Abstract
Epigenetic silencing of tumor suppressor genes in human cancers is associated with aberrant methylation of promoter region CpG islands and local alterations in histone modifications. However, the mechanisms that drive these events remain unclear. Here, we establish an important role for histone H4 lysine 16 acetylation (H4K16Ac) and the histone acetyltransferase hMOF in the regulation of TMS1/ASC, a proapoptotic gene that undergoes epigenetic silencing in human cancers. In the unmethylated and active state, the TMS1 CpG island is spanned by positioned nucleosomes and marked by histone H3K4 methylation. H4K16Ac was uniquely localized to two sharp peaks that flanked the unmethylated CpG island and corresponded to strongly positioned nucleosomes. Aberrant methylation and silencing of TMS1 was accompanied by loss of the H4K16Ac peaks, loss of nucleosome positioning, hypomethylation of H3K4 and hypermethylation of H3K9. In addition, a single peak of histone H4 lysine 20 trimethylation was observed near the transcription start site. Downregulation of hMOF or another component of the MSL complex resulted in a gene-specific decrease in H4K16Ac, loss of nucleosome positioning and silencing of TMS1. Gene silencing induced by H4K16 deacetylation occurred independently of changes in histone methylation and DNA methylation and was reversed upon hMOF re-expression. These results indicate that the selective marking of nucleosomes flanking the CpG island by hMOF is required to maintain TMS1 gene activity, and suggest that the loss of H4K16Ac, mobilization of nucleosomes and transcriptional downregulation may be important events in the epigenetic silencing of certain tumor suppressor genes in cancer.
Keywords: DNA methylation, gene regulation, histone modifications, chromatin, cancer
INTRODUCTION
Epigenetic mechanisms involve DNA methylation at cytosine residues and post-translational modifications of histone tails, both of which regulate gene transcription by altering chromatin structure and DNA-protein interactions (1, 2). In the human genome, most cytosines in CpG dinucleotides are methylated, except those in CpG islands, regions of the genome that contain a high density of CpG sites and encompass the promoter regions of more than half of the known genes (3). Methylation of promoter-associated CpG islands, whether developmentally programmed or occurring aberrantly during carcinogenesis, is associated with gene inactivation (4). Histone modifications can also act synergistically or antagonistically to define the transcription state of genes. Acetylation of histones H3 and H4 is associated with transcriptionally active promoters and an open chromatin configuration (5). Both dimethylation and trimethylation of histone H3 lysine 4 (H3K4me2, H3K4me3) have been linked to actively transcribing genes, although recent studies indicate that CpG island-associated promoters are marked by H3K4me2 regardless of the transcriptional status (6, 7). In contrast, methylation of histone H3 lysine 9 and 27 are associated with transcriptionally inactive promoters and condensed closed chromatin (8, 9). Interplay between the histone modifications and DNA methylation define the transcriptional potential of a particular chromatin domain.
The epigenetic landscape is drastically altered in human cancers. In cancer cells, global hypomethylation occurs at normally methylated CpG sites while hypermethylation occurs at select CpG islands (10). Aberrant CpG island methylation is accompanied by changes in the local histone modification patterns, including the hypoacetylation of histones H3 and H4, hypomethylation of histone H3 lysine 4 and hypermethylation of histone H3 lysine 9 and/or H3 lysine 27, resulting in gene silencing (10). Such epigenetic events contribute to carcinogenesis through the aberrant silencing of tumor suppressor genes. Recently, genome-wide studies showed that cancer cells undergo widespread alterations in histone modifications, including a global loss of acetylation at H4 lysine 16 (H4K16Ac) and trimethylation at H4 lysine 20 (H4K20me3) (11). Though the molecular alterations that occur in cancer cells are well-studied, the precise order and mechanisms by which they precipitate gene silencing are still largely unknown.
Nucleosome positioning, or the preferential association of nucleosomes with specific genomic locations, is another feature with the potential to impact epigenetic regulation. Functionally, nucleosome positioning has been shown to maintain or prohibit gene activity depending on the context (12, 13). The translational positioning of nucleosomes has been attributed to interactions between histone proteins and specific DNA sequences, the binding of non-histone proteins, and neighboring positioned nucleosomes (13-15). DNA methylation does not alter the ability of histone proteins to bind DNA in vitro, but can alter nucleosome positioning in vivo through indirect mechanisms (15, 16). The relationship between histone modifications and positioning of nucleosomes is not well understood.
TMS1 (Target of Methylation-mediated Silencing), also known as ASC, is a proapoptotic signaling factor that is subjected to epigenetic silencing in human cancers (17-21). Originally identified in a screen for genes that were silenced in response to DNMT1 (DNA cytosine-5-methyltransferase-1) over-expression (17), subsequent studies have shown that TMS1 is silenced in conjunction with CpG island hypermethylation in a number of human tumors, including glioblastomas, breast, colorectal and gastric cancers (22-25). Loss of TMS1 function confers resistance to TNFα-induced apoptosis in breast cancer cells and its restoration suppresses cell growth (21). Treatment of cancer cells with the demethylating agent 5-aza-2′-deoxycytidine (5azadC) restores TMS1 expression, suggesting an important role for DNA methylation in TMS1 gene silencing (26).
The TMS1 CpG island is unmethylated in normal cells and breast cancer cells that express TMS1 (26, 27). In the active state, the TMS1 CpG island represents a distinct chromatin domain characterized by acetylated histones and three DNase I hypersensitive sites (HS) that span the CpG island. Two of these, HS1 and HS3, demarcate the 5′ and 3′ boundaries between the unmethylated CpG island DNA and densely methylated flanking DNA (27). Epigenetic silencing of TMS1 in breast cancer is accompanied by a localized remodeling of the CpG island-associated HS sites, hypoacetylation of histones, and hypermethylation of DNA (27). These data have led us to propose that the HS sites act in cis, perhaps by recruiting trans-acting factors, to block or actively oppose the spread of methylation into the CpG island.
To further understand the role of chromatin structure in the epigenetic silencing of tumor suppressor genes in cancer, we have used the TMS1 locus as a model to examine the relationship between histone modifications, nucleosome positioning, and DNA methylation. We found that the TMS1 locus is characterized by distinct histone modification profiles in the active and inactive states. Interestingly, H4K16Ac and H4K20me3 exhibited unique localization across the active and inactive TMS1 locus, respectively. We further pursued the significance of H4K16Ac at TMS1 and determined that abrogation of H4K16 acetylation led to TMS1 silencing, which was accompanied by changes in the local nucleosome architecture. Our findings indicate that H4K16Ac plays a critical role in the maintenance of active gene transcription, and suggest that loss of H4K16Ac and transcriptional downregulation may be important steps in the epigenetic silencing of some tumor suppressor genes in cancer.
MATERIALS AND METHODS
Cell Culture
IMR90 normal diploid fibroblasts and their SV40-transformed derivatives (IMR90/SV40) were obtained from the National Institute of Aging. The generation of IMR90/SV40 cells stably over-expressing DNMT1 (HMT.1E1) has been described (28). MCF7 and MDA-MB231 breast cancer cell lines and 293T embryonic kidney cells were obtained from the American Type Culture Collection and maintained in DMEM containing 2 mM glutamine and 10% FBS.
Chromatin Immunoprecipitation (ChIP)
ChIP was carried out as described in the Acetyl-Histone H3 Immunoprecipitation Assay Kit (Cat. # 17-229) by Millipore. DNA recovered from ChIP was analyzed by real-time PCR. The reaction mixture (25 μL) contained 1 μL of the appropriately diluted DNA sample, 0.2 μM primers and 12.5 μL of IQ SYBR Green Supermix (BioRad). The reaction was subjected to a hot start for 3 minutes at 95°C and 50 cycles of 95°C,10s; 55-65°C, 30s; 72°C, 30s. Melt curve analysis was done to verify a single product species. Starting quantities were determined relative to a common standard curve generated using MCF7 genomic DNA. Percent enrichment in each pulldown was calculated relative to input DNA. Primers pairs used for real-time analysis across the TMS1, CDH1 or ESR1 locus are shown in Figures 1A and 6A. Sequences are available upon request. Antibodies used were: rabbit IgG; (Santa Cruz; #SC-2027), histone H3 acetylated at lysine 9 and 14 (H3K9/14Ac; Millipore; #06-599), H4K16Ac (Abcam; #ab1762), H3K4me2 (Millipore; #07-030), H3K4me3 (Abcam; #ab8580), dimethylated histone H3 lysine 9 (H3K9me2; Abcam; #ab7312), trimethylated histone H3 lysine 9 (H3K9me3; Abcam; #ab8898), trimethylated histone H3 lysine 27 (H3K27me3; Millipore; #07-449), H4K20me3 (Abcam; #ab9053). Antibodies against hMOF and hMSL1 were gifts from Edwin Smith (Emory University).
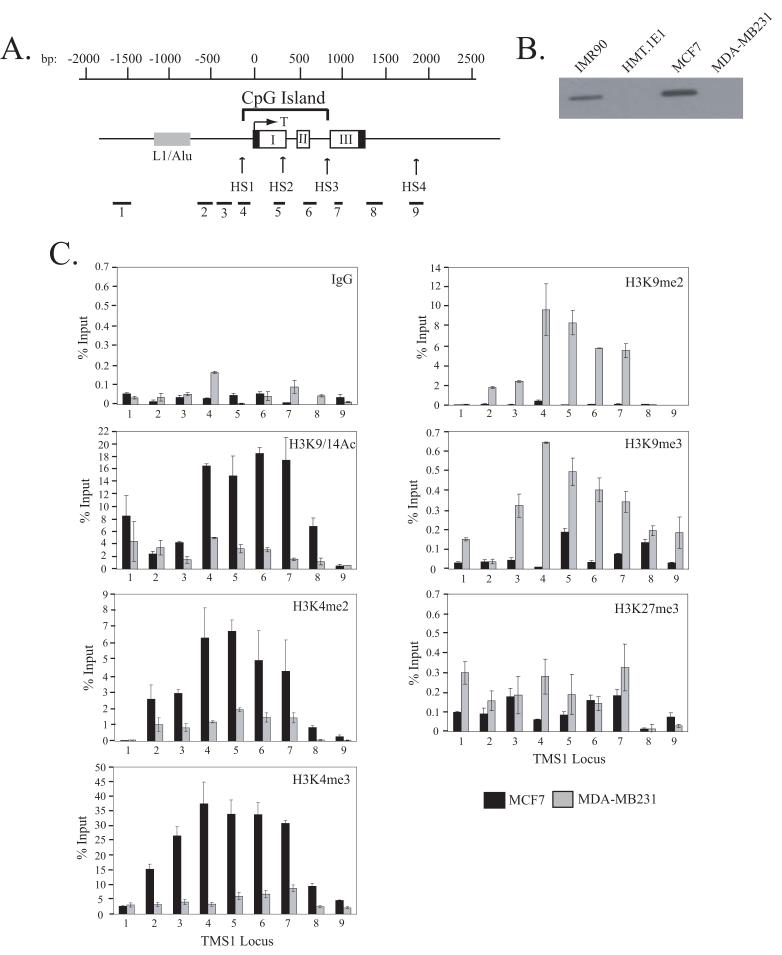
Figure 1.
A) Schematic of the TMS1 genomic locus. The TMS1 gene consists of three exons (I, II and III). Non-coding regions are indicated by black boxes. The nucleotide positions are numbered with respect to the transcription start site (T) and are shown above the gene. The location of the CpG island is marked and spans from approximately -100 to +900 bp. The positions of the hypersensitive sites (HS1-HS4) and an upstream repeat element (L1/Alu) are shown. Primer sets used (1-9) for real-time PCR in ChIP assays are shown below the gene. B) Expression of TMS1. Protein lysates prepared from MCF7, MDA-MB231, HMT.1E1 and IMR90 cells were subject to western blot analysis with antibody against TMS1. C) Distribution of histone H3 modifications across the TMS1 locus. MCF7 and MDA-MB231 cell lines were subjected to ChIP with antibodies against the indicated histone modifications or a rabbit IgG (IgG) control. Immunoprecipitated DNA was amplified by real-time PCR with primer sets indicated in (A). Percent (%) input was determined as the amount of immunoprecipiated DNA relative to input DNA. Each ChIP was repeated at least three times, and although the immunoprecipitation efficiency varied between experiments, the profile of enrichment across the locus was consistent. Shown are the mean +/- standard deviation of triplicate determinations from a representative experiment.
Figure 6.

Role of H4K16Ac in the regulation of the ESR1 and CDH1 loci. A) Schematic of the region encompassing the CpG islands of the ESR1 and CDH1 gene. Nucleotide positions with respect to the transcription start site (T) and the CpG island are indicated above each gene diagram. Exons are shown in grey boxes. The regions amplified by primer sets (1-5) used in real-time PCR are shown below each gene. B) Localization of H3K9/14Ac and H4K16Ac at the ESR1 and CDH1 CpG island. ChIP analyses were performed for MCF7 or MDA-MB231 cells with the indicated antibodies or a negative control (IgG), followed by real-time PCR of regions depicted in A). Each experiment was conducted at least three times and although the immunoprecipitation efficiency varied between experiments, the profile of enrichment across each locus was consistent. Shown are the mean +/- standard deviation of triplicate determinations from a representative experiment. C) MCF7 cells were transfected with siRNA (100 nM) against Lamin A/C (Lamin) (irrelevant control) or hMOF (MOF1 and MOF2) and the expression of ESR1 and CDH1 protein was determined by western blotting. The same blot was also exposed to anti-hMOF and anti-GAPDH antibodies.
Micrococcal Nuclease Digestion and Indirect End-labeling
Isolation of intact nuclei and indirect end labeling was performed as described in (27) with minor modifications. Nuclei from 2 × 106 cells were digested with micrococcal nuclease (MNase) (10 - 200 u) at 25°C for 10 min in RBS (10mM TrisHCl, 5mM MgCl2, 0.5 M DTT, 0.3 mM sucrose, 0.4 mM PMSF). Reactions were stopped by incubation in 1% SDS, 20 mM EDTA, and 10 μg/ml RNaseA for 30 min at 37°C, followed by incubation with 1 mg/ml Proteinase K at 50°C overnight. DNA was recovered by phenol-chloroform extraction and ethanol precipitation. MNase-digested DNA (10 μg) was digested with 2 units each Hind III and Spe I at 37°C overnight, separated on a 1.0% agarose gel and transferred to a nylon membrane. Membranes were hybridized with a random-prime labeled Spe I-Xmn I probe anchored to the downstream Spe I site of TMS1 (Figure 2C). Approximately 100 pg Spe I-Hind III (2765 bp), Spe I-Bam HI (1001 bp) and Spe I-Eco RI (739 bp) fragments from the TMS1 locus were run on each gel as internal size markers. Membranes were washed to high stringency (2X SSC, 0.1% SDS at 65°C), exposed to phosphor storage screens and analyzed by phosphoimage analysis using ImageQuant software (Molecular Dynamics).
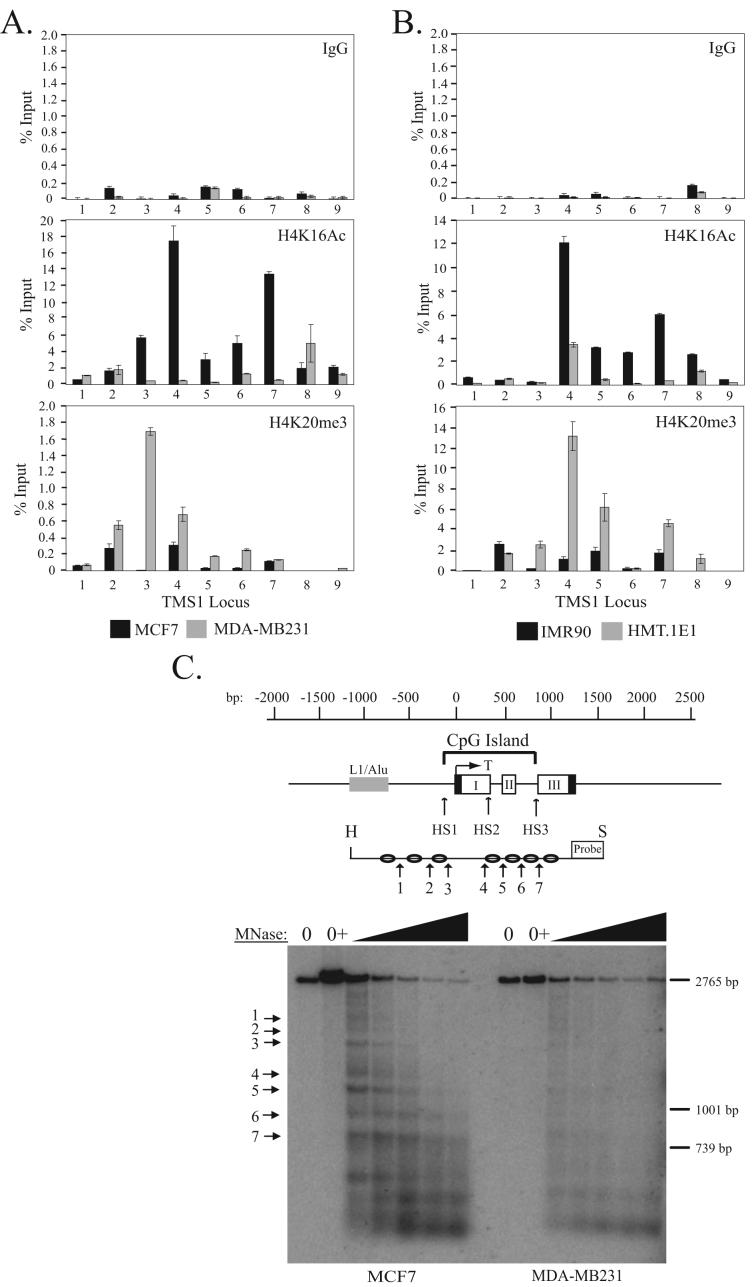
Figure 2.
Localization of H4K16Ac and H4K20me3 across the TMS1 locus. A) MCF7 and MDA-MB231 breast cancer cells or B) IMR90 and HMT.1E1 cell lines were subjected to ChIP with antibodies against rabbit IgG (IgG) or the indicated histone modifications. Immunoprecipitated DNA was amplified by real-time PCR with primer sets indicated in Figure 1A. Percent (%) input was determined as the amount of immunoprecipiated DNA relative to input DNA. Each ChIP was repeated at least three times, and although the immunoprecipitation efficiency varied between experiments, the profile of enrichment across the locus was consistent. Shown are the mean +/- standard deviation of triplicate determinations from a representative experiment. C) Nucleosome positioning at the TMS1 locus. Nuclei from MCF7 (left) or MDA-MB231 (right) cells were incubated in MNase digestion buffer alone (0), digestion buffer plus CaCl2 (0+), or digestion buffer plus CaCl2 and 10-200U MNase. MNase-digested DNA (10 μg) was digested with Hind III (H) and Spe I (S), separated on a 1% agarose gel, and subject to Southern blot analysis using a probe anchored to the 3′ Spe I site. Preferential MNase cut sites are depicted with arrows. Shown is the relative migration of a 2765bp, 1001bp and 739bp Spe I-anchored fragments from the TMS1 locus that were included as internal markers. A representative experiment is shown.
RNAi Silencing
For siRNA transfections, MCF7 cells were transfected with 100 nM siRNA (Dharmacon) against Lamin A/C, hMOF, or hMSL1 using Oligofectamine (Invitrogen). The sequences of hMOF and hMSL1 siRNAs are from Dou et al. (2005) and Smith et al. (2005), respectively. For lentiviral shRNA infections, 293T cells were transfected with 920 ng of pCMV-dR8.91 (viral packaging plasmid), 100 ng pMD2G-VSV-G (viral envelope plasmid) and 1 μg pLKO.1 or pLKO.1 containing hMOF shRNA (Open Biosystems; TRCN0000034875) using Lipofectamine (Invitrogen). Media containing lentiviral particles was collected at 40 and 64 hours post-transfection. Recipient cells were plated in 6 cm plates (1 × 106 cells) or 10 cm plates (2 × 106 cells) and infected with 0.1-0.5 mL supernatant in the presence of 8 μg/mL polybrene. Infected cells were placed under puromycin selection (0.5-1.0 μg/mL) and harvested at given time-points for subsequent analysis.
Western analysis
Cells were lysed in RIPA buffer (50 mM Tris-Cl pH 8.0, 150 mM NaCl, 0.5 mM EDTA, 1% NP-40, 0.5% sodium-deoxycholate, 0.1% SDS) containing protease inhibitors for 10 min on ice. Clarified lysates (100 μg) were electrophoresed on a 12% SDS-PAGE gel and transferred to nitrocellulose membrane. The membrane was incubated with the appropriate 1° antibody and HRP-conjugated 2° antibody and subjected to chemiluminescence detection (Pierce). For detecting histones, cells were washed in PBS containing 5 mM sodium butyrate and resuspended in acid extraction lysis buffer (10 mM HEPES pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT, 1.5 mM PMSF, 0.2 N HCl) for 30 minutes on ice. Lysates (100 μg) were electrophoresed on a 10% SDS-PAGE gel and blotted as described above. Antibodies used were: TMS1 (Protein Tech; #PTG10500-1-AP), GAPDH (Abcam; #ab8245), hMOF, hMSL1, H3K9/14Ac, H4K16Ac, estrogen receptor α (Santa Cruz; #SC-7207) and E-cadherin (BD Biosciences; #C20820).
Reverse Transcription
Total RNA was isolated from MCF7 cells using the RNeasy Mini kit (Qiagen). RNA (6 μg) was pre-treated with DNase I and then reverse-transcribed using random hexamer primers and M-MLV reverse transcriptase. cDNA was amplified with primers against TMS1, hMOF or 18s rRNA using real-time PCR as described for the ChIP assays. Starting quantities were determined relative to a common standard curve generated using MCF7 cDNA. Primer sequences are available upon request.
Methylation-Specific PCR (MSP) and Bisulfite Sequencing
MSP and bisulfite sequencing were performed as previously described (27). Primer sets used for MSP overlap a total of 6 CpG sites in the TMS1 CpG island and have been described (17). Primers used for bisulfite sequencing correspond to primer set B in Stimson and Vertino (27).
RESULTS
The active TMS1 locus is marked by H3K4me while the inactive TMS1 locus is marked by H3K9me
The TMS1 locus consists of 3 exons encompassing approximately 1.5 kb on chromosome 16 (Figure 1A) (17). A 1.0 kb CpG island spans the promoter region as well as exons I and II (Figure 1A). This region is unmethylated in cells expressing TMS1, such as IMR90 human diploid fibroblasts and MCF7 breast cancer cells, but is completely methylated in cells lacking TMS1 expression, such as DNMT1-overexpressing human fibroblasts (HMT.1E1) and MDA-MB231 breast cancer cells (Figure 1B) (17, 26, 27). To determine the relationship between histone modifications and DNA methylation at the TMS1 locus, we examined the distribution of histone H3 marks across a region covering approximately 1.5 kb upstream of the transcription start site to approximately 600 bp downstream of the termination sequence in MCF7 and MDA-MB231 cells by ChIP (Figure 1A, C). We found that the active TMS1 locus in MCF7 cells was enriched in nucleosomes marked by H3K9/14Ac, H3K4me2 and H3K4me3 but lacked H3K9me2 and H3K9me3 (Figure 1C). In contrast, the silent TMS1 locus in MDA-MB231 cells was hypoacetylated at H3K9/14Ac and lacked H3K4me2 and H3K4me3 but was enriched in H3K9me2 and H3K9me3 (Figure 1C). We also examined H3K27me3 at the TMS1 locus and found that there was little enrichment of this mark in either MCF7 or MDA-MB231 cells when compared to a negative control (IgG) and a positive control locus (eg. MYT1) (Figure 1C; data not shown) (29). An inverse relationship between H3K4me and H3K9me was also observed in IMR90 and HMT.1E1 cells (data not shown). These results indicate that distinct histone modification profiles correlate with TMS1 gene activity. Specifically, methylation-associated silencing of TMS1 correlates with a shift from H3K4me to H3K9me.
Discreet Localization of H4K16Ac and H4K20me3 at the TMS1 locus
We next examined the relationship between DNA methylation and histone H4 modifications. H4K16Ac and H4K20me3 are associated with active and constitutive heterochromatic regions, respectively (30-32). Recent studies have shown that cancer cells exhibit reduced levels of H4K16Ac and H4K20me3 overall relative to their normal counterparts (11). It was suggested that this global loss derived primarily from repetitive DNA sequences. However, the relationship between these marks and the epigenetic dysregulation of individual genes in cancer cells has not been addressed. To determine whether histone H4 modifications play a role in the epigenetic regulation of TMS1, we examined the profile of H4K16Ac and H4K20me3 in MCF7 and MDA-MB231 cells. We found that the TMS1 locus was enriched in H4K16Ac in MCF7 cells whereas MDA-MB231 cells were hypoacetylated at H4K16Ac (Figure 2A). Interestingly, the distribution of H4K16Ac in MCF7 cells showed discreet peaks of enrichment that corresponded almost precisely to the 5′ and 3′ boundaries of the unmethylated TMS1 CpG island. These H4K16Ac peaks were completely absent in MDA-MB231 cells (Figure 2A). Furthermore, while TMS1 lacked H4K20me3 in MCF7 cells, there was a prominent peak of H4K20me3 at the TMS1 promoter in MDA-MB231 cells (Figure 2A). To determine whether this pattern was selective for breast cancer cells, we also examined the distribution of H4K16Ac and H4K20me3 in IMR90 normal diploid fibroblasts and HMT.1E1 cells (Figure 2B). Again, two prominent peaks of H4K16Ac flanked the TMS1 CpG island when it is unmethylated and active (IMR90), but were absent in when the locus is hypermethylated and silent (HMT.1E1). As well, a peak of H4K20me3 was observed in the TMS1 promoter region in HMT.1E1, but not in IMR90 cells. These data indicate that the epigenetic silencing of TMS1 is associated with the focal loss of H4K16Ac and the gain of H4K20me3.
Nucleosome positioning at the TMS1 gene
We also examined nucleosome positioning within and around the TMS1 CpG island in MCF7 and MDA-MB231 cells. Isolated nuclei were digested with increasing amounts of MNase followed by digestion of the DNA with Hind III and Spe I to generate a 2.76 kb genomic fragment encompassing the TMS1 CpG island (Figure 2C). Indirect end-labeling analysis with a probe anchored at the 3′ Spe I site allowed mapping of positioned nucleosomes. Nucleosomes were positioned at an approximate 200 bp intervals spanning the CpG island in MCF7 cells (Figure 2C). At similar levels of MNase digestion, MDA-MB231 cells exhibited more of a smear, indicating that nucleosomes are more randomly positioned throughout the CpG island in these cells (Figure 2C, Supplemental Figure 1). One exception to the positioning in MCF7 cells was the distance between MNase cut sites 3 and 4 which flank the transcription start site. This ∼400 bp spacing may correspond to a nucleosome-free region at the transcription start site of TMS1 in actively transcribed cells. Within the regular array of positioned nucleosomes, differences existed in the degree of positioning. Preferential digestion by MNase was observed at cut sites 3, 5 and 7, suggesting that nucleosomes associated with these sites are more strongly positioned (Figure 2C, Supplemental Figure 1). Sites 3 and 7 correspond to the boundaries between the unmethylated CpG island and methylated surrounding DNA as well as the observed peaks of H4K16Ac (27) (Figure 2A). The co-localization of H4K16Ac and strongly positioned nucleosomes suggests a possible link between the two.
Role of H4K16Ac at the TMS1 locus
The finding that there are peaks of H4K16Ac and positioned nucleosomes flanking the CpG island when the TMS1 locus is unmethylated and expressed and that the locus lacks these features when TMS1 is methylated and silent raises the question of whether H4K16Ac plays a role in the regulation of TMS1. The majority of H4K16Ac in humans is mediated by hMOF, a member of the MYST family of histone acetyltransferases (33, 34). Downregulation of hMOF by RNAi has been shown to drastically decrease the global levels of H4K16Ac in human cells (33, 34). Western blot analysis showed no difference in the levels of hMOF between cells that express TMS1 (MCF7 and IMR90) and those that do not express TMS1 (MDA-MB231 and HMT.1E1), indicating that the lack of H4K16Ac at TMS1 in MDA-MB231 and HMT.1E1 cells is not due to differential expression of hMOF (data not shown).
To understand the role of H4K16Ac at the TMS1 locus, we knocked down hMOF in MCF7 cells with two independent siRNA molecules (Figure 3A). Consistent with previous studies (30), we found that downregulation of hMOF led to a global decrease in H4K16Ac, but had no affect on H3K9/14Ac levels (Supplemental Figure 2A). There was also a drastic reduction in the specific association of H4K16Ac, but not H3K9/14Ac, at the TMS1 locus in MCF7 cells downregulated for hMOF. There was a direct correlation between the efficiency of hMOF knock-down by the two siRNAs and loss of H4K16Ac (Figure 3A and B). Knock-down of hMOF in MCF7 cells also led to a loss of nucleosome positioning across the CpG island (Figure 3C, Supplemental Figure 3). In contrast, there was no change in the levels or distribution of H3K4me2, H3K9me2, or H4K20me3 at the TMS1 locus upon hMOF downregulation (Supplemental Figure 4). Thus, hMOF downregulation results in a loss of H4K16Ac and impairs nucleosome positioning at the TMS1 locus, independent of changes in other histone modifications.
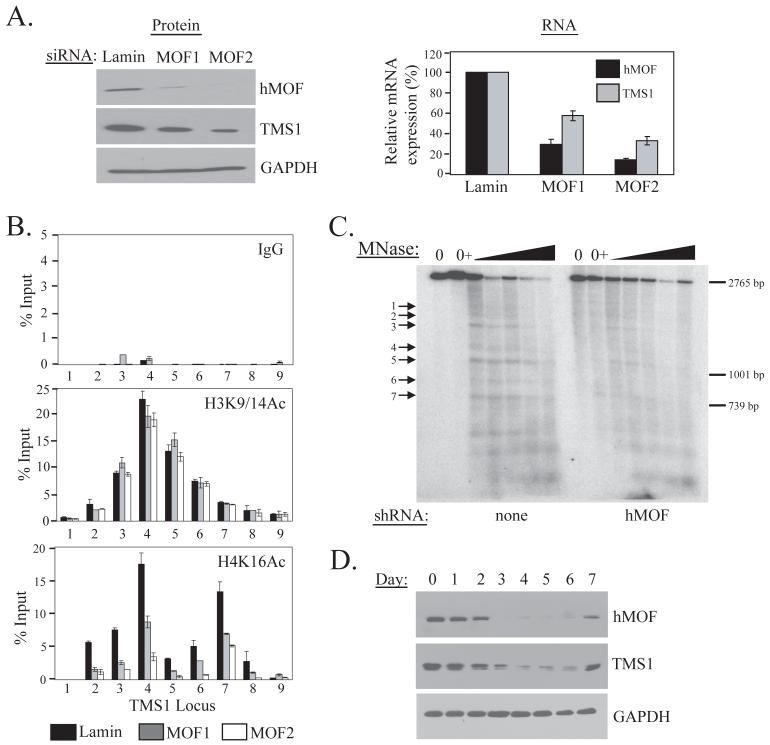
Figure 3.
Effect of hMOF downregulation on the TMS1 locus. A) MCF7 cells were transfected with 100 nM of siRNA targeting hMOF (MOF1 or MOF2) or Lamin A/C (Lamin) (irrelevant control) and harvested four days post-transfection. Cells were analyzed for hMOF, TMS1 or GAPDH (loading control) protein expression by western blot analysis (left) or for hMOF, TMS1 or 18s (internal control) RNA expression by real time PCR (right). The levels of expression of hMOF and TMS1 mRNA are expressed relative to that obtained in cells treated with Lamin siRNA, after normalization to 18s. Shown is the mean +/- standard deviation of three independent experiments. Experiments were also done using scrambled non-targeting siRNA as a control and similar results were observed. B) MCF7 cells were transfected as in A) and ChIP was performed with antibodies against rabbit IgG (IgG), H3K9/14Ac or H4K16Ac. Immunoprecipitated DNA was amplified by real-time PCR with primer sets indicated in Figure 1A. Data represent the percent of input DNA recovered. Each ChIP experiment was repeated at least twice with reproducible results. Shown are the mean +/- standard deviation for triplicate determinations from a representative experiment. C) MCF7 cells infected with an empty pLKO.1 vector (none) or a pLKO.1 expressing hMOF shRNA (hMOF) were analyzed for nucleosome positioning exactly as described in Figure 2C. Preferential MNase cut sites (1-7) are indicated with arrows. Shown is the migration of a 2765bp, 1001bp and 739bp Spe I fragments from the TMS1 locus that were included as internal markers. A representative experiment is shown. D) MCF7 cells were transfected with hMOF siRNA and processed at 0 to 7 days post-transfection for western blot analysis using the indicated antibodies.
We also determined the impact of hMOF knock-down on TMS1 expression. hMOF downregulation led to a concomitant decrease in TMS1 protein and mRNA levels (Figure 3A). Again, there was a direct correlation between the efficiency of hMOF knock-down by the two siRNAs and the magnitude of TMS1 repression (Figure 3A). In time course experiments, a good correlation was observed between the degree and timing of hMOF knock-down and TMS1 downregulation, supporting a direct relationship between the two (Figure 3D). TMS1 silencing followed hMOF downregulation, and was reversed when hMOF expression returned to baseline. Downregulation of hMOF with a lentivirus expressing a shRNA directed against hMOF also led to a decrease in TMS1 expression, in both MCF7 and IMR90 cells (Supplemental Figure 2B, C). As in MCF7 cells, the level of H4K16Ac across the TMS1 locus was drastically reduced in IMR90 cells upon hMOF downregulation (Supplemental Figure 2D). Together, these studies indicate that downregulation of hMOF and the associated loss of H4K16Ac and nucleosome positioning at the TMS1 locus are sufficient to precipitate silencing of TMS1. These studies provide functional relevance to the localization of H4K16Ac at the active TMS1 locus and indicate that hMOF-mediated acetylation of H4K16 positively regulates TMS1 gene expression.
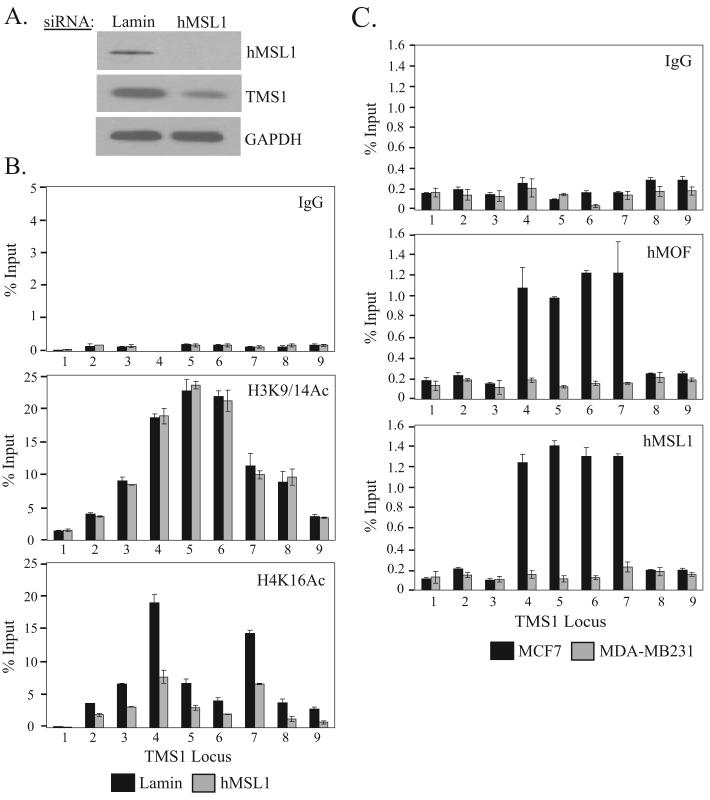
In humans, hMOF is a component of several histone modifying complexes including the hMSL, MLL1-WDR5 and hMSL1v complexes (30, 33). However, hMOF in the hMSL complex is thought to be responsible for the majority of H4K16Ac (33). Indeed, downregulation of hMSL1, another component of the MSL complex, has been shown to drastically reduce global H4K16Ac levels (33). To determine whether the MSL complex played a role at the TMS1 locus, we treated MCF7 cells with siRNA directed against hMSL1. Knock-down of hMSL1 led to a decrease in H4K16Ac levels, loss of the H4K16Ac peaks and suppressed TMS1 expression in a manner similar to that observed after downregulation of hMOF (Figure 4A, B). ChIP experiments showed that both hMSL1 and hMOF were enriched at the TMS1 locus in MCF7 cells, but not in MDA-MB231 cells (Figure 4C). Taken together, these data suggest that hMOF is acting as part of the MSL complex to mediate H4K16 acetylation at the TMS1 locus.
Figure 4.
Effect of hMSL1 silencing and localization of the MSL complex at the TMS1 locus. MCF7 cells transfected with 100 nM of Lamin A/C (Lamin) or hMSL1 siRNA were harvested four days post-transfection and analyzed for (A) hMSL1, TMS1 and GAPDH protein expression by western blot analysis and (B) chromatin immunoprecipitation with antibodies against rabbit IgG (IgG), H3K9/14Ac and H4K16Ac, exactly as described in Figure 3C. Similar results were obtained when a scrambled siRNA was used as a negative control. Shown are the mean +/- standard deviation of triplicate determinations from a representative experiment. C) Chromatin from MCF7 and MDA-MB231 cells were subjected to immunoprecipitation with antibodies against rabbit IgG (IgG), hMOF or hMSL1 as described in Figure 3C. Each ChIP experiment was repeated at least twice with reproducible results. Shown are the mean +/- standard deviation of triplicate determinations from a representative experiment.
Effect of hMOF downregulation and loss of H4K16Ac on DNA methylation
The peaks of H4K16Ac at the boundaries between the unmethylated CpG island and flanking methylated DNA in MCF7 cells, and the absence of these features in MDA-MB231 cells, where the CpG island is methylated suggests that the presence of this histone mark may play an important role in maintaining the integrity of the CpG island domain. To address this question, we determined the impact of hMOF downregulation on the methylation status of TMS1. MCF7 cells transfected with control or hMOF siRNAs were harvested five days post-transfection and the DNA subjected to methylation-specific PCR (MSP) and bisulfite sequencing. Downregulation of hMOF had no affect on the methylation status of the CpG island within a 5 day time frame (Figure 5A). Sequencing of bisulfite-modified DNA showed no difference in the profile of DNA methylation across the TMS1 CpG island in MCF7 cells transfected with control or hMOF siRNA (Figure 5B). Downregulation of hMOF also had no impact on TMS1 CpG island methylation in IMR90 cells (data not shown). These results indicate that, at least within the short time frame analyzed, loss of H4K16Ac leads to TMS1 silencing without accompanying CpG island methylation.
Figure 5.
Effect of hMOF downregulation on TMS1 DNA methylation. A) MCF7 cells were transfected with siRNA (100 nM) against hMOF (MOF1 and MOF2) or an irrelevant control Lamin A/C (Lamin) and harvested 4 days post-transfection. Genomic DNA was isolated, modified by sodium bisulfite and amplified by methylation specific PCR (MSP) with primer sets specific for either methylated (M) or unmethylated (U) DNA. DNA from IMR90 and MCF7 cells served as a control for unmethylated DNA, while that from HMT.1E1 and MDA-MB231 cells served as a control for methylated DNA. The TMS1 region (32-223) amplified by MSP is shown in Part B (MSP). B) DNA from MCF7 cells transfected with Lamin A/C (Lamin) or MOF2 (hMOF) siRNA was modified with bisulfite and amplified with a primer set that spans 53 CpG sites in the TMS1 CpG island (27). Products were subcloned and sequenced. Each row indicates the sequence of an independent clone where methylated (black circles) and unmethylated (white circles) CpG sites are indicated.
To determine whether CpG island methylation would ensue over a longer time period after hMOF knock-down, we also used lentiviral shRNA infections to stably knock-down hMOF in MCF7 cells. However, we found that long-term downregulation of hMOF by this method was lethal. At day 6 post-infection, cells containing lentiviral hMOF shRNA were dead (data not shown). This observation supports previous findings indicating that MOF is required for cell viability (33-36).
H4K16Ac does not regulate all genes silenced by aberrant DNA methylation
The above data indicate that loss of the H4K16Ac peaks at the TMS1 locus, through targeted disruption of hMOF, allows for silencing of TMS1. To determine whether this mechanism is operative at other genes that undergo epigenetic silencing in human cancers, we examined the distribution of H4K16Ac at two other genes, CDH1 and ESR1, which like TMS1, are subject to epigenetic silencing in human breast and other cancers (37, 38). Both CDH1 and ESR1 contain promoter-associated CpG islands and are unmethylated and expressed in MCF7 cells and are methylated and silent in MDA-MB231 cells (Figure 6A) (37, 38). The CDH1 and ESR1 CpG islands were both enriched in H3K9/K14Ac in MCF7 but not MDA-MB231 cells, which is consistent with the tight association of H3Ac mark with actively transcribed genes (Figure 6B). H4K16Ac levels were very low across the CpG islands of both CDH1 and ESR1, as compared to that observed at TMS1 (compare Figure 6B to Figure 2A). More importantly, there was no difference in the levels of H4K16Ac between MCF7 and MDA-MB231 cells at either gene (Figure 6B). Consistent with a lack of association of H4K16Ac with these loci, downregulation of hMOF had no effect on the expression of either ESR1 or CDH1 (Figure 6C). These results indicate that hMOF and H4K16Ac may regulate a specific subset of genes.
DISCUSSION
Alterations in DNA methylation associated with the epigenetic silencing of tumor suppressor genes in human cancers have been well documented. However, the molecular mechanisms underlying this silencing process, including the role of histone modifications and chromatin structural features, remain poorly understood. Here, we have characterized the TMS1 gene, which is silenced in cancers by DNA hypermethylation. Consistent with other studies, we find that the active state is characterized by histones hypermethylated at H3K4 and hypomethylated at H3K9, whereas the inactive state exhibits histones hypomethylated at H3K4 and hypermethylated at H3K9 (39, 40). A recent genome-wide study comparing global histone modifications in normal tissues, cancer cell lines and primary tumors revealed that carcinogenesis is accompanied by a global loss of H4K16Ac and H4K20me3 (11). The study further suggested that this loss was not associated with individual gene loci but rather to alterations at repetitive DNA sequences. Our findings indicate that gene-specific alterations in H4K16Ac and H4K20me3 also accompany carcinogenesis. The active TMS1 locus is characterized by two prominent H4K16Ac peaks and positioned nucleosomes that flank the boundaries of the unmethylated CpG island. Epigenetic silencing of TMS1 is characterized by a loss of these peaks, random positioning of nucleosomes and the appearance of a H4K20me3 peak at the transcription start site. hMOF-mediated acetylation of H4K16, and the hMSL complex, play an integral role in maintaining an open chromatin state at the TMS1 locus, as downregulation of H4K16Ac leads to loss of nucleosome positioning and decreased TMS1 expression.
H4K16Ac has been shown to localize to both heterochromatic and euchromatic regions and has been linked to gene activation (30, 33, 41). Here, we see that H4K16Ac localizes to peaks occurring precisely at the boundaries between the unmethylated CpG island and flanking methylated DNA in the active TMS1 gene. The peaks of H4K16Ac could promote gene expression in two ways. They may act to prevent or oppose the spread of DNA methylation into the regulatory regions contained within the CpG island. In S. cerevisiae, the silencing complex SIR2 binds along the telomeres and at the mating type locus to maintain these regions in a heterochromatic state (42, 43). Acetylation of H4K16 by Sas2p, the yeast ortholog of hMOF, at the boundary of these regions prevents the spreading of the SIR complex and thus heterochromatin, into nearby euchromatic regions. These observations indicate that SIR2 and H4K16Ac act in opposition to define the boundaries between active and inactive chromatin. Though we did not observe methylation of the CpG island upon H4K16Ac downregulation, it is possible that complete deacetylation of H4K16 and/or additional factors is required to trigger CpG island methylation.
Alternatively, the H4K16Ac peaks at the active TMS1 locus may promote the binding of a factor that activates transcription or prevent the docking of a factor that represses TMS1 transcription. Deacetylation of H4K16 in this case would prevent the binding of the activator or promote the association of the repressor, resulting in TMS1 repression. Proteins involved in transcription have been shown to preferentially bind the acetylated or non-acetylated form of H4K16Ac. Bdf1, which associates with the TFIID transcription factor complex to promote transcription, binds H4K16 when it is acetylated (44). ISWI, an ATP-dependent remodeling complex that promotes transcription repression, specifically binds to the non-acetylated form of H4K16 (45).
Our studies indicate that unlike H3Ac and H3K4me, which are more widespread marks among actively transcribing genes, H4K16Ac has a more select set of target genes. The presence of H4K16Ac at these target genes may promote expression, and loss of H4K16Ac may be a pre-requisite for gene silencing. Indeed, the presence of SirT1, which antagonizes H4K16Ac and is the human homolog of yeast SIR2, at certain tumor suppressor genes is associated with gene suppression and ablation of SirT1 leads to a local increase in H4K16Ac levels and activation of these genes (46). While its presence at TMS1 was necessary to maintain TMS1 expression, H4K16Ac was not present at and did not regulate the CDH1 and ESR1 genes, both of which are silenced by aberrant CpG island methylation in cancers. These findings are in contrast to those of Pruitt et al. (46), who showed that inhibition of SirT1 restored CDH1 expression in cancer cells where the gene was methylated and silent, implying a role for H4K16Ac at the CDH1 locus. Genes targeted by H4K16Ac likely include those that are involved in essential cellular functions, since knock-down of hMOF resulted in eventual cell death (35)(data not shown).
We find positioned nucleosomes throughout the CpG island in cells that are unmethylated and express TMS1. On the contrary, epigenetic silencing is associated with a loss of positioning. This is consistent with previous studies showing that CpG islands in the unmethylated and active state are associated with positioned nucleosomes whereas nucleosomes are randomly positioned across the silent locus (12, 13, 47). In the active TMS1 gene, positioning occurred at regular 200 bp intervals with one notable exception at the transcription start site, where there was an ∼400 bp space suggesting the active TMS1 chromatin structure includes a nucleosome-free region at the transcription start site. Genome-wide analysis of nucleosome positioning in yeast has shown that active genes tend to have a nucleosome-free region overlapping the transcription start site that is flanked by two strongly positioned nucleosomes (48). Several mammalian genes have also been shown to exhibit a region thought to be devoid of nucleosomes coincident with transcription factor binding sites (49).
Interestingly, we further show that two strongly positioned nucleosomes flank the unmethylated CpG island and are coincident with the peaks of H4K16Ac (see Figure 2C), suggesting that the marking of these sentinel nucleosomes may play a role in maintaining the integrity of the CpG island domain and the positioning of the remaining nucleosomes in the CpG island. Indeed, knock-down of hMOF and the corresponding decrease in H4K16Ac led to a loss of positioning. Whether this is due to a direct effect of hMOF and associated factors on nucleosome positioning or is an indirect effect of the resulting transcriptional downregulation is not clear. At this point, the factors that dictate preferential nucleosome placement are not well understood, but our data suggest that specific histone modifications, or the complexes that mark them, may play an important role.
H4K20me3 plays an important role in maintaining pericentric heterochromatin (32). Our data indicate that H4K20me3 localizes to a discreet peak upstream of the transcription start site of TMS1 in cases when the gene is methylated and silent, suggesting that H4K20me3 may also play a role in the repression of coding genes in euchromatic regions. Indeed, preliminary data indicates that downregulation of Suv4-20h2, the methyltransferase responsible for H4K20me3, stimulates TMS1 re-expression in cells where it is methylated and silenced, but only when DNA is first demethylated by 5azadC treatment (data not shown). The precise localization of H4K20me3 raises the question of how this mark is directed to the TMS1 locus. At pericentric heterochromatin, a model has been proposed wherein H4K20me3 is directed by pre-exiting H3K9me3 through the binding of HP1 and recruitment of the Suv4-20h enzymes (32). However, what little H3K9me3 was observed at the TMS1 locus was distributed throughout the promoter and coding regions whereas H4K20me3 was localized to a sharp peak near the transcription start (Figures 1B and 2A). The distinct distribution of H4K20me3 and H3K9me3 at the TMS1 locus suggests that additional mechanisms and/or factors may direct H4K20me3 to individual genes.
Recent studies have demonstrated that hMOF is downregulated in primary breast cancers and medullablastomas (36). In addition, loss of hMOF function in human cells leads to genomic instability, defects in cell cycle, chromosomal aberrations and impaired DNA damage response (33, 34, 50). Our findings suggest that an additional consequence may be the epigenetic silencing of certain tumor suppressor genes. Although downregulation of hMOF-mediated H4K16Ac led to local changes in nucleosome positioning and silencing of TMS1 gene expression, it was not sufficient to drive the subsequent demethylation of H3K4 or the acquisition of more ‘heterochromatic’ features (H3K9me2, H4K20me3, DNA methylation) associated with the locus in the stably silent state observed in cancer cells. Thus, while H4K16 deacetylation and transcriptional downregulation may be requisite steps, there must be additional triggers necessary to achieve the more stable and heritable silencing associated with aberrant CpG island DNA methylation. Further studies to understand the precise mechanism by which H4K16 deacetylation mediates gene inactivation will allow development of potential therapeutic agents that prevent silencing of tumor suppressor genes inactivated by this mechanism in tumorigenesis.
ACKNOWLEDGEMENTS
The authors thank Dr. Edwin Smith for the hMOF and hMSL1 antibodies and Dr. John Lucchesi for critical reading of the manuscript.
Footnotes
This work was supported by an NIH Grant (2RO1-CA077337) to PMV, a CHIR Post-Doctoral Fellowship to PKV and a NSF PRISM Fellowship (#DGE0536941 and #DGE0231900) to JDK.
REFERENCES
- 1.Antequera F, Boyes J, Bird A. High levels of de novo methylation and altered chromatin structure at CpG islands in cell lines. Cell. 1990;62:503–14. doi: 10.1016/0092-8674(90)90015-7. [DOI] [PubMed] [Google Scholar]
- 2.Jones PL, Veenstra GJ, Wade PA, et al. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19:187–91. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 3.Bird AP. CpG-rich islands and the function of DNA methylation. Nature. 1986;321:209–13. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- 4.Laird PW. Cancer epigenetics. Hum Mol Genet. 2005;14:R65–76. doi: 10.1093/hmg/ddi113. [DOI] [PubMed] [Google Scholar]
- 5.Davie JR, Spencer VA. Control of histone modifications. J Cell Biochem. 1999;32-33(Suppl):141–8. doi: 10.1002/(sici)1097-4644(1999)75:32+<141::aid-jcb17>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 6.Bernstein BE, Kamal M, Lindblad-Toh K, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–81. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 7.Weber M, Hellmann I, Stadler MB, et al. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39:457–66. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- 8.Heard E, Rougeulle C, Arnaud D, Avner P, Allis CD, Spector DL. Methylation of histone H3 at Lys-9 is an early mark on the X chromosome during X inactivation. Cell. 2001;107:727–38. doi: 10.1016/s0092-8674(01)00598-0. [DOI] [PubMed] [Google Scholar]
- 9.Plath K, Fang J, Mlynarczyk-Evans SK, et al. Role of histone H3 lysine 27 methylation in X inactivation. Science. 2003;300:131–5. doi: 10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- 10.Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet. 2007;8:286–98. doi: 10.1038/nrg2005. [DOI] [PubMed] [Google Scholar]
- 11.Fraga MF, Ballestar E, Villar-Garea A, et al. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat Genet. 2005;37:391–400. doi: 10.1038/ng1531. [DOI] [PubMed] [Google Scholar]
- 12.Patel SA, Graunke DM, Pieper RO. Aberrant silencing of the CpG island-containing human O6-methylguanine DNA methyltransferase gene is associated with the loss of nucleosome-like positioning. Mol Cell Biol. 1997;17:5813–22. doi: 10.1128/mcb.17.10.5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen C, Yang TP. Nucleosomes are translationally positioned on the active allele and rotationally positioned on the inactive allele of the HPRT promoter. Mol Cell Biol. 2001;21:7682–95. doi: 10.1128/MCB.21.22.7682-7695.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Segal E, Fondufe-Mittendorf Y, Chen L, et al. A genomic code for nucleosome positioning. Nature. 2006;442:772–8. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pennings S, Allan J, Davey CS. DNA methylation, nucleosome formation and positioning. Brief Funct Genomic Proteomic. 2005;3:351–61. doi: 10.1093/bfgp/3.4.351. [DOI] [PubMed] [Google Scholar]
- 16.Davey C, Fraser R, Smolle M, Simmen MW, Allan J. Nucleosome positioning signals in the DNA sequence of the human and mouse H19 imprinting control regions. J Mol Biol. 2003;325:873–87. doi: 10.1016/s0022-2836(02)01340-2. [DOI] [PubMed] [Google Scholar]
- 17.Conway KE, McConnell BB, Bowring CE, Donald CD, Warren ST, Vertino PM. TMS1, a novel proapoptotic caspase recruitment domain protein, is a target of methylation-induced gene silencing in human breast cancers. Cancer Res. 2000;60:6236–42. [PubMed] [Google Scholar]
- 18.McConnell BB, Vertino PM. Activation of a caspase-9-mediated apoptotic pathway by subcellular redistribution of the novel caspase recruitment domain protein TMS1. Cancer Res. 2000;60:6243–7. [PubMed] [Google Scholar]
- 19.Ohtsuka T, Ryu H, Minamishima YA, et al. ASC is a Bax adaptor and regulates the p53-Bax mitochondrial apoptosis pathway. Nat Cell Biol. 2004;6:121–8. doi: 10.1038/ncb1087. [DOI] [PubMed] [Google Scholar]
- 20.Masumoto J, Taniguchi S, Ayukawa K, et al. ASC, a novel 22-kDa protein, aggregates during apoptosis of human promyelocytic leukemia HL-60 cells. J Biol Chem. 1999;274:33835–8. doi: 10.1074/jbc.274.48.33835. [DOI] [PubMed] [Google Scholar]
- 21.Parsons MJ, Vertino PM. Dual role of TMS1/ASC in death receptor signaling. Oncogene. 2006;25:6948–58. doi: 10.1038/sj.onc.1209684. [DOI] [PubMed] [Google Scholar]
- 22.Moriai R, Tsuji N, Kobayashi D, et al. A proapoptotic caspase recruitment domain protein gene, TMS1, is hypermethylated in human breast and gastric cancers. Anticancer Res. 2002;22:4163–8. [PubMed] [Google Scholar]
- 23.Virmani A, Rathi A, Sugio K, et al. Aberrant methylation of TMS1 in small cell, non small cell lung cancer and breast cancer. Int J Cancer. 2003;106:198–204. doi: 10.1002/ijc.11206. [DOI] [PubMed] [Google Scholar]
- 24.Yokoyama T, Sagara J, Guan X, et al. Methylation of ASC/TMS1, a proapoptotic gene responsible for activating procaspase-1, in human colorectal cancer. Cancer Lett. 2003;202:101–8. doi: 10.1016/j.canlet.2003.08.027. [DOI] [PubMed] [Google Scholar]
- 25.Stone AR, Bobo W, Brat DJ, Devi NS, Van Meir EG, Vertino PM. Aberrant methylation and down-regulation of TMS1/ASC in human glioblastoma. Am J Pathol. 2004;165:1151–61. doi: 10.1016/S0002-9440(10)63376-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levine JJ, Stimson-Crider KM, Vertino PM. Effects of methylation on expression of TMS1/ASC in human breast cancer cells. Oncogene. 2003;22:3475–88. doi: 10.1038/sj.onc.1206430. [DOI] [PubMed] [Google Scholar]
- 27.Stimson KM, Vertino PM. Methylation-mediated silencing of TMS1/ASC is accompanied by histone hypoacetylation and CpG island-localized changes in chromatin architecture. J Biol Chem. 2002;277:4951–8. doi: 10.1074/jbc.M109809200. [DOI] [PubMed] [Google Scholar]
- 28.Vertino PM, Yen RW, Gao J, Baylin SB. De novo methylation of CpG island sequences in human fibroblasts overexpressing DNA (cytosine-5-)-methyltransferase. Mol Cell Biol. 1996;16:4555–65. doi: 10.1128/mcb.16.8.4555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kirmizis A, Bartley SM, Kuzmichev A, et al. Silencing of human polycomb target genes is associated with methylation of histone H3 Lys 27. Genes Dev. 2004;18:1592–605. doi: 10.1101/gad.1200204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dou Y, Milne TA, Tackett AJ, et al. Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell. 2005;121:873–85. doi: 10.1016/j.cell.2005.04.031. [DOI] [PubMed] [Google Scholar]
- 31.Shogren-Knaak M, Peterson CL. Switching on chromatin: mechanistic role of histone H4-K16 acetylation. Cell Cycle. 2006;5:1361–5. doi: 10.4161/cc.5.13.2891. [DOI] [PubMed] [Google Scholar]
- 32.Schotta G, Lachner M, Sarma K, et al. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev. 2004;18:1251–62. doi: 10.1101/gad.300704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith ER, Cayrou C, Huang R, Lane WS, Cote J, Lucchesi JC. A human protein complex homologous to the Drosophila MSL complex is responsible for the majority of histone H4 acetylation at lysine 16. Mol Cell Biol. 2005;25:9175–88. doi: 10.1128/MCB.25.21.9175-9188.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taipale M, Rea S, Richter K, et al. hMOF histone acetyltransferase is required for histone H4 lysine 16 acetylation in mammalian cells. Mol Cell Biol. 2005;25:6798–810. doi: 10.1128/MCB.25.15.6798-6810.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gupta A, Guerin-Peyrou TG, Sharma GG, et al. Mammalian Ortholog of Drosophila MOF that Acetylates Histone H4 Lysine16 is Essential for Embryogenesis and Oncogenesis. Mol Cell Biol. 2008;28:397–409. doi: 10.1128/MCB.01045-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pfister S, Rea S, Taipale M, et al. The histone acetyltransferase hMOF is frequently downregulated in primary breast carcinoma and medulloblastoma and constitutes a biomarker for clinical outcome in medulloblastoma. Int J Cancer. 2008;122:1207–13. doi: 10.1002/ijc.23283. [DOI] [PubMed] [Google Scholar]
- 37.Graff JR, Herman JG, Myohanen S, Baylin SB, Vertino PM. Mapping patterns of CpG island methylation in normal and neoplastic cells implicates both upstream and downstream regions in de novo methylation. J Biol Chem. 1997;272:22322–9. doi: 10.1074/jbc.272.35.22322. [DOI] [PubMed] [Google Scholar]
- 38.Ottaviano YL, Issa JP, Parl FF, Smith HS, Baylin SB, Davidson NE. Methylation of the estrogen receptor gene CpG island marks loss of estrogen receptor expression in human breast cancer cells. Cancer Res. 1994;54:2552–5. [PubMed] [Google Scholar]
- 39.Nguyen CT, Weisenberger DJ, Velicescu M, et al. Histone H3-lysine 9 methylation is associated with aberrant gene silencing in cancer cells and is rapidly reversed by 5-aza-2′-deoxycytidine. Cancer Res. 2002;62:6456–61. [PubMed] [Google Scholar]
- 40.Kondo Y, Shen L, Issa JP. Critical role of histone methylation in tumor suppressor gene silencing in colorectal cancer. Mol Cell Biol. 2003;23:206–15. doi: 10.1128/MCB.23.1.206-215.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnson CA, O’Neill LP, Mitchell A, Turner BM. Distinctive patterns of histone H4 acetylation are associated with defined sequence elements within both heterochromatic and euchromatic regions of the human genome. Nucleic Acids Res. 1998;26:994–1001. doi: 10.1093/nar/26.4.994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grunstein M. Yeast heterochromatin: regulation of its assembly and inheritance by histones. Cell. 1998;93:325–8. doi: 10.1016/s0092-8674(00)81160-5. [DOI] [PubMed] [Google Scholar]
- 43.Suka N, Luo K, Grunstein M. Sir2p and Sas2p opposingly regulate acetylation of yeast histone H4 lysine16 and spreading of heterochromatin. Nat Genet. 2002;32:378–83. doi: 10.1038/ng1017. [DOI] [PubMed] [Google Scholar]
- 44.Matangkasombut O, Buratowski S. Different sensitivities of bromodomain factors 1 and 2 to histone H4 acetylation. Mol Cell. 2003;11:353–63. doi: 10.1016/s1097-2765(03)00033-9. [DOI] [PubMed] [Google Scholar]
- 45.Corona DF, Clapier CR, Becker PB, Tamkun JW. Modulation of ISWI function by site-specific histone acetylation. EMBO Rep. 2002;3:242–7. doi: 10.1093/embo-reports/kvf056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pruitt K, Zinn RL, Ohm JE, et al. Inhibition of SIRT1 reactivates silenced cancer genes without loss of promoter DNA hypermethylation. PLoS Genet. 2006;2:e40. doi: 10.1371/journal.pgen.0020040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Macleod D, Charlton J, Mullins J, Bird AP. Sp1 sites in the mouse aprt gene promoter are required to prevent methylation of the CpG island. Genes Dev. 1994;8:2282–92. doi: 10.1101/gad.8.19.2282. [DOI] [PubMed] [Google Scholar]
- 48.Yuan GC, Liu YJ, Dion MF, et al. Genome-scale identification of nucleosome positions in S. cerevisiae. Science. 2005;309:626–30. doi: 10.1126/science.1112178. [DOI] [PubMed] [Google Scholar]
- 49.Gal-Yam EN, Jeong S, Tanay A, Egger G, Lee AS, Jones PA. Constitutive nucleosome depletion and ordered factor assembly at the GRP78 promoter revealed by single molecule footprinting. PLoS Genet. 2006;2:e160. doi: 10.1371/journal.pgen.0020160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gupta A, Sharma GG, Young CS, et al. Involvement of human MOF in ATM function. Mol Cell Biol. 2005;25:5292–305. doi: 10.1128/MCB.25.12.5292-5305.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]