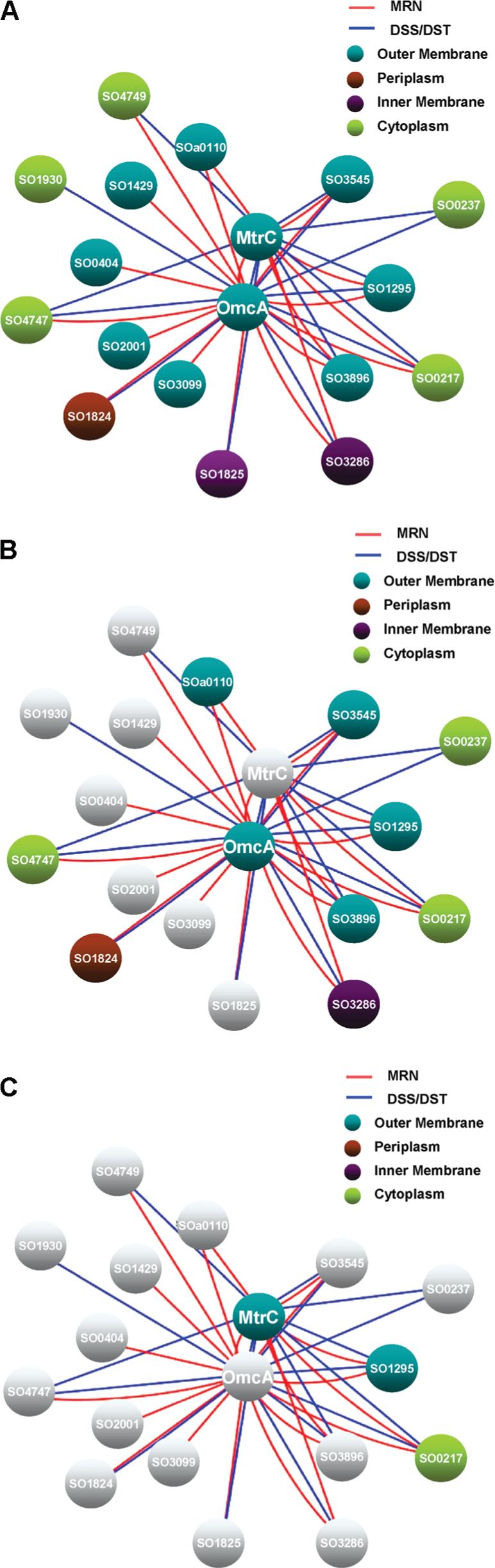
Cytoscape maps of the OmcA–MtrC interaction network based on in vivo cross-linking and IP results: (a) of wild type S. oneidensis MR-1 cells; (b) of mtrC mutant cells; (c) of omcA mutant cells.
In summary, the OmcA–MtrC interaction within S. oneidensis MR-1 cells was identified in vivo with cross-linking and IP methods. The network surrounding the OmcA–MtrC interaction core was further characterized, and identification of other proteins involved in this network could help decipher the unique electron transfer and metal reduction mechanisms associated within S. oneidensis cells. Membrane proteins have presented great challenges for the identification of protein–protein interactions due to their strong hydrophobicity. Our study demonstrates that hydrophobic PIR cross-linkers preferentially label membrane proteins, in particular, OM proteins. Therefore, employing chemical cross-linkers with added hydrophobicity coupled with immunoprecipitation presents a practical strategy for mapping protein–protein interactions in cell membranes.
Acknowledgment
This research was supported by the Office of Science (BER), U.S. Department of Energy, Grant No. DE-FG02-04ER63924; NIH-NCRR, Grant No. 1 S10 RR017805-01; and the Murdock Charitable Trust.
Footnotes
Supporting Information Available: Supplementary Figures 1 to 4 and Supplementary Tables I and II. This material is available free of charge via the Internet at http://pubs.acs.org.
Supplementary Material
References
- 1.Lovley DR, Phillips EJP, Gorby YA, Landa ER. Microbial Reduction of Uranium. Nature. 1991;350:413–416. [Google Scholar]
- 2.Lloyd JR, Macaskie LE. A Novel Phosphorimager-Based Technique for Monitoring the Microbial Reduction of Technetium. Appl. Environ. Microbiol. 1996;62:578–582. doi: 10.1128/aem.62.2.578-582.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Myers CR, Carstens BP, Antholine WE, Myers JM. Chromium(VI) Reductase Activity is Associated with the Cytoplasmic Membrane of Anaerobically Grown Schewanella Putrefaciens MR-1. J. Appl. Microbiol. 2000;88:98–106. doi: 10.1046/j.1365-2672.2000.00910.x. [DOI] [PubMed] [Google Scholar]
- 4.Myers CR, Myers JM. Cell Surface Exposure of the Outer Membrane Cytochromes of Shewanella Oneidensis MR-1. Lett. Appl. Microbiol. 2003;37:254–258. doi: 10.1046/j.1472-765x.2003.01389.x. [DOI] [PubMed] [Google Scholar]
- 5.Meyer TE, Tsapin AI, Vandenberghe I, de Smet L, Frishman D, Nealson KH, Cusanovich MA, van Beeumen JJ. Identification of 42 possible cytochrome C genes in the Shewanella oneidensis genome and characterization of six soluble cytochromes. Omics. 2004;8(1):57–77. doi: 10.1089/153623104773547499. [DOI] [PubMed] [Google Scholar]
- 6.Myers CR, Myers JM. Localization of cytochromes to the outer membrane of anaerobically grown Shewanella putrefaciens MR-1. J. Bacteriol. 1992;174(11):3429–38. doi: 10.1128/jb.174.11.3429-3438.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beliaev AS, Thompson DK, Khare T, Li m. H., Brandt CC, Li G, Murray AE, Heidelberg JF, Giometti CS, Yates J, III, Nealson KH, Tiedje JM, Zhoui J. Gene and protein expression profiles of Shewanella oneidensis during anaerobic growth with different electron acceptors. Omics. 2002;6(1):39–60. doi: 10.1089/15362310252780834. [DOI] [PubMed] [Google Scholar]
- 8.Shi L, Squier TC, Zachara JM, Fredrickson JK. Respiration of metal (hydr)oxides by Shewanella and Geobacter: a key role for multihaem c-type cytochromes. Mol. Microbiol. 2007;65:12–20. doi: 10.1111/j.1365-2958.2007.05783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beliaev AS, Saffarini DA, McLaughlin JL, Hunnicutt D. MtrC, an outer membrane decahaem c cytochrome required for metal reduction in Shewanella putrefaciens MR-1. Mol. Microbiol. 2001;39(3):722–30. doi: 10.1046/j.1365-2958.2001.02257.x. [DOI] [PubMed] [Google Scholar]
- 10.Myers JM, Myers CR. Isolation and sequence of omcA, a gene encoding a decaheme outer membrane cytochrome c of Shewanella putrefaciens MR-1, and detection of omcA homologs in other strains of S.putrefaciens. Biochim. Biophys. Acta. 1998;1373(1):237–51. doi: 10.1016/s0005-2736(98)00111-4. [DOI] [PubMed] [Google Scholar]
- 11.Myers JM, Myers CR. Role for outer membrane cytochromes OmcA and OmcB of Shewanella putrefaciens MR-1 in reduction of manganese dioxide. Appl. Environ. Microbiol. 2001;67(1):260–9. doi: 10.1128/AEM.67.1.260-269.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gorby YA, Yanina S, McLean JS, Rosso KM, Moyles D, Dohnalkova A, Beveridge TJ, Chang IS, Kim BH, Culley DE, Reed SB, Romine MF, Saffarini DA, Hill EA, Shi L, Elias DA, Kennedy DW, Pinchuk G, Watanabe K, Ishii S, Logan B, Nealson KH, Fredrickson JK. Electrically Conductive Bacterial Nanowires Produced by Schewanella Oneidensis Strain MR-1 and Other Microorganisms. PNAS. 2006;103:11358–11363. doi: 10.1073/pnas.0604517103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi L, Chen B, Wang Z, Elias DA, Mayer MU, Gorby YA, Ni S, Lower BH, Kennedy DW, Wunschel DS, Mottaz HM, Marshall MJ, Hill EA, Beliaev AS, Zachara JM, Fredrickson JK, Squier TC. Isolation of high-affinity functional protein complex between OmcA and MtrC: two outer membrane decaheme c-type cytochromes of Shewanella oneidensis MR-1. J. Bacteriol. 2006;188(13):4705–4714. doi: 10.1128/JB.01966-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marshall MJ, Beliaev AS, Dohnalkova AC, Kennedy DW, Shi L, Wang Z, Boyanov MI, Lai B, Kemner KM, McLean JS, Reed SB, Culley DE, Bailey VL, Simonson CJ, Saffarini DA, Romine MF, Zachara JM, Fredrickson JK. c-Type Cytochrome-Dependent Formation of U(IV) Nanoparticles by Shewanella oneidensis. PLOS Biol. 2006;4:1324–1333. doi: 10.1371/journal.pbio.0040268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ross DE, Ruebush SS, Brantley SL, Hartshorne RS, Clarke TA, Richardson DJ, Tien M. Characterization of protein-protein interactions involved in iron reduction by Shewanella oneidensis MR-1. Appl. Environ. Microbiol. 2007;73(18):5797–808. doi: 10.1128/AEM.00146-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anderson NG. Co-immunoprecipitation. Methods Mol. Biol. 1998;88:35–45. doi: 10.1385/0-89603-487-9:35. [DOI] [PubMed] [Google Scholar]
- 17.Lee C. Coimmunoprecipitation assay. Methods Mol. Biol. 2007;362:401–6. doi: 10.1007/978-1-59745-257-1_31. [DOI] [PubMed] [Google Scholar]
- 18.Ceriani MF. Coimmunoprecipitation on Drosophila cells in culture. Methods Mol. Biol. 2007;362:423–7. doi: 10.1007/978-1-59745-257-1_34. [DOI] [PubMed] [Google Scholar]
- 19.Selbach M, Mann M. Protein interaction screening by quantitative immunoprecipitation combined with knockdown (QUICK). Nat. Methods. 2006;3(12):981–3. doi: 10.1038/nmeth972. [DOI] [PubMed] [Google Scholar]
- 20.Su LK. Co-immunoprecipitation of tumor suppressor protein-interacting proteins. Methods Mol. Biol. 2003;223:135–40. doi: 10.1385/1-59259-329-1:135. [DOI] [PubMed] [Google Scholar]
- 21.Masters SC. Co-immunoprecipitation from transfected cells. Methods Mol. Biol. 2004;261:337–50. doi: 10.1385/1-59259-762-9:337. [DOI] [PubMed] [Google Scholar]
- 22.Hari J, Shii K, Roth RA. In vivo association of [125I]-insulin with a cytosolic insulin-degrading enzyme: detection by covalent cross-linking and immunoprecipitation with a monoclonal antibody. Endocrinology. 1987;120(2):829–31. doi: 10.1210/endo-120-2-829. [DOI] [PubMed] [Google Scholar]
- 23.Orlando V, Paro R. Mapping Polycomb-repressed domains in the bithorax complex using in vivo formaldehyde cross-linked chromatin. Cell. 1993;75(6):1187–98. doi: 10.1016/0092-8674(93)90328-n. [DOI] [PubMed] [Google Scholar]
- 24.Orlando V, Strutt H, Paro R. Analysis of chromatin structure by in vivo formaldehyde cross-linking. Methods. 1997;11(2):205–14. doi: 10.1006/meth.1996.0407. [DOI] [PubMed] [Google Scholar]
- 25.Fragoso G, Hager GL. Analysis of in vivo nucleosome positions by determination of nucleosome-linker boundaries in crosslinked chromatin. Methods. 1997;11(2):246–52. doi: 10.1006/meth.1996.0411. [DOI] [PubMed] [Google Scholar]
- 26.Jackson V. Formaldehyde cross-linking for studying nucleosomal dynamics. Methods. 1999;17(2):125–39. doi: 10.1006/meth.1998.0724. [DOI] [PubMed] [Google Scholar]
- 27.Hall DB, Struhl K. The VP16 activation domain interacts with multiple transcriptional components as determined by protein-protein cross-linking in vivo. J. Biol. Chem. 2002;277(48):46043–50. doi: 10.1074/jbc.M208911200. [DOI] [PubMed] [Google Scholar]
- 28.Meunier L, Usherwood YK, Chung KT, Hendershot LM. A subset of chaperones and folding enzymes form multiprotein complexes in endoplasmic reticulum to bind nascent proteins. Mol. Biol. Cell. 2002;13(12):4456–69. doi: 10.1091/mbc.E02-05-0311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim DH, Sarbassov DD, Ali SM, King JE, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110(2):163–75. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- 30.Kametaka S, Shibata M, Moroe K, Kanamori S, Ohsawa Y, Waguri S, Sims PJ, Emoto K, Umeda M, Uchiyama Y. Identification of phospholipid scramblase 1 as a novel interacting molecule with beta -secretase (beta -site amyloid precursor protein (APP) cleaving enzyme (BACE)). J. Biol. Chem. 2003;278(17):15239–45. doi: 10.1074/jbc.M208611200. [DOI] [PubMed] [Google Scholar]
- 31.Alloza I, Martens E, Hawthorne S, Vandenbroeck K. Cross-linking approach to affinity capture of protein complexes from chaotrope-solubilized cell lysates. Anal. Biochem. 2004;324(1):137–42. doi: 10.1016/j.ab.2003.09.017. [DOI] [PubMed] [Google Scholar]
- 32.Schmitt-Ulms G, Hansen K, Liu J, Cowdrey C, Yang J, DeArmond SJ, Cohen FE, Prusiner SB, Baldwin MA. Time-controlled transcardiac perfusion cross-linking for the study of protein interactions in complex tissues. Nat. Biotechnol. 2004;22(6):724–31. doi: 10.1038/nbt969. [DOI] [PubMed] [Google Scholar]
- 33.Kim S, Wong P, Coulombe PA. A keratin cytoskeletal protein regulates protein synthesis and epithelial cell growth. Nature. 2006;441(7091):362–5. doi: 10.1038/nature04659. [DOI] [PubMed] [Google Scholar]
- 34.Guerrero C, Tagwerker C, Kaiser P, Huang L. An integrated mass spectrometry-based proteomic approach: quantitative analysis of tandem affinity-purified in vivo cross-linked protein complexes (QTAX) to decipher the 26 S proteasome-interacting network. Mol. Cell. Proteomics. 2006;5(2):366–78. doi: 10.1074/mcp.M500303-MCP200. [DOI] [PubMed] [Google Scholar]
- 35.Tagwerker C, Flick K, Cui M, Guerrero C, Dou Y, Auer B, Baldi P, Huang L, Kaiser P. A tandem affinity tag for two-step purification under fully denaturing conditions: application in ubiquitin profiling and protein complex identification combined with in vivocross-linking. Mol. Cell. Proteomics. 2006;5(4):737–48. doi: 10.1074/mcp.M500368-MCP200. [DOI] [PubMed] [Google Scholar]
- 36.Vasilescu J, Guo X, Kast J. Identification of protein-protein interactions using in vivo cross-linking and mass spectrometry. Proteomics. 2004;4(12):3845–54. doi: 10.1002/pmic.200400856. [DOI] [PubMed] [Google Scholar]
- 37.Sinz A. Chemical Cross-Linking and Mass Spectrometry to Map Three-Dimensional Protein Structures and Protein-Protein Interactions. Mass Spectrom. Rev. 2006;25:663–682. doi: 10.1002/mas.20082. [DOI] [PubMed] [Google Scholar]
- 38.Vasilescu J, Figeys D. Mapping protein-protein interactions by mass spectrometry. Curr. Opin. Biotechnol. 2006;17(4):394–9. doi: 10.1016/j.copbio.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 39.Gingras AC, Gstaiger M, Raught B, Aebersold R. Analysis of protein complexes using mass spectrometry. Nat. Rev. Mol. Cell. Biol. 2007;8(8):645–54. doi: 10.1038/nrm2208. [DOI] [PubMed] [Google Scholar]
- 40.Tang X, Munske GR, Siems WF, Bruce JE. Mass spectrometry identifiable cross-linking strategy for studying protein-protein interactions. Anal. Chem. 2005;77(1):311–8. doi: 10.1021/ac0488762. [DOI] [PubMed] [Google Scholar]
- 41.Chowdhury SM, Munske GR, Tang X, Bruce JE. Collisionally Activated Dissociation and Electron Capture Dissociation of Several Mass Spectrometry-Identifiable Chemical Cross-Linkers. Anal. Chem. 2006;78:8183–8193. doi: 10.1021/ac060789h. [DOI] [PubMed] [Google Scholar]
- 42.Tang X, Yi W, Munske GR, Adhikari DP, Zakharova NL, Bruce JE. Profiling the Membrane Proteome of Shewanella oneidensis MR-1 with New Affinity Labeling Probes. J. Proteome Res. 2007;6:724–734. doi: 10.1021/pr060480e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gorby YA, Yanina S, McLean JS, Rosso KM, Moyles D, Dohnalkova A, Beveridge TJ, Chang IS, Kim BH, Kim KS, Culley DE, Reed SB, Romine MF, Saffarini DA, Hill EA, Shi L, Elias DA, Kennedy DW, Pinchuk G, Watanabe K, Ishii S, Logan B, Nealson KH, Fredrickson JK. Electrically conductive bacterial nanowires produced by Shewanella oneidensis strain MR-1 and other microorganisms. Proc. Natl. Acad. Sci. U.S.A. 2006;103(30):11358–63. doi: 10.1073/pnas.0604517103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20(18):3551–67. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 45.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pautsch A, Schulz GE. High-resolution structure of the OmpA membrane domain. J. Mol. Biol. 2000;298(2):273–282. doi: 10.1006/jmbi.2000.3671. [DOI] [PubMed] [Google Scholar]
- 47.Saier MH., Jr. A Functional-Phylogenetic Classification System for Transmembrane Solute Transporters. Microbiol. Mol. Biol. Rev. 2000;64(2):354–411. doi: 10.1128/mmbr.64.2.354-411.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Watanabe T, Hayashi S, Wu HC. Synthesis and export of the outer membrane lipoprotein in Escherichia coli mutants defective in generalized protein export. J. Bacteriol. 1988;170(9):4001–7. doi: 10.1128/jb.170.9.4001-4007.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.