Abstract
Cyanobacteria have become a major model system for analyzing clock phenomena. The temporal program in this organism enhances fitness in rhythmic environments and is truly global—essentially all genes are regulated by the circadian system. The topology of the chromosome also oscillates and possibly regulates the rhythm of gene expression. The underlying circadian mechanism appears to consist of both a post-translational oscillator (PTO) and a transcriptional/translational feedback loop (TTFL). The PTO can be reconstituted in vitro with three purified proteins (KaiA, KaiB, and KaiC) and ATP. These three core oscillator proteins have been crystallized and structurally determined, the only full-length circadian proteins to be so characterized. The timing of cell division is gated by a circadian checkpoint, but the circadian pacemaker is not influenced by the status of the cell division cycle. This imperturbability may be due to the presence of the PTO that persists under conditions in which metabolism is repressed. Recent biochemical, biophysical, and structural discoveries bring the cyanobacterial circadian system to the brink of explaining heretofore unexplainable biochemical characteristics of a circadian oscillator: the long time constant, precision, and temperature compensation.
Introduction
Circadian (daily) rhythms are a particularly fascinating example of a biological timing mechanism. When entrained to the 24-h environmental cycle, they provide an intracellular clock that estimates environmental time [1]. However, these self-sustaining oscillators can maintain robust rhythms for years in the absence of any daily cue that the environment provides (such as light/dark or temperature cycles). Under these conditions, they revert to their endogenous periodicity that is usually a little faster or a little slower than 24 h (hence their name, coined from the Latin: circa = about, dies = one day). This endogenous period is remarkably precise from cycle to cycle, especially considering the long ~24 h time constant. Finally, these circadian clocks are “temperature compensated,” meaning that they run at nearly the same rate independently of the average ambient temperature. From an evolutionary perspective a temperature dependent clock is likely to be useless as an internal estimator of environmental time [1]. Therefore, a temperature-compensated daily clock was the product of natural selection, and temperature compensation was conserved–even in cells of homeotherms that can regulate their body temperature [2,3]. These cardinal characteristics of circadian oscillators–the long time constant, precision, and temperature compensation–remain difficult to explain biochemically.
Interactions with the environment–does this clock system do anything useful?
We study circadian organization in the prokaryotic cyanobacterium Synechococcus elongatus PCC7942 [4]. This organism is a unicellular bacterium that depends upon photosynthesis autotrophically. Some cyanobacterial species are able to fix atmospheric nitrogen, but the growth of S. elongatus is dependent upon the availability of fixed nitrogen in the environment. S. elongatus has a genome size of 2.7 Mbp (smaller than that of E. coli), and it grows optimally at 25–35°C. Its division rate varies as a function of light intensity, temperature, and nutrients; under favorable conditions, it can exhibit a doubling time as short as 4–5 h. For many years, circadian biologists were reluctant to believe that organisms as “simple” as prokaryotes could have evolved an elaborate circadian timing mechanism [5]; they reasoned that a rapidly dividing bacterium whose “lifetime” was less than a day had no use for a timing mechanism that extended farther than its “lifetime” [6]. This was “individual-centric” thinking that does not apply appropriately to microbial systems. The circadian system in cyanobacteria clearly enhances fitness for cells that are exposed to environmental cycles. In fact, cyanobacteria are one of the few systems in which the adaptive significance of circadian programs has been rigorously tested [1,7].
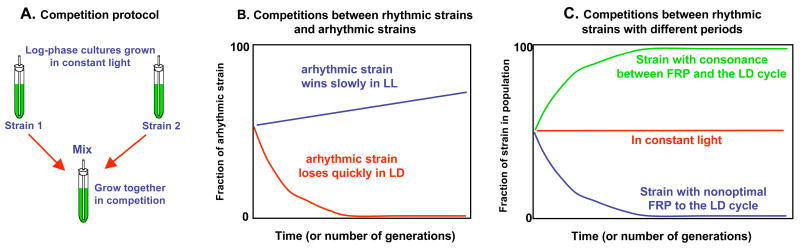
This fitness test was performed in S. elongatus by measuring differential growth of one strain under competition with another strain, which is a good measure of reproductive fitness in asexual microbes [8]. When wild-type cells are competed against a strain in which the circadian clock is inactivated so that it is “arhythmic,” the arhythmic strain is rapidly defeated by wild-type in LD 12:12 cycles (12 h light followed by 12 h darkness). However under competition in LL (constant light), the arhythmic strain grew slightly better than wild-type, as depicted in Fig. 1 [9]. Therefore, the clock system confers a fitness advantage in a rhythmic environment, but not in a constant, non-selective environment. In another series of experiments, mutants with different circadian periods were competed with wild-type and with each other [9,10]. Short-period mutants could out-compete wild-type or long period strains in short-period LD cycles (e.g., LD 11:11 which is a 22-h cycle), whereas long-period mutants were the victors in long-period LD cycles (e.g., LD 15:15, a 30-h cycle). On a 24 h cycle (LD 12:12), wild-type cells could outcompete either mutant as shown in Fig. 1 [7,9,10]. Note that over many cycles, each of these LD conditions have equal amounts of light and dark (which is important since photosynthetic cyanobacteria derive their energy from light); it is only the frequency of light/dark cycling that differs. These data clearly show that in rhythmic environments, the strain whose period most closely matches that of the LD cycle defeats the competitors. Therefore, an intact clock system whose free-running period is consonant with the environment significantly enhances the fitness of cyanobacteria in rhythmic environments.
Figure 1. Competition between circadian strains in different LD cycles.
(A) Different strains of S. elongatus were mixed together in batch cultures and grown in competition under different LD cycles. Every 8 days, the cultures were diluted with fresh medium. At various times during the competition, aliquots were plated as single colonies and their luminescence rhythms were monitored to determine the frequency distribution of the different circadian phenotypes.
(B) In competitions between wild-type and arhythmic strains, the arhythmic strain is rapidly out-competed in LD cycles, but slowly defeats wild-type in constant light (LL, nonselective conditions).
(C) In competitions among strains that are rhythmic, the strain whose endogenous free-running period (FRP) most closely matches that of the environmental LD cycle is able to out-compete strains with a nonoptimal FRP. In LL, all the strains were able to maintain their initial fraction in the population. (Modified from refs. 9 & 10).
How do cyanobacterial cells sense the environmental cycles? Although the endogenous clock is not driven by the environmental cycles, they are entrained to those cycles. As a result of this entrainment, the circadian clock takes on the 24 h period of the environment with a phase relationship that is determined by the biological clock’s endogenous period and its resetting characteristics [1]. Even a 5 h dark pulse can strongly reset the cyanobacterial clock when the cells are otherwise in constant light (LL)[11]. The input pathway by which this entrainment is accomplished is a subject of active research, implicating numerous candidate genes [12,13]. The first such gene to be identified was cikA, which encodes a histidine kinase whose disruption causes interruption of light/dark resetting [11]. Recently four new proteins have been found to interact with CikA (NhtA, PrkE, IrcA, and CdpA), and they may contribute to the input pathway [13]. Two other proteins, Pex and LdpA, also affect the ability of S. elongatus cells to respond to external light/dark cues [14,15]. LdpA contains iron-sulfur clusters that may enable it to sense the redox state of the cell [15], which could be an important daily signal in photosynthetic cyanobacteria. Light drives large changes in redox potential via photosynthesis, and this could be an entraining cue for the cyanobacterial clock [12].
Rhythms in cyanobacteria
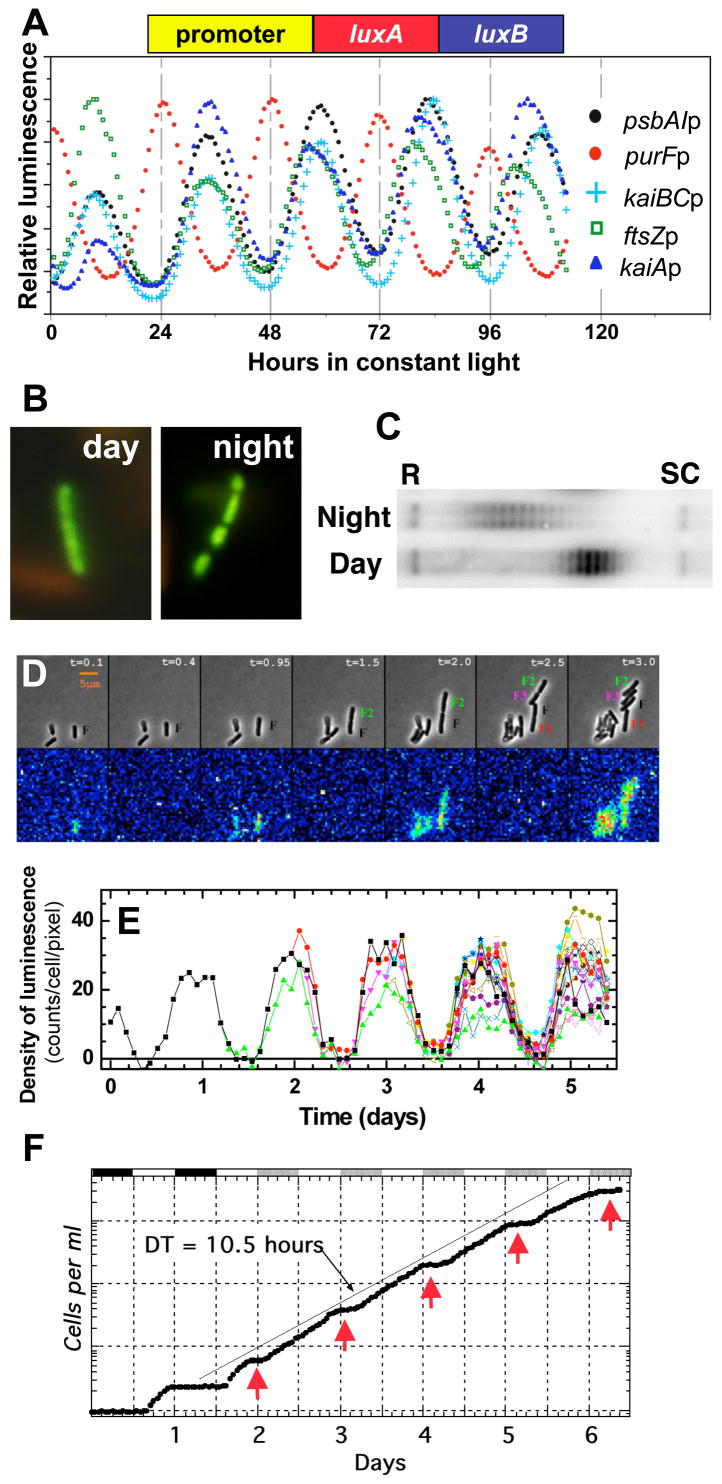
The first circadian rhythm discovered in a cyanobacterium was that of nitrogen fixation, a process whose rate is regulated by the circadian clock to be maximal in the night phase [16]. This nocturnal regulation is potentially of adaptive significance because nitrogenase, the enzyme that reduces atmospheric nitrogen to ammonia during nitrogen fixation, is oxygen sensitive and photosynthesis produces oxygen throughout the daytime. Therefore, the circadian system orchestrates a temporal separation of incompatible metabolic events: photosynthesis in the day, nitrogen fixation at night. In searching for a cyanobacterium that was genetically malleable, we and our collaborators focused upon S. elongatus PCC 7942 (which does not fix nitrogen); we were enabled by the excellent genetics of this species to discover that gene expression was globally regulated by a circadian timekeeper. Initially, we used bacterial luciferase as a reporter of the activity of the promoter for the psbAI gene [4], but we subsequently found by applying the luciferase reporter system that essentially every promoter in the S. elongatus genome was regulated by the circadian system [17]. Rhythms from a few selected promoter::reporter constructs are shown in Fig. 2A, and include the cyanobacterial promoters for the psbAI, kaiA, kaiBC, purF, and ftsZ genes. Note that most of the promoters are activated in the subjective day phase, but the purF promoter is activated in the nocturnal phase, which is interesting because its gene product is involved in an oxygen sensitive pathway and therefore this might be another example of temporal separation [17,18]. These daily rhythms of gene activity may be regulated–at least in part–via the putative transcriptional factor RpaA that is coupled to the KaiABC oscillator described below by the two-component system kinase SasA [19].
Figure 2. Circadian rhythms in S. elongatus.
(A) Circadian rhythms of gene expression monitored as rhythms of luminescence emanating from cells transformed with bacterial luciferase (luxAluxB) fused to the promoters for the psbAI, kaiA, kaiBC, purF, and ftsZ genes.
(B) Circadian rhythm of chromosomal compaction as visualized by a fluorescent DNA-binding dye. The chromosome is more compacted in the subjective night than in the subjective day [22]; S. elongatus has several copies of the same chromosome [28] that appear as fluorescent balls when compacted as seen in the night-phase cells in this panel.
(C) Chromosomal topology shows a circadian rhythm as assayed by supercoiling of an endogenous plasmid. Topoisomers of the plasmid are more relaxed (R) in the subjective night and are more supercoiled (SC) in the subjective day [23].
(D,E) Circadian rhythms of luminescence in single cyanobacterial cells. Panel D shows micrographs of cyanobacterial cells at different times in constant light—upper panel is the brightfield images showing growth and cell division as a function of approximate circadian time, lower panel is the luminescence emanating from these cells (luminescence reporter was the psbAI promoter driving expression of bacterial luciferase). Panel E shows the quantification of bioluminescence from a single cell as it divides into multiple cells as a function of time in constant light; starting at day 1.5, there are two differently colored traces as a result of cell division, the next division occurs at day 2.0, and so on (panels D & E are courtesy of Dr. Irina Mihalcescu).
(F) Cell division of a population of S. elongatus cells. The first 36 h are in a light/dark cycle (phases of darkness are indicated by the black bars at the top of the panel), thereafter the cells are in constant light. Cell number is plotted on the ordinate and the phases in constant light when the cells stop dividing are indicated by the red arrowheads. The average doubling time of this culture was 10.5 h, as indicated by the diagonal line [28].
However, the fact that heterologous promoters from E. coli such as conIIp [20] and trcp [21] exhibit circadian activity in S. elongatus suggested the global regulation is not mediated solely by transcriptional factors that have co-evolved with cis elements of S. elongatus promoters. Perhaps an even more global process is underway that can modulate the activity of any promoter that is introduced into the genome? Such a global process has been found–the circadian system in S. elongatus regulates pervasive changes in the compaction/topology of the entire chromosome! Both compaction/decompaction as visualized by DNA-binding dyes [22] and DNA topology as indicated by plasmid supercoiling [23] undergo dramatic circadian changes (Fig. 2B,C). Because transcriptional rates are known to be sensitive to DNA topology/torsion, these circadian changes in chromosomal topology could be partially responsible for daily modulation of promoter activity [22–25]. While the two available explanations for the global expression patterns (oscillating chromosomal topology vs. transcription factors such as RpaA) seem to exclude each other, a recent analysis of stochastic gene expression in cyanobacteria [26] supports an amalgamative hypothesis that circadian gene expression in cyanobacteria is regulated by multiple factors, for example by changes in both DNA topology and transcriptional factor activity.
The rhythms shown in Fig. 2A are from populations of cells, but a tour de force imaging investigation showed that single isolated cyanobacterial cells also display luminescence rhythms from the luciferase reporter fused to the psbAI promoter [27]. The rhythms from single cells are shown in Fig. 2D, and the quantification of these images revealed that cell division did not perturb the circadian oscillator (Fig. 2E); that is, the daughter cells retained the circadian phase of mother cells. Studies of circadian timing in populations of dividing cells also indicate that the circadian clock is not disturbed by cell division–whether S. elongatus cells are dividing rapidly, dividing slowly, or not dividing at all, the circadian system ambles sedately at its intrinsic ~24 h period [28–30]. However, not only does the circadian oscillator tick independently of cell division, it specifies a checkpoint for division so that the circadian system regulates the timing in which cell division is allowed. For example, Fig. 2F depicts an experiment in which a population of cells is rapidly dividing in constant light (average doubling time of 10.5 h), but in which cell division is restrained every ~24 h by the circadian system (red arrows in Fig. 2F). Therefore, circadian timing is unperturbed by the pervasive changes in metabolism, structure, and gene expression that accompany cell division, but on the other side, the timing of cell division operates at the behest of the circadian program. Recent studies suggest that the circadian-implicated genes cikA and cdpA are involved in the regulation of cell division in S. elongatus [13,31,32].
The underlying clockwork: molecular rhythms in vivo and in vitro
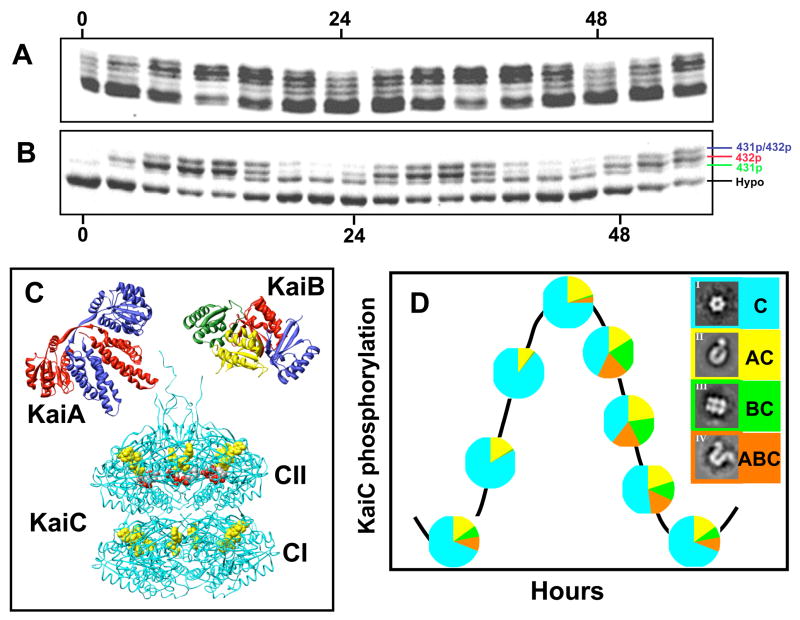
What is the clockwork mechanism that underlies these remarkable properties? A mutation/complementation analysis identified three essential clock genes in S. elongatus: kaiA, kaiB, and kaiC, that are clustered together on the chromosome [33]. The initial studies of expression from the kaiABC cluster supported the hypothesis that the cyanobacterial clockwork was a transcriptional/translational feedback loop (TTFL) oscillator that was similar in principle to that proposed for the circadian mechanism of eukaryotic organisms [34,35]. This hypothesis was based on evidence from cyanobacteria that was similar to that used to support a TTFL oscillator in eukaryotes: (i) rhythms of abundance for mRNAs and proteins encoded by “clock genes,” (ii) negative feedback of clock proteins on their gene’s transcription, (iii) phase setting by overexpression of clock genes, (iv) interaction among the Kai proteins that modulates their activity, and other evidence [33,36–39]. For example, the abundances of KaiB and KaiC proteins and transcripts oscillate [33,38,39] and overexpression of KaiC protein abundance will repress the expression of the kaiC gene and reset the phase of the clock [33,38]. Another similarity to eukaryotic clock systems is that KaiC is rhythmically phosphorylated in vivo [37], as shown in Fig. 3A.
Figure 3. Rhythms of KaiC proteins in vivo and in vitro.
(A) Rhythmic KaiC phosphorylation in vivo at different times in constant light. Samples were collected every 4 h in constant light and processed for SDS PAGE and immunoblotting. The lowest band is hypo-phosphorylated KaiC and the upper bands are various forms of phosphorylated KaiC. Hyper-phosphosphorylated KaiC is most prevalent at hours 8–16 and 32–40.
(B) Rhythmic KaiC phosphorylation in the in vitro reaction. Purified KaiA, KaiB, and KaiC are combined with ATP in vitro and samples collected every 3 h and processed for SDS PAGE and staining. Four bands are obvious in these samples: hypo-phosphorylated KaiC and KaiC phosphorylated at the S431, T432, or S431/T432 residues (see labels on the right side of the panel).
(C) 3-D structures of the KaiA dimer (monomer MW = 30 kD; PDB: 1R8J [45]), the KaiB tetramer (monomer MW = 11 kD; PDB: 1WWJ [46]), and the KaiC hexamer (monomer MW = 58 kD; PDB: 2GBL [48]). In the cases of KaiA and KaiB, various subunits are distinguished by different colors. In the case of KaiC, all the subunits are shown in the same color, but the bound ATP molecules are shown in yellow and the S431/T432 phosphorylation sites are shown in red. Each monomer of KaiC has a CI and CII domain, leading to a CI “end” and a CII “end” of KaiC. Available evidence indicates that KaiA and KaiB interact with KaiC at the CII end. In the case of KaiA, this interaction is probably via the C-terminal peptides shown as tentacles extending upwards from the CII end.
(D) Rhythms of KaiA/B/C complexes during the in vitro rhythm as visualized by electron microscopy of the protein mixtures. The complexes themselves are shown in the boxes to the right, with color coding that corresponds with the pie charts along the KaiC phosphorylation rhythm. For example, only free KaiC & KaiA·KaiC complexes are present during the rising phase of KaiC phosphorylation, while free KaiC, KaiA·KaiC, KaiB·KaiC, and KaiA·KaiB·KaiC are all present during the dephosphorylation phase [56].
Astonishingly, the group of Dr. Takao Kondo in Japan was able to reconstitute the rhythm of KaiC phosphorylation by combining purified KaiA, KaiB, and KaiC proteins together with ATP in a test tube [40], as shown in Fig. 3B. This in vitro rhythm was even temperature compensated, so a temperature compensation mechanism is encoded in the characteristics of the three Kai proteins and the nature of their interactions. Mutations of KaiC that alter the circadian period in vivo similarly altered the period of this post-translational oscillator (PTO) in vitro. Therefore, the Kondo group concluded that the “oscillation of KaiC phosphorylation is the molecular timer for the circadian rhythm of Synechococcus” [40]. Subsequent studies from the Kondo lab discovered that KaiC has a weak but stable ATP hydrolytic activity of ~16 ATPs per day per monomer [41]. The rate of this activity oscillates over the PTO cycle; a few of these ATPs are used to phosphorylate key residues on KaiC. The function of the remaining ATP hydrolysis is unknown but may provide the energy to drive conformational changes and/or monomer exchange (see below). This rhythm of ATP hydrolytic activity is also temperature compensated [41,42].
The availability of the in vitro PTO system conjoined with the ability to crystallize KaiA, KaiB, and KaiC to make cyanobacteria the first circadian system in which the techniques of structural biology and biophysics could first be fully applied. These capabilities enable truly molecular analyses, including the interpretation and prediction of the effect of mutations on the structure and function of the clock proteins. The structure of cyanobacterial KaiA has been studied by several groups, and solved by solution NMR [43] and X-ray crystallography [44,45]. The crystal structure of KaiA from S. elongatus revealed a dimer that is composed of swapped domains (Figure 3C)[45]. The crystal structure of cyanobacterial KaiB revealed an α-β meander motif [44,46] and KaiB has been found as both dimers and tetramers (the tetrameric form is illustrated in Fig. 3C). The fold of KaiB closely resembles the α-β motif of thioredoxin. While the folds of KaiA vs. KaiB are clearly different, some surface features of the physiologically relevant oligomers are similar, suggesting that KaiA and KaiB may compete for a potential common binding site on KaiC [44,46]. Indeed, recent structural studies support the interpretation that KaiA and KaiB both interact with the same “end” of KaiC, namely the CII domain as depicted in Fig. 3C [47].
KaiC is the largest of the Kai proteins and it forms a “double doughnut” hexamer with a central pore that is partially sealed at one end (Figure 3C) [48]. The crystal structure revealed ATP binding sites, inter-subunit organization, a scaffold for Kai-protein complex formation, the location of critical KaiC mutations, and evolutionary relationships to other proteins. The KaiCI and KaiCII domains of each subunit adopt similar conformations, but their ATP-binding pockets exhibit significant differences that are likely of functional importance. The most important binding site of KaiA to KaiC appears to be on the surface of the CII terminus and to 22-amino acid “tentacles” that extend from CII (Fig. 3C)[49,50]. In addition to facilitating our understanding of KaiA-KaiC interaction, the KaiC structure also shed light on the mechanism of rhythmic phosphorylation of KaiC by identifying threonine and serine residues at positions 426, 431, and 432 (T426, S431, & T432) in KaiCII as potential sites at which KaiC is phosphorylated [51]. A crucial role for these residues was shown by the loss of rhythmicity in T426A, S431A, and T432A single alanine mutants [51,52]. Mass spectrometry studies also found that S431 and T432 are key phosphorylation sites [52], while the role of the T426 is still unknown. Recent investigations established that the S431 and T432 residues are phosphorylated in an ordered sequence over the cycle of the in vitro PTO: first T432 is phosphorylated (432p), then S431 is phosphorylated to form a doubly phosphorylated KaiC (431p/432p), then T432 is dephosphorylated to form a singly phosphorylated KaiC at the S431 residue (431p), and finally KaiC is completely dephosphorylated [53,54]. These various phosphoforms can be visualized by SDS PAGE, as shown in Fig. 3B. Furthermore, the singly phosphorylated 431p appears to be the form of KaiC that KaiB binds [53]. This ordered sequence of phosphorylation and dephosphorylation was interpreted to be the mechanism that serves as the basis for cyanobacterial circadian rhythm generation [53,54].
The knowledge that the Kai proteins interact [36] and the availability of the in vitro system led to investigations of Kai interaction and formation of complexes by both structural and biochemical methods that visualized the ticking of the PTO [55,56]. For example, we and our collaborators used electron microscopy (EM) of samples from the in vitro reaction to analyze and quantify the time dependent interaction among the three proteins (KaiA, KaiB, and KaiC) to elucidate the timing of the formation of complexes among the Kai proteins [56]. We could visualize by EM all the possible combinations of complexes between KaiC and KaiA/KaiB, as shown in Fig. 3D. During the course of the in vitro oscillation, KaiC existed in any of all the possible combinations, namely, free KaiC hexamers, KaiA·KaiC complexes, KaiB·KaiC complexes, and KaiA·KaiB·KaiC complexes, but the proportions of these complexes vary in a phase-dependent manner: free KaiC hexamers predominate at all phases, ~10% of KaiC hexamers appear as KaiA·KaiC complexes at all phases, and KaiB·KaiC & KaiA·KaiB·KaiC complexes are clearly rhythmic with a peak in the KaiC dephosphorylation phase (Fig. 3D). Similar results were observed by chromatography and 2-D native gel electrophoresis [55,56]. Structural, biochemical, and biophysical methods have also shown that KaiC can exchange monomers among its hexamers, and this exchange is maximally active during the dephosphorylation phase of the in vitro rhythm [55–57].
Modeling the in vitro PTO, including robustness and synchrony
Mathematical modeling of the in vitro oscillator has been attempted by many groups [54,56,58–66]. Most of these mathematical models share a similar core KaiC hexamer model in which KaiC can exist in two (or more) conformational states over the course of the cycle. The cyclic phosphorylation status of KaiC is considered by these models as both an indicator of the oscillation as well as a regulator of KaiC conformational changes and interactions with KaiB and KaiA. The various models differ in a number of respects, including the role of KaiA which figures more prominently in some models [59,62,63] than in other models.
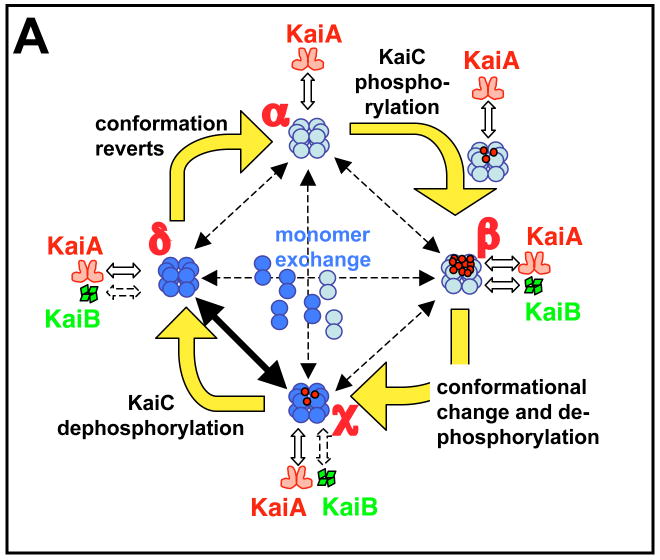
We will briefly describe one of these models [56] as an illustrative example (Fig. 4A). This particular model stochastically simulates the kinetics of KaiC hexamers and the degree of phosphorylation of each monomer in every hexamer. Starting from a hypophosphorylated state of KaiC (state α), rapid association and disassociation of KaiA facilitates phosphorylation until the KaiC hexamer is hyperphosphorylated (state β). In this model, KaiB is assumed to bind with hyperphosphorylated KaiC, but we now know that KaiB associates with the 431p state of KaiC [53,54]. The association of KaiB with KaiC induces a conformational change to a new state (KaiC*, state χ). The KaiC* hexamer (state χ) dephosphorylates to a relatively hypophosphorylated status (state δ) and relaxes to the original conformation (state α). Simultaneous with the phosphorylation cycle of a hexamer is the possibility of the exchange of monomers between any two hexamers in any of the states. The rate of this exchange is highest during the KaiC dephophorylation phase when KaiC is associated with KaiB [57]. This model assumes that KaiA stochastically binds and unbinds rapidly from KaiC hexamers and that KaiB associates/disassociates with the KaiC hexamer when the total degree of phosphorylation of the hexamer exceeds a threshold that places KaiC in state β [56].
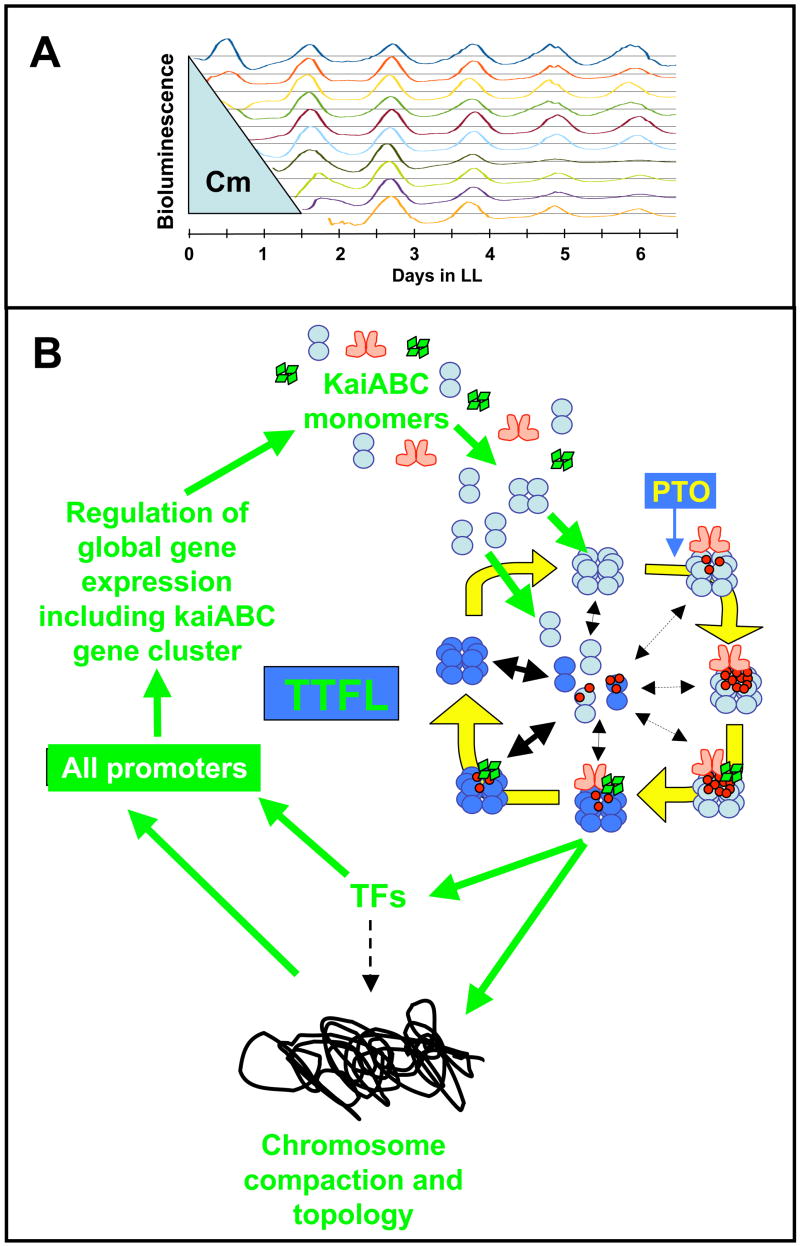
Figure 4. Models for the in vitro and in vivo oscillators.
(A) The diagram represents a mathematical model for the phosphorylation cycle of a KaiC hexamer and its association with KaiA and KaiB [56]. A KaiC monomer is shown as a double circle that can form a hexamer. KaiC hexamers can associate/dissociate with KaiA and/or KaiB. KaiC hexamers are depicted in two conformational states: a default KaiC status (light blue color) and an altered KaiC* state that has undergone a conformational change (darker blue color). Red dots are phosphates attached to the S431 and T432 phosphorylation sites on KaiC. Starting from a hypo-phosphorylated state (α), rapid binding and unbinding of KaiA facilitates autophosphorylation until the KaiC hexamers are hyper-phosphorylated (state β). In this model, KaiB is assumed to preferentially associate with hyper-phosphorylated KaiC; there is a simultaneous conformational change of KaiC to a new state (KaiC* depicted as a darker blue hexamer). The KaiC* hexamer de-phosphorylates through states χ and δ until it is no longer phosphorylated, at which time it reverts to the original conformation (state α). Robustness is maintained by synchronization of KaiC hexameric status via monomer exchange [55–57], depicted by “dumbbell” KaiC monomers exchanging with KaiC hexamers in the central region of the figure. The rate of this exchange is highest during the dephosphorylation phase [57], as indicated by the thicker exchange arrow in this phase.
(B) Inhibiting protein synthesis does not perturb the phase of the clock. Cyanobacterial cells were grown in batch culture prior to addition of the translational inhibitor chloramphenicol. Chloramphenicol (50 μg/ml) was applied to cyanobacterial cultures for various durations, then removed. The chloramphenicol treatment began immediately after a 12 h exposure to darkness that synchronizes the cells in the population. Filled circles indicate the phases of the troughs of the rhythm of psbAI promoter activity after wash-out of chloramphenicol. If the clock were stopped or otherwise perturbed by the chloramphenicol treatment, the phase of the subsequent rhythms would be expected to follow the diagonal line of the wash-out treatments. In contrast, we found that the phase of the subsequent rhythms align vertically, indicating that the cyanobacterial clock’s phase was not affected by the chloramphenicol treatments (in the first cycle after chloramphenicol wash-out [~hours 24–36], there was a transient impact upon the phase of psbAIp activity, but thereafter the steady-state phase of the rhythm was unaffected by the chloramphenicol treatment) [38].
(C) A self-sustained post-translational oscillator (PTO) embedded within a transcription and translation feedback loop (TTFL). The PTO is shown in greater detail in panel A. New synthesis of KaiC monomers feeds into the KaiABC oscillator as non-phosphorylated hexamers (panel C) or as monomers that exchange into pre-existing hexamers (panel C). If the new synthesis of KaiC occurs at a phase when hexamers are predominantly hypo-phosphorylated, the oscillation of KaiC phosphorylation is reinforced (enhanced amplitude). If on the other hand, new synthesis of unphosphorylated KaiC happens at a phase when hexamers are predominantly hyper-phosphorylated, this could lead to an overall decrease in the KaiC phosphorylation status, thereby altering the phase of the KaiABC oscillator (phase shift) and/or reducing its amplitude. Some configuration of KaiC (here shown as the dephosphorylating phase, but it could be any phase of the PTO) mediates rhythmic DNA torsion/compaction and transcriptional factor activity to control global transcription of all promoters, including those driving expression of the essential clock genes kaiA, kaiB, and kaiC (“TFs” = transcriptional factors such as the putative transcriptional factor RpaA).
How does the in vitro oscillator maintain a robust, high amplitude oscillation for more than 10 cycles without apparent damping [57] ? The first published mathematical model for the in vitro oscillation [58] assumed the existence of monomer exchange among KaiC hexamers, which was experimentally confirmed shortly thereafter [55]. The model depicted in Fig. 4A incorporated KaiC monomer exchange as a mechanism for keeping the phosphorylation state of hexamers synchronized in the population, thereby maintaining a high-amplitude oscillation [56], and these predictions were experimentally confirmed [57]. Therefore, the in vitro oscillator is an excellent candidate for a PTO in vivo that can maintain a robust oscillation in the face of metabolic perturbations by virtue of biochemical mass-action kinetics among the 1500–2000 KaiC hexamers present in a single cyanobacterial cell [67]. Despite the fact that KaiC hexamers synchronize to each other intracellularly, however, cyanobacterial cells do not appear to communicate phase information intercellularly. Both from observations of the rhythms of single cells in colonies on agar plates (Fig. 2D [27]) and of populations of cells in liquid cultures [68], cyanobacteria cells appear to be independently oscillating entities and therefore the exquisite rhythms observed from populations of cells (Fig. 2A) must be due to highly precise oscillators residing within cells that have nearly the same period.
Relationship between the TTFL and the PTO in cyanobacteria
As summarized above, there was substantial early evidence in favor of a TTFL pacemaker in cyanobacteria [33,38] that was similar to that proposed for circadian clocks in eukaryotes [34,35]. However, even before the discovery of the in vitro KaiC phosphorylation rhythm that corresponds to the PTO in vivo, there were indications that the usual formulation of a circadian TTFL oscillator was inadequate to explain the circadian system of cyanobacteria. For example, the promoters driving kaiABC gene expression can be replaced with non-specific heterologous promoters without disturbing the circadian rhythm [21,69]. If the TTFL model as proposed for eukaryotic clocks were correct for cyanobacteria, the Kai proteins would be expected to inhibit their own transcription through direct or indirect feedback upon their specific promoters. If that hypothesis were true, replacement of the kaiABC promoters with non-specific heterologous promoters should interrupt the central feedback loop. But that result was not obtained in S. elongatus; heterologous promoters driving KaiABC expression were competent to support robust rhythmicity in vivo [21,69].
Studies of cyanobacterial cells under metabolic repression also failed to support a simple TTFL model. For example, we showed in 2000 that prolonged treatments with the protein synthesis inhibitor chloramphenicol did not perturb the phase of the circadian system after return to normal conditions (Fig. 4B [38]), and Tomita and coworkers found that the PTO was similarly unaffected by chloramphenicol or by constant darkness (which also represses transcription and translation, [39]). Since a TTFL oscillator will be inoperative in the absence of translation, these results supported the hypothesis that a translation-independent PTO was acting as an uninterruptible oscillator that could keep track of time, even when metabolism was repressed. It was therefore natural for the Kondo lab to assume with the discovery of the in vitro rhythm of KaiC phosphorylation [40] that seemed to operate as a PTO in vivo [39], the identity of a circadian oscillator had been found that did not involve a TTFL.
Clearly, the PTO shows that a circadian oscillator can be built without transcriptional and translational feedback. Does this mean that transcriptional and translational feedback plays no role in the larger circadian system in cyanobacteria, and that the early studies that found phase resetting by KaiC expression [33,38] were misleading? A re-evaluation of the relative roles of TTFL and PTO in the cyanobacterial circadian system in vivo is underway. In particular, the Kondo lab has recently reported that interfering with the KaiC phosphorylation rhythm in vivo does not abolish the rhythm of circadian gene expression [70]. These authors disrupted the KaiC phosphorylation rhythm by (i) constitutive KaiC hyperphosphorylation (by overexpression of KaiA) and (ii) the mutation of the S431/T432 residues of KaiC to glutamate (431E/432E) which should mimic constitutive phosphorylation. The authors reasoned that the persisting rhythms of gene expression they observed while KaiC was locked into a constitutive phosphorylation status (i.e., a “clamped” PTO) argues for another oscillator in the cyanobacteria. Therefore, despite their earlier conclusion that the PTO is the core circadian pacemaker [40], the Kondo lab now favors the interpretation that the only other described candidate for an alternative pacemaker—the TTFL—can oscillate independently and therefore that the cyanobacterial clock is a multiple oscillator system composed of hierarchically equal coupled oscillators, the PTO and the TTFL [70].
What is the relationship between the PTO and TTFL? The Kondo group now suggests that the two oscillating systems are both self-sustained oscillators that provide additional robustnessness when coupled, especially at lower temperatures [70]. As mentioned above, the PTO could act as an “uninterruptible” oscillator that operates under metabolic repression/activation. More significantly, the PTO could provide the imperturbability observed when cells are actively dividing [27–30]. A simple TTFL would be sensitive to the consequences of cell division: chromosomes condense (and are therefore less accessible for transcription) and there are changes in the ratio between DNA and transcriptional factors. Indeed, the period of synthetic TTFL oscillators in bacteria are strictly dependent upon doubling time [71]. An alternative to the “dual coupled oscillator” hypothesis of the Kondo lab [70] that we favor is a self-sustained PTO that is embedded within a damped oscillator; this damped oscillator regulates a transcription and translation feedback loop (Fig. 4C). By this interpretation, the PTO acts as the core circadian oscillator that runs under all conditions. When metabolism is active, a damped oscillator additionally elaborates rhythmic transcription of all promoters in the cell and mediates a TTFL of central Kai proteins. By this scenario, the TTFL could behave as a damped oscillator and indeed, the rhythms observed when KaiC is mutated to the constitutively active form (431E/432E) suggest a damped oscillation [70].
Resolving the relationship between the PTO and the TTFL will ultimately require the elucidation of KaiC’s most fundamental biochemical activity in regulating the clock. At this time, the most elemental activity known for KaiC is its rhythm of ATP hydrolysis [41]. However, this activity probably reflects a rhythm of ATP-dependent KaiABC conformational changes and/or oligomeric associations that drive the essential oscillating circuit by a more fundamental enzymatic activity. What is this primary activity that mediates the outputs of KaiC? An early idea about KaiC’s enzymatic activity that was based upon its sequence similarity to RecA and DnaB [72] was that KaiC might have helicase activity [73]. However, despite the observation that KaiC can bind forked DNA substrates [73], there is no direct evidence that KaiC has a helicase activity. What is known, however, is that KaiC appears to influence the phosphorylation status of the putative transcriptional factor RpaA via the two-component system kinase SasA [19], and perhaps this is the most relevant enzymatic activity of KaiC in mediating downstream activities.
Implications for eukaryotic clocks and unanswered questions
The hypothesis of a TTFL in cyanobacteria has come full circle, from its first proposal [33,38] to marginalization [39,40,69] and back again to prominence [70]. Now that a TTFL is once again considered to be significant for the cyanobacterial clock system, cyanobacterial and eukaryotic clocks appear to be more similar than we thought three years ago. Noteworthy is that the early evidence in support of a core TTFL oscillator in cyanobacteria that was summarized above is the same as that which currently supports a TTFL core in eukaryotic circadian systems, including the setting of circadian phase by overexpression of core clock proteins such as PER or FRQ [34,35]. If a PTO is embedded in the TTFL of cyanobacteria, perhaps a similar organization exists for eukaryotic clocks? Indeed, there was early evidence for circadian rhythms in enucleated eukaryotic cells [74–76]—obviously a TTFL cannot operate in cells without nuclei. But the quality of this old evidence might not satisfy the standards of contemporary research.
A re-evaluation of the role of the TTFL in eukaryotes is underway [77–79]. Can the cyanobacterial clock system tell us anything about clocks in eukaryotes? Eukaryotic circadian genes have no detectable homology to kaiABC sequences, so if there is an evolutionary relationship between the bacterial and eukaryotic systems, it is so diverged as to be genetically invisible. But what about the possibility of convergence to a fundamentally similar biochemical mechanism? It might seem implausible that independent origins for clocks would converge upon an essentially similar core post-translational oscillator made more robust by an overlying TTFL. However, the advantages that accrue to the cyanobacterial system by having a post-translational mechanism at its core are also relevant to eukaryotic clocks. For example, individual mammalian fibroblasts express cell-autonomous, self-sustained circadian oscillations of gene expression that are largely unperturbed by cell division [80,81] in a fashion reminiscent of cyanobacteria [27–30,68]. Could the necessity for imperturbability even when buffeted by the massive intracellular changes provoked by cell division provide an evolutionary driving force for circadian clock mechanisms to converge on a relatively similar core mechanism? The results from cyanobacteria combined with recent results from eukaryotic systems that do not easily fit into the original TTFL formulation [77–79] embolden such speculations.
Circadian timekeeping is common in organisms from bacteria to humans. We now appreciate many features underlying the cyanobacterial system, which has a PTO embedded within a larger transcriptional/translational control loop. A variety of structural and biophysical methods including X-ray crystallography, NMR, EM, FRET, and mathematical modeling in addition to biochemical assays are enabling us to appreciate the inner workings of the in vitro oscillator. Nevertheless, unanswered questions abound. How does the KaiC-based oscillator regulate global gene expression? How is the central oscillator linked to the environment cycle (i.e., what is the input pathway) ? What is the relationship/coupling between the PTO and the TTFL that results in the overall emergent circadian program? How is this program temperature compensated from the truly molecular perspective? Finally, what is the mechanism whereby strains with clock properties that resonate with the environment are able to outcompete strains with less favorable clock properties? The objectives of the research on cyanobacterial clocks are to take advantage of this system’s capabilities and tools to delve deeply into its molecular nature, to examine the interaction of the core oscillator with its input and output mediators, and to discover if our own clocks have attributes that are similar to those of cyanobacteria.
Acknowledgments
We thank our colleagues and co-authors at Vanderbilt University who have helped to bring our understanding of the S. elongatus circadian system to its current level, especially Drs. Mark Byrne, Martin Egli, Ximing Qin, Rekha Pattanayek, Sabuj Pattanayek, Phoebe Stewart, Dewight Williams, and Mark Woelfle. We also thank Drs. Takao Kondo, Susan Golden, Masahiro Ishiura, and their laboratory members; these labs together with ours initiated the study of circadian clocks in S. elongatus and continue to make seminal contributions that make this subject fascinatingly unpredictable. Work in our lab is supported by funds from the National Institutes of Health (GM067152).
References
- 1.Dunlap JC, Loros JJ, DeCoursey PJ, editors. Chronobiology: Biological Timekeeping. Sinauer; Sunderland, MA: 2004. [Google Scholar]
- 2.Ruby NF, Burns DE, Heller HC. Circadian rhythms in the suprachiasmatic nucleus are temperature-compensated and phase-shifted by heat pulses in vitro. J Neurosci. 1999;19:8630–6. doi: 10.1523/JNEUROSCI.19-19-08630.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Izumo M, Johnson CH, Yamazaki S. Circadian gene expression in mammalian fibroblasts revealed by real-time luminescence reporting: temperature compensation and damping. Proc Natl Acad Sci USA. 2003;100:16089–16094. doi: 10.1073/pnas.2536313100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kondo T, Strayer CA, Kulkarni RD, Taylor W, Ishiura M, Golden SS, Johnson CH. Circadian rhythms in prokaryotes: luciferase as a reporter of circadian gene expression in cyanobacteria. Proc Natl Acad Sci USA. 1993;90:5672–5676. doi: 10.1073/pnas.90.12.5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnson CH, Golden SS, Ishiura M, Kondo T. Circadian clocks in prokaryotes. Mol Microbiol. 1996;21:5–11. doi: 10.1046/j.1365-2958.1996.00613.x. [DOI] [PubMed] [Google Scholar]
- 6.Pittendrigh CS. Temporal organization: reflections of a Darwinian clock-watcher. Annu Rev Physiol. 1993;55:17–54. doi: 10.1146/annurev.ph.55.030193.000313. [DOI] [PubMed] [Google Scholar]
- 7.Johnson CH. Testing the adaptive value of circadian systems. Methods in Enzymology. 2005;393:818–837. doi: 10.1016/S0076-6879(05)93043-7. [DOI] [PubMed] [Google Scholar]
- 8.Lenski RE, Travisano M. Dynamics of adaptation and diversification: a 10,000 generation experiment with bacterial populations. Proc Natl Acad Sci USA. 1994;91:6808–6814. doi: 10.1073/pnas.91.15.6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Woelfle MA, Ouyang Y, Phanvijhitsiri K, Johnson CH. The adaptive value of circadian clocks: An experimental assessment in cyanobacteria. Current Biol. 2004;14:1481–1486. doi: 10.1016/j.cub.2004.08.023. [DOI] [PubMed] [Google Scholar]
- 10.Ouyang Y, Andersson CR, Kondo T, Golden SS, Johnson CH. Resonating circadian clocks enhance fitness in cyanobacteria. Proc Natl Acad Sci USA. 1998;95:8660–8664. doi: 10.1073/pnas.95.15.8660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmitz O, Katayama M, Williams SB, Kondo T, Golden SS. CikA, a bacteriophytochrome that resets the cyanobacterial circadian clock. Science. 2000;289:765–768. doi: 10.1126/science.289.5480.765. [DOI] [PubMed] [Google Scholar]
- 12.Mackey SR, Golden SS. Winding up the cyanobacterial circadian clock. Trends Microbiol. 2007;15:381–8. doi: 10.1016/j.tim.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 13.Mackey SR, Choi JS, Kitayama Y, Iwasaki H, Dong G, Golden SS. Proteins found in a CikA-interaction assay link the circadian clock, metabolism, and cell division in Synechococcus elongatus. J Bacteriol. 2008 doi: 10.1128/JB.01721-07. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takai N, Ikeuchi S, Manabe K, Kutsuna S. Expression of the circadian clock-related gene pex in cyanobacteria increases in darkness and is required to delay the clock. J Biol Rhythms. 2006;21:235–244. doi: 10.1177/0748730406289400. [DOI] [PubMed] [Google Scholar]
- 15.Ivleva NB, Bramlett MR, Lindahl PA, Golden SS. LdpA: a component of the circadian clock senses redox state of the cell. EMBO J. 2005;24:1202–1210. doi: 10.1038/sj.emboj.7600606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grobbelaar N, Huang T-C, Lin HY, Chow TJ. Dinitrogen-fixing endogenous rhythm in Synechococcus RF-1. FEMS Microbiol Lett. 1986;37:173–177. [Google Scholar]
- 17.Liu Y, Tsinoremas NF, Johnson CH, Lebedeva NV, Golden SS, Ishiura M, Kondo T. Circadian orchestration of gene expression in cyanobacteria. Genes Dev. 1995;9:1469–1478. doi: 10.1101/gad.9.12.1469. [DOI] [PubMed] [Google Scholar]
- 18.Liu Y, Tsinoremas NF, Golden SS, Kondo T, Johnson CH. Circadian expression of genes involved in the purine biosynthetic pathway of the cyanobacterium Synechococcus sp strain PCC 7942. Mol Microbiol. 1996;20:1071–1081. doi: 10.1111/j.1365-2958.1996.tb02547.x. [DOI] [PubMed] [Google Scholar]
- 19.Takai N, Nakajima M, Oyama T, Kito R, Sugita C, Sugita M, Kondo T, Iwasaki H. A KaiC-associating SasA-RpaA two-component regulatory system as a major circadian timing mediator in cyanobacteria. Proc Natl Acad Sci USA. 2006;103:12109–12114. doi: 10.1073/pnas.0602955103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Katayama M, Tsinoremas NF, Kondo T, Golden SS. cpmA, a gene involved in an output pathway of the cyanobacterial circadian system. J Bacteriol. 1999;181:3516–3524. doi: 10.1128/jb.181.11.3516-3524.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakahira Y, Katayama M, Miyashita H, Kutsuna S, Iwasaki H, Oyama T, Kondo T. Global gene repression by KaiC as a master process of prokaryotic circadian system. Proc Natl Acad Sci USA. 2004;101:881–885. doi: 10.1073/pnas.0307411100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith RM, Williams SB. Circadian rhythms in gene transcription imparted by chromosome compaction in the cyanobacterium Synechococcus elongatus. Proc Natl Acad Sci USA. 2006;103:8564–8569. doi: 10.1073/pnas.0508696103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Woelfle MA, Xu Y, Qin X, Johnson CH. Circadian rhythms of superhelical status of DNA in cyanobacteria. Proc Natl Acad Sci USA. 2007;104:18819–18824. doi: 10.1073/pnas.0706069104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mori T, Johnson CH. Circadian programming in cyanobacteria. Semin Cell Dev Biol. 2001;12:271–278. doi: 10.1006/scdb.2001.0254. [DOI] [PubMed] [Google Scholar]
- 25.Min H, Liu Y, Johnson CH, Golden SS. Phase determination of circadian gene expression in Synechococcus elongatus PCC 7942. J Biol Rhythms. 2004;19:103–112. doi: 10.1177/0748730403262056. [DOI] [PubMed] [Google Scholar]
- 26.Chabot JR, Pedraza JM, Luitel P, van Oudenaarden A. Stochastic gene expression out-of-steady-state in the cyanobacterial circadian clock. Nature. 2007;450:1249–52. doi: 10.1038/nature06395. [DOI] [PubMed] [Google Scholar]
- 27.Mihalcescu I, Hsing W, Leibler S. Resilient circadian oscillator revealed in individual cyanobacteria. Nature. 2004;430:81–85. doi: 10.1038/nature02533. [DOI] [PubMed] [Google Scholar]
- 28.Mori T, Binder B, Johnson CH. Circadian gating of cell division in cyanobacteria growing with average doubling times of less than 24 hours. Proc Natl Acad Sci USA. 1996;93:10183–10188. doi: 10.1073/pnas.93.19.10183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kondo T, Mori T, Lebedeva NV, Aoki S, Ishiura M, Golden SS. Circadian rhythms in rapidly dividing cyanobacteria. Science. 1997;275:224–227. doi: 10.1126/science.275.5297.224. [DOI] [PubMed] [Google Scholar]
- 30.Mori T, Johnson CH. Independence of circadian timing from cell division in cyanobacteria. J Bacteriol. 2001;183:2439–2444. doi: 10.1128/JB.183.8.2439-2444.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miyagishima SY, Wolk CP, Osteryoung KW. Identification of cyanobacterial cell division genes by comparative and mutational analyses. Mol Microbiol. 2005;56:126–143. doi: 10.1111/j.1365-2958.2005.04548.x. [DOI] [PubMed] [Google Scholar]
- 32.Zhang X, Dong G, Golden SS. The pseudo-receiver domain of CikA regulates the cyanobacterial circadian input pathway. Mol Microbiol. 2006;60:658–668. doi: 10.1111/j.1365-2958.2006.05138.x. [DOI] [PubMed] [Google Scholar]
- 33.Ishiura M, Kutsuna S, Aoki S, Iwasaki H, Andersson CR, Tanabe A, Golden SS, Johnson CH, Kondo T. Expression of a gene cluster kaiABC as a circadian feedback process in cyanobacteria. Science. 1998;281:1519–1523. doi: 10.1126/science.281.5382.1519. [DOI] [PubMed] [Google Scholar]
- 34.Hardin PE, Hall JC, Rosbash M. Feedback of the Drosophila period gene product on circadian cycling of its messenger RNA levels. Nature. 1990;343:536–540. doi: 10.1038/343536a0. [DOI] [PubMed] [Google Scholar]
- 35.Dunlap JC. Molecular bases for circadian clocks. Cell. 1999;96:271–290. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- 36.Iwasaki H, Taniguchi Y, Ishiura M, Kondo T. Physical interactions among circadian clock proteins KaiA, KaiB and KaiC in cyanobacteria. EMBO J. 1999;18:1137–1145. doi: 10.1093/emboj/18.5.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iwasaki H, Nishiwaki T, Kitayama Y, Nakajima M, Kondo T. KaiA-stimulated KaiC phosphorylation in circadian timing loops in cyanobacteria. Proc Natl Acad Sci USA. 2002;99:15788–15793. doi: 10.1073/pnas.222467299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu Y, Mori T, Johnson CH. Circadian clock-protein expression in cyanobacteria: rhythms and phase setting. EMBO J. 2000;19:3349–3357. doi: 10.1093/emboj/19.13.3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tomita J, Nakajima M, Kondo T, Iwasaki H. No transcription-translation feedback in circadian rhythm of KaiC phosphorylation. Science. 2005;307:251–254. doi: 10.1126/science.1102540. [DOI] [PubMed] [Google Scholar]
- 40.Nakajima M, Imai K, Ito H, Nishiwaki T, Murayama Y, Iwasaki H, Oyama T, Kondo T. Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science. 2005;308:414–415. doi: 10.1126/science.1108451. [DOI] [PubMed] [Google Scholar]
- 41.Terauchi K, Kitayama Y, Nishiwaki T, Miwa K, Murayama Y, Oyama T, Kondo T. ATPase activity of KaiC determines the basic timing for circadian clock of cyanobacteria. Proc Natl Acad Sci USA. 2007;104:16377–16381. doi: 10.1073/pnas.0706292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Murakami R, Miyake A, Iwase R, Hayashi F, Uzumaki T, Ishiura M. ATPase activity and its temperature compensation of the cyanobacterial clock protein KaiC. Genes to Cells. 2008;13:387–395. doi: 10.1111/j.1365-2443.2008.01174.x. [DOI] [PubMed] [Google Scholar]
- 43.Vakonakis I, Sun J, Wu T, Holzenburg A, Golden SS, LiWang AC. NMR structure of the KaiC-interacting C-terminal domain of KaiA, a circadian clock protein: Implications for the KaiA-KaiC Interaction. Proc Natl Acad Sci USA. 2004;101:1479–1484. doi: 10.1073/pnas.0305516101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Garces RG, Wu N, Gillon W, Pai EF. Anabaena circadian clock proteins KaiA and KaiB reveal potential common binding site to their partner KaiC. EMBO J. 2004;23:1688–1698. doi: 10.1038/sj.emboj.7600190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ye S, Vakonakis I, Ioerger TR, LiWang AC, Sacchettini JC. Crystal structure of circadian clock protein KaiA from Synechococcus elongatus. J Biol Chem. 2004;279:20511–20518. doi: 10.1074/jbc.M400077200. [DOI] [PubMed] [Google Scholar]
- 46.Hitomi K, Oyama T, Han S, Arvai AS, Getzoff ED. Tetrameric architecture of the circadian clock protein KaiB. A novel interface for intermolecular interactions and its impact on the circadian rhythm. J Biol Chem. 2005;280:19127–19135. doi: 10.1074/jbc.M411284200. [DOI] [PubMed] [Google Scholar]
- 47.Pattanayek R, Williams DR, Pattanayek S, Mori T, Johnson CH, Stewart PL, Egli M. Structural model of the circadian clock KaiB-KaiC complex and mechanism for modulation of KaiC phosphorylation. EMBO J. 2008 doi: 10.1038/emboj.2008.104. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pattanayek R, Wang J, Mori T, Xu Y, Johnson CH, Egli M. Visualizing a circadian clock protein: crystal structure of KaiC and functional insights. Mol Cell. 2004;15:375–388. doi: 10.1016/j.molcel.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 49.Vakonakis I, LiWang AC. Structure of the C-terminal domain of the clock protein KaiA in complex with a KaiC-derived peptide: implications for KaiC regulation. Proc Natl Acad Sci USA. 2004;101:10925–10930. doi: 10.1073/pnas.0403037101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pattanayek R, Williams DR, Pattanayek S, Xu Y, Mori T, Johnson CH, Stewart PL, Egli M. Analysis of KaiA-KaiC protein interactions in the cyano-bacterial circadian clock using hybrid structural methods. EMBO J. 2006;25:2017–2028. doi: 10.1038/sj.emboj.7601086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu Y, Mori T, Pattanayek R, Pattanayek S, Egli M, Johnson CH. Identification of key phosphorylation sites in the circadian clock protein KaiC by crystallographic and mutagenetic analyses. Proc Natl Acad Sci USA. 2004;101:13933–13938. doi: 10.1073/pnas.0404768101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nishiwaki T, Satomi Y, Nakajima M, Lee C, Kiyohara R, Kageyama H, Kitayama Y, Temamoto M, Yamaguchi A, Hijikata A, Go M, Iwasaki H, Takao T, Kondo T. Role of KaiC phosphorylation in the circadian clock system of Synechococcus elongatus PCC 7942. Proc Natl Acad Sci USA. 2004;101:13927–13932. doi: 10.1073/pnas.0403906101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nishiwaki T, Satomi Y, Kitayama Y, Terauchi K, Kiyohara R, Takao T, Kondo T. A sequential program of dual phosphorylation of KaiC as a basis for circadian rhythm in cyanobacteria. EMBO J. 2007;26:4029–4037. doi: 10.1038/sj.emboj.7601832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rust MJ, Markson JS, Lane WS, Fisher DS, O’Shea EK. Ordered phosphorylation governs oscillation of a three-protein circadian clock. Science. 2007;318:809–812. doi: 10.1126/science.1148596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kageyama H, Nishiwaki T, Nakajima M, Iwasaki H, Oyama T, Kondo T. Cyanobacterial circadian pacemaker: Kai protein complex dynamics in the KaiC phosphorylation cycle in vitro. Mol Cell. 2006;23:161–171. doi: 10.1016/j.molcel.2006.05.039. [DOI] [PubMed] [Google Scholar]
- 56.Mori T, Williams DR, Byrne MO, Qin X, Egli M, McHaourab HS, Stewart PL, Johnson CH. Elucidating the ticking of an in vitro circadian clockwork. PLoS Biol. 2007;5:e93. doi: 10.1371/journal.pbio.0050093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ito H, Kageyama H, Mutsuda M, Nakajima M, Oyama T, Kondo T. Autonomous synchronization of the circadian KaiC phosphorylation rhythm. Nat Struct Mol Biol. 2007;14:1084–1088. doi: 10.1038/nsmb1312. [DOI] [PubMed] [Google Scholar]
- 58.Emberly E, Wingreen NS. Hourglass model for a protein-based circadian oscillator. Phys Rev Lett. 2006;96:38303. doi: 10.1103/PhysRevLett.96.038303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mehra A, Hong CI, Shi M, Loros JJ, Dunlap JC, Ruoff P. Circadian rhythmicity by autocatalysis. PLoS Comput Biol. 2006;2:e96. doi: 10.1371/journal.pcbi.0020096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kurosawa G, Aihara K, Iwasa Y. A model for the circadian rhythm of cyanobacteria that maintains oscillation without gene expression. Biophys J. 2006;91:2015–2023. doi: 10.1529/biophysj.105.076554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Takigawa-Imamura H, Mochizuki A. Predicting regulation of the phosphorylation cycle of KaiC clock protein using mathematical analysis. J Biol Rhythms. 2006;21:405–416. doi: 10.1177/0748730406291329. [DOI] [PubMed] [Google Scholar]
- 62.Clodong S, Duhring U, Kronk L, Wilde A, Axmann I, Herzel H, Kollmann M. Functioning and robustness of a bacterial circadian clock. Mol Syst Biol. 2007;3:90. doi: 10.1038/msb4100128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.van Zon JS, Lubensky DK, Altena PR, ten Wolde PR. An allosteric model of circadian KaiC phosphorylation. Proc Natl Acad Sci USA. 2007;104:7420–7425. doi: 10.1073/pnas.0608665104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yoda M, Eguchi K, Terada TP, Sasai M. Monomer-shuffling and allosteric transition in KaiC circadian oscillation. PLoS ONE. 2007;2:e408. doi: 10.1371/journal.pone.0000408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Miyoshi F, Nakayama Y, Kaizu K, Iwasaki H, Tomita M. A mathematical model for the Kai-protein-based chemical oscillator and clock gene expression rhythms in cyanobacteria. J Biol Rhythms. 2007;22:69–80. doi: 10.1177/0748730406295749. [DOI] [PubMed] [Google Scholar]
- 66.Li S, Fang YH. Modelling circadian rhythms of protein KaiA, KaiB and KaiC interactions in cyanobacteria. Biol Rhythm Res. 2007;38:43–53. [Google Scholar]
- 67.Kitayama Y, Iwasaki H, Nishiwaki T, Kondo T. KaiB functions as an attenuator of KaiC phosphorylation in the cyanobacterial circadian clock system. EMBO J. 2003;22:2127–2134. doi: 10.1093/emboj/cdg212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Amdaoud M, Vallade M, Weiss-Schaber C, Mihalcescu I. Cyanobacterial clock, a stable phase oscillator with negligible intercellular coupling. Proc Natl Acad Sci USA. 2007;104:7051–7056. doi: 10.1073/pnas.0609315104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xu Y, Mori T, Johnson CH. Cyanobacterial circadian clockwork: roles of KaiA, KaiB and the kaiBC promoter in regulating KaiC. EMBO J. 2003;22:2117–2126. doi: 10.1093/emboj/cdg168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kitayama Y, Nishiwaki T, Terauchi K, Kondo T. Dual KaiC-based oscillations constitute the circadian system of cyanobacteria. Genes & Development. 2008 doi: 10.1101/gad.1661808. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Atkinson MR, Savageau MA, Myers JT, Ninfa AJ. Development of genetic circuitry exhibiting toggle switch or oscillatory behavior in Escherichia coli. Cell. 2003;113:597–607. doi: 10.1016/s0092-8674(03)00346-5. [DOI] [PubMed] [Google Scholar]
- 72.Leipe DD, Aravind L, Grishin NV, Koonin EV. The bacterial replicative helicase DnaB evolved from a RecA duplication. Genome Res. 2000;10:5–16. [PubMed] [Google Scholar]
- 73.Mori T, Saveliev SV, Xu Y, Stafford WF, Cox MM, Inman RB, Johnson CH. Circadian Clock Protein KaiC forms ATP-dependent Hexameric Rings and Binds DNA. Proc Natl Acad Sci USA. 2002;99:17203–17208. doi: 10.1073/pnas.262578499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sweeney BM, Haxo FT. Persistence of a photosynthetic rhythm in enucleated Acetabularia. Science. 1961;134:1361–1363. doi: 10.1126/science.134.3487.1361. [DOI] [PubMed] [Google Scholar]
- 75.Ashkenazi IE, Hartman H, Strulovitz B, Dar O. Activity rhythms of enzymes in human red blood cell suspensions. J Interdiscipl Cycle Res. 1975;6:291–301. [Google Scholar]
- 76.Hartman H, Ashkenazi I, Epel BL. Circadian changes in membrane properties of human red blood cells in vitro, as measured by a membrane probe. FEBS Lett. 1976;67:161–163. doi: 10.1016/0014-5793(76)80356-0. [DOI] [PubMed] [Google Scholar]
- 77.Lakin-Thomas PL. Transcriptional feedback oscillators: maybe, maybe not... J Biol Rhythms. 2006;21:83–92. doi: 10.1177/0748730405286102. [DOI] [PubMed] [Google Scholar]
- 78.Merrow M, Roenneberg T. Circadian clock: time for a phase shift of ideas? Current Biol. 2007;17:R636–R638. doi: 10.1016/j.cub.2007.06.041. [DOI] [PubMed] [Google Scholar]
- 79.Hastings MH, Maywood ES, O’Neill JS. Cellular circadian pacemaking: biology’s loops without end. This volume of Current Biology. 2008 doi: 10.1016/j.cub.2008.07.021. [DOI] [PubMed] [Google Scholar]
- 80.Nagoshi E, Saini C, Bauer C, Laroche T, Naef F, Schibler U. Circadian gene expression in individual fibroblasts: cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell. 2004;119:693–705. doi: 10.1016/j.cell.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 81.Welsh DK, Yoo SH, Liu AC, Takahashi JS, Kay SA. Bioluminescence imaging of individual fibroblasts reveals persistent, independently phased circadian rhythms of clock gene expression. Curr Biol. 2004;14:2289–95. doi: 10.1016/j.cub.2004.11.057. [DOI] [PMC free article] [PubMed] [Google Scholar]