Abstract
In order to assess the mission and strategic direction in an academic department of biomedical informatics, we used social network analysis to identify patterns of common interest among the department's multidisciplinary faculty. Data representing faculty and their self-identified research methods and expertise were analyzed by applying a network modularity algorithm to detect community structure. Three distinct communities of practice emerged: empirical discovery and prediction; human and organizational factors; and information management. This analysis made intuitive sense and served the goal of stimulating discussion from new perspectives. The findings will guide future direction and faculty recruitment efforts. Communities of practice present a novel view of interdisciplinarity in biomedical informatics.
Introduction
Traditional categories that are used to define biomedical informatics center on application areas (e.g., clinical informatics, bioinformatics, public health informatics). While useful and informative, application areas can be an imperfect fit for multidisciplinary academic researchers whose expertise, inquiry and practice cut across categories. A research faculty that is engaged in a range of multi-dimensional activities and that use a range of methods constitutes a complex human system. To understand the relationships within this system we explored the interests of the faculty using social network analysis, an empirical technique for understanding complexity. Our goal was to gain insight to inform an updated mission and to direct future actions such as hiring new faculty.
Social Network Analysis
Social network analysis is grounded in the systematic analysis of empirical data using formal theory organized in mathematical terms. 1 The goal is to capture patterns of human interactions. A network is represented as graph consisting of a finite set of nodes or vertices linked by lines or edges. A “social” network is defined as a group of collaborating (or competing) entities that have some type of relationship and interact within a shared environment often referred to as a community. 2 Network graphs are notated in the form of an adjacency matrix that allows use of operations from matrix and linear algebra to mathematically define characteristics of the network. 3 The social network can be used to characterize and describe the community structure of its members. Some communities have a densely connected core of members with less central members scattered around the periphery. In others there is fairly uniform distribution of links across people, and still others have obvious divisions into sub-groups or “modules.” One approach to quantifying such patterns is a modularity algorithm developed by Newman. 4 The algorithm reflects the degree to which a network contains community structure by calculating the number of edges that fall within sub-areas of the network minus the expected number in an equivalent network with edges placed at random. The modularity can be either positive or negative on a −1 to +1 scale. Positive values greater than 0.3 indicate the likely presence of a community structure. 5
Case Description
A transition of leadership and a planned recruitment initiative in an academic department of biomedical informatics was the occasion for reviewing the department's mission and strategic direction. As part of that process faculty members were asked to self identify their areas of expertise, research, and practice during the department's annual retreat. The methodological areas and the scope of practice that emerged challenged the faculty to develop a cohesive description of “who we are.” To explore the structure of the department we turned to information presented during the retreat which consisted of a single slide prepared by each faculty member summarizing their current research methods and areas of expertise.
Example of Network Analysis Used to Describe Department Structure
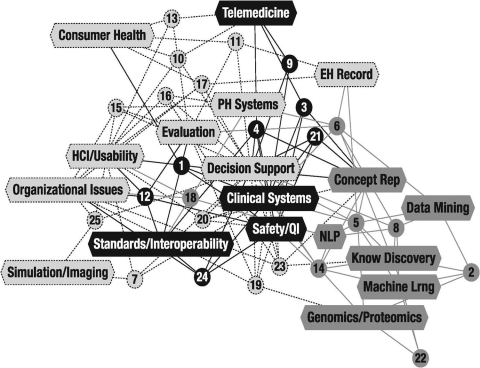
We extracted terms or short strings describing the self-identified methods and areas of expertise from the content of the faculty-prepared slides. Similar or closely related items were clustered and de-duplicated within broader terms. For example “evidence-based practice” and “guideline-based care” and “bring scientific evidence into clinical practice” were clustered within decision support. The term “electronic patient charting” was clustered within “electronic health record.” A total of 18 terms were extracted from slides prepared by 25 faculty members. These were arranged in a rectangular adjacency matrix wherein each faculty member who was associated with a term received a score of “1” and all others received a score of “0.” These data were entered into the Organization Risk Analyzer (ORA) computer program developed at Carnegie Mellon University. 6 We used the Newman modularity algorithm to detect sub-groups in the network. Three sub-groups or “communities” were identified within the department. The modularity score of 0.32 suggested that the groups are relatively distinct, without a high degree of integration. A graphical display of the network colored by Newman groups is shown in ▶.
Figure 1.
- Light Grey A community around methods and expertise related to human and organizational factors
- Dark Grey A community around methods and expertise related to empirical discovery and prediction
- Black A community around methods and expertise related to information management
Three communities clustered around activities broadly related to empirical discovery and prediction, human and organizational factors, and information management. Visualization of the data showed clearly not only which people and activities got grouped together, but how three distinct groups emerged. The results resonated strongly with the faculty. Bioinformatics faculty and clinical faculty emerged as a group sharing practice in computational methods such as data mining and knowledge discovery. While there is co-authorship between these faculties, the analysis led us to consider how the two groups may move the department forward by sharing resources, particularly in the area of curriculum development and training. A series of discussions held via email and during two monthly faculty meetings served to refine the results more inclusively. This process focused the faculty's energies on defining what the department is, and gleaning insights on future direction and hiring new faculty. The dialogue clarified the relationship of the three communities of practice to the Department's overarching mission of research, education and service in the context of four recognized areas of biomedical informatics application. 7 The methods and areas of expertise associated with the communities of practice are displayed in ▶. These results are already guiding faculty and student recruitment efforts and redesign of the Department's website.
Table 1.
Table 1 Methods and Expertise Associated with Three Communities of Practice with the Corresponding Label on the Nodes in ▶
| Community of Practice | Method or Area of Expertise Associated with Community | Label in ▶ |
|---|---|---|
| Empirical discovery and prediction | Computational simulation of biological systems focused on genomics and proteomics | Genomics/Proteomics |
| Data mining | Data Mining | |
| Knowledge discovery | Know Discovery | |
| Natural language processing | NLP | |
| Concept representation | Concept Rep | |
| Machine learning and intelligent systems | Machine Lrng | |
| Human and organizational factors | Decision support | Decision Support |
| Health technology evaluation | Evaluation | |
| Consumer health | Consumer Health | |
| Electronic health records | EH Record | |
| Public health systems | PH Systems | |
| Cognition, human computer interface, and usability | HCI/usability | |
| Visual simulation of human biological systems | Simulation/Imaging | |
| Information Management | Clinical information systems | Clinical Systems |
| Safety and quality improvement | QI | |
| Information standards and interoperability | Standards/Interoperability | |
| Telemedicine | Telemedicine |
Discussion
Social network analysis is a method for unraveling complexity. It was applied to gain insight during a period of transition within an academic department of biomedical informatics. When an environment changes, unexamined assumptions may become out-of-date and the actions based on them may become ineffective. 8 Insight into the complex nature of the department has helped faculty to critique and reconstruct ideas about who we are, and to generate a renewed and shared direction. The process itself assisted in transitioning the department toward a stronger identity by providing insight into how the scope and range of faculty expertise interacts with the mission of the department. The groups listed in ▶ are logical and might be grouped similarly if identified by any expert observer. Perhaps the more interesting outcome of the analysis is that the Newman algorithm identified three communities of practice as opposed to some other number. That is, an observer operating without the aid of a computational technique might potentially cluster the methods into two, four, or more groups.
The results of a network analysis can be judged by how accurately the network reflects the real world situation, i.e., face validity. 9 In this case we used self-identified and subjectively categorized research methods and expertise of faculty. Yet when these empirically identified groupings were presented to the departmental faculty for interpretation they just “felt” right, thus satisfying the face validity criteria. The faculty easily came up with labels for the communities of practice represented by the groupings. In the past, the department had loosely organized itself around application areas: clinical, imaging, public health, and bioinformatics. Due in part to external forces like a separate physical location and dedicated funding 10 the bioinformatics faculty constitute a formal division within the department. The other application areas have not formally separated into divisions. The current findings help to explain why this separation has not fully emerged: communities of practice span application areas, preventing any simple separation into divisions.
The groupings produced in this analysis reflect a single point in time. As the department evolves the methods and expertise applied in communities of practice may shift, particularly in areas of cross-cutting practice. For example, electronic health records (EHRs) is grouped within “Human and Organizational Factors” because several faculty were actively working on that aspect of EHRs at the time of the study. Future funding or faculty recruitment may result in more of those within the “Information Management” community applying their energies to EHRs.
Application areas remain an important way to characterize biomedical informatics. The method-oriented communities of practice identified here supply an alternative “fit” that acknowledges the multi-disciplinary nature of the field. The clustering of methods and expertise associated with these communities of practice have helped faculty in our department understand competencies required for training scientists and have informed curriculum development to improve the quality of our academic training program. The insight gained by identifying communities of practice can help us to meet the challenge of integrating the skills and perspectives of multiple disciplines to build knowledge and to translate that knowledge to solve relevant problems. We studied overlapping areas of methods and expertise rather than actual collaboration among faculty. It would be interesting to compare the two networks to see how communities of practice interact internally and with each other.
Acknowledgments
The authors thank the faculty of the Department of Biomedical Informatics at Columbia University who contributed to the development of the concepts presented here.
References
- 1.Freeman LC. The Development of Social Network Analysis: A Study in the Sociology of ScienceVancouver: Booksurge Publishing; 2004.
- 2.Aviv RE, Zippy, Ravid, Gilad. Geva, Aviva Network analysis of knowledge construction in asynchronous learning networks J Asynchronous Learning Networks 2003;7(3):1-23. [Google Scholar]
- 3.Scott JG, Tallia A, Crosson J, et al. Social Network Analysis as an Analytic Tool for Interaction Patterns in Primary Care Practices Ann Fam Med 2005;3(5):443-4482005 September 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Newman MEJ. Fast algorithm for detecting community structure in networks Physics Review 2004;69. [DOI] [PubMed]
- 5.Newman MEJ. From the Cover: Modularity and community structure in networks Proceedings of the National Academy of Sciences 2006;103(23):8577-85822006 June 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carley KM. ORA: Organizational Risk AnalyzerPittsburgh, PA: Center for Analysis of Social and Organizational Systems; 2007.
- 7.National Library of Medicine. National Library of Medicine Awards $75 Million for Informatics Research Training. 2006http://www.nlm.nih.gov/news/press_releases/award_info_res_training06.html 2007. Accessed September 18, 2008.
- 8.Lane D, Maxfield R. Strategy Under Complexity: Fostering Generative Relationships Long Range Planning 1996;29:215-231. [Google Scholar]
- 9.Wasserman S, Faust K. Social Network Analysis: Methods and Applications. Cambridge: Cambridge University Press; 1994.
- 10.Columbia University Center for Computational Biology & Bioinformaticswww.c2b2.columbia.edu 2005. Accessed September 18, 2008.