Abstract
Precise control of Wnt/β-catenin signaling is critical for animal development, stem cell renewal, and prevention of disease. In the fruit fly Drosophila melanogaster, the naked cuticle (nkd) gene limits signaling by the Wnt ligand Wingless (Wg) during embryo segmentation. Nkd is an intracellular protein that is composed of separable membrane- and nuclear-localization sequences (NLS) as well as a conserved EF-hand motif that binds the Wnt receptor-associated scaffold protein Dishevelled (Dsh), but the mechanism by which Nkd inhibits Wnt signaling remains a mystery. Here we identify a second NLS in Nkd that is required for full activity and that binds to the canonical nuclear import adaptor Importin-α3. The Nkd NLS is similar to the Importin-α3-binding NLS in the Drosophila heat shock transcription factor (dHSF), and each Importin-α3-binding NLS required intact basic residues in similar positions for nuclear import and protein function. Our results provide further support for the hypothesis that Nkd inhibits nuclear step(s) in Wnt/β-catenin signaling and broaden our understanding of signaling pathways that engage the nuclear import machinery.
Keywords: Wnt, Wingless, Naked cuticle, Dishevelled, Importin-α3, Nuclear transport
Introduction
Wnts comprise a family of protein signals that govern morphogenesis and cell inductive events throughout the animal life cycle (Clevers, 2006). Depending on the organism, tissue, and type of cell, a Wnt signal can act locally to regulate binary cell fate decisions or over longer distances as a morphogen to orchestrate differential gene expression. Many Wnts elicit a well-conserved chain of intracellular events termed the canonical Wnt/β-catenin pathway (see http://www.stanford.edu/~rnusse/wntwindow.html and references therein). In the absence of Wnt, phosphorylation of β-catenin by a “destruction complex” consisting of the Axin and Apc proteins as well as the CK1 and GSK3 kinases triggers ubiquitin-dependent proteasomal degradation of β-catenin. Association of Wnt with Lrp5-6/Arrow (Arr) and Frizzled (Fz) family co-receptors promotes Axin-Lrp5/6 interactions and inhibits β-catenin degradation via linkages with the Dishevelled (Dsh) family of scaffold proteins (Bilic et al., 2007; Wallingford and Habas, 2005). β-catenin then binds transcription factors of the TCF/Lef family to regulate Wnt target genes {reviewed in (Willert and Jones, 2006)}. Altered canonical Wnt signaling is associated with a growing list of human diseases, including cancer, osteoporosis, and type 2 diabetes (Clevers, 2006). Therefore, understanding the mechanisms by which human cells and tissues normally control Wnt signaling has become an urgent priority of biomedical science and the biopharmaceutical industry alike.
Although much effort has been expended to understand how Wnt/β-catenin signals are activated, much less is known about how signals are normally terminated. Naked cuticle (Nkd) is a family of intracellular proteins that antagonize Wnt signaling (Van Raay et al., 2007; Wharton et al., 2001; Zeng et al., 2000). In the fruit fly Drosophila melanogaster, transcription of the sole nkd gene is Wnt inducible, suggesting that Nkd acts in a feedback loop (Zeng et al., 2000). Known vertebrate genomes encode two dynamically expressed nkd paralogs, nkd1 and nkd2 (Katoh, 2001; Van Raay et al., 2007; Wharton et al., 2001). Our current understanding of how Nkd proteins inhibit Wnt signaling derives largely from studies in Drosophila. Fly and mammalian Nkd proteins use an EF-hand motif (“EFX”) to associate with Dsh (Rousset et al., 2002; Wharton et al., 2001), but conserved motifs that confer membrane association and - at least in Drosophila - mediate nuclear entry are also important for function (Chan et al., 2007; Waldrop et al., 2006). How each motif in Nkd acts in concert within a single protein to inhibit signaling in vivo remains unclear. A recent report suggests that vertebrate Dsh proteins act in the nucleus (Itoh et al., 2005), but what Dsh does in the nucleus, whether fly Dsh acts in the nucleus, or whether the action of Nkd in the nucleus requires Dsh association is not known.
Nkd attenuates signaling by the Wnt ligand Wingless (Wg) during early segmentation of the Drosophila embryo (Jürgens et al., 1984; Zeng et al., 2000). In the cellular blastoderm embryo, Wg is produced by a single transverse row of cells per segmental anlage. As the germ band extends, Wg-dependent accumulation of the β-catenin homolog Armadillo (Arm) in nearby cells activates the localized transcription of target genes including hedgehog (hh), engrailed (en), and nkd (Baker, 1988; DiNardo et al., 1988; Lee et al., 1992; Martinez Arias et al., 1988; Riggleman et al., 1990; Tabata et al., 1992; Zeng et al., 2000). In nkd-mutant embryos, the quantity and distribution of Wg is initially similar to wild type, but Arm accumulates to higher levels and genes activated by Wg are expressed in broader domains than in wild type, indicating that nkd mutant cells are hypersensitive to Wg (Bejsovec and Wieschaus, 1993; Dougan and DiNardo, 1992; Martinez Arias et al., 1988; Waldrop et al., 2006; Zeng et al., 2000). Later, Wg signaling instructs epidermal cell fate: cells beyond the influence of Wg secrete ventral cuticle with actin-based apical cell processes termed denticles, whereas most cells in close proximity to Wg producers suppress denticle synthesis and remain “naked” (Bejsovec and Martinez Arias, 1991; Dougan and DiNardo, 1992). In embryos homozygous for null or strongly hypomorphic nkd alleles, an ectopic stripe of Wg is induced in most segments, creating mirror-image pattern duplications, increased cell death, and extra naked cuticle, whereas weaker nkd alleles give rise to milder (yet still lethal) cuticle phenotypes due to variable ectopic Wg (Bejsovec and Wieschaus, 1993; Dougan and DiNardo, 1992; Pazdera et al., 1998; Waldrop et al., 2006).
In order for intercellular signals to impinge upon gene transcription in the nucleus of responding cells, key signal transducers typically engage a nuclear transport machinery that is shared by a myriad of intracellular proteins. In the canonical nuclear import paradigm, the adaptor protein Importin-α links nuclear localization sequence (NLS)-containing cargo proteins to the nuclear import receptor Importin-β1 {reviewed in (Chook and Blobel, 2001; Goldfarb et al., 2004)}. {A notable exception is β-catenin itself, which, due to its structural similarity to Importin-α is able to translocate to the nucleus in an Importin-α/β- and NLS-independent fashion (Fagotto et al., 1998).} D. melanogaster has three Importin-α paralogs: Importin-α1 (Kap-α1) (Mason et al., 2002), Importin-α2 (Pendulin/Kap-α2) (Kussel and Frasch, 1995; Torok et al., 1995), and Importin-α3 (Kap-α3) (Dockendorff et al., 1999; Mason et al., 2003; Mathe et al., 2000); and a single Importin-β1 paralog: Fs(2)Ketel (Ket) (Lippai et al., 2000). We previously identified a 30 amino acid (aa) NLS in Drosophila Nkd (Waldrop et al., 2006), but how Nkd enters the nucleus is not known. Here we identify an additional NLS in Nkd and show that its interaction with Importin-α3 is crucial for nuclear localization and function of Nkd. Our findings provide further support for the hypothesis that Nkd acts, in part, within the nucleus to inhibit Wnt signaling, and broaden our understanding of signaling pathways that engage the nuclear import machinery.
Materials and Methods
DNA constructs
Nkd constructs were built in pBSII-KS+ (Stratagene) with C-terminal enhanced-GFP (Clontech). NkdGFPC, NkdΔ30aa/GFPC, NkdΔ30aaNLS/GFPC, and NkdΔR1S/GFPC have been described (Waldrop et al., 2006). Mutant/junctional regions were synthesized by Pfu-PCR, subcloned/sequenced, then cloned into pUAS-T (Brand and Perrimon, 1993). Residues deleted: NkdΔD6/GFPC 424–466; NkdΔR1SΔ30aa/GFPC 179–370, 543–572; NkdΔR1SΔD6Δ30aa/GFPC 179–370, 424–466, and 543–572; NkdΔD6Δ30aa/GFPC 424–466, 543–572. Point mutations: NkdR441A/GFPC Arg441 to Ala; NkdK445A/GFPC Lys445 to Ala; NkdRAKA/GFPC Arg441 and Lys445 to Ala. NkdΔD6-HSFNLS/GFPC substituted Nkd aa 424–466 with dHSF aa 392–435 (Genbank AAA28642); NkdΔR1SΔ30aaNLS/GFPC deleted aa 179–370 and replaced aa 543–572 with the SV40-NLS (APKKKRKVGST) (Kalderon et al., 1984). NkdD6/GFPC consisted of aa 424–466 fused to GFP. For Y2H, Importin-α1/-α2/-α3 cDNAs (Genbank AAC26055, AAA85260, and AAD37442) were amplified by PCR from a fly embryo cDNA library, sequenced, and cloned into the pAS2-1 bait and pAct2 prey plasmids (Clontech). pAS2-Nkd and pAct-Dsh constructs have been described (Rousset et al., 2001). For GST-pulldown, the following residues of each Importin-α lacking its N-terminal auto-inhibitory Importin-β-binding (IBB) domain (Harreman et al., 2003) (ΔIBB) were cloned into pGEX-4T-1 (Amersham): Importin-α1ΔIBB 118–543; Importin-α2ΔIBB 93–522; Importin-α3ΔIBB 65−514. Nkd or NkdΔD6 cDNAs were cloned into pBSKS(−) (Stratagene) for in vitro transcription/translation. For Drosophila Kc cell transfections, the D6 motif (aa 424–466), as well as NkdD6-R441A, NkdD6-K445A, and NkdD6-R441A,K445A(RAKA) point mutant constructs fused to GFP were cloned into pAc5 (Invitrogen).
Yeast two-hybrid
The pAS2-Nkd bait plasmid was transformed into the yeast strain PJ69-4A (Rousset et al., 2001). A 0–24 hour Drosophila embryo cDNA library fused to the GAL4 transcriptional activation domain was transformed into the pAS2-Nkd-carrying yeast using a variation of the lithium acetate method (Clontech). Sequencing of one of the clones whose growth on minimal medium lacking Leu, Trp, His and Ade (4D) required the presence of pAS2-Nkd plasmid revealed the complete open reading frame of Importin-α3. To confirm the interaction, competent yeast strain AH109 was transformed using the EZ Yeast Transformation II kit (ZymoResearch). Co-transformed yeast was grown under 2D (double dropout: -Leu-Trp) and, for stringent selection, 4D conditions. Western blot of yeast lysates showed that lack of growth of yeast strains under 4D were not due to bait or prey protein instability {data not shown and (Waldrop et al., 2006)}.
GST pulldown
Lysates containing each GST-fusion protein were prepared from E. coli strain BL21pLys as described by Amersham Biosciences, except that MTPBS buffer (Rousset et al., 2002) was used for washing. Each lysate was incubated with glutathione-Sepharose 4B beads for 1 hour at 4°C, and then washed 3X with DT80 buffer (Rousset et al., 2002). Nkd proteins were labeled with 35S-Methionine using the TNT T7 coupled reticulocyte lysate system (Promega) and incubated with the beads for 2 hours at 4°C in DT80 buffer. Beads were washed 4X with DT300 buffer, and labeled proteins were eluted in SDS-PAGE sample buffer, boiled for 5 minutes, and then separated by 8% SDS-PAGE. Gels were vacuum dried, and protein bands were detected using a Typhoon phosphorimager with ImageQuant software (Molecular Dynamics).
Fly stocks and genetics
Fly culture and P-element transformation were performed according to standard procedures. All fly crosses were performed at 25°C. In rescue assays, the strongest known nkd allele, nkd7H16, which encodes a predicted truncated 59 aa protein with no nkd activity (Jürgens et al., 1984; Zeng et al., 2000; Waldrop et al., 2006), was used in the following cross: UAS-Nkd/UAS-Nkd or CyO;nkd7H16/TM3-hb-lacZ X nkd7H16da-Gal4/TM3-hb-lacZ. In immunofluorescence experiments, all nkd mutant embryos were unambiguously identified by the absence of head-specific β-galactosidase staining from hb-lacZ on the balancer chromosome. Number of independent UAS lines of each construct that were examined in nkd rescue assays are as follows: GFP 1; NkdGFPC 2; NkdΔR1S/GFPC 2; NkdΔ30aa/GFPC 3; NkdΔ30aaΔR1S/GFPC 2; NkdΔ30aaNLS/GFPC 3; NkdΔR1SΔ30aaNLS/GFPC 3; NkdΔD6/GFPC 3; NkdΔD6Δ30aa/GFPC 2; NkdΔR1SΔD6/GFPC 2; NkdΔR1SΔD6Δ30aa/GFPC 2; NkdR441A/GFPC 2; NkdK445A/GFPC 2; NkdR441A,K445A/GFPC 2; NkdΔD6-HSFNLS/GFPC 3. FRT82B importin-a3D93 was provided by Robert Fleming and David Goldfarb (Mason et al., 2003). FRT82B D-axinP was provided by Jin Jiang (Hamada et al., 1999). UAS-importin-α3 RNAi flies (ID#36103 and #36104) were provided by the Vienna Drosophila RNAi Center (VDRC) (Dietzl et al., 2007). Fly stocks/chromosome: UAS-lacZ (II) (Brand and Perrimon, 1993), da-Gal4 (III) (Wodarz et al., 1995), prd-Gal4 (III) (Yoffe et al., 1995), 71B-Gal4 (III) (Brand and Perrimon, 1993).
Cuticle preparations
Cuticles were prepared and scored as described (Chan et al., 2007; Waldrop et al., 2006; Zeng et al., 2000). Briefly, each nkd cuticle was scored as “strong” if it had two or fewer complete denticle bands and severe head involution and tail defects, “moderate” if it had partial head involution and three or more complete denticle bands, or “weak” if it had denticle bands in every segment, complete or nearly complete head involution and tail morphogenesis, but patchy naked cuticle due to focal ectopic Wg production. 125–600 cuticles were scored for each rescue experiment.
Germ line clones
Females with germlines mutant for importin-α3D93 were generated by heat-shock of hs-FLP;FRT82B ovoD/FRT82B α3D93 larvae twice a day for 1.5 hours on three consecutive days at 37°C during early larval stages (Chou et al., 1993). Female progeny were crossed to α3D93/TM3-GFP males. The recovery of stage 11 embryos with uniformly elevated Arm and wide En stripes from females with germlines mutant for the Wg antagonist D-axin (Hamada et al., 1999), which lies on the same chromosomal arm as importin-α3, indicated that our procedure for making germline clones was successful (data not shown). D-axin clones were generated by heat-shock in hs-FLP; FRT82B ovoD/FRT82B D-axinP larvae. The resulting adult females were then crossed to D-axinP/TM6B males.
Computer programs
Drawings were prepared in Canvas (ACD Systems) and Powerpoint (Microsoft), and composite figures were prepared in Photoshop (Adobe). Graphs were made in Deltagraph (Red Rock Software). Selected confocal image channel spectra were maximized below saturation, with the exception that anti-Arm channels were not altered. ImageJ (NIH) was used to quantitate mean greyscale pixel intensity of raw confocal images of Arm-stained embryos as described (Waldrop et al., 2006). Statistical calculations were performed using Prism (GraphPad Software). ClustalW analysis and linked boxshade output were obtained at http://dot.imgen.bcm.tmc.edu:9331/multi-align/multi-align.html.
Immunocytochemistry and Microscopy
All embryo collection, salivary gland dissection, fixation, staining procedures (including antibodies and their dilutions), and confocal microscopy were performed as described (Chan et al., 2007; Waldrop et al., 2006). Mouse monoclonal anti-Importin-α3 5E3 (Fang et al., 2001) was kindly provided by Carl Parker.
Cell culture, dsRNA feeding, and transfections
Drosophila Kc167 (Kc) cells were grown in Schneider's Drosophila medium (Invitrogen) supplemented with 10% FBS. Wild type or point mutant pAc-NkdD6 or pArm-GFP were transfected with Effectene (Qiagen) into cells grown on coverslips. Forty-eight hours post-transfection, coverslips were fixed in 4% paraformaldehyde and counterstained with DRAQ5 (Biostatus Limited). GFP/DRAQ5 were imaged by confocal microscopy as previously described (Chan et al., 2007).
To deplete Importin-α3 in Kc cells with RNAi, 37 nM double-stranded RNA (dsRNA) – synthesized according to the manufacturer’s instructions using the MEGAscript in vitro transcription kit (Ambion) – was fed to Kc cells once per day (6 hour feeding in serum-free medium, followed by 18 hours with 10% FBS) for three consecutive days prior to transfection (Clemens et al., 2000). To synthesize dsRNA, the following primer pairs were used to PCR-amplify two independent importin-α3 DNA templates with flanking T7 polymerase promoter sequences (5’-TAATACGACTCACTATAGGG): dsRNA#1 (520 bp): 5’-(T7)TCATCACACCGACACGAACATCCT and 5’-(T7)ATCCTGGCATGAAAGGAGGTCACA; dsRNA#2 (429 bp): 5’-(T7)AGGATGAAATGCGACGTCGGAGAA and 5’-(T7)TGAGAAGTTGAAGGAAGAGCGGCA. Each importin-α3 dsRNA reduced Importin-α3 protein levels and resulted in uniform distribution of NkdD6/GFPC (Fig. 5C and data not shown).
Fig. 5.
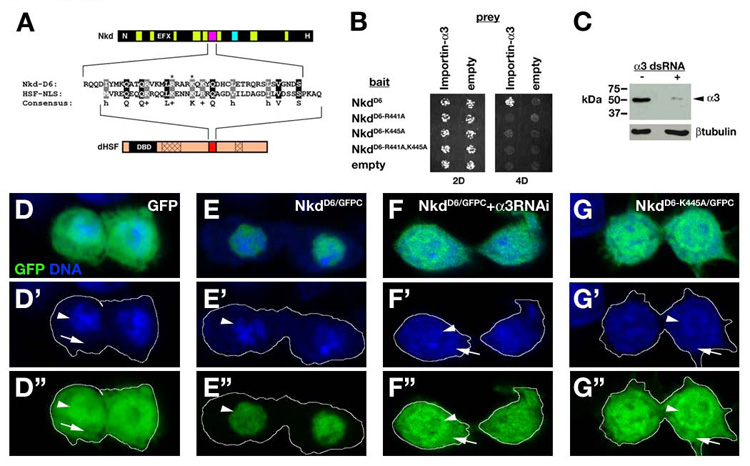
The Nkd D6 motif is an importin-α3-dependent NLS. (A) Alignment of Nkd D6 motif (magenta) and HSF-NLS (red). dHSF has a DNA binding domain (DBD) and two oligomerization domains (cross-hatch). Asterisks mark Nkd basic residues R441/K445. (B) Y2H showing that D6 motif with R441A and/or K445A mutations eliminated D6/Importin-α3 binding. (C) Western blots of Drosophila Kc cells minus (left lane) or plus (right lane) two independent importin-α3 dsRNAs (right lane) probed with anti-Importin-α3 antibody (top) or anti- βtubulin (bottom) (see Materials and methods). Quantitation of Importin-α3 bands, normalized to the βtubulin loading control, indicates a 92.5% reduction in Importin-α3 protein levels relative to the control. (D-G) Kc cells expressing GFP (D), NkdD6/GFPC (E,F) or NkdD6-K445A/GFPC (G), and imaged for GFP (green) and nucleic acids (blue). D’-G’ shows DNA channel, and D”-G” shows GFP channel, with cell outlines indicated by the white line. Peripheral nucleic acid staining corresponds to RNA in cytoplasm (arrow), while central staining (DNA) is the nucleus (arrowhead). GFP - at 27 kDa small enough to freely diffuse through nuclear pores - is uniformly distributed in nucleus and cytoplasm, while NkdD6/GFP (32 kDa – also below the nuclear pore diffusion limit of ~60 kDa) is exclusively nuclear (E-E”). Feeding either or both independent importin-α3 dsRNAs resulted in uniform localization of NkdD6/GFPC (e.g. F-F”), similar to the uniform localization of NkdD6-K445A/GFPC (G-G”). NkdD6-R441A/GFPC and NkdD6-R441A,K445A/GFPC were also uniformly localized (data not shown).
Western blots
Yeast lysates were prepared following the manufacturer’s protocol (Clontech), and Kc cell lysates were prepared as described (Chan et al., 2007), with 10 µg supernatant resolved per lane. Antibodies/dilutions were as follows: Anti-Gal4 DNA-BD sc510 (Santa Cruz Biotechnology) 1:1000; Anti-Porin (Molecular Probes) 1:1000; Anti-Importin-α3 5E3 1:1,000); Anti-βtubulin (Covance) TU27 1:2,500. Signals were visualized with SuperSignal West Dura Extended Duration Substrate (Pierce Biotechnology) followed by autoradiography.
Results
Requirement of Nkd’s Dsh binding sequences for function of a nuclear localized Nkd
In the germband-extended Drosophila embryo, Wg signaling promotes en transcription in the 2–3 adjacent rows of epidermal cells posterior to Wg-producing cells (DiNardo et al., 1988; Martinez Arias et al., 1988). In nkd mutant embryos, Wg activates en during stage 9 in further posterior cells, such that by stage 10–11 approximately half of the cells in each segment express en (Fig. 1A) (Martinez Arias et al., 1988). Production of Nkd fused to a C-terminal GFP (NkdGFPC), but not GFP alone, via the ubiquitous da-Gal4 driver in a strong nkd7H16 mutant (nkd da>NkdGFPC) narrowed the width of the En stripes - revealed by a monoclonal antibody against En - to 2–3 cells by stage 10–11, restored wild type cuticle pattern, and rescued some mutants to adulthood (Fig. 1A) (Waldrop et al., 2006; Zeng et al., 2000). Misexpression of NkdGFPC in otherwise wild type embryos had no effect on Wg signaling or segmentation, but overexpression during larval development produced adult phenotypes characteristic of wg loss-of-function (Waldrop et al., 2006; Zeng et al., 2000). The subcellular localization of NkdGFPC – as revealed by an antibody against GFP - was very similar to that of endogenous Nkd: diffuse and punctate cytoplasmic staining as well as a lower level of nuclear staining (Fig. 1A) (Waldrop et al., 2006; Zeng et al., 2000).
Fig. 1.
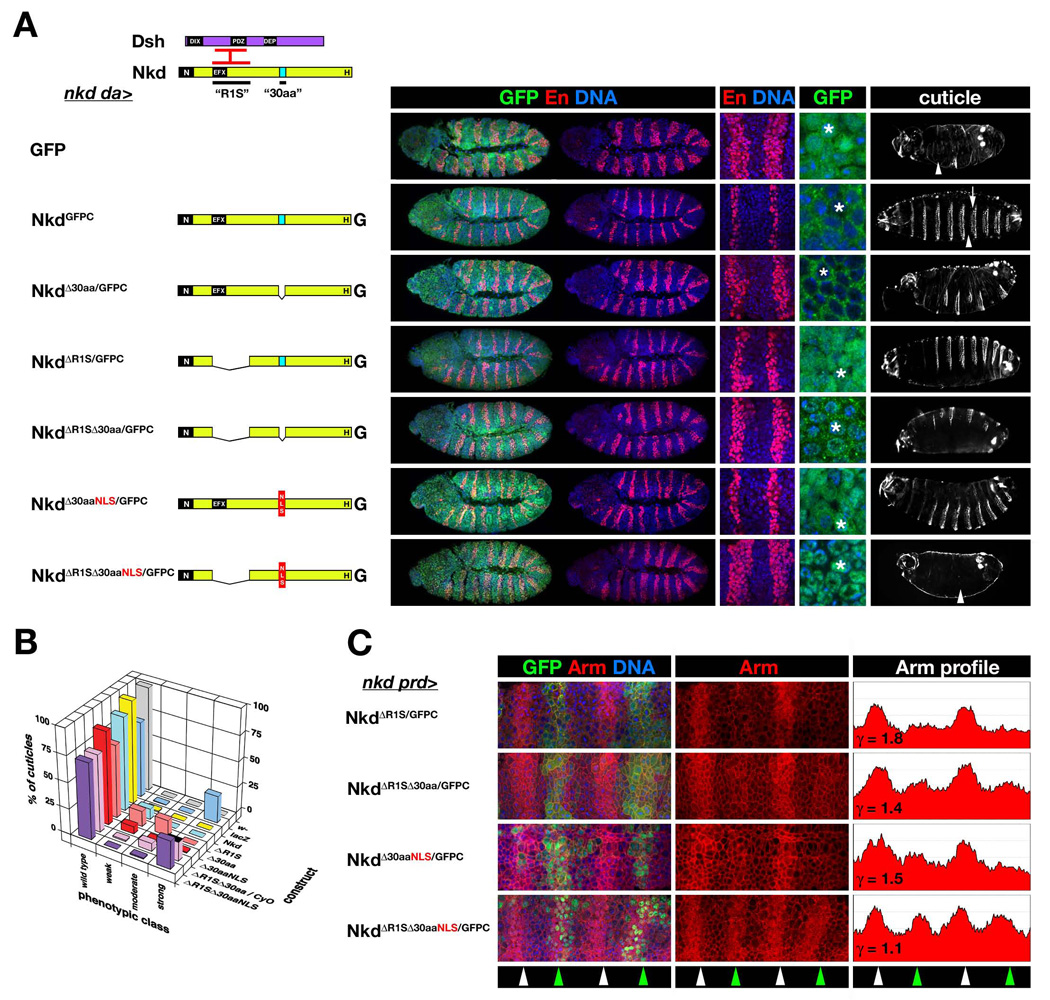
Requirement of Nkd’s Dsh-binding sequences for activity of nuclear-localized Nkd. (A) nkd rescue via ubiquitous expression of UAS-Nkd constructs driven by da-Gal4 (nkd da>). Nkd (yellow) has N-terminal (N), EFX, 30aa NLS (“30aa” - light blue), and histidine-rich (H) motifs; Dsh (purple) has DIX, PDZ, and DEP domains. Dsh basic/PDZ region binds EFX and adjacent region of Nkd (“R1S” - interacting regions in red bars). For each rescue construct is shown, from left to right, a representative stage 11 rescued embryo stained for GFP (green), En (red) and DNA (blue), with higher power images showing En stripes in two segments and GFP subcellular localization. Rescued nkd cuticle is in right column, and anterior is to the left in all columns. GFP has no rescue activity, resulting in nkd embryos with wide En stripes that occupy half of each segmental anlage, and near uniform naked cuticle (arrowhead). NkdGFPC {Nkd with C-terminal GFP (G)} restored En stripes to 2–3 cells, localized predominantly to the cytoplasm but also in epidermal nuclei (asterisk), and restored wild type denticle bands in each abdominal segment (arrow). NkdΔ30aa/GFPC only slightly narrowed nkd-mutant En stripes, was predominantly cytoplasmic during stage 11, and partly rescued the nkd cuticle phenotype, whereas NkdΔR1S/GFPC narrowed En stripes, localized predominantly to the nucleus, and restored mostly wild type cuticle pattern (Waldrop et al., 2006). NkdΔR1SΔ30aa/GFPC accessed the nucleus and did not narrow En stripes but restored some denticle bands. NkdΔR1SΔ30aaNLS/GFPC, lacking R1S and replacing the 30aa NLS with the SV40 NLS (red box), localized to the nucleus but had no rescue activity. (B) Percent of wild type and nkd cuticle phenotypes {weak, moderate, strong - described in Materials and methods)} for each nkd rescue cross with indicated UAS-Nkd construct. For lethal UAS-Nkd inserts on chromosome II, the persistence of the CyO balancer in the rescue cross results in ~12.5% of cuticles remaining unrescued and hence “strong” (black-topped bar in “strong” column). w- is negative control, with 100% wild type. da>lacZ rescue gives the expected Mendelian ratio of ~75%/25% wild type/strong nkd mutant, while da>Nkd rescued nearly all nkd mutants to wild type. NkdΔR1SΔ30aaNLS/GFPC had no activity, but NkdΔR1SΔ30aa/GFPC produced some moderate-class cuticles indicating weak Nkd activity. (C) Rescue of elevated Arm in alternate segments of nkd mutants via expression of UAS-Nkd driven by prd-Gal4 (nkd prd>). Four segmental anlagen of indicated stage 10 nkd prd>Nkd embryo stained for GFP (green), Arm (red) and DNA (blue) are shown. Since Arm/β-catenin also links E-cadherin to the actin cytoskeleton, Arm staining is a composite of plasma membrane-associated Arm and Wg-dependent accumulation of cytoplasmic and nuclear Arm. Middle column shows Arm channel, and right panel is mean greyscale pixel intensity of Arm channel along anterior/posterior axis (ordinate axis 0–200). Green arrowheads indicate segments with prd-Gal4 expression, white arrowheads alternate unrescued segments. Previously, we defined γ as the ratio of peak Arm intensity in non-rescued to rescued segments, with γ=~1.0 indicating no rescue, and for NkdGFPC γ=~1.8 (Waldrop et al., 2006). While the activity of NkdΔR1S/GFPC was comparable to NkdGFPC, the activities of NkdΔ30aa/GFPC or NkdΔR1SΔ30aa/GFPC (γ=~1.4) were reduced relative to NkdGFPC {NkdGFPC and NkdΔ30aa/GFPC are not pictured but γvalues are from (Waldrop et al., 2006)}. NkdΔR1SΔ30aaNLS/GFPC had nearly absent activity (γ=1.1) as compared to NkdΔ30aaNLS/GFPC (γ=1.5) (Waldrop et al., 2006). Images of embryos and cuticles rescued by NkdGFPC, NkdΔ30aa/GFPC, and NkdΔ30aaNLS/GFPC in panel A, as well as the nkd rescue data in panel B for NkdΔR1S/GFPC, NkdΔ30aa/GFPC, and NkdΔ30aaNLS/GFPC have been previously published and have been used with permission (Waldrop et al., 2006).
We previously showed that NkdΔR1S/GFPC, which lacks the two adjacent Dshbinding regions (termed “R1S”), retained a significant amount of nkd rescue activity but was mostly nuclear (Fig. 1A,B) (see Materials and methods for nkd cuticle scoring system) (Rousset et al., 2002; Waldrop et al., 2006). In contrast, NkdΔ30aa/GFPC, with a deleted 30aa NLS, had substantially reduced rescue activity and was predominantly cytoplasmic (Fig. 1A,B) (Waldrop et al., 2006). To determine whether the 30aa NLS is responsible for the nuclear localization of NkdΔR1S/GFPC, we deleted both regions in construct NkdΔR1SΔ30aa/GFPC. nkd da>NkdΔR1SΔ30aa/GFPC embryos retained wide En stripes (Fig. 1A) and developed a moderate to strong nkd cuticle phenotype more severe than nkd da>NkdΔ30aa/GFPC embryos (Fig. 1B) (Waldrop et al., 2006; Zeng et al., 2000). In embryos, NkdΔR1SΔ30aa/GFPC accessed the nucleus (Fig. 1A), indicating that the 30aa NLS is not necessary for nuclear localization in the absence of Nkd’s Dsh-binding sequences.
In nkd prd>NkdGFPC embryos, in which Nkd was expressed by the pair-rule gene driver prd-Gal4 in alternate segments, Arm protein was reduced to wild-type levels in rescued segments by stage 10 (Waldrop et al., 2006). NkdΔR1S/GFPC lowered Arm levels in nkd mutants (Fig. 1C) as well as did NkdGFPC (Waldrop et al., 2006), but, like the latter construct, did not reduce Arm levels in otherwise wild-type embryos (data not shown). Despite the inability of NkdΔR1SΔ30aa/GFPC to narrow nkd-mutant En stripes, nkd prd>NkdΔR1SΔ30aa/GFPC embryos had partially reduced Arm levels in alternate segments (Fig. 1C), indicating that NkdΔR1SΔ30aa/GFPC retains a low but detectable level of Nkd activity.
In embryos, Dsh is predominantly cytoplasmic but can also be observed in nuclear puncta, and Nkd/Dsh colocalization can be observed in cytoplasm, and rarely, in the nucleus (Waldrop et al., 2006; Yanagawa et al., 1995). In support of the hypothesis that Nkd acts, in part, within the nucleus, NkdΔ30aaNLS/GFPC – in which the 30aa NLS was replaced by a strong SV40 NLS – rescued nkd embryos to a greater extent than did NkdΔ30aa/GFPC (Fig. 1A,B) (Waldrop et al., 2006). If Nkd/Dsh association is important for NkdΔ30aaNLS/GFPC to antagonize Wg signaling, then deletion of Dsh binding regions (to make NkdΔR1SΔ30aaNLS/GFPC) might eliminate Nkd activity. Conversely, if NkdΔ30aaNLS/GFPC activity is Dsh-independent, then NkdΔR1SΔ30aaNLS/GFPC might retain activity. As shown in Fig. 1A–C, NkdΔR1SΔ30aaNLS/GFPC was exclusively nuclear yet had no ability to reduce Arm, narrow En stripes, or rescue nkd cuticles (Fig. 1A–C), in further support of the hypothesis that Nkd/Dsh interactions contribute to Nkd’s ability to inhibit Wg signaling.
Nkd associates with the nuclear import adaptor Importin-α 3 via a motif conserved in D. pseudoobscura
Since NkdΔR1SΔ30aa/GFPC is partly nuclear, Nkd must have NLS(s) in addition to the 30aa NLS. In a yeast-two-hybrid (Y2H) screen we recovered the nuclear import adaptor Importin-α3 as a Nkd-binding protein (see Materials and methods). The Nkd/Importin-α3 interaction is specific, because Nkd did not bind to the other two Drosophila Importins, -α1 or -α2 (Fig. 2A,B). To identify the Importin-α3-binding region of Nkd, we first assembled a putative nkd cDNA sequence from the genome of D. pseudoobscura (Dp) (Richards et al., 2005), a Drosophilid whose common ancestor with D. melanogaster (Dm) lived ~25–50 Myr ago. DpNkd is predicted to be 1009aa and Mr=111.8×103 (Fig. 2D). Dm/Dp Nkd alignment revealed nine regions of sequence similarity that included the four regions we previously noted to be conserved in mosquito Nkd (Fig. 2C,D) (Waldrop et al., 2006). Region #6 (designated “D6”) of conservation was necessary and sufficient for Importin-α3 interaction (Fig. 2E,F). The 30aa NLS - part of conserved region #7 - did not bind any of the three Importin-αs by Y2H (data not shown).
Fig. 2.
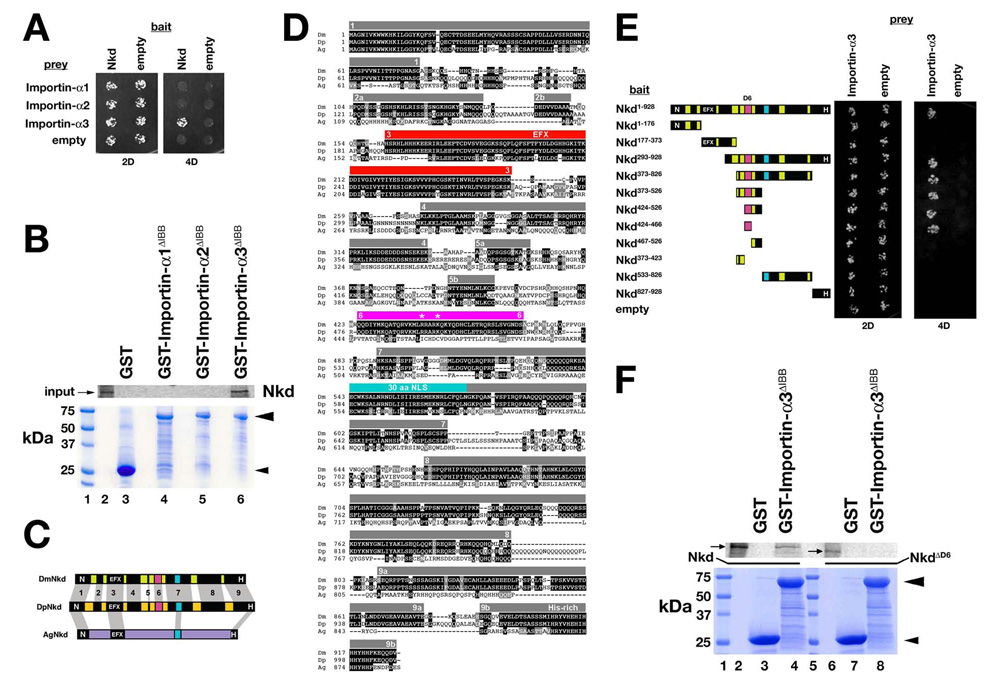
Drosophila Nkd binds Importin-α3. (A) Y2H of strains expressing indicated bait (left) and prey (top) constructs under double (2D) or quadruple (4D) dropout conditions. Only strains with Nkd+Importin-α3 plasmids grew under 4D. (B) GST-pulldown assay showing that in vitro translated Nkd was retained on Glutathione agarose bound to GST-Importin-α3ΔIBB (Importin-α3 residues 118–543 lacking the N-terminal Importin-β-binding (IBB) domain; see Materials and methods) in lane 6 but not to GST (lane 3), GST-Importin-α1ΔIBB (lane 4) or GST-Importin-α2ΔIBB (lane 5). Coomassie stained gel below confirmed that each recombinant protein (arrowheads) was bound to glutathione agarose. Size markers are in lane 1, and 35S-Methionine labeled Nkd input is in lane 2. (C) Schematic of D. melanogaster Nkd (DmNkd), D. pseudoobscura Nkd (DpNkd), and A. gambiae Nkd (AgNkd). Grey bars show sequence similarity. DmNkd/DpNkd have nine regions of similarity, numbered 1–9. Region #6 (D6; magenta) binds Importin-α3. (D) ClustalW alignment of insect Nkd proteins. Amino acid identities are dark-shaded, similarities light-shaded. Grey bars indicate regions of Dm/Dp sequence similarity (numbered). EFX–red; 30aa NLS-light blue; D6-magenta. Asterisks designate basic residues mutated in this work. (E) NkdD6 (residues #424–466) binds Importin-α3 by Y2H. Yeast expressing indicated Nkd deletion constructs were assayed for growth as in panel A. (F) GST-pulldown assay showing that in vitro translated Nkd (lanes 2–4), but not NkdΔD6 (lanes 6–8), bound to glutathione agarose to which GST-Importin-α3ΔIBB but not GST was bound. Coomassie-stained gel shows that equal amounts of each GST fusion protein were bound to the glutathione agarose. Size markers are in lanes 1 and 5, and Nkd or NkdΔD6 inputs are in lanes 2 and 6, respectively.
To investigate whether Importin-α3 is required for Nkd activity, we attempted to generate embryos that lacked Importin-α3. Importin-α3 mRNA and protein have been detected during oogenesis (Dockendorff et al., 1999; Mason et al., 2003; Mathe et al., 2000), but Importin-α3 protein only becomes detectable in the embryo after cell division cycle 12 (stage 4), correlating with the onset of responsiveness to heat shock-dependent transcription by dHSF (Fang et al., 2001). importin-α3 mRNA is also synthesized zygotically, as mutants homozygous for the null importin-α3 allele α3D93 survive until the transition between first and second larval instars (Mason et al., 2003). Stage 11 homozygous α3D93 embryos (n > 50) had En stripes indistinguishable from wild type and produced wild type cuticles (n > 50; data not shown), consistent with Importin-α3 protein translated from maternally deposited mRNA being responsible for postembryonic survival of the zygotic α3D93 mutants (Mason et al., 2003). To eliminate the maternal importin-α3 contribution, we used the ovoD technique to generate germ-line clones of the α3D93 allele (Chou et al., 1993) (see Materials and Methods); unfortunately, α3D93-mutant females (n > 30) did not lay eggs when mated with either wild type or α3D93/TM3 males, and examination of their ovarioles revealed only the ovoD phenotype. Our results suggest that importin-α3 has essential roles in oogenesis (Mathe et al., 2000), thereby precluding examination of embryos that lack maternal and zygotic importin-α3.
Importin-α3 is required for Nkd nuclear localization
Next we used RNA-interference (RNAi) to deplete Importin-α3. Expression of either of two UAS-importin-α3 RNAi lines (Dietzl et al., 2007) with da-Gal4 resulted in larval lethality, but no obvious cuticle abnormalities were observed (data not shown). Similarly, prd-Gal4-driven expression of UAS-importin-α3 RNAi did not affect En, Arm, or Importin-α3 staining in stage 10–11 embryos (data not shown), the latter perhaps due to an inability of zygotically synthesized double-stranded RNA produced by the snapback construct to fully deplete maternal and zygotic importin-α3 mRNA stores. We therefore examined the effect of Importin-α3 depletion on NkdGFPC distribution in postembryonic cells. When synthesized in larval salivary gland, NkdGFPC is detected at low levels at the plasma membrane and cytoplasm and at higher levels in cytoplasmic/perinuclear puncta and in the nucleus (Fig. 3A). Importin-α3 is also enriched in salivary gland nuclei, in the presence or absence of NkdGFPC (Fig. 3A’ and data not shown). However, co-expression of NkdGFPC and UAS-importin-α3 RNAi in salivary gland (see Materials and methods) dramatically reduced Importin-α3 immunoreactivity and resulted in complete exclusion of NkdGFPC from the nucleus (Fig. 3B-B”). These data demonstrate that Importin-α3 is required for nuclear localization of Nkd.
Fig. 3.
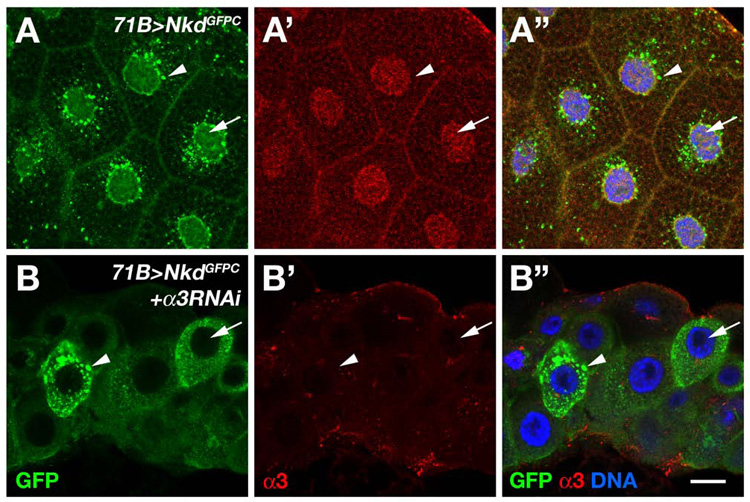
Nkd requires importin-α3 for nuclear localization. (A-A”) 71B>NkdGFPC third instar salivary gland stained with Importin-α3 antibody (red channel in panel A’) and imaged for GFP (green channel in panel A) and DNA (blue in merged image of panel A”). NkdGFPC localizes to the plasma membrane, in cytoplasmic/perinuclear puncta (arrowhead), and diffusely in the nucleus (arrow), whereas Importin-α3 is enriched in the nucleus. (B-B”) 71B>NkdGFPC+importin-α3 RNAi salivary gland stained as in A. Note that NkdGFPC is completely excluded from nuclei (arrow) but cytoplasmic puncta (arrowhead) remain, and that Importin-α3 immunoreactivity is reduced to absent in nuclei. Scale bar in B”: 30 µm in A-A”; 18 µm in B-B”
D6 motif is required for nuclear localization and function of Nkd
Next we examined the rescue activity of NkdΔD6/GFPC, a mutant construct that lacks the Importin-α3-binding motif. Stage 11 nkd da>NkdΔD6/GFPC embryos had En stripes of intermediate width (~3–5 cells) and produced weak and moderate-class nkd cuticles (Fig. 4A,B), indicating that D6 motif is required for full Nkd activity. The subcellular localization of NkdΔD6/GFPC was very similar to NkdGFPC, likely due to the intact 30aa NLS in the latter construct (Fig. 4A). However, in contrast to NkdGFPC, NkdΔD6/GFPC was predominantly cytoplasmic in larval salivary glands (cf. Fig. 3A and Fig 4D). NkdΔD6/GFPC reduced Arm levels in nkd mutants as well as did NkdGFPC (Fig. 4C), which is perhaps not surprising given the sufficiency of Nkd N-terminal and 30aa motifs, when substituted for the homologous regions of mouse Nkd1, for significant Nkd rescue activity (Chan et al., 2007).
Fig. 4.
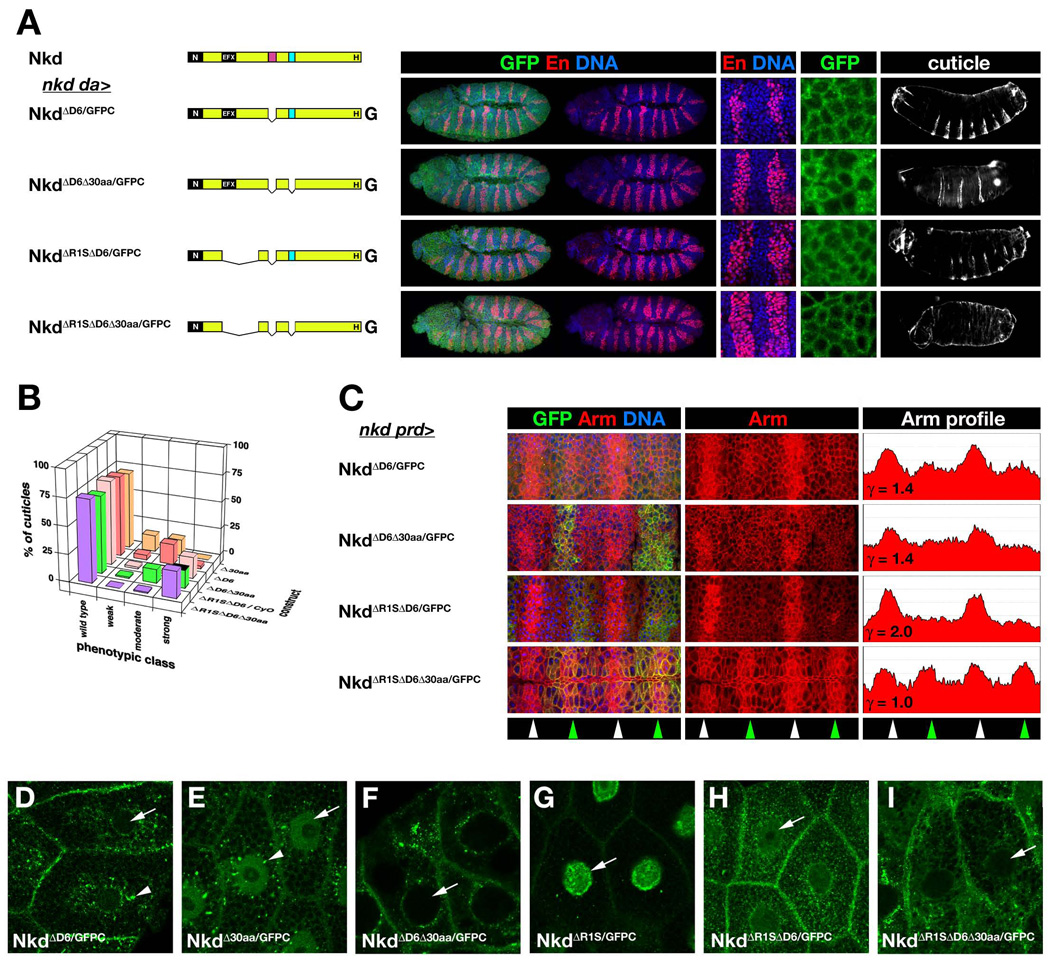
D6 is required for Nkd function and nuclear localization. (A) nkd da>NkdGFP rescue and localization as in Fig. 1A. (B) Distribution of wild type and nkd cuticle phenotypes for rescue by indicated constructs as in Fig. 1B. (C) Rescue of elevated Arm in alternate segments of nkd mutants via prd-Gal4 (nkd prd>) as in Fig. 1C (ordinate axis 0–250 pixels). (D–I) 71B>NkdGFP third instar salivary gland imaged for GFP. NkdΔD6/GFPC (D) is in perinuclear aggregates (arrowhead) but has reduced nuclear localization (arrow) compared to NkdGPFC in Fig. 3A. NkdΔ30aa/GFPC (E) localizes to cytoplasm and nucleus in a distribution very similar to NkdGFPC, but NkdΔD6Δ30aa/GFPC (F) is excluded from nuclei. NkdΔR1S/GFPC (G) is almost exclusively nuclear, with enrichment at the nuclear membrane (arrow). NkdΔR1SΔD6/GFPC (H) exhibits reduced nuclear localization relative to NkdΔR1S/GFPC, while NkdΔR1SΔD6Δ30aa/GFPC (I) is excluded from the nucleus.
Next we deleted both NLSs (NkdΔD6Δ30aa/GFPC). Although embryos rescued with either NkdΔ30aa/GFPC or NkdΔD6Δ30aa/GFPC had wide En stripes and intermediate levels of Arm, cuticles rescued by the former construct were mostly weak and moderate nkd, while the majority of cuticles rescued by the latter construct were strong nkd, indicating that each NLS contributes to Nkd function (Fig. 4A–C) (Waldrop et al., 2006). In contrast to the marked difference in NkdGFPC vs. NkdΔ30aa/GFPC localizations during embryonic stage 11 {Fig. 1A and (Waldrop et al., 2006)}, NkdΔ30aa/GFPC localized to larval salivary gland nuclei to the same extent as did NkdGFPC (cf. Fig. 1A and Fig. 4E). In contrast, NkdΔD6Δ30aa/GFPC was excluded from embryonic and salivary gland nuclei (Fig. 4A,F), indicating that each NLS makes distinct contributions to nuclear localization in embryonic vs. postembryonic stages.
We previously showed that two constructs that lack the EFX motif (NkdΔEFX/GFPC and NkdΔR1S/GFPC) had substantial nkd rescue activity but localized predominantly to embryonic nuclei, consistent with Dsh or other EFX-binding proteins anchoring Nkd in the cytoplasm (Waldrop et al., 2006). We assessed the requirement of the D6 motif for NkdΔR1S/GFPC activity/localization by deleting D6 to make NkdΔR1SΔD6/GFPC. As shown in Fig. 4A–C, NkdΔR1SΔD6/GFPC reduced Arm levels but did not narrow En stripes and gave rise to moderate and strong nkd cuticles. As in the embryo, NkdΔR1S/GFPC was predominantly nuclear in salivary gland (Fig. 4G), but NkdΔR1SΔD6/GFPC localized at the cell membrane and in the cytoplasm, with low levels in the nucleus (Fig. 4H), indicating that the D6 motif contributes to NkdΔR1S/GFPC nuclear localization. NkdΔR1SΔD6Δ30aa/GFPC, lacking Dsh-binding sequences and both NLSs, had no Nkd activity in any of the assays and was cytoplasmic in embryos and in salivary gland, indicating that the D6 motif is necessary for both the nuclear localization and weak activity of NkdΔR1SΔ30aa/GFPC (Fig. 4A–C,I).
The Nkd D6 NLS is similar to the dHSF Importin-α3-binding NLS
The three Drosophila Importin-αs serve both unique and shared roles in development {reviewed by (Goldfarb et al., 2004)}, but sequence features that confer specificity between NLSs and each Importin-α are not known. In Drosophila, Importin-α3 has been shown to bind DNA polymerase-α 180 (DNApol-α180) (Mathe et al., 2000), the transcriptional regulator Germ-cell-less (Gcl) (Dockendorff et al., 1999), and Heat-Shock Transcription Factor (dHSF) (Fang et al., 2001). We observed no obvious sequence similarity between the Nkd D6 motif and DNApol-α180 or Gcl, with the exception of a few basic residues characteristic of NLSs (data not shown), but sequence alignment with dHSF-NLS revealed 33% (13/39 amino acids) similarity (Fig. 5A). Importantly, basic residues critical for dHSF/Importin-α3 binding and dHSF nuclear localization are also basic in similar positions of the Nkd-D6 motif (Fang et al., 2001; Zandi et al., 1997). Mutation of either or both of the Nkd-D6 basic residues to alanine (R441A, K445A) eliminated the D6/Importin-α3 interaction by Y2H (Fig. 5B). D6 is also sufficient to function as a NLS: when transiently produced in Drosophila Kc cells, the D6 motif fused to GFP (NkdD6/GFPC) was exclusively nuclear, in contrast to GFP which was uniformly distributed in nucleus and cytoplasm {cf. Fig. 5D,E; at 27 kDa, GFP is smaller than the size – typically ~60 kDa or less - at which protein diffusion between nucleus and cytoplasm is restricted by the nuclear pore (Wang and Brattain, 2007)}. RNAi-mediated depletion of Importin-α3 (Fig. 5C; see Materials and methods) resulted in a uniform distribution of NkdD6/GFPC similar to GFP or each D6 point mutant construct fused to GFP (Fig. 5F,G and data not shown). Thus, D6 basic residues required for Importin-α3 interaction are also required for the exclusive nuclear localization of NkdD6/GFPC.
To test the biological significance of the basic residues required for Nkd/Importin-α3 interaction, we introduced the point mutations into NkdGFPC. Point mutation of either or both conserved D6 basic residues eliminated the Nkd/Importin-α3 interaction, and D6 deletion did not affect Nkd/Dsh interactions by Y2H (Fig. 6A). As shown in Fig. 6B, En stripes in embryos rescued by each point mutant construct were ~1–3 cells wider than those rescued by wild type Nkd, with focal areas of extreme widening indicative of ectopic Wg synthesized by further posterior cells (Chan et al., 2007). Consequently, the majority of nkd embryos rescued by each point mutant construct developed a moderate nkd cuticle phenotype similar to that of several lethal nkd alleles with nonsense mutations in the vicinity of the D6 motif (Fig. 6B,C) (Waldrop et al., 2006). Although the subcellular distributions of each point mutant construct appeared similar to that of NkdGFPC in stage 10–11 embryos (Fig. 6B; like NkdΔD6/GFPC presumably due to an intact 30 aa NLS), each point mutant construct was excluded from third instar salivary gland nuclei to the same extent as NkdΔD6/GFPC (data not shown). The inability of the point mutant constructs to rescue nkd mutants was not due to their reduced expression levels relative to NkdGFPC or to an early defect each construct’s ability to lower Arm levels, because each of the three point mutant constructs lowered Arm levels during stage 10 to an extent comparable to NkdGFPC (Fig. 6D).
Fig. 6.
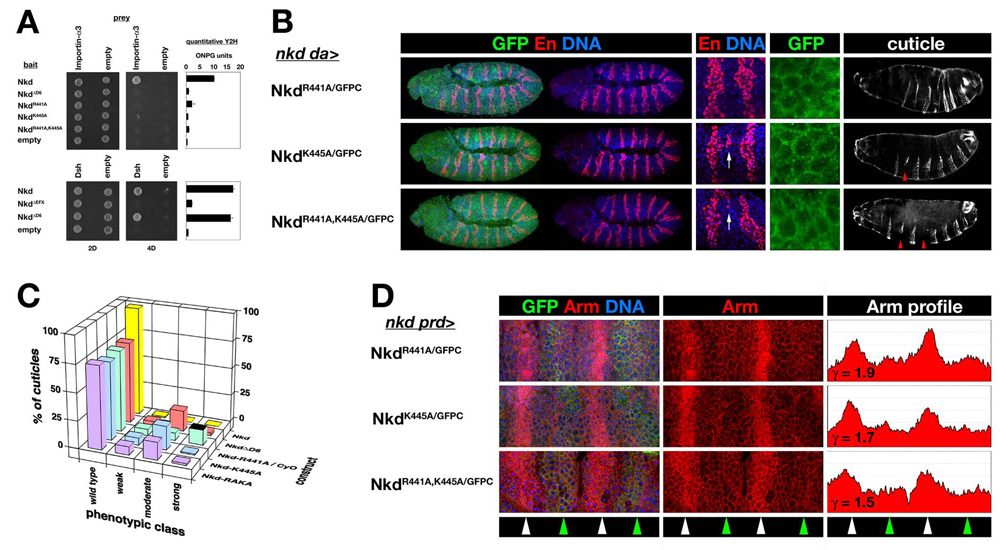
D6 NLS basic residues are required for Nkd function. (A) Y2H showing that Nkd R441A and/or K445A mutations disrupted Nkd/Importin-α3 interactions, but unlike EFX deletion, D6 deletion did not affect the Nkd/Dsh interaction. Quantitative Y2H assay is depicted in bar graphs on the right. (B) nkd da>NkdGFP rescue and localization as in Fig. 5A. Note that rescue by each point mutant construct results in En stripes widened by 1–3 cells relative to embryos rescued by NkdGPFC (cf. Fig. 1A), with focally ectopic En indicative of ectopic Wg production in cells just posterior to the En+ cells (arrow) (Chan et al., 2007), and moderate-class rescued nkd cuticle with deleted denticle bands (arrowheads). Each point mutant protein localized in a pattern – cytoplasmic and weakly nuclear – similar to wild type NkdGFPC (cf. Fig. 1A). (C) Distribution of wild type and nkd cuticle phenotypes for rescue by indicated NkdGFPC point mutant constructs as in Fig. 1B. (D) Representative stage 10 nkd prd>NkdGFP (point mutant) embryos stained for Arm as in Fig. 1C (ordinate axis=0–250 pixels). Note that each construct lowered Arm to an extent comparable to NkdGFPC (γ=~1.8) (Waldrop et al., 2006).
dHSF NLS restores Importin-α3 binding and nuclear localization but not function to Nkd
If Importin-α3-dependent nuclear import is necessary for Nkd function, then replacing the D6-NLS with the dHSF-NLS (to make the construct NkdΔD6-HSFNLS/GFPC) might restore nuclear localization and function to NkdΔD6/GFPC. The dHSF-NLS, either by itself or when placed in NkdΔD6, bound to Importin-α3, and replacement of the Nkd D6 motif with the dHSF-NLS did not affect Nkd/Dsh interactions by Y2H (Fig. 7A). Although NkdΔD6-HSFNLS/GFPC entered the nucleus and reduced Arm levels of stage 10 nkd mutants, by stage 11 the rescued embryos had wide En stripes and gave rise to moderate-strong nkd cuticles (Fig. 7B–E), indicating that Importin-α3-dependent nuclear localization is not sufficient to restore NkdΔD6/GFPC activity. We hypothesize that the dHSF-NLS compromises NkdΔD6/GFPC activity through its mislocalization in the nucleus, perhaps similar to that observed when Nkd’s N-terminal membrane-anchoring motif was deleted (Chan et al., 2007).
Fig. 7.
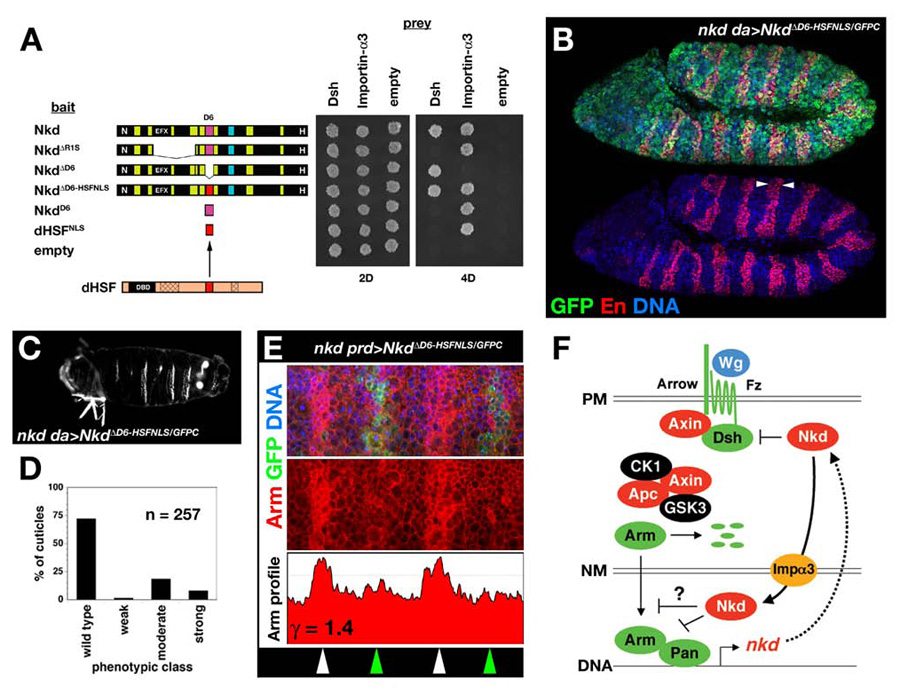
Restoration of Importin-α3-association by the dHSF-NLS does not restore Nkd function. (A) Y2H showing that replacement of the D6-NLS (magenta) with the dHSF-NLS (red) restored the Nkd/Importin-α3 interaction but did not affect Nkd/Dsh interactions. (B) Stage 11 nkd da>NkdΔD6-HSFNLS/GFPC stained for GFP (green), En (red), and DNA (blue) with wide En stripes (arrowheads). (C) Representative nkd da>NkdΔD6-HSFNLS/GFPC cuticle, with 3 complete denticle belts indicative of moderate-class nkd cuticle. (D) Distribution of nkd cuticle phenotypes for NkdΔD6-HSFNLS/GFPC rescue cross (n=number of cuticles scored). (E) Stage 10 nkd prd>NkdΔD6-HSFNLS/GFPC embryo stained for GFP (green), Arm (red), and DNA (blue) in top panel, with Arm-only channel and Arm greyscale intensity profile (ordinate axis 0–200 pixels) in middle and lower panels, similar to Fig. 1C. (F) Model for fly Nkd function. Wg signaling at the plasma membrane (PM) leads to Arm/Pan-dependent transcription of target genes, including nkd (see Introduction for details). Nkd targets Dsh in the cytoplasm and/or plasma membrane, and binds Importin-α3 to traverse the nuclear membrane (NM). In the nucleus, whether Nkd inhibits nucleocytoplasmic transport of Arm or another molecule, or whether Nkd directly regulates target gene transcription, remains unknown.
Discussion
A growing body of evidence indicates that the traditionally “cytoplasmic” Wnt signal transducers Axin, Apc, and Dsh also act in the nucleus {reviewed by (Willert and Jones, 2006)}. In the cytoplasm, Apc promotes β-catenin degradation and regulates the cytoskeleton, but nuclear Apc can recruit transcriptional corepressors to Wnt target genes and, like Axin, escort β-catenin from the nucleus {(Cong and Varmus, 2004; Sierra et al., 2006; Wiechens et al., 2004); reviewed in (Aoki and Taketo, 2007)}. In light of recent evidence that Axin/Dsh oligomers crosslink Wnt-bound receptors at the plasma membrane during signal activation (Bilic et al., 2007), it remains unclear whether the nuclear roles for Axin or Dsh are similar to their cytoplasmic functions or whether they, like Apc, have novel nuclear functions.
Nkd is also a conserved Wnt signal regulator whose subcellular localization initially suggested a cytoplasmic site of action (Zeng et al., 2000). Our studies have shown that fly Nkd is composed of discrete motifs that confer membrane localization and binding to Dsh, as well as two NLSs {(Chan et al., 2007) and this work}. In addition to Nkd targeting an uncharacterized fraction of Dsh in the cytoplasm and/or at the plasma membrane, our data strongly support a nuclear role for Nkd, but whether Nkd inhibits Wnt signaling by altering nucleo-cytoplasmic transport of critical signaling components, such as Dsh or Arm, or by acting on the chromatin of Wnt target genes remains to be elucidated (Fig. 7F).
Genetic epistasis can be a powerful method to infer the regulatory logic of signal transduction cascades. Because Nkd is a “side-regulator” whose loss-of-function phenotype is dependent on intact Wg signaling, double-mutants between nkd and dsh or arm did not help us to discern at which level Nkd inhibits the linear Wg signaling pathway (Rousset et al., 2001). However, Nkd overexpression suppressed the gain-of-Wg signaling phenotype caused by overexpression of Dsh but not that caused by overexpression of an N-terminally deleted and hence degradation-resistant Arm/β-catenin (Pai et al., 1997; Rousset et al., 2001; Waldrop et al., 2006); taken together with the observation that Nkd binds Dsh, our previous epistasis experiments allowed us to conclude that Nkd acted at the level of Dsh and not “downstream” of Arm/β-catenin in Wg signaling (Rousset et al., 2001). However, in view of the present data, Nkd might also act in the nucleus at or above the level of Arm/β-catenin. Unfortunately, overproduction of wild type Arm is without phenotypic consequence (Pai et al., 1997), presumably because of an excess capacity of the β-catenin “destruction complex” to degrade ectopic Arm, thus preventing us from making further conclusions at present about the epistatic relationship between Nkd and endogenous, degradation-sensitive Arm/β-catenin.
Despite the deletion of Dsh-binding sequences in otherwise wild type Nkd having only a minor effect on cuticle rescue activity (Waldrop et al., 2006), the present experiments further support the hypothesis that the Nkd/Dsh interaction is important for Nkd to inhibit Wg signaling. However, our experiments thus far do not clarify how the interaction is regulated in vivo or whether it must occur in the cytoplasm, nucleus, or both locations. Nevertheless, several lines of evidence indicate that a Nkd/Dsh interaction in the cytoplasm and/or near the plasma membrane is important for Nkd function: First, both proteins are predominantly cytoplasmic and/or membrane-associated (Axelrod, 2001; Chan et al., 2007; Waldrop et al., 2006; Yanagawa et al., 1995). Second, punctate cytoplasmic Nkd/Dsh colocalization can be observed in embryos and in salivary gland (Rousset et al., 2001; Waldrop et al., 2006). Third, the Dsh-binding EFX motif fused to GFP was predominantly cytoplasmic (Waldrop et al., 2006). Fourth, deletion of Dsh-binding sequences in Nkd promoted nuclear localization, consistent with Dsh anchoring Nkd in the cytoplasm (Waldrop et al., 2006). Fifth, deletion of both Nkd NLSs eliminated nuclear localization whether or not Dsh-binding sequences were present, but Dsh-binding sequences were required for Nkd activity.
How Nkd acts on Dsh in the cytoplasm to inhibit Wg signaling is not known. One possibility is that Nkd sequesters Dsh away from Fz and/or Axin during signal activation, freeing Axin to regenerate β-catenin destruction complexes. Alternatively, Nkd might target “activated” Dsh, possibly the pool of Dsh bound to the Wnt receptor complex, for degradation; consistent with Nkd targeting only a fraction of Dsh is the minimal colocalization of the two proteins in embryos as well as the lack of any obvious changes in Dsh levels in nkd mutants (Waldrop et al., 2006). Since Nkd can block the gain-of-Wg signaling phenotypes caused by overexpression of the Dsh kinase CK1 (Zhang et al., 2006), a third possibility is that Nkd blocks CK1-dependent phosphorylation of Dsh via a steric mechanism, although the relationship between Dsh phosphorylation status and activity remains unclear. Future experiments should clarify this issue, because each of these hypotheses makes distinct predictions about phosphorylation status and associated proteins in a Nkd/Dsh complex.
Our studies also provide several lines of evidence that the Nkd/Dsh interaction is not sufficient for Nkd to inhibit Wg signaling (Rousset et al., 2001; Waldrop et al., 2006), and that binding in the nucleus might also be required to fully antagonize Wg signaling. First, the Dsh-binding regions of Nkd when overexpressed blocked phenotypes induced by Dsh overexpression but had no nkd rescue activity (Waldrop et al., 2006). Second, (fly) Nkd and (vertebrate) Dsh have NLSs, although it is not yet known whether fly Dsh acts in the nucleus. Third, rare punctate Nkd/Dsh nuclear colocalization can be observed by confocal microscopy in fly embryos (Waldrop et al., 2006). Fourth, the SV40-NLS increased NkdΔ30aa/GFPC activity when Dsh-binding sequences were intact but reduced activity when Dsh-binding sequences were deleted {(Waldrop et al., 2006) and this study}. We cannot rule out the possibility that the activity of NkdΔ30aaNLS/GFPC, some of which remains outside the nucleus despite the strong heterologous NLS, is due to cytoplasmic Nkd/Dsh interactions. Similarly, NkdΔR1SΔ30aaNLS/GFPC, which was exclusively nuclear in embryos, might lack activity because of its inability to bind and be retained by Dsh in the cytoplasm. While one must be cautious when inferring site(s) of protein action from subcellular localizations, our studies collectively suggest that fly Nkd is required at multiple locations in Wg-receiving cells.
The Nkd-D6 motif has been subject to intense selection pressure, as it is identical in Nkd from D. pseudoobscura, a fly species that diverged from D. melanogaster approximately one billion generations ago. Similarly, the 30aa NLS is part of a 58 aa motif, and the EFX is part of a 91 aa motif, which are also identical in the two Drosophila species. Using Y2H, we have identified Nkd-EFX residues that are either dispensable or critical for NkdEFX/DshbPDZ interactions (K.W. and C.-C. C., unpublished data), suggesting that interactions between the EFX motif and proteins other than Dsh might enforce strict motif conservation. Although each NLS contributes to Nkd activity and nuclear localization, heterologous NLSs did not fully replace the function of each Nkd NLS in rescue assays, and in both cases in this work a heterologous NLS was deleterious to protein function. Absolute conservation of each of these motifs implies that both the tertiary structure and every square angstrom of each motif’s surface are necessary for species survival. Taken together with our previous work (Chan et al., 2007; Waldrop et al., 2006), our experiments also suggest that each Nkd motif is required for distinct thresholds and/or duration of Wg signal inhibition: the N-terminal and 30 aa motifs were required for reduction of Arm levels by stage 10, whereas the Dsh-binding EFX and Importin-α3-binding D6 motifs were dispensable for Arm reduction but were required, either directly or indirectly, to fully repress en and/or wg transcription by stage 11. Since the deletion of two highly conserved motifs (EFX and D6) preserved the mutant Nkd protein’s ability to reduce Arm levels during stage 10, it seems unlikely that these motifs will be shown to possess an intrinsic catalytic activity. We therefore favor the hypothesis that Nkd acts as an inducible protein scaffold, with each of the conserved motifs able to bind additional protein(s). Perhaps there exist distinct Nkd-complexes depending on the subcellular compartment, state of signal activation, or time following signal initiation.
Alignment of the Importin-α3-binding NLSs in Nkd and dHSF revealed several conserved residues. Interestingly, the dHSF-NLS has been shown to be bifunctional, suppressing dHSF trimerization in the absence of heat shock, and in response to heat or other stress conferring Importin-α3-dependent dHSF nuclear translocation and transcriptional induction of heat-responsive genes such as hsp70 (Voellmy, 2004; Wu, 1995; Zandi et al., 1997). Our data suggest that the Nkd D6-NLS is also bifunctional, conferring Importin-α3-dependent nuclear localization as well as possibly binding nuclear protein(s) that repress Wg target gene transcription in some cells through stages 10–11. While non-import – presumably scaffolding - functions for Importin-αs have been inferred from phenotypes observed with importin-α deficiency in flies and worms {reviewed in (Goldfarb et al., 2004)}, all of our experiments support the hypothesis that the Nkd/Importin-α3 interaction promotes nuclear localization. The central region of Importin-α consists of ten alpha-helical “Arm” repeats - so named because they were first identified in the Drosophila Arm protein (Peifer et al., 1994) - stacked to form a banana-shaped molecule, the concave side of which harbors a groove that binds basic residues within NLSs {reviewed in (Lange et al., 2007)}. At present, it is not possible based on primary sequence to predict which Importin-α a given NLS will bind, although both the NLS and its three dimensional context (i.e. adjacent sequence) have been demonstrated to contribute to NLS/Importin-α specificity (Friedrich et al., 2006). Future experiments will investigate whether the residues conserved between Nkd and dHSF represent a consensus Importin-α3-specific binding motif.
Vertebrate Nkds have a conserved 30aa motif between the EFX and C-terminal histidine-rich regions (Waldrop et al., 2006), but whether the vertebrate proteins act in the nucleus like fly Nkd is not known. In this regard, we observed no obvious difference between the subcellular localizations of mouse Nkd1 fused to C-terminal GFP (mNkd1GFPC) vs. a similar construct that lacks the 30aa motif (mNkd1Δ30aa/GFPC) when either protein was produced in cultured mammalian cells (T. Cagatay and K. W., unpublished data) (Waldrop et al., 2006). However, it deserves mention that we also observed no obvious difference between fly Nkd vs. NkdΔ30aa localizations in Drosophila S2 cells (T. Cagatay and K. W., unpublished data), but the differences in localization and function of these two constructs when produced in nkd mutant embryos were dramatic. Our findings illustrate the importance of investigating the subcellular localizations of mutant proteins in a native environment that lacks the endogenous wild-type protein. It might therefore be interesting to examine the subcellular localization of vertebrate Nkd constructs in nkd-mutant mice or zebrafish just as we have done in Drosophila. More importantly, future experiments must address the critical question of how Nkd antagonizes Wnt/β-catenin signaling in each of the compartments to which it localizes.
Acknowledgements
Importin-α3 was identified as a Nkd binding partner by R.R. in the laboratory of Matthew Scott, whom K.W. acknowledges for generosity and encouragement. Thanks to Tolga Cagatay, Beatriz Fontoura, and Yuh Min Chook for advice and discussion of Importin-αs; Robert Fleming, David Goldfarb, Jin Jiang, Joseph Park and the Vienna RNAi center for fly stocks; Joseph Park for advice regarding the ovoD technique; Carl Parker for anti-Importin-α3 antibody and Nipam Patel for anti-En antibody; and Tolga Cagatay, Yuh Min Chook, Robert Fleming, Beatriz Fontoura, Jin Jiang, and Duojia Pan for comments on the manuscript. The Genetics Society of America has granted permission to reproduce previously published panels as described in Fig. 1. K.W. is a W.W. Caruth, Jr. Scholar in Biomedical Research at UT Southwestern and acknowledges support by NIH R-01-GM065404.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aoki K, Taketo MM. Adenomatous polyposis coli (APC): a multifunctional tumor suppressor gene. J Cell Sci. 2007;120:3327–3335. doi: 10.1242/jcs.03485. [DOI] [PubMed] [Google Scholar]
- Axelrod JD. Unipolar membrane association of Dishevelled mediates Frizzled planar cell polarity signaling. Genes Dev. 2001;15:1182–1187. doi: 10.1101/gad.890501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker NE. Localization of transcripts from the wingless gene in whole Drosophila embryos. Development. 1988;103:289–298. doi: 10.1242/dev.103.2.289. [DOI] [PubMed] [Google Scholar]
- Bejsovec A, Martinez Arias A. Roles of wingless in patterning the larval epidermis of Drosophila. Development. 1991;113:471–485. doi: 10.1242/dev.113.2.471. [DOI] [PubMed] [Google Scholar]
- Bejsovec A, Wieschaus E. Segment polarity gene interactions modulate epidermal patterning in Drosophila embryos. Development. 1993;119:501–517. doi: 10.1242/dev.119.2.501. [DOI] [PubMed] [Google Scholar]
- Bilic J, Huang YL, Davidson G, Zimmermann T, Cruciat CM, Bienz M, Niehrs C. Wnt induces LRP6 signalosomes and promotes dishevelled-dependent LRP6 phosphorylation. Science. 2007;316:1619–1622. doi: 10.1126/science.1137065. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Chan C-C, Zhang S, Cagatay T, Wharton KA., Jr Cell-autonomous, myristyl-independent activity of the Drosophila Wnt/Wingless antagonist Naked cuticle (Nkd) Dev Biol. 2007;311:538–553. doi: 10.1016/j.ydbio.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chook YM, Blobel G. Karyopherins and nuclear import. Curr Opin Struct Biol. 2001;11:703–715. doi: 10.1016/s0959-440x(01)00264-0. [DOI] [PubMed] [Google Scholar]
- Chou TB, Noll E, Perrimon N. Autosomal P[ovoD1] dominant female-sterile insertions in Drosophila and their use in generating germ-line chimeras. Development. 1993;119:1359–1369. doi: 10.1242/dev.119.4.1359. [DOI] [PubMed] [Google Scholar]
- Clemens JC, Worby CA, Simonson-Leff N, Muda M, Maehama T, Hemmings BA, Dixon JE. Use of double-stranded RNA interference in Drosophila cell lines to dissect signal transduction pathways. Proc Natl Acad Sci U S A. 2000;97:6499–6503. doi: 10.1073/pnas.110149597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H. Wnt/β-Catenin Signaling in Development and Disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- Cong F, Varmus H. Nuclear-cytoplasmic shuttling of Axin regulates subcellular localization of β-catenin. Proc Natl Acad Sci U S A. 2004;101:2882–2887. doi: 10.1073/pnas.0307344101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, Couto A, Marra V, Keleman K, Dickson BJ. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- DiNardo S, Sher E, Heemskerk-Jongens J, Kassis JA, O'Farrell PH. Two-tiered regulation of spatially patterned engrailed gene expression during Drosophila embryogenesis. Nature. 1988;332:604–609. doi: 10.1038/332604a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dockendorff TC, Tang Z, Jongens TA. Cloning of karyopherin-α3 from Drosophila through its interaction with the nuclear localization sequence of germ cell-less protein. Biol Chem. 1999;380:1263–1272. doi: 10.1515/BC.1999.161. [DOI] [PubMed] [Google Scholar]
- Dougan S, DiNardo S. Drosophila wingless generates cell type diversity among engrailed expressing cells. Nature. 1992;360:347–350. doi: 10.1038/360347a0. [DOI] [PubMed] [Google Scholar]
- Fagotto F, Gluck U, Gumbiner BM. Nuclear localization signal-independent and importin/karyopherin-independent nuclear import of β-catenin. Curr Biol. 1998;8:181–190. doi: 10.1016/s0960-9822(98)70082-x. [DOI] [PubMed] [Google Scholar]
- Fang X, Chen T, Tran K, Parker CS. Developmental regulation of the heat shock response by nuclear transport factor karyopherin-α3. Development. 2001;128:3349–3358. doi: 10.1242/dev.128.17.3349. [DOI] [PubMed] [Google Scholar]
- Friedrich B, Quensel C, Sommer T, Hartmann E, Kohler M. Nuclear localization signal and protein context both mediate importin α specificity of nuclear import substrates. Mol Cell Biol. 2006;26:8697–8709. doi: 10.1128/MCB.00708-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldfarb DS, Corbett AH, Mason DA, Harreman MT, Adam SA. Importin α: a multipurpose nuclear-transport receptor. Trends Cell Biol. 2004;14:505–514. doi: 10.1016/j.tcb.2004.07.016. [DOI] [PubMed] [Google Scholar]
- Hamada F, Tomoyasu Y, Takatsu Y, Nakamura M, Nagai S, Suzuki A, Fujita F, Shibuya H, Toyoshima K, Ueno N, Akiyama T. Negative regulation of Wingless signaling by D-axin, a Drosophila homolog of axin. Science. 1999;283:1739–1742. doi: 10.1126/science.283.5408.1739. [DOI] [PubMed] [Google Scholar]
- Harreman MT, Hodel MR, Fanara P, Hodel AE, Corbett AH. The auto-inhibitory function of importin α is essential in vivo. J Biol Chem. 2003;278:5854–5863. doi: 10.1074/jbc.M210951200. [DOI] [PubMed] [Google Scholar]
- Itoh K, Brott BK, Bae GU, Ratcliffe MJ, Sokol SY. Nuclear localization is required for Dishevelled function in Wnt/β-catenin signaling. J Biol. 2005;4:3. doi: 10.1186/jbiol20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jürgens G, Wieschaus E, Nüsslein-Volhard C, Kluding H. Mutations affecting the pattern of the larval cuticle in Drosophila melanogaster. II. Zygotic loci on the third chromosome. Wilhelm Roux Arch. Dev. Biol. 1984;193:283–295. doi: 10.1007/BF00848157. [DOI] [PubMed] [Google Scholar]
- Kalderon D, Roberts BL, Richardson WD, Smith AE. A short amino acid sequence able to specify nuclear location. Cell. 1984;39:499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- Katoh M. Molecular cloning, gene structure, and expression analyses of NKD1 and NKD2. Int J Oncol. 2001;19:963–969. [PubMed] [Google Scholar]
- Kussel P, Frasch M. Pendulin, a Drosophila protein with cell cycle-dependent nuclear localization, is required for normal cell proliferation. J Cell Biol. 1995;129:1491–1507. doi: 10.1083/jcb.129.6.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange A, Mills RE, Lange CJ, Stewart M, Devine SE, Corbett AH. Classical nuclear localization signals: definition, function, and interaction with importin α. J Biol Chem. 2007;282:5101–5105. doi: 10.1074/jbc.R600026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JJ, von Kessler DP, Parks S, Beachy PA. Secretion and localized transcription suggest a role in positional signaling for products of the segmentation gene hedgehog. Cell. 1992;71:33–50. doi: 10.1016/0092-8674(92)90264-d. [DOI] [PubMed] [Google Scholar]
- Lippai M, Tirian L, Boros I, Mihaly J, Erdelyi M, Belecz I, Mathe E, Posfai J, Nagy A, Udvardy A, Paraskeva E, Gorlich D, Szabad J. The Ketel gene encodes a Drosophila homologue of importin-β. Genetics. 2000;156:1889–1900. doi: 10.1093/genetics/156.4.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez Arias A, Baker NE, Ingham PW. Role of segment polarity genes in the definition and maintenance of cell states in the Drosophila embryo. Development. 1988;103:157–170. doi: 10.1242/dev.103.1.157. [DOI] [PubMed] [Google Scholar]
- Mason DA, Fleming RJ, Goldfarb DS. Drosophila melanogaster importin α1 and α3 can replace importin α2 during spermatogenesis but not oogenesis. Genetics. 2002;161:157–170. doi: 10.1093/genetics/161.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason DA, Mathe E, Fleming RJ, Goldfarb DS. The Drosophila melanogaster importin α3 locus encodes an essential gene required for the development of both larval and adult tissues. Genetics. 2003;165:1943–1958. doi: 10.1093/genetics/165.4.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathe E, Bates H, Huikeshoven H, Deak P, Glover DM, Cotterill S. Importin-α3 is required at multiple stages of Drosophila development and has a role in the completion of oogenesis. Dev Biol. 2000;223:307–322. doi: 10.1006/dbio.2000.9743. [DOI] [PubMed] [Google Scholar]
- Pai LM, Orsulic S, Bejsovec A, Peifer M. Negative regulation of Armadillo, a Wingless effector in Drosophila. Development. 1997;124:2255–2266. doi: 10.1242/dev.124.11.2255. [DOI] [PubMed] [Google Scholar]
- Pazdera TM, Janardhan P, Minden JS. Patterned epidermal cell death in wild-type and segment polarity mutant Drosophila embryos. Development. 1998;125:3427–3436. doi: 10.1242/dev.125.17.3427. [DOI] [PubMed] [Google Scholar]
- Peifer M, Berg S, Reynolds AB. A repeating amino acid motif shared by proteins with diverse cellular roles. Cell. 1994;76:789–791. doi: 10.1016/0092-8674(94)90353-0. [DOI] [PubMed] [Google Scholar]
- Richards S, Liu Y, Bettencourt BR, Hradecky P, Letovsky S, Nielsen R, Thornton K, Hubisz MJ, Chen R, Meisel RP, Couronne O, Hua S, Smith MA, Zhang P, Liu J, Bussemaker HJ, van Batenburg MF, Howells SL, Scherer SE, Sodergren E, Matthews BB, Crosby MA, Schroeder AJ, Ortiz-Barrientos D, Rives CM, Metzker ML, Muzny DM, Scott G, Steffen D, Wheeler DA, Worley KC, Havlak P, Durbin KJ, Egan A, Gill R, Hume J, Morgan MB, Miner G, Hamilton C, Huang Y, Waldron L, Verduzco D, Clerc-Blankenburg KP, Dubchak I, Noor MA, Anderson W, White KP, Clark AG, Schaeffer SW, Gelbart W, Weinstock GM, Gibbs RA. Comparative genome sequencing of Drosophila pseudoobscura: chromosomal, gene, and cis-element evolution. Genome Res. 2005;15:1–18. doi: 10.1101/gr.3059305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggleman B, Schedl P, Wieschaus E. Spatial expression of the Drosophila segment polarity gene armadillo is posttranscriptionally regulated by wingless. Cell. 1990;63:549–560. doi: 10.1016/0092-8674(90)90451-j. [DOI] [PubMed] [Google Scholar]
- Rousset R, Mack JA, Wharton KA, Jr, Axelrod JD, Cadigan KM, Fish MP, Nusse R, Scott MP. naked cuticle targets dishevelled to antagonize Wnt signal transduction. Genes Dev. 2001;15:658–671. doi: 10.1101/gad.869201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousset R, Wharton KA, Jr, Zimmermann G, Scott MP. Zinc-dependent interaction between dishevelled and the Drosophila Wnt antagonist Naked cuticle. J Biol Chem. 2002;277:49019–49026. doi: 10.1074/jbc.M203246200. [DOI] [PubMed] [Google Scholar]
- Sierra J, Yoshida T, Joazeiro CA, Jones KA. The APC tumor suppressor counteracts β-catenin activation and H3K4 methylation at Wnt target genes. Genes Dev. 2006;20:586–600. doi: 10.1101/gad.1385806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata T, Eaton S, Kornberg TB. The Drosophila hedgehog gene is expressed specifically in posterior compartment cells and is a target of engrailed regulation. Genes Dev. 1992;6:2635–2645. doi: 10.1101/gad.6.12b.2635. [DOI] [PubMed] [Google Scholar]
- Torok I, Strand D, Schmitt R, Tick G, Torok T, Kiss I, Mechler BM. The overgrown hematopoietic organs-31 tumor suppressor gene of Drosophila encodes an Importin-like protein accumulating in the nucleus at the onset of mitosis. J Cell Biol. 1995;129:1473–1489. doi: 10.1083/jcb.129.6.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Raay TJ, Coffey RJ, Solnica-Krezel L. Zebrafish Naked1 and Naked2 antagonize both canonical and non-canonical Wnt signaling. Dev Biol. 2007;309:151–168. doi: 10.1016/j.ydbio.2007.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voellmy R. On mechanisms that control heat shock transcription factor activity in metazoan cells. Cell Stress Chaperones. 2004;9:122–133. doi: 10.1379/CSC-14R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldrop S, Chan C-C, Cagatay T, Zhang S, Rousset R, Mack J, Zeng W, Fish M, Zhang M, Amanai M, Wharton KA., Jr An unconventional nuclear localization motif is crucial for function of the Drosophila Wnt/wingless antagonist Naked cuticle. Genetics. 2006;174:331–348. doi: 10.1534/genetics.106.061853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallingford JB, Habas R. The developmental biology of Dishevelled: an enigmatic protein governing cell fate and cell polarity. Development. 2005;132:4421–4436. doi: 10.1242/dev.02068. [DOI] [PubMed] [Google Scholar]
- Wang R, Brattain MG. The maximal size of protein to diffuse through the nuclear pore is larger than 60kDa. FEBS Lett. 2007;581:3164–3170. doi: 10.1016/j.febslet.2007.05.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wharton KA, Jr, Zimmermann G, Rousset R, Scott MP. Vertebrate proteins related to Drosophila Naked Cuticle bind Dishevelled and antagonize Wnt signaling. Dev Biol. 2001;234:93–106. doi: 10.1006/dbio.2001.0238. [DOI] [PubMed] [Google Scholar]
- Wiechens N, Heinle K, Englmeier L, Schohl A, Fagotto F. Nucleo-cytoplasmic shuttling of Axin, a negative regulator of the Wnt-β-catenin pathway. J Biol Chem. 2004;279:5263–5267. doi: 10.1074/jbc.M307253200. [DOI] [PubMed] [Google Scholar]
- Willert K, Jones KA. Wnt signaling: is the party in the nucleus? Genes Dev. 2006;20:1394–1404. doi: 10.1101/gad.1424006. [DOI] [PubMed] [Google Scholar]
- Wodarz A, Hinz U, Engelbert M, Knust E. Expression of crumbs confers apical character on plasma membrane domains of ectodermal epithelia of Drosophila. Cell. 1995;82:67–76. doi: 10.1016/0092-8674(95)90053-5. [DOI] [PubMed] [Google Scholar]
- Wu C. Heat shock transcription factors: structure and regulation. Annu Rev Cell Dev Biol. 1995;11:441–469. doi: 10.1146/annurev.cb.11.110195.002301. [DOI] [PubMed] [Google Scholar]
- Yanagawa S, van Leeuwen F, Wodarz A, Klingensmith J, Nusse R. The dishevelled protein is modified by wingless signaling in Drosophila. Genes Dev. 1995;9:1087–1097. doi: 10.1101/gad.9.9.1087. [DOI] [PubMed] [Google Scholar]
- Yoffe KB, Manoukian AS, Wilder EL, Brand AH, Perrimon N. Evidence for engrailed-independent wingless autoregulation in Drosophila. Dev Biol. 1995;170:636–650. doi: 10.1006/dbio.1995.1243. [DOI] [PubMed] [Google Scholar]
- Zandi E, Tran TN, Chamberlain W, Parker CS. Nuclear entry, oligomerization, and DNA binding of the Drosophila heat shock transcription factor are regulated by a unique nuclear localization sequence. Genes Dev. 1997;11:1299–1314. doi: 10.1101/gad.11.10.1299. [DOI] [PubMed] [Google Scholar]
- Zeng W, Wharton KA, Jr, Mack JA, Wang K, Gadbaw M, Suyama K, Klein PS, Scott MP. naked cuticle encodes an inducible antagonist of Wnt signalling. Nature. 2000;403:789–795. doi: 10.1038/35001615. [DOI] [PubMed] [Google Scholar]
- Zhang L, Jia J, Wang B, Amanai K, Wharton KA, Jr, Jiang J. Regulation of wingless signaling by the CK1 family in Drosophila limb development. Dev Biol. 2006;299:221–237. doi: 10.1016/j.ydbio.2006.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]


