Abstract
The fluorescent intercalator displacement assay using thiazole orange has been adapted to the study of RNA-binding helix-threading peptides (HTPs). This assay is highly sensitive with HTP-binding RNAs and provides binding affinity data in good agreement with quantitative ribonuclease footprinting without the need for radiolabeling or gel electrophoresis. The FID assay was used to define structure activity relationships for a small library of helix-threading peptides. Results of these studies indicate their RNA binding is dependent on peptide sequence, α-amino acid stereochemistry, and cyclization (versus linear peptides), but independent of macrocyclic ring size for the penta-, tetra- and tri peptides analyzed.
Keywords: RNA recognition, cyclic peptides, threading intercalation, high throughput screening
Introduction
RNA-binding small molecules have been studied in the past for their potential as drug leads.1, 2 This area of research continues to hold promise for the development of new therapeutics, particularly given the recent advances in defining novel regulatory roles for RNA and advances in determining high-resolution structures of RNA drug targets.3–5 However, small molecule/RNA interactions are also now recognized as important for controlling gene expression through both natural and synthetic riboswitches, RNA elements within mRNA that bind small molecules. For instance, advances from the labs of Smolke, Mulligan, Gallivan and Yokobayashi have shown how small molecule-binding RNAs can be used as synthetic riboswitches to regulate the function of mRNAs into which these RNA structures have been inserted.6–9 Although small molecule control of a variety of intracellular RNAs has been demonstrated in this way, relatively few small molecules have been described for this purpose. To fully exploit the potential of RNA-binding small molecules either as drug candidates or as chemical biology tools, new classes of RNA-binding compounds are needed. However, the design and development of new ligands that can bind RNA inside living cells remains a challenging task.1 Several factors need to be considered including RNA target affinity and selectivity, cell permeability, and metabolic stability. It is often difficult to predict what structural features can result in the desired properties for different classes of molecules. In such cases, it can be useful to generate libraries of compounds with structural variations and screen for these properties.10–12 Diversity-oriented synthesis techniques aid in rapidly generating large numbers of molecules.13 However, rapid, high-throughput assays must be available to evaluate specific properties of the library members. When considering assaying the RNA-binding properties of library components, some of the most commonly used techniques (e.g. ribonuclease footprinting, affinity cleaving, gel-mobility shift assays, isothermal titration calorimetry, and surface plasmon resonance) are time consuming and, in some cases, require the use of radioactively or fluorescently labeled RNA.2, 14 For libraries of ligands generated on solid-phase, on-bead screening techniques have been used to screen for tight binders.15, 16 Often, however, on-bead screening techniques result in false-positive hits that can arise as a result of non-specific interactions of the RNA probe to the solid support or with the library members.17 To circumvent these potential problems, we have adapted a spectroscopic, solution phase nucleic acid binding assay to the screening of libraries of helix-threading peptides (HTPs) prepared in our laboratory.18–21 HTPs contain a heterocyclic chromophore embedded in a peptide backbone and can selectively bind stem-loop RNA structures that contain a 5’-PyPu-3’ intercalation site flanked by at least a single bulge on each strand, 3’ to this site.18, 20, 22 We recently demonstrated that a macrocyclic variant binds a naturally occurring pre-miRNA containing such a binding site.21 In this paper, we describe the use of the fluorescent intercalator displacement assay to analyze the RNA binding properties of a small collection of HTPs.23, 24 This assay is highly sensitive, simple, does not required labeled RNA and can be carried out in homogeneous solutions in multi-well plates.25 Moreover, the assay appears particularly well suited for screening HTPs, given the large increase (up to 2300-fold) in fluorescence signal observed when the intercalator thiazole orange binds RNA containing a preferred HTP site. This assay was used to evaluate structure activity relationships for HTP/RNA binding by varying HTP sequence, amino acid stereochemistry, macrocyclic vs linear HTPs, and macrocycle size. We found that all of these structural parameters except ring size influence the RNA binding properties of HTPs to the RNA target chosen. We also report an efficient synthesis of a new 2-phenylquinoline carboxylic acid for incorporation into macrocyclic HTPs.
Results and Discussion
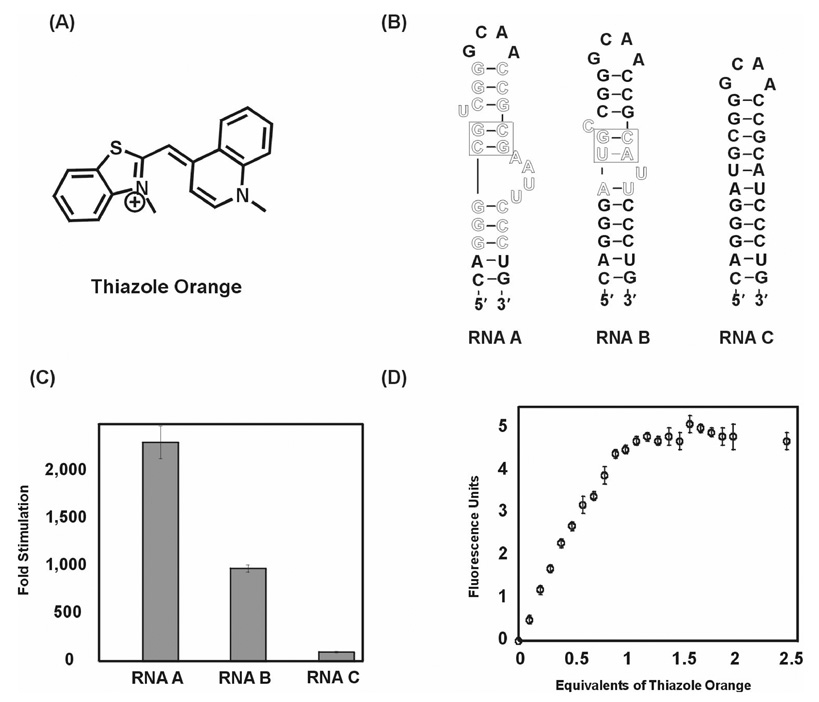
We adapted the fluorescent intercalator displacement (FID) assay to the study of HTP/RNA binding. Boger and coworkers described this assay previously for establishing the affinity and selectivity of DNA binding compounds.23, 25–28 This assay involves the displacement of an intercalator, such as ethidium bromide or thiazole orange, from a nucleic acid target upon addition of a binding ligand. When a double stranded nucleic acid is treated with certain fluorescent intercalators, there is an increase in the observed fluorescence signal. Addition of a binding ligand to the complex results in the displacement of the intercalator, causing a decrease in the fluorescence. Recently, Pei and Stojanovic screened a series of RNA and DNA aptamers in a thiazole orange displacement assay and found that the assay was reliable for the study of ligand binding to some, but not all, of the aptamers investigated.29 We tested three RNA targets in our assay, namely, RNA A, RNA B, and RNA C (Figure 1). RNA A is a 29-mer stem-loop RNA construct based on an in vitro selected RNA aptamer that is predisposed to bind HTPs.18 RNA B is a 28-mer hairpin stem structure containing sequence found in human pre-miRNA 23b and also has a binding site for HTPs.21 RNA C is a control RNA duplex that does not contain an HTP site.
Figure 1.
(A) Structure of thiazole orange; (B) Secondary structures of RNA A, B with putative intercalation site boxed and control RNA C used for the displacement assay, nucleotides highlighted in outlined text are present in the in vitro selected aptamer (RNA A) and human pre-miRNA 23b (RNA B); (C) Fold stimulation of thiazole orange (2.6 µM) fluorescence with RNA A, B, and C at 2 µM concentration each; (D) Titration of RNA A (5 µM) with thiazole orange.
We found that when thiazole orange and RNA are present at 2.6 µM and 2 µM concentrations respectively, the thiazole orange fluorescence is enhanced by the presence of RNA A by 2308-fold (Figure 1). RNA B, which contains a lower affinity HTP site, caused a 977-fold increase in thiazole orange fluorescence at 2.6 µM concentration. Importantly, under these same conditions, the binding of thiazole orange to control RNA C caused a much smaller fluorescence increase (96-fold). Thus, thiazole orange binding to RNAs with intercalation "hot spots" (e.g. RNAs A and B) leads to approximately three orders of magnitude enhancement in fluorescence signal, suggesting the possibility of a highly sensitive RNA binding assay. Thiazole orange binding to RNA A was analyzed further. Titration of the fluorophore into a 5 µM solution of RNA A resulted in pronounced increase in fluorescence with each addition until one equivalent of thiazole orange had been added (Figure 1). No additional RNA-dependent fluorescence enhancement was observed upon further addition, suggesting the large fluorescence enhancement is a result of the formation of a 1:1 complex.
The large fluorescence enhancement observed when thiazole orange binds RNAs A and B suggested that competition experiments could be carried out at low thiazole orange concentrations. After screening several different concentrations and thiazole orange:RNA ratios, the combination of 50 nM thiazole orange and 100 nM RNA A was chosen for the displacement experiments. Briefly, in the displacement assays, the RNA-thiazole orange complex was pre-formed and added to increasing HTP concentrations. The solutions were then transferred to a 96-well plate and scanned for fluorescence changes. As described above, binding of thiazole orange to the stem-loop RNA structure enhances the fluorescence of thiazole orange (Figure 2a). Addition of HTP to this complex results in the displacement of the bound thiazole orange causing a decrease in the fluorescence. The extent of fluorescence decrease can be correlated to the binding affinity of the peptide. The IC50 for the decrease in the fluorescence of thiazole orange was calculated by plotting the relative fluorescence decrease as a function of HTP concentration. In each case, background fluorescence of thiazole orange alone was subtracted. These IC50 values were converted to apparent dissociation constants (KD) assuming a competitive binding mode and using a thiazole orange KD for RNA A of 3.1 ± 1.8 µM as determined by quantitative RNase V1 footprinting (see Experimental Section and Supplementary Information).30 It should be noted that given the amount of RNA required to observe the thiazole orange fluorescence enhancement, accurately measuring Kd's for very tight binders (low nM or better) may not be possible with this approach.
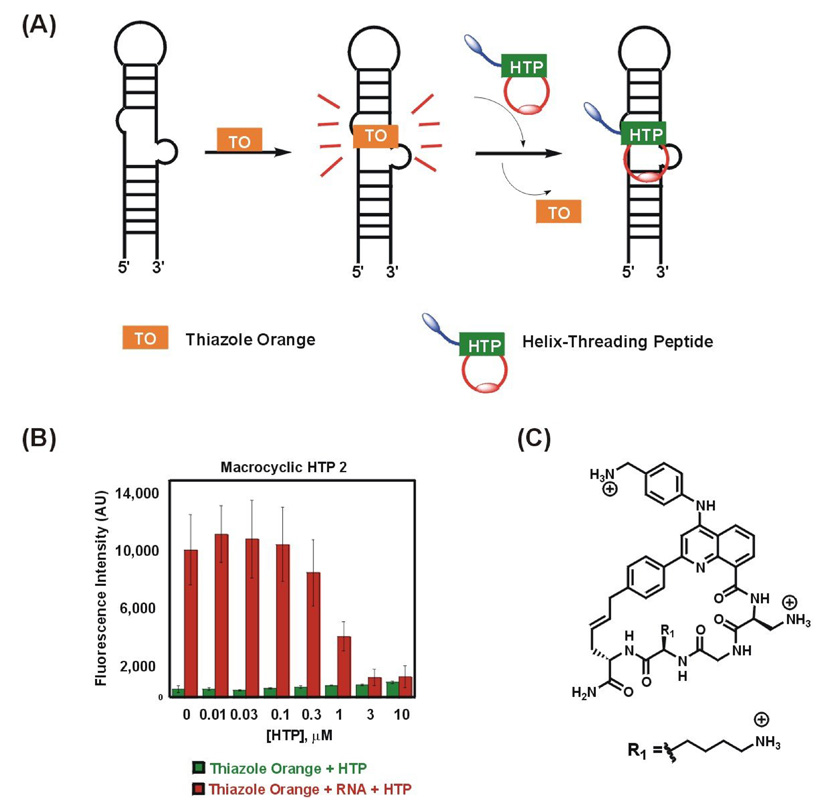
Figure 2.
(a) Schematic representation of thiazole orange displacement assay; (b) Binding of HTP 2 to RNA A causes displacement of thiazole orange resulting in decrease in fluorescence; (c) Structure of macrocyclic HTP 2.
Shown in Figure 2 is the thiazole orange displacement for a representative macrocyclic HTP. At 50 nM thiazole orange and 100 nM RNA concentrations, binding of thiazole orange to RNA A resulted in an approximate 20-fold stimulation of fluorescence signal. Titration of macrocyclic HTP 2 (Figure 2c) resulted in 96% reduction of this initial fluorescence at the highest concentration of HTP as a result of HTP binding and displacement of thiazole orange (Figure 2b).
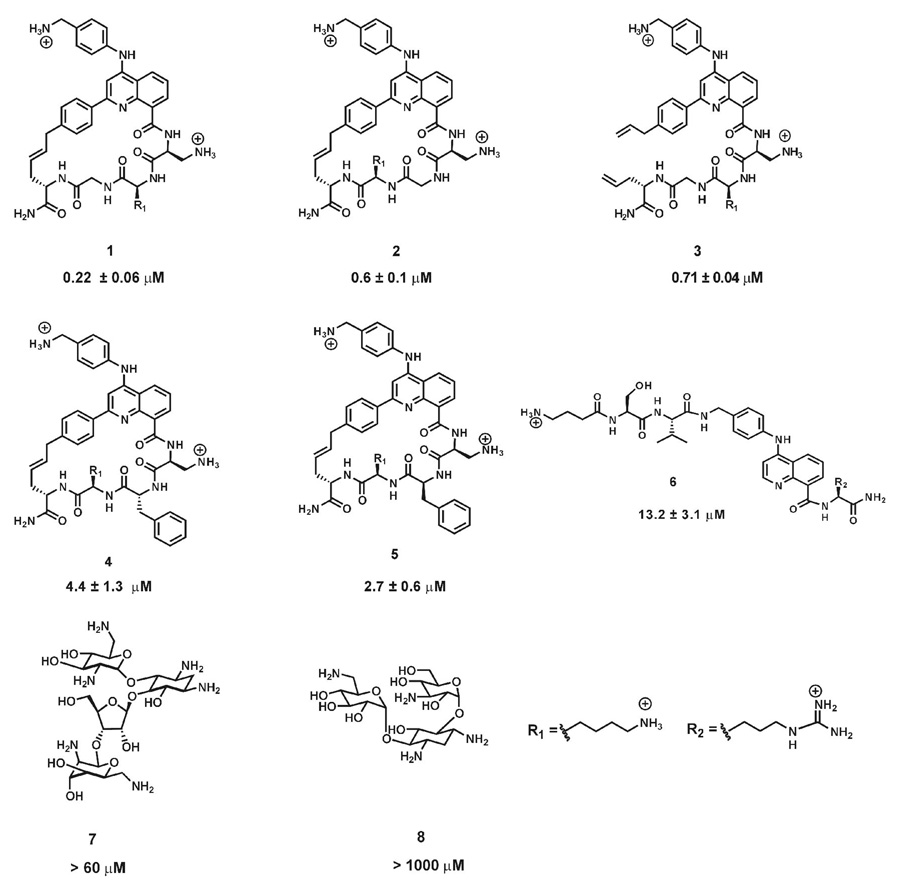
We initially tested this assay for a small set of HTPs that had been previously shown to bind RNA A using RNase V1 footprinting (Figure 3). We found that the trends in the dissociation constants from the FID assay correlated well with those obtained from quantitative footprinting experiments carried out previously with larger deviations observed for a tight binder (HTP1) and a weaker binding compound (HTP6) (Table 1).20, 21 This indicated that the FID assay can be used reliably for screening HTPs to determine RNA binding affinities without the investment in time and effort necessary for the footprinting experiments.
Figure 3.
Chemical structures of HTPs and aminoglycosides analyzed by thiazole orange displacement assay. The KD values obtained from the displacement assay are indicated below the structures for each ligand.
Table 1.
Comparison of dissociation constants for HTPs measured in two different assays.
| HTP | Dissociation constants (footprinting) (µM)[a] | Dissociation constants (FID) (µM)[b] |
|---|---|---|
| 1 | 0.05 ± 0.01 | 0.22 ± 0.06 |
| 2 | 0.27 ± 0.11 | 0.6 ± 0.1 |
| 3 | 0.33 ± 0.06 | 0.70 ± 0.04 |
| 4 | 8.6 ± 4.3 | 4.4 ± 1.3 |
| 5 | 2.3 ± 0.9 | 2.7 ± 0.6 |
| 6 | 3.3 ± 1.8 | 13.2 ± 3.1* |
| 7 | nd | > 60 |
| 8 | nd | > 100 |
Conditions: 50 mM Bis-Tris·HCl, pH 7.0, 100 mM NaCl, 10 mM MgCl2, and 10 µg/mL of yeast tRNAPhe, 25 °C, 15 min. equilibration prior to digestion with ribonuclease V1.
Conditions: 50 mM Bis-Tris·HCl, pH 7.0, 100 mM NaCl, 10 mM MgCl2, 50 nM thiazole orange and 100 nM RNA A, 25 °C. Dissociation constants are reported as the average of three independent measurements ± standard deviation. nd not determined.
Average of two independent measurements.
In order to further test its scope, we performed the assay with aminoglycosides neomycin (7) and kanamycin (8) (Figure 3). Interestingly, complete displacement was not observed with these compounds even at high micromolar concentrations, conditions likely to cause them to bind these RNAs nonspecifically (Table 1, Supplementary Figure 2 and Supplementary Figure 3). The more efficient displacement by HTPs may be the result of a shared binding mode with thiazole orange (intercalation). Groove binding by the aminoglycosides could be compatible with the thiazole orange-RNA complex and result in poor displacement.
Since macrocyclic scaffolds provide the possibility of orienting different functional groups on the cyclic backbone in specific positions in space, we were interested in studying the structure activity relationships of macrocyclic HTPs and their binding affinity to target RNA using this assay. Encouraged by the results from the displacement assay, we synthesized a small macrocyclic HTP library containing a 2-phenylquinoline intercalating unit, in order to test the effect of changing the structure of amino acid side chains in the peptide, the α-amino acid stereochemistry, backbone cyclization as well as ring size (Figure 4). In the design of our new library, our goal was to reduce non-selective RNA binding in the HTPs as much as possible. Since highly positively charged compounds are capable of nonselective interactions with the negatively charged RNA backbone, we chose to alter the core HTP structure to reduce the number of positive charges by replacing the 4-aminobenzylamine unit with aniline. We therefore synthesized a new 2-phenylquinoline carboxylic acid with anilino substitution at the C4 position. The carboxylic acid required for synthesis of the library members was generated using a short and efficient scheme (Scheme 1).
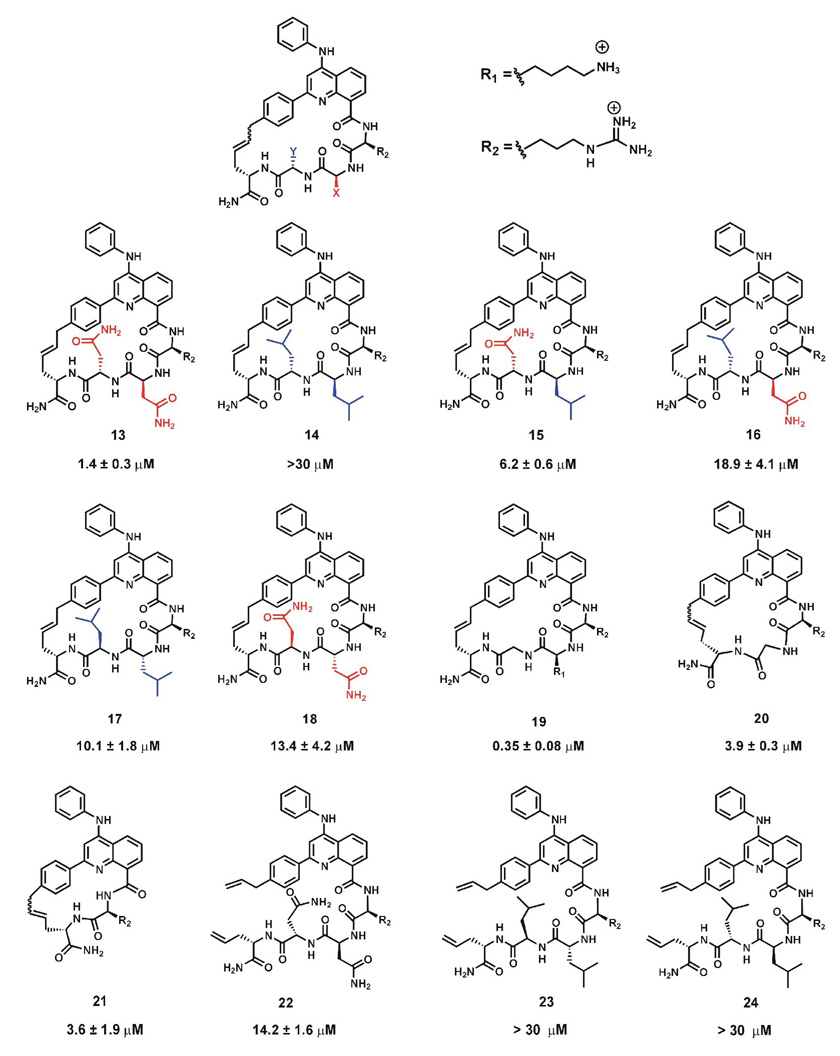
Figure 4.
Chemical structures of macrocyclic HTPs used in the displacement assay. The KD values obtained from the displacement assay are indicated below the structures for each HTP.
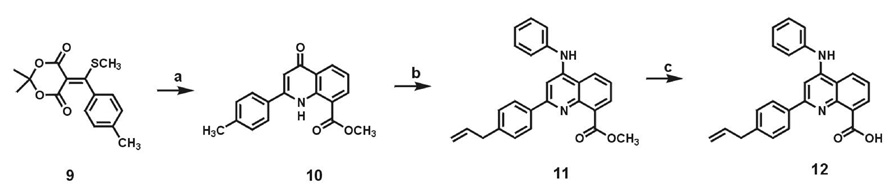
Scheme 1.
Synthesis of 2-phenylquinoline carboxylic acid 12. (a) Methyl anthranilate, Ph2O, MW, 240 °C, 20 min, 83%.; (b) (i) POCl3, reflux,1 h, 87%; (ii) NBS, AIBN, cyclohexane, reflux, 50 min, 77%; (iii) Vinyl tributyltin, Pd(PPh3)4, DMF, 45 °C, 2 h, 73%; (iv) aniline, SnCl4, CH3CN, rt , 1 h and 86%; (c) LiOH.H2O, THF-H2O, rt, 24 h, 87%.
2-Phenylquinolone 10 was synthesized from Meldrum’s acid derivative, 5-(p-methyl-α-thiomethoxybenzylidene)-2,2-dimethyl-1,3-dioxane-4,6-dione (9) using microwave conditions in very good yields. As compared to our previously reported synthesis of 10 using standard heating conditions, we observed that the reaction time decreased considerably by using microwave conditions and resulted in greater yields of the quinolone.21 Amination with a 4-chloro-2-p allylphenylquinoline derivative reported earlier by us, using aniline in the presence of tin tetrachloride gave ester 11 in good yield.21 This methyl ester was hydrolyzed to obtain carboxylic acid 12 in overall good yield. The acid was used for solid-phase peptide synthesis of the macrocyclic HTP library as previously described.21 The library represented members that differed in the structure of amino acid side chains, α-carbon stereochemistry, cyclic vs. linear HTP as well as macroccycle ring size (Figure 4). Dissociation constants for the binding to RNA A determined using the FID assay as discussed above are shown below each HTP structure in Figure 4.
To test the effect of changing amino acid side chain structure, we decided to focus initially on two amino acids present in the macrocyclic pentapeptides (positions X and Y, Figure 4). Comparing asparagines at these positions to leucines would allow us to determine if a preference for hydrophilic or hydrophobic side chains existed, since the asparagine and leucine side chains are of similar size and steric differences would be minimized. RNA A was chosen as the target for these initial studies as a representative HTP-binding RNA. It is clear from our results that asparagine is preferred at these positions when L amino acids are used. For instance, in HTP 13, replacement of asparagine residues at X and Y with leucine (HTP 14) dramatically reduced the binding affinity (KD = 1.4 µM vs KD > 30 µM). We also found that the position of the asparagine residue in the ring is important as can be seen comparing HTP 13 to 15 or 16. Mutation of residue X in 13 from asparagine to leucine to give HTP 15, resulted in a four-fold loss in affinity. However, mutation of residue Y in HTP 13 to give 16 was more detrimental resulting in a 13-fold reduction in binding affinity. Although complete understanding of the affinity changes observed will require high resolution structural data, we can speculate that asparagines engage in productive interactions with RNA functional groups around the intercalation site not possible with leucines. Our previous work with structurally related linear peptides indicated that the quinoline 8-carboxamide substitutent is localized in the major groove of the RNA duplex.20, 22 Therefore, asparagine side chains at X and Y may be hydrogen bonding within the major groove at the binding site. Regardless of the basis for this effect, these results indicate that the structure of the amino acid side chain at the X and Y positions of the macrocyclic pentapeptides controls RNA affinity, signifying that libraries of these molecules where X and Y are varied contain compounds with distinct RNA recognition properties.
To determine if the observed effect arose simply from a general preference for the more polar side chain of asparagine, we switched the α-amino acid stereochemistry to D at the variable positions. The stereochemistry at the α- carbon affects the binding affinity as well, as seen by comparisons to HTPs 17 and 18, which have D-amino acids at positions X and Y. Replacing the L-asparagine with D-asparagine at both positions reduces binding affinity by 10- fold (compare HTP 13 and HTP 18). Surprisingly, replacing the two L-leucine residues (HTP 14) with D-leucine residues (HTP 17) causes an improvement in binding affinity. The overall effect is that with D stereochemistry at the X and Y positions, macrocycle binding is largely independent of side chain structure, since 17 and 18 bind with nearly the same affinity.
Importantly, comparison of macrocyclic HTPs 13 and 17 with their linear precursors HTPs 22 and 23 respectively, indicated that cyclization is beneficial for both the sequences. This result is similar to that observed previously when comparing linear vs cyclic pentapeptide HTPs binding to the binding site found on RNA A.21 We also explored the effect of reducing the macrocycle ring size. Macrocyclic tetrapeptide HTP 20 and tripeptide HTP 21 were prepared for this purpose. As observed with the other macrocycles in this study, ring-closing metathesis (RCM) was efficient with HTP 20 yielding the cyclic compound as the major reaction product. However, a complex mixture of products was observed for RCM reactions to generate the smaller HTP 21, requiring extensive HPLC purification and mass spectrometry to identify 21 as a minor component. Interestingly, 20 and 21 bound RNA with nearly the same affinity and only slightly reduced from pentapeptide 13 bearing the same overall charge, suggesting that ring size is not a major determinant for binding to RNA A when comparing these tri-, tetra-, and pentapeptides. However, the poor cyclization reaction for the tripeptide suggests preparing libraries of macrocyclic HTPs of this size will be problematic.
Conclusion
In summary, we have adapted the fluorescent intercalator displacement assay using thiazole orange to the study of RNA-binding helix-threading peptides. This assay is highly sensitive and provides binding affinity data in good agreement with other methods. This assay was used to define structure activity relationships for a small library of compounds. Results of these studies indicate their RNA binding is dependent on peptide sequence, α-amino acid stereochemistry, and cyclization (versus linear peptides), but independent of macrocyclic ring size. These studies indicate that libraries of macrocyclic, helix-threading peptides with different amino acids at variable positions contain members with distinct RNA binding properties.
Experimental
Preparation of 8-(methoxycarbonyl)-2-(4’-methylphenyl)-4(1H)-quinolone (10)
To a solution of 9 (0.150 g, 0.514 mmol) in phenyl ether (3.5 mL) was added methyl anthranilate (0.1 mL, 0.770 mmol) in a microwave vial. The vial was capped and subjected to MW irradiation at 240 °C for 20 min. The reaction mixture was cooled to room temperature. Purification by silica gel column chromatography (initially 100% hexanes to remove phenyl ether, 10–40% EtOAc/hexanes to remove impurities, 70–80% EtOAc/hexanes to elute product) afforded 0.121 g (83%) of the product as a white solid. The characterization of the product by NMR and mass spectroscopy has been reported previously.21
Preparation of methyl-2-(4’-allylphenyl)-4-(anilino)quinoline-8-carboxylate (11)
To a solution of methyl-2-(4’-allylphenyl)-4-chloro-quinoline-8-carboxylate (0.060 g, 0.179 mmol) (synthesized according to our previously reported procedure) in CH3CN (5 mL) was added aniline (0.048 mL, 0.358 mmol) and SnCl4 (0.021 mL, 0.179 mmol).21 The resulting solution was stirred at room temperature for 1 h. The reaction mixture was quenched with saturated NaHCO3, passed through a celite pad and extracted with dichloromethane. The organic layer was dried over anhydrous Na2SO4, and the solvent was removed under reduced pressure. Purification by silica gel column chromatography (initially 1% MeOH/CHCl3 followed by 2–3 % MeOH/CHCl3) afforded 0.060 g (86 %) of the product as a yellow solid. 1H NMR (600 MHz, CDCl3) δ (ppm): 7.84 (brs, 4H), 7.35–7.03 (m, 8H), 6.79 (brs, 1H), 5.87 (m, 1H), 5.01 (d, J = 0.9 Hz, 1H), 5.00 (d, J = 0.9 Hz, 1H), 3.94 (s, 3H), 3.32 (d, J = 6.5 Hz, 2H) 13C NMR (100 MHz, CDCl3) δ (ppm): 169.37 142.11, 137.19, 129.82, 129.31, 128.72, 127.30, 123.96, 123.43, 122.20, 116.39, 100.03, 52.72, 40.18, 29.94, 21.59. High resolution CI-MS calculated mass for C26H22N2O2: 394.1676, found [M + H]+ 394.1680 m/z.
Preparation 2-(4’-allylphenyl)-4-(anilino)–quinoline-8-carboxylic acid (12)
To a solution of 11 (0.250 g, 0.638 mmol) in THF/H2O (13.5 mL, 2:1) was added lithium hydroxide monohydrate (0.059 g, 1.423 mmol) and the resulting solution stirred at room temperature for 24 h. The reaction mixture was quenched with saturated NH4Cl, when a yellow precipitate formed. The precipitate was filtered, washed with water and dried under vacuum to afford 0.209 g (87%) of the product as a yellow solid. 1H NMR (500 MHz, CDCl3/CD3OD) δ (ppm): 8.42 (d, J = 7.3 Hz, 1H), 8.37 (dd, J = 8.5, 1.2 Hz, 1H), 7.50 (d, J = 7.9 Hz, 2H), 7.47–7.42(m, 2H), 7.40–7.37 (m, 3H), 7.32–7.31 (m, 3H), 7.26–7.23( m, 2H), 7.15 (d, J = 7.9 Hz, 3H), 6.87 (s, 1H), 5.79–5.72 (m, 1H), 4.95–4.91 (m, 2H), 3.27 (d, J = 6.5 Hz, 2H). 13C NMR (150 MHz, CDCl3/CD3OD) δ (ppm): 170.44, 155.12, 151.45, 145.10, 143.17, 140.33, 137.53, 136.39, 136.09, 130.51, 130.18, 130.06, 127.62, 127.20, 126.99, 126.19, 125.33, 125.29, 124.75, 117.06, 116.78, 97.02, 39.88, 21.31. High resolution FAB calculated mass for C25H20N2O2: 380.1519, found [M + H]+ 380.1535 m/z.
Preparation of linear HTPs 22 and 23
Linear HTPs were synthesized using 9-fluoronylmethoxycarbonyl (Fmoc)-protected amino acids according to standard SPPS protocols using Novagel Rink Amide resin (NovaBiochem, 0.64 mmol/g loading, 45 mg, 0.028 mmol) as described previously.21 Following coupling of carboxylic acid 12 (40 mg, 3.7 eq), the resin was washed and cleaved using TFA/TIS/H2O (90:8:2, 3 mL, 3.5 h). The resulting solution was collected and concentrated under reduced pressure. Ether precipitation of the residue, followed by extraction with water gave the crude product, which was purified by HPLC.
Cyclization via ring-closing metathesis
For the synthesis of macrocyclic HTPs 13–21, the linear peptides were first assembled as described above using Novagel Rink Amide resin (NovaBiochem, 0.64 mmol/g loading, 55 mg, 0.035 mmol). Following the coupling of carboxylic acid 12 (52.5 mg, 4 eq), the resin was washed with CH2Cl2 (3 × 2 mL), DMF (3 × 2 mL), MeOH (3× 2 mL) and diethyl ether (3 × 2 mL) and lyophilized for 12 h. The dried resin was transferred to a flame-dried round-bottomed flask and preswelled with 1,2-dichloroethane (4 mL) for 45 min. Hoveyda-Grubbs catalyst (5.5 mg, 25 mol%) was weighed in a flame-dried vial and to this 1,2-dichloroethane (1 mL) was added and transferred to the flask containing the suspended resin. The flask was then heated at reflux for ~ 30 h. After cooling to room temperature, the resin was transferred to a polypropylene column (Bio-Rad) and drained. It was then washed with CH2Cl2 (3 × 2 mL), 10% 1,3-bis(diphenylphosphino)propane in CH2Cl2 (1 × 2 mL) and MeOH (3 × 2 mL) and lyophilized for 12 h. The dry resin was incubated with TFA/TIS/PhOH/H2O (88:5:5:2, 3 mL) and shaken for 3.5 h to effect cleavage from the resin and amino acid side chain deprotection. The resulting solution was collected and concentrated under reduced pressure. Ether precipitation of the residue, followed by extraction with water, gave the crude product. After the standard work-up procedures, the linear and cyclic peptides were purified using reverse-phase HPLC (Waters Corporation) equipped with an auto sampler unit (2767 Sample Manager, Waters). The crude peptides were analyzed using a reverse phase analytical C-18 column (4.6 × 50 mm, SunFire, Waters). The purification was carried out using a reverse phase preparative C-18 column (19 × 50 mm, SunFire, Waters). For analytical runs, a flow-rate of 1.5 mL/min was used and for the preparative runs, a flow-rate of 25 mL/min was used. Typically, 15–20 µL of the sample was injected as an aqueous solution for analytical runs, while 500–1000 µL of sample was injected when the preparative column was used. HTPs 13–21 were injected as 5% AcOH solutions. HTP 22 was injected as a solution in 50% ACN and HTP 23 was injected as a 50% MeOH solution to address solubility issues. ESI-MS analyzed and calculated mass for the macrocyclic peptides are given in Supplementary Table 1. For macrocyclic pentapeptides 13–19, olefin stereochemistry is assumed to be trans based on the structures of HTPs 1 and 2 prepared under these conditions.21 The olefin stereochemistry for macrocyclic tetrapeptide HTP 20 or tripeptide HTP 21 was not determined.
Preparation of RNA for experiments
RNA A was purchased from Dharmacon RNA Technologies and used directly for the FID experiments. RNA B and C were purchased from Dharmacon RNA Technologies as 2’-bis(2-acetoxyethoxy)methyl orthoester (2’-ACE)® protected RNA. RNA was prepared for the experiments by deprotecting the 2’-ACE group according to the manufacturer’s protocol. Briefly, 400 µL of 2’-deprotection buffer (100 mM acetic acid, adjusted to pH 3.8 with TEMED) was added to the RNA pellet and mixed well. The solution was incubated at 60 ° C for 30 min, following which the RNA was extracted with phenol/chloroform, ethanol precipitated, dissolved in nanopure water and stored at − 20 ° C.
Quantitative footprinting of thiazole orange with RNA A
A dissociation constant for thiazole orange on RNA A was obtained using RNase V1 footprinting experiments. For the preparation of 5’-32P RNA, 60 pmol of RNA A in T4 Polynucleotide Kinase buffer (70 mM Tris-HCl, 10 mM MgCl2, 5 mM dithiothreitol, pH 7.6) was treated with T4 Polynucleotide Kinase (2.0 U/µL), [γ-32P]-ATP (2 mCi/mL) and incubated for 45 min at 37 °C followed by heat inactivation at 65 °C for 20 min. Labeled RNA was purified on a 19% denaturing polyacrylamide gel, visualized by storage phosphor autoradiography, excised from the gel and eluted overnight via the crush and soak method. After filtration, the resulting solution was extracted with phenol/chloroform, ethanol precipitated, and stored in water. Ligand/protein–RNA complexes were formed by incubating increasing concentrations of the ligand with 5’-32P labeled RNA A (~ 5 nM) for 15 min in reaction buffer (50 mM Bis-Tris·HCl, pH 7.0, 100 mM NaCl, 10 mM MgCl2) at ambient temperature. Enzymatic digestions with RNase V1 (1 × 10−6 U) were carried out for 30 min at ambient temperature and quenched with hot formamide loading buffer. Cleaved RNA was heat denatured and analyzed by 19% denaturing polyacrylamide gel electrophoresis. The cleavage data for the RNA was converted to binding data for the ligand, assuming that the maximum cleavage efficiency corresponds to 0% occupancy and the minimum cleavage efficiency corresponds to 100% occupancy. The fraction of RNA bound by the ligand was plotted as a function of concentration, and the data were fitted to the equation: fraction RNA bound = [ligand]/([ligand] + KD(TO)). The dissociation constant (KD(TO) = 3.1 ± 1.8 µM) is reported as the average and standard deviation for three different experiments (see Supplementary Figure 1).
RNA-thiazole orange stoichiometry determination
RNA stock solutions were prepared in nanopure water and thiazole orange stock solutions were prepared in DMSO. A reaction mixture (98 µL) consisting of RNA A (5 µM) in reaction buffer (50 mM Bis-Tris·HCl, pH 7.0, 100 mM NaCl, 10 mM MgCl2) was added to 2 µL of increasing concentration of thiazole orange (0.1 to 2.5 equivalents) in a 96-well black OptiPlate (Perkin Elmer) at 25 °C. The mixture was thoroughly mixed with a pipet and incubated for ~ 15 min at 25 °C. For the control wells, 98 µL of a reaction mixture consisting of reaction buffer (50 mM Bis-Tris·HCl, pH 7.0, 100 mM NaCl, 10 mM MgCl2) was added to 2 µL of increasing concentration of thiazole orange (0.1 to 2.5 equivalents). The 96-well plate was then read in Victor 3 fluorescent plate reader (Perkin Elmer, excitation 485 nm, emission 535 nm). The background fluorescence of the control wells at each concentration was subtracted from the wells containing the RNA-thiazole orange complex at that concentration. The fluorescence change was plotted as a function of equivalents of thiazole orange added. The stoichiometry was determined using Microsoft Excel by simultaneously solving the equations representing the pre- and post-saturation regions of the curve.
Thiazole orange displacement assay
RNA stock solutions were prepared in nanopure water and thiazole orange stock solutions were prepared in DMSO. HTP stocks were usually prepared in water except for HTPs 17 and 23, which were prepared in DMSO. RNA-thiazole orange complex was formed by adding RNA (100 nM) to a solution of thiazole orange (50 nM) in reaction buffer (50 mM Bis-Tris·HCl, pH 7.0, 100 mM NaCl, 10 mM MgCl2) and incubated for 15 min. This solution (98 µL) was added to increasing concentrations of the desired HTP (2 µL) such that the total reaction volume was 100 µL. The mixture was thoroughly mixed with a pipet and incubated for ~ 15 min at 25 °C. After incubation for 15 min, 100 µL of the solution was transferred to a 96-well black OptiPlate (Perkin Elmer). For HTP 17, the total reaction volume was 50 µL. For the control wells, a solution of thiazole orange (50 nM) in reaction buffer (50 mM Bis-Tris·HCl, pH 7.0, 100 mM NaCl, 10 mM MgCl2) was added to increasing concentrations of the HTP (2 µL) such that the total reaction volume was 100 µL. The 96-well plate was then read in Victor 3 fluorescent plate reader (Perkin Elmer, excitation 485 nm, emission 535 nm). The background fluorescence of the control wells at each concentration was subtracted from the wells containing the RNA-HTP-thiazole orange complex at that concentration. In order to calculate the IC50 of fluorescence displacement, the fluorescence data was converted to binding data assuming that maximum fluorescence corresponds to 0% displacement. The fluorescence of the other wells was normalized to the well with 0% displacement (100 % fluorescence) to obtain the relative fluorescence decrease. Using Kaleidagraph 4.0 software, the above data were plotted as a function of HTP concentration and fitted to the equation: Percentage decrease in fluorescence = F/(1+IC50/[ligand]) where F = Normalized relative fluorescence of RNA-thiazole orange complex at each HTP concentration subtracted from background. With aminoglycosides 7 and 8, there was no displacement of thiazole orange up to the highest concentrations (see Supplementary Figure 2). The apparent dissociation constants of the HTPs (KHTP) were calculated using a competitive model using the KD of thiazole orange with RNA A (KTO = 3.1 ± 1.8 □M) as determined by RNase V1 footprinting. The equation, KHTP = IC50 /(1 + ([TO]/KTO))30, was used to determine the apparent dissociation constants, where [TO] = 50 nM and is the concentration of thiazole orange in the FID assay. The KHTP values are reported in Table 1 and Table 2.
Table 2.
Dissociation constants of HTPs from thiazole orange displacement.
| HTP | Dissociation constants (FID) (M)[a] |
|---|---|
| 13 | 1.4 ± 0.3 |
| 14 | > 30 |
| 15 | 6.2 ± 0.6 |
| 16 | 18.9 ± 4.1 |
| 17* | 10.1 ± 1.8 |
| 18 | 13.4 ± 4.2 |
| 19 | 0.35 ± 0.08 |
| 20 | 3.9 ± 0.3 |
| 21 | 3.6 ± 1.9 |
| 22 | 14.2 ± 1.6 |
| 23 | > 30 |
| 24 | > 30 |
Conditions: 50 mM Bis-Tris·HCl, pH 7.0, 100 mM NaCl, 10 mM MgCl2, 50 nM thiazole orange and 100 nM RNA A, 25 °C. Dissociation constants are reported as the average of three independent measurements ± standard deviation.
Average of two independent measurements.
Supplementary Material
Acknowledgements
P.A.B. acknowledges NIH for financial support GM061115.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gallego J, Varani G. Acc. Chem. Res. 2001;34:836. doi: 10.1021/ar000118k. [DOI] [PubMed] [Google Scholar]
- 2.Thomas JR, Hergenrother P. J. Chem. Rev. 2008;108:1171. doi: 10.1021/cr0681546. [DOI] [PubMed] [Google Scholar]
- 3.Bartel DP. Cell. 2004;116:281. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 4.Winkler WC, Breaker RR. Annu. Rev. Microbiol. 2005;59:487. doi: 10.1146/annurev.micro.59.030804.121336. [DOI] [PubMed] [Google Scholar]
- 5.Noller HF. Science. 2005;309:1508. doi: 10.1126/science.1111771. [DOI] [PubMed] [Google Scholar]
- 6.Win MN, Smolke CD. Proc. Natl. Acad. Sci. U.S.A. 2007;104:14283. doi: 10.1073/pnas.0703961104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nomura Y, Yokobayashi Y. J. Am. Chem. Soc. 2007;129:13814. doi: 10.1021/ja076298b. [DOI] [PubMed] [Google Scholar]
- 8.Topp S, Gallivan JP. J. Am. Chem. Soc. 2007;129:6807. doi: 10.1021/ja0692480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yen L, Svendsen J, Lee JS, Gray JT, Magnier M, Baba T, D'Amato RJ, Mulligan RC. Nature. 2004;431:471. doi: 10.1038/nature02844. [DOI] [PubMed] [Google Scholar]
- 10.Murray JK, Sadowsky JD, Scalf M, Smith LM, Tomita Y, Gellman SH. J. Comb. Chem. 2008;10:204. doi: 10.1021/cc700153z. [DOI] [PubMed] [Google Scholar]
- 11.Kwon YU, Kodadek T. Chem. Biol. 2007;14:671. doi: 10.1016/j.chembiol.2007.05.006. [DOI] [PubMed] [Google Scholar]
- 12.Yu P, Liu B, Kodadek T. Nat. Biotechnol. 2005;23:746. doi: 10.1038/nbt1099. [DOI] [PubMed] [Google Scholar]
- 13.Tan DS. Nat. Chem. Biol. 2005;1:74. doi: 10.1038/nchembio0705-74. [DOI] [PubMed] [Google Scholar]
- 14.Chaltin P, Borgions F, Van Aerschot A, Herdewijn P. Bioorg. Med. Chem. Lett. 2003;13:47. doi: 10.1016/s0960-894x(02)00836-3. [DOI] [PubMed] [Google Scholar]
- 15.Hwang S, Tamilarasu N, Kibler K, Cao H, Ali A, Ping YH, Jeang KT, Rana TM. J. Biol. Chem. 2003;278:39092. doi: 10.1074/jbc.M301749200. [DOI] [PubMed] [Google Scholar]
- 16.Hwang S, Tamilarasu N, Ryan K, Huq I, Richter S, Still WC, Rana TM. Proc. Natl. Acad. Sci. U.S.A. 1999;96:12997. doi: 10.1073/pnas.96.23.12997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He XG, Gerona-Navarro G, Jaffrey SR. J. Pharmacol. Exp. Ther. 2005;313:1. doi: 10.1124/jpet.104.076943. [DOI] [PubMed] [Google Scholar]
- 18.Carlson CB, Vuyisich M, Gooch BD, Beal PA. Chem. Biol. 2003;10:663. doi: 10.1016/s1074-5521(03)00147-9. [DOI] [PubMed] [Google Scholar]
- 19.Krishnamurthy M, Gooch BD, Beal PA. Org. Lett. 2004;6:63. doi: 10.1021/ol036094+. [DOI] [PubMed] [Google Scholar]
- 20.Krishnamurthy M, Gooch BD, Beal PA. Org. Biomol. Chem. 2006;4:639. doi: 10.1039/b513591e. [DOI] [PubMed] [Google Scholar]
- 21.Krishnamurthy M, Simon K, Orendt AM, Beal PA. Angew. Chem. Int. Ed. 2007;46:7044. doi: 10.1002/anie.200702247. [DOI] [PubMed] [Google Scholar]
- 22.Gooch BD, Beal PA. J. Am. Chem. Soc. 2004;126:10603. doi: 10.1021/ja047818v. [DOI] [PubMed] [Google Scholar]
- 23.Tse WC, Boger DL. Acc. Chem. Res. 2004;37:61. doi: 10.1021/ar030113y. [DOI] [PubMed] [Google Scholar]
- 24.Monchaud D, Allain C, Bertrand H, Smargiasso N, Rosu F, Gabelica V, De Cian A, Mergny JL, Teulade-Fichou MP. Biochimie. 2008;90(8):1207–1223. doi: 10.1016/j.biochi.2008.02.019. [DOI] [PubMed] [Google Scholar]
- 25.Boger DL, Fink BE, Brunette SR, Tse WC, Hedrick MP. J. Am. Chem. Soc. 2001;123:5878. doi: 10.1021/ja010041a. [DOI] [PubMed] [Google Scholar]
- 26.Boger DL, Fink BE, Hedrick MP. J. Am. Chem. Soc. 2000;122:6382. [Google Scholar]
- 27.Boger DL, Dechantsreiter MA, Ishii T, Fink BE, Hedrick MP. Bioorg. Med. Chem. 2000;8:2049. doi: 10.1016/s0968-0896(00)00137-1. [DOI] [PubMed] [Google Scholar]
- 28.Boger DL, Tse WC. Bioorg. Med. Chem. 2001;9:2511. doi: 10.1016/s0968-0896(01)00243-7. [DOI] [PubMed] [Google Scholar]
- 29.Pei R, Stojanovic MN. Anal. Bioanal. Chem. 2008;390:1093. doi: 10.1007/s00216-007-1773-2. [DOI] [PubMed] [Google Scholar]
- 30.Chang Y, Covey DF, Weiss DS. Mol. Pharmacol. 2000;58:1375. doi: 10.1124/mol.58.6.1375. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.