Abstract
Hepatocellular carcinoma (HCC) is the fifth most common cancer in the world, and has a wide geographical variation. Eighty per cent of HCC is attributed to hepatitis B virus (HBV). The predominant carcinogenic mechanism of HBV associated HCC is through the process of liver cirrhosis, but direct oncogenic effects of HBV may also contribute. Prevention of HBV infections as well as effective treatment of chronic hepatitis B is still needed for the global control of HBV associated HCC. Continued investigation of the mechanisms of hepatocarcinogenesis will refine our current understanding of the molecular and cellular basis for neoplastic transformation in the liver.
Keywords: hepatocellular carcinoma, hepatitis b virus, hepatitis B x protein (HBx), transactivation
Hepatits B virus (HBV) is a serious public health problem worldwide and important cause of chronic hepatitis, cirrhosis, and hepatocellular carinoma (HCC).1 Of the approximately two billion people who have been infected worldwide, more than 400 million are chronic carriers of HBV.2 There is a noticeable difference in geographical distribution of the carrier state in the world population ranging from 10% to 20% in South East Asia and sub‐Sahara Africa to less than 1% in Northern Europe and America.1,3 About 75% of chronic carriers live in Asia and the Western Pacific.1,3 In low endemic areas, most HBV infections acquire by horizontal transmission in adolescents and young adults in comparatively well defined high risk groups, including injection drug users, homosexual men, health care workers, patients who require regular blood transfusion or haemodialysis. In areas of high endemicity, the most common route of transmission is perinatal or the infection is acquired during the preschool years.4 The risk of becoming a virus carrier after infection ranges from 90% in perinatal infection to 25% to 30% in infants and children under 5 and less than 10% in adults.2 In addition, immunosuppressed persons are more likely to develop chronic HBV infection after acute infection.2 Exposure to other hepatotoxins, such as alcohol and aflatoxins, in the person with HBV infection hastens both the development of cirrhosis and HCC.5 Coinfection with hepatitis C virus and HBV also increases the risk of developing HCC by at least twofold.5 About 15%–40% of infected patients will develop cirrhosis, liver failure, or HCC.3 Presently, more than 80% of HCC are found in HBV infected people.3 HCC is now the fifth most frequent cancer in the world, killing 300 000–500 000 people each year.3,5 Overall, the outcome of HCC is poor. Unfortunately, many patients at presentation already have advanced and unresectable disease, which is associated with a median survival of less than six months. A minority of patients are eligible for liver resection, which results in a five year survival of about 40%.1
The details of how HBV causes carcinogenic changes in the liver are still not apparent.
Epidemiological studies have clearly shown that chronic HBV infection is an important aetiological factor in the development of HCC. The incidence of HCC and the prevalence of HBV serological markers follow the same general geographical pattern of distribution.3 HCC is common in regions where HBV is endemic, but is much less common than other types of cancer in regions where HBV infection is uncommon.3,5 The highest rates are in South East Asia and sub‐Saharan Africa, with the HCC incidence >50/100 000 population.5 Moreover, HCC can occur even in convalescent HBV infection.3 Patients found to be hepatitis B core antibody positive or hepatitis B surface antibody positive are also at a several‐fold increased risk for HCC over HBV naive controls, although the risk is much lower than in those with active infection.6,7 Importantly, a study from Taiwan showed a decline in the incidence of HCC in children after a universal hepatitis B vaccination.8 Another study in Korea suggested that the immunisation with hepatitis B vaccine, even in adulthood, could reduce the risk of HCC.9 Regardless of vaccination, large numbers of persons are infected with HBV or will become infected. Several studies strongly supported the relation between viral replication and the risk of HCC.3,10 Relative risk of HCC among hepatitis B e antigen (HBeAg) positive patients is much higher than that among inactive hepatitis B surface antigen (HBsAg) carries.10 Furthermore, HBV DNA has been identified as the most important predictor of the development of HCC in HBsAg positive patients with different clinical conditions.11 Therefore, treatment for the underlying liver disease may be the best approach for prevention of HCC.
The study of HBV has been difficult because of the lack of cell culture systems permitting virus propagation in vitro. Much of what we know about the replication and expression of HBV is derived from the study of other closely related hepadnaviruses, such as the woodchuck hepatitis virus and the Beechey ground squirrel hepatitis virus.12 As with most viral diseases, HBV infected host cells are exposed to multiple opposing signals mediated by growth hormones, immune cytokines, and the effects of adjacent cells such as lymphocytes or Kupffer cells. Hepatocytes must convert these signals into a defined response such as proliferation, differentiation, or death, through signal transduction. Moreover, the infected hepatocytes must modulate signal transduction pathways leading to growth, inflammation, or cell death to maximise the symbiotic survival of both the virus and the cell, in a process, which often progresses to cirrhosis and HCC.2 Furthermore, early HCC progress to advanced HCC, which have increased tumour size and malignant potential, are moderately or poorly differentiated.2 The development and progression of HCC are believed to be caused by the accumulation of genetic changes as well as genes involved in different regulatory pathways, such as cell cycle control, apoptosis, adhesion, and angiogenesis.1
This review will summarise the current knowledge on the mechanisms involved in HBV associated hepatocarcinogenesis.
Structure of HBV genome
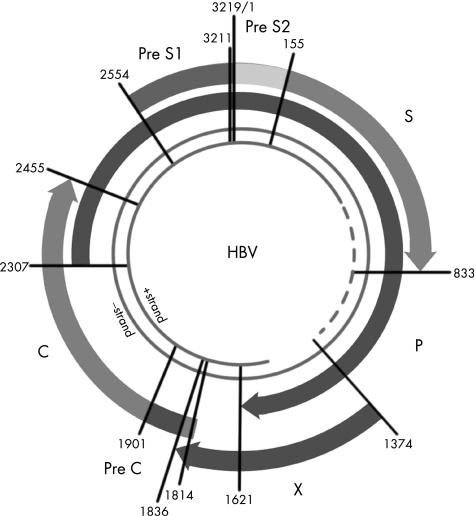
HBV belongs to the family of hepadnaviruses. The HBV genome is a relaxed circular, partially double stranded DNA of about 3200 base pairs (fig 1). There are four partially overlapping open reading frames encoding the envelope (pre‐S/S), core (precore/core), polymerase (P), and X proteins (X).13 The pre‐S/S open reading frame encodes the large (L), middle (M), and small (S) surface glycoproteins. The precore/core open reading frame is translated into a precore polypeptide, which is modified into a soluble protein, HBeAg and the nucleocapsid protein, hepatitis B core antigen. The polymerase protein functions as a reverse transcriptase as well as a DNA polymerase. The X protein is a potent transactivator and may play a crucial part in hepatocarcinogenesis.13
Figure 1 Schematic organisation of the hepatitis B virus genome. The HBV genome is a relaxed circular, partially double stranded DNA of about 3200 base pairs. There are four partially overlapping open reading frames encoding the envelope (pre‐S/S), core (precore/core), polymerase (P), and X proteins (X).
The mechanism of liver injury is still unclear. However, it is believed that host immune responses accompanying HBV infection induce tissue damages, as HBV is not directly cytotoxic.2 Chronic HBV infection is associated with a weak virus specific response by the cytotoxic T lymphocytes (CTL). A strong CTL response may be essential for viral clearance and a weak CTL response may be responsible for the hepatocyte damage.2
Current concepts of HBV associated hepatocarcinogenesis
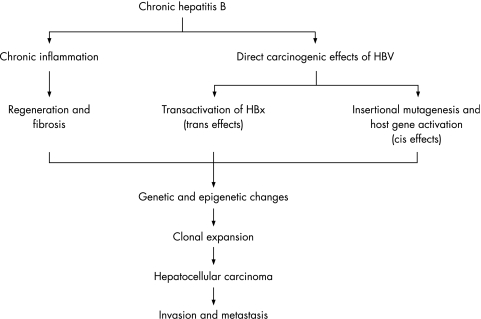
The exact mechanisms by which HBV infection causes HCC are unclear. HCC exhibits a high degree of genetic heterogeneity, suggesting that multiple molecular pathways may be involved in the genesis of subsets of HCC. Two pathways have been proposed (fig 2).6 One involves chronic necroinflammation of hepatocytes, cellular injury, mitosis, and hepatocyte regeneration. Continuous necrosis of hepatocytes during chronic HBV infection accompanied by rapid regeneration, may lead to accumulation of mutations and selection of cells with a malignant phenotype.12 Regenerative hepatocytes in chronic liver diseases give rise to hyperplastic hepatocyte nodules, and these progresses to dysplastic nodules, which are thought to be the direct precursor of HCC.2 Several facts support this theory, such as the fact that the extension of the inflammation in HBV infection is proportional to the risk of developing HCC. Patients with cirrhosis are more prone to develop HCC than those that have active chronic hepatitis or HBV infection without cirrhosis.6,7 Chronic HBV leading to cirrhosis remains the most important precancerous aetiological factor, with 70% to 90% of HBV associated HCC developing on a background of cirrhosis.3,5 However, HBV associated HCC may also develop without a background of cirrhosis.3,5 The other pathway evokes direct oncogenic potential of HBV through chromosomal integration (cis‐activation) or transactivation of cellular genes.14 There are several lines of evidence that suggest that HBV may lead directly to HCC. Persistent HBV replication is associated with a high frequency of integration of HBV sequences into the human host genome.6,14 However, the insertion is random and is not usually near other important genes.15 Twenty per cent of the patients with HBV associated HCC do not integrate the DNA of the virus.16 This concept cannot also explain the clinical finding that asymptomatic HBV carriers with extensive viral replication rarely develop HCC.3,6 Furthermore, some patients develop HCC during occult HBV infection that is not even detectable in the serum.17 That is, the cause of HBV associated HCC could be a combination of generalised processes that lead to a chronic hepatic disease, and those specific ones related to HBV infection.
Figure 2 Mechanism of hepatitis B virus associated hepatocarcinogenesis. The predominant carcinogenic mechanism of HBV associated HCC is through the process of liver cirrhosis, but direct oncogenic effects of HBV may also contribute.
Integration of HBV DNA
HBV shares a replication strategy that includes the reverse transcription of a RNA intermediate.13 HBV DNA sequences have been shown to be integrated into cellular DNA in HCC tissue and can also be identified in non‐tumorous tissue from patients with chronic hepatitis.6 Moreover, at least in some cases, integration of the viral DNA occurs during the acute or early stages of infection.18 When HBV integrations occur they produce a wide range of genetic changes within the host genome, including deletions, translocations, the production of fusion transcripts, and generalised genomic instability.1 Several reports have investigated the chromosomal integrity of HCC and have identified deletions in portions of chromosomes. Chromosomal losses are present in 25%–45% of patients in 1p, 4q, 5q, 6q, 8p, 9p, 13q, 16p, 16q, and 17p while gains occur 30%–55% of the time at 1p, 6p, 8q, and 17q.1,19 Many of these chromosomal segments contain known tumour suppressor genes such as p53, Rb, cyclin D1, p16.1
Research over the past decade has unravelled that HBx protein seems to have a pivotal part in hepatocarcinogenesis related to HBV. Transgenic mice have provided insight into the mechanisms of hepatocarcinogenesis but the results obtained so far have not been consistent. HBx has been shown to promote liver tumour formation in some transgenic mice, but not in others.12 However, HBx at least acted as a cofactor in carcinogen induced as well as in c‐Myc induced hepatocarcinogenesis.12 More than 95% of patients with HBV associated cirrhosis and dysplasia are positive for HBx and 70% of patients with HBV associated HCC produce HBx protein.5,20 Therefore, it seems probable that HBx protein contributes to the initiation of tumour formation in the liver during the processes of chronic active hepatitis and cirrhosis. Integrated HBV sequences frequently have a carboxy‐terminal deleted X gene, resulting in translation of a truncated HBx protein.15 These deletion mutants seem to lose their capacity for oncogene induced apoptosis and cell cycle arrest, leading to an increased tendency to unregulated cell proliferation that increases the transforming capacity of the protein.21
Another HBV gene product that has been reported to possess transactivational properties is a truncated form of the pre‐S2/S gene.6 Truncated pre‐S2/S sequences are frequently found in HBV DNA integration sites in HCC.6 Specific activation of mitogen activated protein kinases (MAPK/ERK) signalling by the truncated pre‐S2/S protein has been shown to result in an activation of transcription factors such as AP‐1 and nuclear factor‐κ B (NF‐κB). Furthermore, by activation of this signalling cascade, the Pre‐S2/S activators cause an increased proliferation rate of hepatocytes.22 Cytologically, overproduction of HBV envelope proteins (pre‐S2/S), particularly L and possibly M, results in their intracellular accumulation and may predispose the cell to stress, which in turn may lead to the development of HCC.23 HBV envelope protein (pre‐S2/S) mutants that overaccumulate envelope polypeptides within the cell have been also seen to be associated with advancing liver disease and may be, in part, responsible for ground glass hepatocytes and perhaps even HCC lesions.24 In addition, mutations of core promoter region of the virus have also been described in integrated HBV sequences.25
Roles of HBX on hepatocarcinogenesis
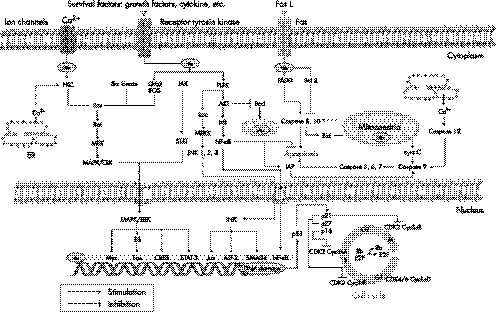
Cumulative data suggest that HBx is a multifunctional regulatory protein, which communicates directly or indirectly with a variety of host targets and mediates many opposing cellular functions, including function in cell cycle regulation, transcriptional regulation, signalling, encoding of cytoskeleton and cell adhesion molecules, as well as oncogenes and tumour suppressor genes(fig 3).14,26
Figure 3 Important molecular pathways in hepatitis B virus associated hepatocarcinogenesis. HBx is a multifunctional regulatory protein, which communicates directly or indirectly with a variety of host targets and mediates many opposing cellular functions, including function in apoptosis, cell cycle regulation, transcriptional regulation, as well as cell signalling pathways. HBx transactivates gene expression by acting on different cellular regulatory pathways, depending on its subcellular localisation. HBx activates components of the mitogen activated protein kinases (MAPK/ERK), stress activated protein kinase/Jun N‐terminal kinase (SAPK/JNK), protein kinase B (AKT/PKB), protein kinase C (PKC), phosphatidylinositol 3‐kinase (PI‐3‐K), and Janus kinase (JAK)/STAT kinase pathways in the cytoplasm. HBx also behaves as a transcriptional transactivator that up‐regulates the expression of proto‐oncogenes such as c‐Myc and c‐Jun, transcriptional factors like nuclear factor‐κ B (NF‐κB), AP‐1, AP‐2, and ATF/cAMP‐response element binding protein (CREB) in the nucleus.
Transcriptional transactivation
Increasing experimental evidence exists that HBx transactivates gene expression by acting on different cellular regulatory pathways, depending on its subcellular localisation. Intracellular localisation studies have shown that HBx protein is present predominantly in the cytoplasm but can also be found at lesser amounts in the nucleus.14 Thus, HBx stimulates signal transduction pathways like MAPK/ERK pathway in the cytoplasm, and HBx also behaves as a transcriptional transactivator that up‐regulates the expression of proto‐oncogenes such as c‐Myc and c‐Jun, transcriptional factors like NF‐κB, AP‐1, AP‐2, RPB5 subunit of RNA polymerase II, TATA binding protein and ATF/cAMP‐response element binding protein (CREB), as well as other viral genes such as HBV enhancers in the nucleus.14 The identification of target proteins that directly interact with HBx is increasing, including Smad4 and nuclear factor activated T cells (NF‐AT) in TNFα signalling, and COX‐2,27 retinoid X receptor in phosphoenolpyruvate carboxykinase expression.28 Moreover, several host genes have been also reported to be indirectly affected by HBx, such as the upregulation of IL6, IL8, the induction of nitric oxide synthetase, and Fas ligand.28
NF‐κB and AP‐1 have been reported to be directly or indirectly affected by HBx. NF‐κB activation by HBx includes direct interaction with inhibitory proteins, such as the p105 precursor and IκB kinase (IKK), and induction of oxidative stress by mitochondrial associated HBx protein.14,29 In addition, HBx activates components of the MAPK/ERK, stress activated protein kinase/ Jun N‐terminal kinase (SAPK/JNK), and protein kinase C (PKC) signalling pathways to regulate NF‐κB and AP‐1 dependent transcription.14 Regardless of the mechanism, HBx induction of NF‐κB and AP‐1 activity ultimately leads to the acceleration of cell cycle progression, increased proliferation, and may contribute to repression of apoptosis.
Regulation of signal pathways
Several reports have investigated that the effects of the HBx on signal pathways such as, MAPK/ERK, protein kinase B (AKT/PKB), PKC, SAPK/JNK, phosphatidylinositol 3‐kinase (PI‐3‐K), and Janus kinase (JAK)/STAT kinase pathways. These pathways can overlap or interact with one another to generate a unique response, causing their constitutive or prolonged activation, resulting in increased cell proliferation and survival. Aberrant expression of MAPK and associated proteins are involved in the development of HCC. HBx activates Ras as an intracellular cytoplasmic activator of the Src family of tyrosine kinases and the downstream MAPK pathway such as Raf, MAPKK, MEK1/2, and ERK1/2.14,30 Activated ERK1/2 translocates to the nucleus to activate a variety of transcription factors such as ELK‐1, c‐Fos, c‐Jun, c‐Myc, STAT‐3 etc, involved in regulating cell proliferation and differentiation.30 HBx increases the levels of epidermal growth factor receptor (EGFR) by transactivating the EGFR promoter, and subsequently activates the Raf/MEK/ERK cascade.30 Furthermore, HBx protein activates AP‐1 and NF‐κB transcription factors through the endogenous PKC.30,31 HBx could also activate the JAK/STAT pathway, leading to activation of STAT regulated genes.30 Because of its association with mitochondria and through oxidative stress, HBx constitutively induces the transcription factors STAT‐3 and NF‐κB.30
Recent studies showed that HBx could activate Wnt/β‐catenin signalling by stabilising cytoplasmic β‐catenin in HCC.32,33 An alternative mechanism for Wnt activation in HCC is that HBx repressed E‐cadherin expression at the transcriptional level through hypermethylation of E‐cadherin promoter.34 Mutations of AXIN1, another factor in the Wnt/β‐catenin signalling pathway, have been found in a substantial proportion of HCC with β‐catenin accumulation in the absence of mutation of the β‐catenin gene.35 Furthermore, upregulation of frizzled‐7(FZD‐7) receptors in association with activation of the Wnt/β‐catenin pathway is a common in HCC.36 After all, HBx is associated with decreased expression of E‐cadherin, accumulation of β‐catenin in the cytoplasm and nucleus, and increased cell migration, which may contribute importantly to hepatocarcinogenesis
Regulation of apoptosis
HBx is involved in apoptosis, including p53, retinoblastoma protein (Rb), Fas associated death domain protein (FADD), tumour necrosis factor (TNF) receptor associated death domain protein (TRADD), and NF‐κB in HBV associated HCC.26 Hepatocyte apoptosis can be induced through the death receptor dependent pathway (extrinsic pathway) or the mitochondrial dependent pathway (intrinsic pathway). The extrinsic pathway is started in the liver by death ligands like TNF, Fas ligand (FasL, CD95L), and TNF related apoptosis inducing ligand (TRAIL), after binding to their relevant death receptors. In contrast, intrinsic pathway is triggered by a variety of intracellular stressors such as DNA damage, growth factor deprivation, metabolic disturbances, and detachment from matrix and/or surrounding cells.37
Survival or apoptosis of cells depends upon the balance of various extracellular and/or intracellular stimuli. HBx is located in mitochondria and causes loss of mitochondrial membrane potential and subsequently induces mitochondria dependent cell death.38 Hepatitis viruses also modulate other antiapoptotic signals such as STAT‐3 and NF‐κB through ER stress and the generation of reactive oxygen species (ROS).39 Therefore, ROS ultimately leads to the activation of STAT‐3 and NF‐κB.40 HBx protein may modulate calcium homoeostasis, by inhibiting cytosolic calcium dependent proline rich tyrosine kinase‐2 (Pyk2), a Src kinase activator, or calcium signalling mediated by mitochondrial calcium channels.41 HBx also inhibits TGFβ and FasL induced apoptosis through the activation of PI‐3‐K pathway.42,43 In addition, the activation of PI‐3‐K by HBx, and the phosphorylation of the downstream factors, AKT and Bad, and downregulation of caspase 3, are seen.44,45
Inhibition of apoptosis may be a consequence of the failure of p53, in the presence of HBx, to upregulate genes, such as p21WAF1, Bax, or Fas, that are involved in the apoptotic pathway. A p53 mutation is present in 30%–60% of patients with HCC.1 HBx can bind to the C‐terminus of p53 forming a protein‐protein complex, thereby inactivating several critical p53 dependent activities, including p53 mediated apoptosis.46 Additional studies show that HBx sequesters p53 in the cytoplasm and prevents it from entering the nucleus.47,48 Moreover, HBV achieves protection from apoptotic death through a HBx‐PI3K‐AKT‐Bad pathway and by inactivating caspase 3 activity that is at least partially p53 independent in liver cells.45,49
The proapoptotic activity of HBx overcomes or bypasses the inhibitory effect of Bcl‐2 against Fas cytotoxicity.50 HBx also upregulates survivin, member of the inhibitor of apoptosis (IAP) family, expression.51 The reduction of proapoptotic protein, Bid, may result from a mechanism associated with HBx.52 However, HBx promotes the apoptosis of hepatocytes by regulating the expressions of Fas/FasL, Bax/Bcl‐2, Bcl‐xL, and c‐Myc gene in a dose dependent.14,53,54,55 Accordingly, HBx can modulate both preapoptotic and antiapoptotic pathways, dependent upon different cell settings. The imbalance of increased antiapoptosis and decreased pro‐apoptosis seen in HCC is a critical mechanism leading to the uncontrolled growth of tumour cells.
Regulation of cell cycle
Several studies have shown that various types of changes in cell cycle regulators are found in HCC. Control of the cell cycle is regulated by activity of cyclins, cyclin dependent kinases (CDKs), and cyclin dependent kinase inhibitors (CDKIs). CDKs, cyclins, and CDKIs generally function within several defined pathways, including the p21WAF1‐p27KIP1‐cyclinE‐CDK2 pathway, the p16INK4A‐cyclinD1‐CDK4/6‐Rb‐E2F pathway, and the p14ARF‐MDM2‐p53 pathway.56 HBx increases the rate and level of activation of the CDK2, and their respective active association with cyclins E and A or cyclin B.57 HBx also causes cyclin D1 overexpression.58 Rb gene is a negative regulator of cell proliferation and plays a critical part in cell proliferation control. Rb suppresses the G1 to S transition.59 Previous studies have shown that Rb is deleted or mutated in HCC.60,61 Furthermore, HBx represses the transcription of p21WAF1.62 Interestingly, the reduction or loss by methylation of the p16INK4A, p14ARF, p15INK4b, and p27KIP1 promoters are detected in HCC.63,64,65,66,67 Ultimately, HBx has been shown to stimulate cell cycle progression by accelerating transit through the G1/S and G2/M checkpoints.
Key points
More than 80% of HCC are caused by hepatitis B infection.
HBV infection is preventable with a vaccine.
Vaccination decreases the incidence of HCC.
Integration of HBV and/or the long term inflammatory changes by chronic hepatitis B infection has an important part in hepatocarcinogenesis.
Treatment of hepatitis B infection reduces the risk of developing HCC
Regulation of angiogenesis
HBx is strongly implicated in angiogenesis and metastasis during hepatocarcinogenesis. HBx induces angiogenesis through up‐regulation of the potent angiogenic factor, vascular endothelial growth factor (VEGF) transcription.68 More recent studies have supported that HBx could induce the expression of VEGF through stabilisation of hypoxia inducible factor‐1α (HIF‐1α) by increasing its association with CREB binding protein (CBP), thereby leading to angiogenesis during hepatocarcinogenesis.69,70 Additional factors, such as angiopoietin‐2,71 seem to be involved as well.
Regulation of telomere function
Activation of telomerase is considered as the important episode for HCC. Recent evidences suggest that telomere dysfunction, leading to telomere based chromosomal instability, may be operative during the early stages of hepatocarcinogenesis while telomerase activation occurs during HCC progression.72,73 Telomerase reverse transcriptase (hTERT) is the rate limiting determinant for regulating telomerase activity. HBV genome integrated in the hTERT promoter region and HBV enhancer could cis‐activate the transcription of hTERT gene.74 In addition, the telomeric repeat binding factor 1 (TRF1), TRF2, and the TRF1 interacting nuclear protein 2 (TIN2) are involved in telomere maintenance.74 The expression of TRF1, TRF2, TIN2 mRNA, and TRF1 protein was gradually increased according to the progression of hepatocarcinogenesis.75
Inhibition of DNA mismatch repair
DNA mismatch repair gene defects seem to play a part in HCC. DNA changes resulting from exogenous genotoxic factors or normal replication processes are corrected by several repair pathways. HBx inhibits the repair of damaged hepatocyte DNA.14 This effect may be mediated by interaction with p53 or through binding to the damaged DNA binding protein (DDB), a highly conserved protein implicated in DNA repair and cell cycle regulation, which plays an accessory part in nucleotide excision repair.76,77 HBx also represses the transcription repair factor TFIIH, impairing TFIIH function in DNA repair mechanisms.78 Therefore, HBx acts as a cofactor in hepatocarcinogenesis by preventing the cell from efficiently repairing damaged DNA, thus leading to an accumulation of DNA mutations and, eventually, cancer.
Modulation of adhesion‐deadhesion balance
HBx may play a part in tumour spreading by modulating the adhesion‐deadhesion balance of the cells in the primary tumour site and favouring integrin mediated cell migration. HBx induces adherens junction disruption and decreases cellular adhesion to ECM, resulting in modulation of cell adhesion and cell motility.79 HBx also increases the migratory phenotype of hepatoma cells through the up‐regulation of matrix metalloproteinases‐1 (MMP‐1) and MMP‐9.80,81 Moreover, HBx represses several cell adhesion molecules and cytoskeleton proteins, including E‐cadherin,34 integrin,79 fibronectin,82 CD47,83 and hyaluronan receptor (CD44).84 Genes regulating the composition of the extra cellular matrix and the cytoskeleton such as tubulin α1, MMP14, osteonectin SPARC, RhoA are also found to be up‐regulated in HCC.85 These genes play important parts in cell motility and invasion.
In addition, HBx takes part in regulation of proteasome and, thus, controls degradation of cellular and viral proteins.86 Expressions of HSP27, HSP70, and HSP90 are also commonly up‐regulated in HBV associated HCC.87
Genetic and epigenetic changes
Numerous genetic and epigenetic changes have been identified to be responsible for the activation of carcinogenic pathways in HCC. In addition to above mentioned p53, p21, Rb, AXIN1, and cyclin D1, several genes are involved in hepatocarcinogenesis.14,88 These include insulin growth factor (IGF) activation through IGF2 overexpression and M6P/IGF2R inactivated mutations.89,90 Other genetic and epigenetic changes include mutations of Smad2/4,28 HCC suppressor 1 (HCCS1),28 phosphatase and tensin homolog deleted on chromosome ten (PTEN),28,91 WWOX (WW‐domain containing oxidoreductase),92 and T cell factor 1 (TCF1),28 and gene silencing by hypermethylation of GSTP1,28,93 and suppressor of cytokine signalling‐1 (SOCS‐1).28 Moreover, the expression of IGF binding protein 3 (IGFBP3) down‐regulates in HCC. In the IGF receptor dependent pathway, IGFBP‐3 mediates a wide variety of growth suppression signals such as TGFβ, retinoic acid, TNFα and p53.94,95 Transcriptional silencing by hypermethylation of deleted in liver cancer‐1 (DLC‐1), a putative tumour suppressor gene mapped at 8p21.3–22, is frequent in HCC, suggesting a possible role of the inactivation of DLC‐1 in hepatocarcinogenesis.28 DLC‐2, one of the most frequently deleted chromosome arms at 13q, has reported to be significantly under‐expressed in HCC.96 Recently, brain derived neutrophilic factor (BDNF) has also been found to be involved in hepatocarcinogenesis.97
The principal growth factors and cytokines stimulate liver growth, inhibition of apoptosis, and development of HCC. These include hepatocyte growth factor (HGF), EGF, TGFβ, IL6, TNFα.88
Fibrogenesis and chronic HBV infection
Chronic stellate cell activation induced by HBV replication could contribute to fibrogenesis and increased proliferation of hepatocytes. This increased production of extracellular matrix and hepatocyte turnover coupled with activation of MAPK pathway may ultimately lead to HCC. The Wnt/β‐catenin pathways and MMT could be directly associated with hepatocarcinogenesis.98 Other functions that could contribute to disease that are known to be increased during cirrhosis are TGFβ, increased gelatinases, fibroblast activating proteins, and members of the interferon response pathway.98 Thus, several factors, directly or indirectly and alone or in combination, can lead to HCC.
Five key references for further reading
Ganem D, Prince AM. Hepatitis B virus infection—natural history and clinical consequences. N Engl J Med 2004;350:1118–29.
Brechot C. Pathogenesis of hepatitis B virus‐related hepatocellular carcinoma: old and new paradigms. Gastroenterology 2004;127:S56–61.
Thomas MB, Zhu AX. Hepatocellular carcinoma: the need for progress. J Clin Oncol 2005;23:2892–9.
Lok AS. Prevention of HBV‐related hepatocellular carcinoma. Gastroenterology 2004;127:S303–9.
Cha C, Dematteo RP. Molecular mechanisms in hepatocellular carcinoma development. Best Pract Res Clin Gastroenterol 2005;19:25–37.
Conclusion
HBV is strongly associated with HCC by its presence in the tumour cell and by the striking role of persistent HBV infection as a risk factor for the development of HCC. It is generally shown that vaccination significantly decreases the incidence of HCC. Moreover, preventing the most severe HBV disease consequences in infected people, such as cirrhosis and HCC, will require appropriate therapeutic agents and reduces the risk of developing HCC.
The past decade has shown the important mechanism of action of oncogenes and tumour suppressor genes in hepatocarcinogenesis, together with the molecular pathways of control through which their effects on proliferation are mediated. Recent studies have delineated many different oncogenic pathways involved in hepatocarcinogenesis. Accordingly, a large number of novel molecular therapeutics targeting these signalling components is in clinical development. Recently, genetic information of new genomic technologies and approaches is accumulated at a rapid pace. The establishment of microarray methods for large scale analysis of gene expression has made it possible to seek molecular markers for cancer classification and outcome prediction and to identify molecules involved in hepatocarcinogenesis.
Multiple choice questions (true (T)/false (F); answers at the end of references)
1. Regarding the epidemiology HBV
HBV is a serious public health problem worldwide
HBV is an important cause of chronic hepatitis, cirrhosis, and HCC
About two billion people who have been infected worldwide
Less than 400 million are chronic carriers of HBV
About 75% of chronic carriers live in Asia and the Western countries
2. In geographical distribution of the carrier state
There is a noticeable difference in geographical distribution of the carrier state in the world
Ranging from 10% to 20% in South East Asia and sub‐Sahara Africa
Less than 1% in Southern Europe and America
In low endemic areas, most HBV infections are acquired in adolescents and young adults
In areas of high endemicity, the most common route of transmission is preschool years
3. In the risk of becoming a virus carrier
In low endemic areas, most HBV infections are acquired by horizontal transmission
In areas of high endemicity, the most common route of transmission is vertical
Ranges from 90% in perinatal infection
More than 10% in adults
Immunosuppressed persons are more likely to develop chronic HBV infection
4. In the incidence of HCC
HCC is the fifth most frequent cancer in the world, killing 300 000–500 000 people each year
Exposure to other hepatotoxins, such as alcohol and aflatoxins, hastens the development of HCC
Coinfection with HCV and HBV also increases the risk of developing HCC by at least twofold
About 15%–40% of infected patients will develop HCC
The outcome of HCC is good
5. How HBV causes carcinogenic changes in the liver
HCC is common in regions where HBV is endemic
The highest rates are in South East Asia and sub‐Saharan Africa
Does not occur in convalescent HBV infection
A decline in the incidence of HCC in children after a universal hepatitis B vaccination
Relative risk of HCC among HBeAg positive patients is much higher than that among inactive HBsAg carries
6. In the structure of HBV genome
HBV belongs to the family of hepadnaviruses
The HBV genome is a relaxed circular, completely double stranded DNA of about 3200 base pairs
There are four partially overlapping open reading frames encoding the envelope (pre‐S/S), core (precore/core), polymerase, and X proteins
The pre‐S/S open reading frame encodes the large (L), middle (M), and small (S) surface glycoproteins
The precore/core open reading frame functions as a reverse transcriptase as well as a DNA polymerase.
7. In the mechanism of HBV associated HCC
One entails chronic necroinflammation of hepatocytes, cellular injury, mitosis, and hepatocyte regeneration
Cirrhosis are more prone to develop HCC than those that have active chronic hepatitis or HBV infection without cirrhosis
40% to 50% of HBV associated HCC developing on a background of cirrhosis
Does not develop without a background of cirrhosis
The other pathway evokes direct oncogenic potential of HBV through chromosomal integration (cisactivation) or transactivation of cellular genes
8. In the integration of HBV DNA
HBV DNA sequences have been shown to be integrated into cellular DNA in HCC tissue and can also be identified in non‐tumorous tissue from patients with chronic hepatitis
At least in some cases, integration of the viral DNA occurs during the acute or early stages of infection
Eighty per cent of the patients with HCC associated to HBV do not integrate the DNA of the virus
The insertion site is uniform
Is usually near other important genes
Abbreviations
HCC - hepatocellular carcinoma
HBV - hepatitis B virus
CREB - cAMP response element binding protein
NF‐κB - nuclear factor κ B
TNF - tumour necrosis factor
CDK - cyclin dependent kinase
VEGF - vascular endothelial growth factor
MMP - metalloproteinase
IGF - insulin growth factor
Answers
1. (A) T, (B) T, (C) T, (D) F, (E) F; 2. (A) T, (B) T, (C) F, (D) T, (E) T ; 3. (A) T, (B) T, (C) T, (D) F, (E) T; 4. (A) T, (B) T, (C) T, (D) T, (E) F; 5. (A) T, (B) T, (C) F, (D) T, (E) T; 6. (A) T, (B) F, (C) T, (D) T, (E) F ; 7. (A) T, (B) T, (C) F, (D) F, (E) T; 8. (A) T, (B) T, (C) F, (D) F, (E) F.
Footnotes
Funding: none.
Conflicts of interest: none declared.
References
- 1.Cha C, Dematteo R P. Molecular mechanisms in hepatocellular carcinoma development. Best Pract Res Clin Gastroenterol 20051925–37. [DOI] [PubMed] [Google Scholar]
- 2.Ganem D, Prince A M. Hepatitis B virus infection—natural history and clinical consequences. N Engl J Med 20043501118–1129. [DOI] [PubMed] [Google Scholar]
- 3.Lok A S. Prevention of hepatitis B virus‐related hepatocellular carcinoma. Gastroenterology 2004127S303–S309. [DOI] [PubMed] [Google Scholar]
- 4.Lok A S, Heathcote E J, Hoofnagle J H. Management of hepatitis B: 2000—summary of a workshop. Gastroenterology 20011201828–1853. [DOI] [PubMed] [Google Scholar]
- 5.Fattovich G, Stroffolini T, Zagni I.et al Hepatocellular carcinoma in cirrhosis: incidence and risk factors. Gastroenterology 2004127S35–S50. [DOI] [PubMed] [Google Scholar]
- 6.Brechot C. Pathogenesis of hepatitis B virus‐related hepatocellular carcinoma: old and new paradigms. Gastroenterology 2004127S56–S61. [DOI] [PubMed] [Google Scholar]
- 7.Tanaka H, Iwasaki Y, Nouso K.et al Possible contribution of prior hepatitis B virus infection to the development of hepatocellular carcinoma. J Gastroenterol Hepatol 200520850–856. [DOI] [PubMed] [Google Scholar]
- 8.Chang M ‐ H, Chen C ‐ J, Lai M ‐ S.et al Universal hepatitis B vaccination in Taiwan and the incidence of hepatocellular carcinoma in children. N Engl J Med 19973361855–1859. [DOI] [PubMed] [Google Scholar]
- 9.Lee M S, Kim D H, Kim H.et al Hepatitis B vaccination and reduced risk of primary liver cancer among male adults: a cohort study in Korea. Int J Epidemiol 199827316–319. [DOI] [PubMed] [Google Scholar]
- 10.Yang H I, Lu S N, Liaw Y F.et al Hepatitis B e antigen and the risk of hepatocellular carcinoma. N Engl J Med 2002347168–174. [DOI] [PubMed] [Google Scholar]
- 11.Ikeda K, Arase Y, Kobayashi M.et al Consistently low hepatitis B virus DNA saves patients from hepatocellular carcinogenesis in HBV‐related cirrhosis. A nested case‐control study using 96 untreated patients. Intervirology 20034696–104. [DOI] [PubMed] [Google Scholar]
- 12.Singh M, Kumar V. Transgenic mouse models of hepatitis B virus‐associated hepatocellular carcinoma. Rev Med Virol 200313243–253. [DOI] [PubMed] [Google Scholar]
- 13.Seeger C, Mason W S. Hepatitis B virus biology. Microbiol Mol Biol Rev 20006451–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pang R, Tse E, Poon R T. Molecular pathways in hepatocellular carcinoma. Cancer Lett. (in press) [DOI] [PubMed]
- 15.Wang Y, Lau S H, Sham J S.et al Characterization of HBV integrants in 14 hepatocellular carcinomas: association of truncated x gene and hepatocellular carcinogenesis. Oncogene 200423142–148. [DOI] [PubMed] [Google Scholar]
- 16.Brechot C, Gozuacik D, Murakami Y.et al Molecular bases for the development of hepatitis B virus (HBV)‐related hepatocellular carcinoma (HCC). Semin Cancer Biol 200010211–231. [DOI] [PubMed] [Google Scholar]
- 17.Pollicino T, Squadrito G, Cerenzia G.et al Hepatitis B virus maintains its pro‐oncogenic properties in the case of occult HBV infection. Gastroenterology 2004126102–110. [DOI] [PubMed] [Google Scholar]
- 18.Murakami Y, Minami M, Daimon Y.et al Hepatitis B virus DNA in liver, serum, and peripheral blood mononuclear cells after the clearance of serum hepatitis B virus surface antigen. J Med Virol 200472203–214. [DOI] [PubMed] [Google Scholar]
- 19.Zhang S H, Cong W M, Xian Z H.et al Clinicopathological significance of loss of heterozygosity and microsatellite instability in hepatocellular carcinoma in China. World J Gastroenterol 2005113034–3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murakami Y, Saigo K, Takashima H.et al Large scaled analysis of hepatitis B virus (HBV) DNA integration in HBV related hepatocellular carcinomas. Gut 2005541162–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tu H, Bonura C, Giannini C.et al Biological impact of natural COOH‐terminal deletions of hepatitis B virus x protein in hepatocellular carcinoma tissues. Cancer Res 2001617803–7810. [PubMed] [Google Scholar]
- 22.Pagano J S, Blaser M, Buendia M‐A.et al Infectious agents and cancer: criteria for a causal relation. Semin Cancer Biol 200414453–471. [DOI] [PubMed] [Google Scholar]
- 23.Xu Z, Jensen G, Yen T S. Activation of hepatitis B virus S promoter by the viral large surface protein via induction of stress in the endoplasmic reticulum. J Virol 1997717387–7392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tai P‐C, Suk F‐M, Gerlich W H.et al Hypermodification and immune escape of an internally deleted middle‐envelope (M) protein of frequent and predominant hepatitis B virus variants. Virology 200229244–58. [DOI] [PubMed] [Google Scholar]
- 25.Momosaki S, Hsia C C, Nakashima Y.et al Integration of hepatitis B virus containing mutations in the core promoter/X gene in patients with hepatocellular carcinoma. Dig Liver Dis 200335795–800. [DOI] [PubMed] [Google Scholar]
- 26.Peng Z, Zhang Y, Gu W.et al Integration of the hepatitis B virus X fragment in hepatocellular carcinoma and its effects on the expression of multiple molecules: a key to the cell cycle and apoptosis. Int J Oncol 200526467–473. [PubMed] [Google Scholar]
- 27.Cheng A S, Chan H L, Leung W K.et al Expression of HBx and COX‐2 in chronic hepatitis B, cirrhosis and hepatocellular carcinoma: implication of HBx in upregulation of COX‐2. Mod Pathol 2004171169–1179. [DOI] [PubMed] [Google Scholar]
- 28.Pang R, Yuen J, Yuen M F.et al PIN1 overexpression and beta‐catenin gene mutations are distinct oncogenic events in human hepatocellular carcinoma. Oncogene 2004234182–4186. [DOI] [PubMed] [Google Scholar]
- 29.Chan C‐F, Yau T‐O, Jin D‐Y.et al Evaluation of nuclear factor‐{kappa}B, urokinase‐type plasminogen activator, and HBx and their clinicopathological significance in hepatocellular carcinoma. Clin Cancer Res 2004104140–4149. [DOI] [PubMed] [Google Scholar]
- 30.Panteva M, Korkaya H, Jameel S. Hepatitis viruses and the MAPK pathway: is this a survival strategy? Virus Res 200392131–140. [DOI] [PubMed] [Google Scholar]
- 31.Wen‐Sheng W, Jun‐Ming H. Activation of protein kinase C alpha is required for TPA‐triggered ERK (MAPK) signaling and growth inhibition of human hepatoma cell HepG2. J Biomed Sci 200512289–296. [DOI] [PubMed] [Google Scholar]
- 32.Cha M Y, Kim C M, Park Y M.et al Hepatitis B virus X protein is essential for the activation of Wnt/beta‐catenin signaling in hepatoma cells. Hepatology 2004391683–1693. [DOI] [PubMed] [Google Scholar]
- 33.Ding Q, Xia W, Liu J‐C.et al Erk associates with and primes GSK‐3beta for its inactivation resulting in upregulation of beta‐catenin. Mol Cell 200519159–170. [DOI] [PubMed] [Google Scholar]
- 34.Lee J O, Kwun H J, Jung J K.et al Hepatitis B virus X protein represses E‐cadherin expression via activation of DNA methyltransferase 1. Oncogene 2005246617–6625. [DOI] [PubMed] [Google Scholar]
- 35.Satoh S, Daigo Y, Furukawa Y.et al AXIN1 mutations in hepatocellular carcinomas, and growth suppression in cancer cells by virus‐mediated transfer of AXIN1. Nat Genet 200024245–250. [DOI] [PubMed] [Google Scholar]
- 36.Merle P, Kim M, Herrmann M.et al Oncogenic role of the frizzled‐7/beta‐catenin pathway in hepatocellular carcinoma. J Hepatol 200543854–862. [DOI] [PubMed] [Google Scholar]
- 37.Yin X M, Ding W X. Death receptor activation‐induced hepatocyte apoptosis and liver injury. Curr Mol Med 20033491–508. [DOI] [PubMed] [Google Scholar]
- 38.Shirakata Y, Koike K. Hepatitis B virus X protein induces cell death by causing loss of mitochondrial membrane potential. J Biol Chem 200327822071–22078. [DOI] [PubMed] [Google Scholar]
- 39.Lee Y I, Hwang J M, Im J H.et al Human hepatitis B virus‐X protein alters mitochondrial function and physiology in human liver cells. J Biol Chem 200427915460–15471. [DOI] [PubMed] [Google Scholar]
- 40.Waris G, Huh K ‐ W, Siddiqui A. Mitochondrially associated hepatitis B virus X protein constitutively activates transcription factors STAT‐3 and NF‐kappa B via oxidative stress. Mol Cell Biol 2001217721–7730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bouchard M J, Wang L H, Schneider R J. Calcium signaling by HBx protein in hepatitis B virus DNA replication. Science 20012942376–2378. [DOI] [PubMed] [Google Scholar]
- 42.Sirma H, Giannini C, Poussin K.et al Hepatitis B virus X mutants, present in hepatocellular carcinoma tissue abrogate both the antiproliferative and transactivation effects of HBx. Oncogene 1999184848–4859. [DOI] [PubMed] [Google Scholar]
- 43.Shih W‐L, Kuo M‐L, Chuang S‐E.et al Hepatitis B virus X protein inhibits transforming growth factor‐beta ‐induced apoptosis through the activation of phosphatidylinositol 3‐kinase pathway. J Biol Chem 200027525858–25864. [DOI] [PubMed] [Google Scholar]
- 44.Shin E C, Shin J S, Park J H.et al Expression of fas ligand in human hepatoma cell lines: role of hepatitis‐B virus X (HBX) in induction of Fas ligand. Int J Cancer 199982587–591. [DOI] [PubMed] [Google Scholar]
- 45.Lee Y I, Kang‐Park S, Do S‐I.et al The hepatitis B virus‐X protein activates a phosphatidylinositol 3‐kinase‐dependent survival signaling cascade. J Biol Chem 200127616969–16977. [DOI] [PubMed] [Google Scholar]
- 46.Lin Y, Nomura T, Yamashita T.et al The transactivation and p53‐interacting functions of hepatitis B virus X protein are mutually interfering but distinct. Cancer Res 1997575137–5142. [PubMed] [Google Scholar]
- 47.Wang X W, Gibson M K, Vermeulen W.et al Abrogation of p53‐induced apoptosis by the hepatitis B virus X gene. Cancer Res 1995556012–6016. [PubMed] [Google Scholar]
- 48.Shintani Y, Yotsuyanagi H, Moriya K.et al Induction of apoptosis after switch‐on of the hepatitis B virus X gene mediated by the Cre/loxP recombination system. J Gen Virol 1999803257–3265. [DOI] [PubMed] [Google Scholar]
- 49.Kim Y C, Song K S, Yoon G.et al Activated ras oncogene collaborates with HBx gene of hepatitis B virus to transform cells by suppressing HBx‐mediated apoptosis. Oncogene 20012016–23. [DOI] [PubMed] [Google Scholar]
- 50.Terradillos O, de La Coste A, Pollicino T.et al The hepatitis B virus X protein abrogates Bcl‐2‐mediated protection against Fas apoptosis in the liver. Oncogene 200221377–386. [DOI] [PubMed] [Google Scholar]
- 51.Zhang X, Dong N, Yin L.et al Hepatitis B virus X protein upregulates survivin expression in hepatoma tissues. J Med Virol 200577374–381. [DOI] [PubMed] [Google Scholar]
- 52.Chen G G, Lai P B S, Chan P K S.et al Decreased expression of Bid in human hepatocellular carcinoma is related to hepatitis B virus X protein. Eur J Cancer 2001371695–1702. [DOI] [PubMed] [Google Scholar]
- 53.Zhang S J, Chen H Y, Chen Z X.et al Possible mechanism for hepatitis B virus X gene to induce apoptosis of hepatocytes. World J Gastroenterol 2005114351–4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim W H, Hong F, Jaruga B.et al Hepatitis B virus X protein sensitizes primary mouse hepatocytes to ethanol‐and TNF‐alpha‐induced apoptosis by a caspase‐3‐dependent mechanism. Cell Mol Immunol 2005240–48. [PubMed] [Google Scholar]
- 55.Miao J, Chen G G, Chun S Y.et al Hepatitis B virus X protein induces apoptosis in hepatoma cells through inhibiting Bcl‐xL expression. Cancer Lett 2006236115–124. [DOI] [PubMed] [Google Scholar]
- 56.Semczuk A, Jakowicki J A. Alterations of pRb1‐cyclin D1‐cdk4/6‐p16(INK4A) pathway in endometrial carcinogenesis. Cancer Lett 20042031–12. [DOI] [PubMed] [Google Scholar]
- 57.Bouchard M, Giannakopoulos S, Wang E H.et al Hepatitis B virus HBx protein activation of cyclin A‐cyclin‐dependent kinase 2 complexes and G1 transit via a Src kinase pathway. J Virol 2001754247–4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Klein A, Guhl E, Tzeng Y J.et al HBX causes cyclin D1 overexpression and development of breast cancer in transgenic animals that are heterozygous for p53. Oncogene 2003222910–2919. [DOI] [PubMed] [Google Scholar]
- 59.Singh R P, Agarwal C, Agarwal R. Inositol hexaphosphate inhibits growth, and induces G1 arrest and apoptotic death of prostate carcinoma DU145 cells: modulation of CDKI‐CDK‐cyclin and pRb‐related protein‐E2F complexes. Carcinogenesis 200324555–563. [DOI] [PubMed] [Google Scholar]
- 60.Niketeghad F, Decker H J, Caselmann W H.et al Frequent genomic imbalances suggest commonly altered tumour genes in human hepatocarcinogenesis. Br J Cancer 200185697–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Edamoto Y, Hara A, Biernat W.et al Alterations of RB1, p53 and Wnt pathways in hepatocellular carcinomas associated with hepatitis C, hepatitis B and alcoholic liver cirrhosis. Int J Cancer 2003106334–341. [DOI] [PubMed] [Google Scholar]
- 62.Kwun H J, Jang K L. Natural variants of hepatitis B virus X protein have differential effects on the expression of cyclin‐dependent kinase inhibitor p21 gene. Nucleic Acids Res 2004322202–2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li X, Hui A M, Sun L.et al p16INK4A hypermethylation is associated with hepatitis virus infection, age, and gender in hepatocellular carcinoma. Clin Cancer Res 2004107484–7489. [DOI] [PubMed] [Google Scholar]
- 64.Pascale R M, Simile M M, Calvisi D F.et al Role of HSP90, CDC37, and CRM1 as modulators of P16(INK4A) activity in rat liver carcinogenesis and human liver cancer. Hepatology 2005421310–1319. [DOI] [PubMed] [Google Scholar]
- 65.Lei P P, Zhang Z J, Shen L J.et al Expression and hypermethylation of p27 kip1 in hepatocarcinogenesis. World J Gastroenterol 2005114587–4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fukai K, Yokosuka O, Imazeki F.et al Methylation status of p14ARF, p15INK4b, and p16INK4a genes in human hepatocellular carcinoma. Liver Int 2005251209–1216. [DOI] [PubMed] [Google Scholar]
- 67.Qin Y, Liu J Y, Li B.et al Association of low p16INK4a and p15INK4b mRNAs expression with their CpG islands methylation with human hepatocellular carcinogenesis. World J Gastroenterol 2004101276–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lee S‐W, Lee Y M, Bae S‐K.et al Human hepatitis B virus X protein is a possible mediator of hypoxia‐induced angiogenesis in hepatocarcinogenesis. Biochem Biophys Res Commun 2000268456–461. [DOI] [PubMed] [Google Scholar]
- 69.Yoo Y‐G, Cho S, Park S.et al The carboxy‐terminus of the hepatitis B virus X protein is necessary and sufficient for the activation of hypoxia‐inducible factor‐1 (alpha). FEBS Lett 2004577121–126. [DOI] [PubMed] [Google Scholar]
- 70.Moon E‐J, Jeong C‐H, Jeong J‐W.et al Hepatitis B virus X protein induces angiogenesis by stabilizing hypoxia‐inducible factor‐1alpha. FASEB J 200418382–384. [DOI] [PubMed] [Google Scholar]
- 71.Mitsuhashi N, Shimizu H, Ohtsuka M.et al Angiopoietins and Tie‐2 expression in angiogenesis and proliferation of human hepatocellular carcinoma. Hepatology 2003371105–1113. [DOI] [PubMed] [Google Scholar]
- 72.Farazi P A, Glickman J, Jiang S.et al Differential impact of telomere dysfunction on initiation and progression of hepatocellular carcinoma. Cancer Res 2003635021–5027. [PubMed] [Google Scholar]
- 73.Satyanarayana A, Manns M P, Rudolph K L. Telomeres and telomerase: a dual role in hepatocarcinogenesis. Hepatology 200440276–283. [DOI] [PubMed] [Google Scholar]
- 74.Zhang X, Dong N, Zhang H.et al Effects of hepatitis B virus X protein on human telomerase reverse transcriptase expression and activity in hepatoma cells. J Lab Clin Med 200514598–104. [DOI] [PubMed] [Google Scholar]
- 75.Oh B K, Kim Y J, Park C.et al Up‐regulation of telomere‐binding proteins, TRF1, TRF2, and TIN2 is related to telomere shortening during human multistep hepatocarcinogenesis. Am J Pathol 200516673–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mathonnet G, Lachance S, Alaoui‐Jamali M.et al Expression of hepatitis B virus X oncoprotein inhibits transcription‐coupled nucleotide excision repair in human cells. Mutat Res 2004554305–318. [DOI] [PubMed] [Google Scholar]
- 77.Leupin O, Bontron S, Schaeffer C.et al Hepatitis B virus X protein stimulates viral genome replication via a DDB1‐dependent pathway distinct from that leading to cell death. J Virol 2005794238–4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jaitovich‐Groisman I, Benlimame N, Slagle B L.et al Transcriptional regulation of the TFIIH transcription repair components XPB and XPD by the hepatitis B virus x protein in liver cells and transgenic liver tissue. J Biol Chem 200127614124–14132. [DOI] [PubMed] [Google Scholar]
- 79.Lara‐Pezzi E, Majano P L, Yanez‐Mo M.et al Effect of the hepatitis B virus HBx protein on integrin‐mediated adhesion to and migration on extracellular matrix. J Hepatol 200134409–415. [DOI] [PubMed] [Google Scholar]
- 80.Lara‐Pezzi E, Gomez‐Gaviro M V, Galvez B G.et al The hepatitis B virus X protein promotes tumor cell invasion by inducing membrane‐type matrix metalloproteinase‐1 and cyclooxygenase‐2 expression. J Clin Invest 20021101831–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chung T W, Lee Y C, Kim C H. Hepatitis B viral HBx induces matrix metalloproteinase‐9 gene expression through activation of ERK and PI‐3K/AKT pathways: involvement of invasive potential. FASEB J 2004181123–1125. [DOI] [PubMed] [Google Scholar]
- 82.Matsuo M, Sakurai H, Ueno Y.et al Activation of MEK/ERK and PI3K/Akt pathways by fibronectin requires integrin alpha‐mediated ADAM activity in hepatocellular carcinoma: A novel functional target for gefitinib. Cancer Sci 200697155–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yoshida H, Tomiyama Y, Oritani K.et al Interaction between Src homology 2 domain bearing protein tyrosine phosphatase substrate‐1 and CD47 mediates the adhesion of human B lymphocytes to nonactivated endothelial cells. J Immunol 20021683213–3220. [DOI] [PubMed] [Google Scholar]
- 84.Lara‐Pezzi E, Serrador J M, Montoya M C.et al The hepatitis B virus X protein (HBx) induces a migratory phenotype in a CD44‐dependent manner: possible role of HBx in invasion and metastasis. Hepatology 2001331270–1281. [DOI] [PubMed] [Google Scholar]
- 85.Delpuech O, Trabut J B, Carnot F.et al Identification, using cDNA macroarray analysis, of distinct gene expression profiles associated with pathological and virological features of hepatocellular carcinoma. Oncogene 2002212926–2937. [DOI] [PubMed] [Google Scholar]
- 86.Cui F, Wang Y, Wang J.et al The up‐regulation of proteasome subunits and lysosomal proteases in hepatocellular carcinomas of the HBx gene knockin transgenic mice. Proteomics 20066498–504. [DOI] [PubMed] [Google Scholar]
- 87.Lim S O, Park S G, Yoo J H.et al Expression of heat shock proteins (HSP27, HSP60, HSP70, HSP90, GRP78, GRP94) in hepatitis B virus‐related hepatocellular carcinomas and dysplastic nodules. World J Gastroenterol 2005112072–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Thomas M B, Zhu A X. Hepatocellular carcinoma: the need for progress. J Clin Oncol 2005232892–2899. [DOI] [PubMed] [Google Scholar]
- 89.Fan Z R, Yang D H, Cui J.et al Expression of insulin like growth factor II and its receptor in hepatocellular carcinogenesis. World J Gastroenterol 20017285–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Oka Y, Waterland R A, Killian J K.et al M6P/IGF2R tumor suppressor gene mutated in hepatocellular carcinomas in Japan. Hepatology 2002351153–1163. [DOI] [PubMed] [Google Scholar]
- 91.Ma D Z, Xu Z, Liang Y L.et al Down‐regulation of PTEN expression due to loss of promoter activity in human hepatocellular carcinoma cell lines. World J Gastroenterol 2005114472–4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Park S W, Ludes‐Meyers J, Zimonjic D B.et al Frequent downregulation and loss of WWOX gene expression in human hepatocellular carcinoma. Br J Cancer 200491753–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Niu Z S, Wang M. Expression of c‐erbB‐2 and glutathione S‐transferase‐pi in hepatocellular carcinoma and its adjacent tissue. World J Gastroenterol 2005114404–4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hanafusa T, Shinji T, Shiraha H.et al Functional promoter upstream p53 regulatory sequence of IGFBP3 that is silenced by tumor specific methylation. BMC Cancer 200559–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yumoto E, Nakatsukasa H, Hanafusa T.et al IGFBP‐3 expression in hepatocellular carcinoma involves abnormalities in TGF‐beta and/or Rb signaling pathways. Int J Oncol 2005271223–1230. [PubMed] [Google Scholar]
- 96.Ching Y P, Wong C M, Chan S F.et al Deleted in liver cancer (DLC) 2 encodes a RhoGAP protein with growth suppressor function and is underexpressed in hepatocellular carcinoma. J Biol Chem 200327810824–10830. [DOI] [PubMed] [Google Scholar]
- 97.Yang Z F, Ho D W, Lam C T.et al Identification of brain‐derived neurotrophic factor as a novel functional protein in hepatocellular carcinoma. Cancer Res 200565219–225. [PubMed] [Google Scholar]
- 98.Xu X‐R, Huang J, Xu Z‐G.et al Insight into hepatocellular carcinogenesis at transcriptome level by comparing gene expression profiles of hepatocellular carcinoma with those of corresponding noncancerous liver. Proc Natl Acad Sci 20019815089–15094. [DOI] [PMC free article] [PubMed] [Google Scholar]