Abstract
This study investigates the effects of digital image compression on automatic quantification of immunohistochemical nuclear markers. We examined 188 images with a previously validated computer-assisted analysis system. A first group was composed of 47 images captured in TIFF format, and other three contained the same images converted from TIFF to JPEG format with 3×, 23× and 46× compression. Counts of TIFF format images were compared with the other three groups. Overall, differences in the count of the images increased with the percentage of compression. Low-complexity images (≤100 cells/field, without clusters or with small-area clusters) had small differences (<5 cells/field in 95–100% of cases) and high-complexity images showed substantial differences (<35–50 cells/field in 95–100% of cases).
Compression does not compromise the accuracy of immunohistochemical nuclear marker counts obtained by computer-assisted analysis systems for digital images with low complexity and could be an efficient method for storing these images.
Introduction
Quantification of immunohistochemistry with computer-assisted analysis systems is becoming a useful tool in pathology for research and clinical practice. 1–5 However, the archiving of digital images (DIs) continues to be one of the biggest problems. 6 Very large DI databases require a huge digital storage space and, as a result, some forms of data compression have become necessary. Image-compression techniques can greatly reduce the volume of storage or transmission time required per image 7 and has been investigated in several fields of medicine. 8–11
Tagged Image File Format (TIFF) and Joint Photographic Experts Group (JPEG) are two of the most frequently used image formats in medicine. 7,12–14 Tagged Image File Format (TIFF) is an example of a lossless format, which has the advantage that no data are lost, but with the disadvantage that files sizes are quite large. Joint Photographic Experts Group (JPEG) is an example of lossy format, which has the advantage that it can reduce file sizes but with the disadvantage that some detail may be lost in the compression. Acquired images in TIFF format could be transformed in a JPEG format with different degree of compression. At low and moderate compression levels, it is very difficult for the human eye to discern any difference from the original, even at extreme magnification. However, JPEG inevitably introduces digital artefacts and variations in several image variables, and may modify the grey values of each pixel for every one of the three channels of RGB 24-bit true color images. 12
Case Description
In the field of anatomical and surgical pathology, computerized image analysis software is used to detect and quantify the number of positively stained markers and combine different tools that evaluate standard morphometric and densitometric features (area, diameter, roundness, light intensity) of these markers. Previous results obtained with automated or semi-automated segmentation of immunohistochemically stained DIs have used available commercial programs such as Photoshop®, 15 Image-Pro® Plus, 14,16,17 Visilog® 18 and ACIS®. 1 Although JPEG transformation produces changes in Feulgen-stained samples, especially for the standard densitometric features, 12 the effects of compression in the computer-assisted analysis of immunohistochemical digital images have not yet been thoroughly evaluated.
In looking for a more efficient way to store immunohistochemical DIs, this study attempted to determine the influence of different JPEG image compression rates on the immunohistochemical digital quantification compared with the original TIFF images. We determined the rates of JPEG compression levels that did not compromise the accuracy of the automatic nucleus count of immunohistochemical markers for diagnostic or research purposes.
Methods
Selection of Histopathological Material and Immunohistochemistry
Slides stained with DAB(3,3'Diaminobenzidine) from routinely formalin-fixed and paraffin-embedded histopathological tissues were selected from the archives of the Department of Pathology of the Hospital de Tortosa Verge de la Cinta. Representative slides immunostained with monoclonal antibodies directed against the nuclear protein Ki67 (clone MIB-1, Dako, Carpinteria, CA, USA) and FOXP3 (clone FOXP3-236A/E7, CNIO, Spain) were selected. All slides were prepared, processed and immunohistochemically stained in our laboratory using previously described standard protocols. 3
Digitalization Procedures
Samples were vizualized with a standard Leica DM LB2 light microscope (Leica Microsystems Wetzlar, Germany) with ×40 magnification. We acquired DIs of representative fields with a Leica DFC320 digital camera (3.3 Mpixels) connected to a Compaq Professional Workstation computer (2 GHz Pentium IV CPU, 750 mB RAM). Photographs were saved as TIFF files, with a resolution of 2088 × 1550 pixels in RGB 24-bit true color format, with the LEICA IM50 v4.0 program. To avoid differences in the illumination that might produce significant differences in the quantification of the image, the same range of values of illumination was used to ensure maximum reproducibility.
Image Processing and Quantification
Initially, 47 DIs were captured and saved in the uncompressed TIFF format. ACDSee software uses a 0-100 scale of JPEG compression that represents the compression parameters that can be adjusted from the best compression to the best quality. Under these conditions and taken into consideration the size of the original TIFF images (around 9.27 Mbytes), the images with the lowest compression and the highest quality that the software allows have 3× compression (around 3.25 Mbytes), intermediate quality images have 23× compression (around 400 Kbytes) and images with maximum compression and poor quality have 46× compression (around 200 Kbytes).
One hundred eighty-eight images were analysed using an automatic computer-assisted procedure, 47 from original uncompressed TIFF DIs and the other 141 from different JPEG compressed images (47 with 3×, 47 with 23× and 47 with 46× JPEG compression). This automatic process combined the analysis of images by the Image-Pro® Plus 5.0 program and mathematical algorithms developed as a macro in Excel®, as previously tested and validated. 19
Statistical Analysis
Computer-assisted counts of TIFF DIs were taken as the reference. Differences in the nuclear count obtained between paired results (TIFF versus each group of compressed JPEG images with 3×, 23× and 46× compression) were evaluated using Student's t-test, intra-class correlation coefficients (ICC), Bland-Altman and Kaplan-Meier analysis with their corresponding graphical illustration. All statistical analyses were carried out using SPSS 11.0.
Example
The paired t-test was carried out in order to establish how the formats with JPEG compression were likely to differ from the TIFF format so that the different compressed formats could be interchangeable with TIFF. Globally and for high-complexity images, the paired t-test demonstrated significant differences between the TIFF and the JPEG images of all three compression levels (▶; p < 0.001 for TIFF vs. 3×, 23× and 46× compressed JPEG images). These significance disappear in low-complexity images with the exception of TIFF vs. JPEG 3× counts (p = 0.002). However, these findings must be interpreted with caution since the significance was largely due to the fact that variance of differences was very small. Therefore, when comparing date of different measurement methods, the uses of correlation or mean comparison (t-test) are misleading. On the other hand, the ICC coefficients showed excellent agreement but the condition of equality of variances was no satisfied.
Table 1.
Table 1 Probability Values from Student's t-Tests Comparing Mean Counts in TIFF and JPEG Images of Different Compression Rates
| Differences | TIFF vs. 3× JPEG | TIFF vs. 23× JPEG | TIFF vs. 46× JPEG |
|---|---|---|---|
| Globally | <0.001 | <0.001 | <0.001 |
| No cluster/Low area cluster | 0.002 | 0.059 | 0.125 |
| Fewer than 100 nuclei | 0.125 | 0.052 | 0.175 |
| Large-area cluster | <0.001 | <0.001 | <0.001 |
| More than 100 nuclei | <0.001 | <0.001 | <0.001 |
JPEG = joint photographic experts group; TIFF = tagged image file format.
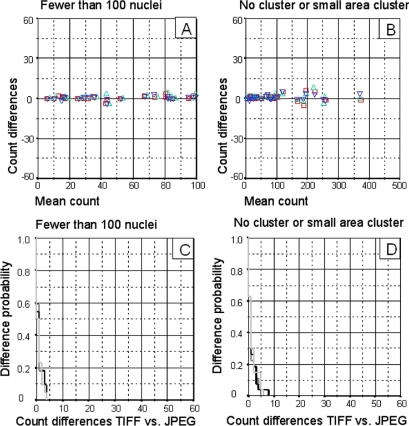
As demonstrated in a previous study, 19 the complexity of the images may affect the results of the automated quantification. The density of positively stained cells in each image and the presence of large clusters of these positive cells is the biggest problem in the automated immunohistochemical quantification process. Taking these points into consideration, the degree of JPEG compression did not appear to affect the automatic quantification in images of low complexity (fewer than 100 nuclei/field, with no clusters or small-area clusters). In such images, at any level of compression, there were very small differences (▶A and ▶B). Furthermore, the Kaplan-Meier procedure indicates that in these DIs the probability of observing differences of less than 5 nuclei/image is around of 95–100% (▶C and ▶D).
Figure 2.
Results obtained by automatic quantification from low-complexity images (fewer than 100 nuclei/image, with no cluster or small-area cluster). A. Superimposed Bland-Altman graphs comparing count differences between TIFF and JPEG images. TIFF vs. 3× compression JPEG (□), TIFF vs. 23× compression JPEG (▵), TIFF vs. 46× compression JPEG (▿). B. Superimposed Kaplan-Meier curves comparing the probability of difference between TIFF and JPEG image counts. TIFF vs. 3× compression JPEG (▩), TIFF vs. 23× compression JPEG (▪), TIFF vs. 46× compression JPEG (□).
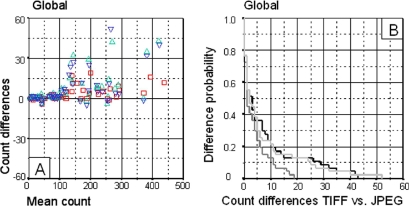
An alternative approach, based on graphical techniques and simple calculations such as the Bland-Altman representations and the Kaplan-Meier curves is recommended in this context. The nuclei count differences which were considered acceptable (independently of statistical significance) were used as values to create both a Bland-Altman plot and a Kaplan-Meier curve. The Bland-Altman method described the extent of agreement of the nucleus counts obtained with TIFF vs. JPEG 3×, 23×, and 46× compression; the Kaplan-Meier method described the conditional probability of observing differences between the counts. Overall, these graphical representations indicate that count differences become higher as compression increases (▶A and ▶B).
Figure 1.
Global results obtained by automatic quantification. The Bland-Altman graph (A) represents the difference between each pair of results (Y axis) and the mean of both counts (X axis) for each comparison. The Kaplan-Meier curves (B) represent the conditional probability of observing differences between the TIFF and the JPEG images at different levels of compression. The X axis represents count differences between the number of nuclei in TIFF and JPEG images at 3×, 23× and 46× compression. The Y axis represents the probability of observing these count differences. A. Superimposed Bland-Altman graphs comparing count differences between TIFF and JPEG images. TIFF vs. 3× compression JPEG (□), TIFF vs. 23× compression JPEG (▵), TIFF vs. 46× compression (▿). B. Superimposed Kaplan-Meier curves comparing the probability of difference between TIFF and JPEG image counts. TIFF vs. 3× compression JPEG (▩), TIFF vs. 23× compression JPEG (▪), TIFF vs. 46× compression JPEG (□).
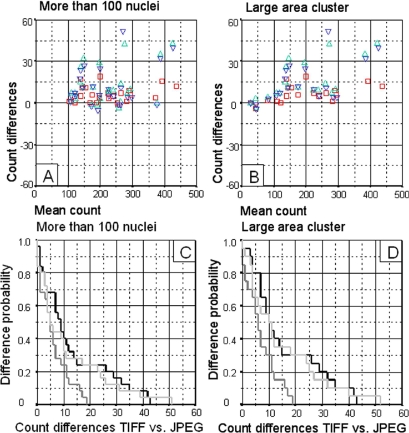
On the other hand, when images were more complex, with more than 100 cells/images (▶A) or of large clusters (▶B), there was greater dispersion of the difference in the Bland-Altman representations. Count differences were less than 15–20 nuclei in 95–100% of cases for JPEG images with 3× compression. Otherwise, in images with 23× or 46× compression count differences were less than 40–50 nuclei in 95–100% of cases (▶C and ▶D).
Figure 3.
Results obtained with automatic quantification from high-complexity images (more than 100 nuclei/field or with large-area clusters). A. Superimposed Bland-Altman graphs comparing count differences between TIFF and JPEG images. TIFF vs. 3× compression JPEG (□), TIFF vs. 23x compression JPEG (▵), TIFF vs. 46× compression JPEG (▿). B. Superimposed Kaplan-Meier curves comparing the probability of difference between TIFF and JPEG image counts. TIFF vs. 3× compression JPEG (▩), TIFF vs. 23× compression JPEG (▪), TIFF vs. 46× compression JPEG (□).
Discussion
Manual (non-automated) counting of immunohistochemical stained markers is a tedious and not absolutely objective process that ideally should be replaced, when possible, by automatic systems (computerized image-analysis systems) which are faster, more reproducible and objective. 1,20 Previous results indicated that the accuracy and precision of our automated process for the detection and quantification of the immunostained nuclei in TIFF format images have been validated for diagnostic and prognostic purposes in haematological malignancies including follicular lymphoma 4,14 and Hodgkin lymphoma. 3,21
An important part of computer-assisted image analysis depends on image segmentation. 4,5,14,17,18 Segmentation is the partitioning of a DI into various meaningful regions by identifying regions of an image that have common properties and separating those that are dissimilar. 22 Changes produced during JPEG image compression affect the grey values of each pixel 12 so that some positive pixels altered by the compression fall outside our selected range of positive color and some negative pixels appearing to fall within the positive range of values. Changes produced during JPEG image compression affect the grey values of pixel 12 in each of the three RGB channels in the 24-bit images. This could result in some positive pixels being altered by the compression so that they fall outside the range of positive color values selected by us, and in some negative pixels appearing to fall within the positive range of values. This variability would reduce the efficiency of segmentation algorithms used in the computer-assisted image-analysis procedure, leading to serious and unacceptable inaccuracy in the final nucleus count.
Globally, the efficiency of the image-analysis algorithms decreases with the degree of compression. The choice of compression degree also depends on the magnitude of the nucleus count differences allowed or considered acceptable. The most important aim of this study was to determine in which cases the count differences were lowest, and the most useful tools to quantify these differences were the Kaplan-Meier and Bland-Altman plots. The results indicate that the variability produced by the automated analysis of these immunohistochemical JPEG compressed DIs does not compromise the accuracy of nuclear marker quantification and that the method could be an efficient way to store DIs for diagnostic and research purposes in images with low complexity.
When JPEG compression does not compromise the accuracy of the results, the advantage of the compression is to reduce the storage capacity. One of our several studies, have 200 samples and consisted in the quantification of a total amount of 5600 photographies. Considering that all the TIFF images on this study could be analyzed with JPEG 46× compression, images could be stored in only 2 DVDs (2.2 Gbytes including a second security copy) in comparison to the 24 DVDs needed for the TIFF format images (100 Gbytes with the second copy). Three other studies of the same magnitude (same number of images) could be also stored in these DVDs. If we made these three other studies in TIFF format these would take up 72 DVDs and 300 Gbytes more. This represents that four studies in TIFF format would occupy 400 Gbytes and 96DVDs versus 8,8 Gbytes and 2DVDs in JPEG 46× compression. Other potential uses of images compression are nowadays being investigated in other automated quantification procedures in order to generalize counts to cytoplasmatic and membrane immunohistochemical markers.
This study demonstrated that a high degree of compression can be used in computer-assisted quantification of low complexity DIs, since the same levels of accuracy and reproducibility are obtained as with the original TIFF format images. The use of compression in these images allows reduction of up to 46× the original size of the file, the hard disks space, the economical costs, and the time spent making security copies.
Acknowledgments
The authors thank María del Mar Barbera, Bárbara Tomás, Ainhoa Monteserrat, Vanesa Gestí, Ana Suñé, and Marc Iniesta for their skilful technical assistance, Anna Carot and Rosa Cabrera for their excellent secretarial work.
Footnotes
This work was supported by grants FIS 04/1440, 04/1467 and 05/1527 from the Ministerio de Sanidad y Consumo, Spain.
References
- 1.Wang S, Saboorian MH, Frenkel EP, et al. Assessment of HER-2/neu status in breast cancer. Automated Cellular Imaging System (ACIS)-assisted quantitation of immunohistochemical assay achieves high accuracy in comparison with fluorescence in situ hybridization assay as the standard. Am J Clin Pathol 2001;116:495-503. [DOI] [PubMed] [Google Scholar]
- 2.Pierga JY, Bonneton C, Vincent-Salomon A, et al. Clinical significance of immunocytochemical detection of tumor cells using digital microscopy in peripheral blood and bone marrow of breast cancer patients Clin Cancer Res 2004;10:1392-1400. [DOI] [PubMed] [Google Scholar]
- 3.Alvaro-Naranjo T, Lejeune M, Salvado-Usach MT, et al. Tumor-infiltrating cells as a prognostic factor in Hodgkin's lymphoma: a quantitative tissue microarray study in a large retrospective cohort of 267 patients Leuk Lymphoma 2005;46:1581-1591. [DOI] [PubMed] [Google Scholar]
- 4.Alvaro T, Lejeune M, Camacho FI, et al. The presence of STAT1-positive tumor-associated macrophages and their relation to outcome in patients with follicular lymphoma Haematologica 2006;91:1605-1612. [PubMed] [Google Scholar]
- 5.Zhang K, Prichard JW, Yoder S, et al. Utility of SKP2 and MIB-1 in grading follicular lymphoma using quantitative imaging analysis Hum Pathol 2007. [DOI] [PubMed]
- 6.Leong FJ, Leong AS. Digital imaging in pathology: theoretical and practical considerations, and applications Pathology 2004;36:234-241. [DOI] [PubMed] [Google Scholar]
- 7.Slone RM, Foos DH, Whiting BR, et al. Assessment of visually lossless irreversible image compression: comparison of three methods by using an image-comparison workstation Radiology 2000;215:543-553. [DOI] [PubMed] [Google Scholar]
- 8.Gurdal P, Hildebolt CF, Akdeniz BG. The effects of different image file formats and image-analysis software programs on dental radiometric digital evaluations Dentomaxillofac Radiol 2001;30:50-55. [DOI] [PubMed] [Google Scholar]
- 9.Li F, Sone S, Takashima S, et al. Effects of JPEG and wavelet compression of spiral low-dose ct images on detection of small lung cancers Acta Radiol 2001;42:156-160. [DOI] [PubMed] [Google Scholar]
- 10.Ohgiya Y, Gokan T, Nobusawa H, et al. Acute cerebral infarction: effect of JPEG compression on detection at CT Radiology 2003;227:124-127. [DOI] [PubMed] [Google Scholar]
- 11.Belair ML, Fansi AK, Descovich D, et al. The effect of compression on clinical diagnosis of glaucoma based on non-analyzed confocal scanning laser ophthalmoscopy images Ophthalmic Surg Lasers Imaging 2005;36:323-326. [PubMed] [Google Scholar]
- 12.Atalag K, Sincan M, Celasun B, et al. Effects of lossy image compression on quantitative image analysis of cell nuclei Anal Quant Cytol Histol 2004;26:22-27. [PubMed] [Google Scholar]
- 13.Singh SS, Kim D, Mohler JL. Java Web Start based software for automated quantitative nuclear analysis of prostate cancer and benign prostate hyperplasia Biomed Eng Online 2005;4:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alvaro T, Lejeune M, Salvado MT, et al. Immunohistochemical patterns of reactive microenvironment are associated with clinicobiologic behavior in follicular lymphoma patients J Clin Oncol 2006;24:5350-5357. [DOI] [PubMed] [Google Scholar]
- 15.Lehr HA, van der Loos CM, Teeling P, et al. Complete chromogen separation and analysis in double immunohistochemical stains using Photoshop-based image analysis J Histochem Cytochem 1999;47:119-126. [DOI] [PubMed] [Google Scholar]
- 16.Carai A, Diaz G, Santa Cruz R, et al. Computerized quantitative color analysis for histological study of pulmonary fibrosis Anticancer Res 2002;22:3889-3894. [PubMed] [Google Scholar]
- 17.Alvaro T, Lejeune M, Salvado MT, et al. Outcome in Hodgkin's lymphoma can be predicted from the presence of accompanying cytotoxic and regulatory T cells Clin Cancer Res 2005;11:1467-1473. [DOI] [PubMed] [Google Scholar]
- 18.Loukas CG, Wilson GD, Vojnovic B, et al. An image analysis-based approach for automated counting of cancer cell nuclei in tissue sections Cytometry A 2003;55:30-42. [DOI] [PubMed] [Google Scholar]
- 19.Alvaro T, Lejeune M, Garcia JF, et al. Tumor-infiltrated immune response correlates with alterations in the apoptotic and cell cycle pathways in Hodgkin and Reed-Sternberg cells Clin Cancer Res 2008;14:685-691. [DOI] [PubMed] [Google Scholar]
- 20.Lejeune M, Jaen J, Pons L, et al. Quantification of diverse subcellular immunohistochemical markers with clinicobiological relevancies: validation of a new computer-assisted image analysis procedure J Anat 2008;212:868-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lopez C, Lejeune M, Salvado MT, et al. Automated quantification of nuclear immunohistochemical markers with different complexity Histochem Cell Biol 2008;129:379-387. [DOI] [PubMed] [Google Scholar]
- 22.Levine MD. Vision in Man and MachinesNew York: McGraw-Hill; 1985.