Abstract
Systemic lupus erythematosus (SLE) is the prototype of complex autoimmune diseases. Studies have suggested that genetic, hormonal, and environmental factors contribute to the development of the disease. Interestingly, several recent studies involving SLE patients and mouse models of the disease have suggested a role for interferon (IFN)-stimulated genes (ISGs) in the development of SLE. One family of ISGs is the Ifi200-family, which includes mouse (Ifi202a, Ifi202b, Ifi203, Ifi204, and Ifi205) and human (IFI16, MNDA, AIM2, and IFIX) genes. The mouse genes cluster between serum amyloid P-component (Apcs) and α-spectrin (Spna-1) genes on chromosome 1 and the human genes cluster in syntenic region 1q23. The Ifi200-family genes encode structurally and functionally-related proteins (the p200-family proteins). Increased expression of certain p200-family proteins in cells is associated with inhibition of cell proliferation, modulation of apoptosis, and cell differentiation. Our studies involving generation of B6.Nba2 congenic mice, coupled with gene expression analyses, identified the Ifi202 as a candidate lupus-susceptibility gene. Importantly, recent studies using different mouse models of SLE have suggested that increased expression of Ifi202 gene (encoding p202 protein) in immune cells contributes to lupus susceptibility. Consistent with a functional role for the p202 protein in lupus susceptibility, increased levels of IFI16 protein in human SLE patients are associated with the diseases. This review summarizes recent findings concerning the regulation and role of p200-family proteins in the development of SLE.
Keywords: Interferons, Ifi200-family, SLE, Gender, Transcriptional regulation
1. Introduction
Interferons (IFNs) are a family of cytokines [1–3]. The family includes Type-I (IFN-α and IFN-β) and Type-II (IFN-γ) IFNs among others [1, 2]. Infection of cells with certain viruses or treatment of cultured cells with polyinosinic: polycytidylic acid (polyI: C) results in production of IFNs as a part of anti-viral cellular response [4]. The IFNs exhibit multiple biological activities both in vitro and in vivo [1–4]. These activities include well-characterized antiviral and immunomodulatory activities, and relatively less understood cell growth-regulatory activities, such as inhibition of cell proliferation and modulation of cell survival [1, 2].
Binding of an IFN (α, β, or γ) to the corresponding cell surface receptor results in activation of the Janus family of tyrosine kinases and activation of signal transducer and activator of transcription (STAT) proteins [1, 4]. Importantly, transcriptional activation of IFN-stimulated genes (ISGs) by the activated STATs results in induction of IFN-inducible proteins that mediate the biological activities of IFNs [1, 5]. One family of structurally-related IFN-inducible proteins is the p200-family [6, 7].
Systemic lupus erythematosus (SLE) is the prototypical autoimmune disease of unknown etiology [8–11]. The disease predominantly affects women between the age of 15 and 40-years [10, 11]. The disease is a heterogeneous syndrome with a complex immunopathogenesis. The disease has potential to affect many organ systems, including the skin, kidneys, and nervous system [10, 12]. Elevated levels of antinuclear autoantibodies (ANA) are a hallmark of SLE. Interestingly, recent studies have indicated that SLE patients with active disease have elevated levels of interferon-α/β in their serum [13–20]. Moreover, consistent with increased serum levels of IFN-α in SLE patients, studies [13, 15, 19, 20] have demonstrated that peripheral blood mononuclear cells from SLE patients exhibit a gene expression profile indicative of an active IFN-α signaling. Additionally, a strong correlation between the expression levels of IFN-α-inducible genes and renal disease has been reported [15].
Several mouse models of SLE have been used [8, 21–24]. For example, (NZB × NZW)F1 mice spontaneously develop an autoimmune glomerulonephritis associated with anti-dsDNA antibody deposits [8, 10, 21–24]. MLR/lpr mice also produce high amounts of autoantibodies. The lpr (lpr: lymphoproliferation) trait results from an autosomal recessive mutation in Fas gene leading to the accumulation of non-deleted autoimmune T and B lymphocytes [24]. Notably, defects in the Fas gene are not found in human SLE patients. In BXSB mice, Yaa (Y-linked autoimmune accelerator) induces severe disease in males, but not in females [24, 25]. In all these mouse models of SLE, mice develop increased levels of anti-nuclear antibodies and kidney disease, which are also seen in patients with SLE [21–25].
Several studies [26–32] involving mouse models of lupus have provided evidence for the IFN-signaling in the development of lupus-like disease. For example, lupus-prone mice that cannot signal through the IFN-α/β receptor fail to develop lupus-like disease [26, 27] and treatment of (NZB × NZW)F1 lupus susceptible mice with IFN-α accelerates disease development and death. Importantly, a recent study has provided a direct proof that increased levels of IFN-α induce early lethal lupus in pre-autoimmune (NZB × NZW)F1 but not in Balb/c mice [28]. The study also demonstrated that the prolonged expression of IFN-α in vivo leads to dramatic acceleration of lupus manifestation in young lupus-prone mice. Moreover, the study also revealed that increased levels of IFN-α in mice are not sufficient to induce lupus disease and the genetic background appears to play a critical role. Consistent with a role for genetic background of mice in lupus susceptibility, it has been reported that type I IFNs suppress autoimmunity in MRL/lpr mice [33]. Moreover, antibody-mediated blockade of type I interferon activity in B6.Sle2 mice and C57BL/6 control mice augmented serum autoantibody levels and boosted B1a cell numbers [34], indicating that increased serum levels of the type I interferons have different role in some mouse models of SLE.
There are excellent reviews [6, 7, 35–40] concerning the structural similarities among the p200-family proteins and their role in the regulation of cell growth and differentiation. Therefore, focus of this review is on the recent advances in our understanding of the regulation and role of the p200-family proteins in the development of autoimmune diseases. It is anticipated that recent genetic and molecular studies concerning the Ifi200-family genes and encoded proteins will advance our understanding of the role of IFNs in the development of autoimmune diseases, such as SLE
2. Interferon-inducible Ifi200-family genes
The p200-family proteins are encoded by Ifi200-family genes [6, 7]. The family includes mouse ISGs (for example, Ifi202a, Ifi202b, Ifi203, Ifi204 and Ifi205) and human ISGs (for example, IFI16, MNDA, AIM2, and IFIX). The murine Ifi200-family genes cluster between Apcs (encoding serum amyloid P-component or Sap protein) and Spna-1 (encoding α-spectrin protein) genes on chromosome 1 and the human Ifi200-family genes cluster in syntenic region 1q23 [6, 7, 38, 39]. Based on genetic studies using various mouse models of lupus disease and human SLE patients, the human 1q23 region is predicted [41, 42] to harbor autoimmunity susceptibility genes.
3. Mouse Ifi200-family genes in lupus susceptibility
New Zealand Black (NZB) mice develop a lupus-like autoimmune disease that is characterized by production of autoantibodies and mild glomerulonephritis that develops late in life [8–10, 24]. Offspring (NZB × NZW)F1 of the cross between NZB and New Zealand White (NZW) mice develop a progressive immune complex-mediated glomerulonephritis and high titer anti-dsDNA antibodies. These mice are considered to be excellent models of SLE [8, 24]. Importantly, characterization of immune defects in (NZB × NZW)F1 mice has contributed to our understanding of the human lupus disease.
Mapping studies [9, 10] have revealed that susceptibility to lupus disease in NZB and (NZB × NZW)F1 mice is polygenic, with multiple genes contributing to the generation of autoantibodies, kidney disease, and mortality. In particular, genetic loci on NZB chromosome 1 have been shown in multiple crosses to play a prominent role in disease susceptibility [10]. In particular, an interval extending from ~79 to 109 cM, which is termed Nba2 (NZB autoimmunity 2) has emerged as a major genetic contribution for the development of lupus-like disease [10, 43].
Our studies [36, 44] involving generation of congenic mice (designated as B6.Nba2) on C57BL/6 background, which contains the Nba2 interval, revealed that the congenic female mice develop splenomegaly and produce high titer IgG anti-nuclear antibodies. Interestingly, the NZB-derived Nba2 interval contains the Ifi200-gene cluster [44]. Furthermore, in an independent study, Wither et al. reported [45] that mice that are congenic for the NZB derived interval extending from 85–106 cM (this interval includes the Ifi200-gene cluster) on C57BL/6 background also develop splenomegaly, anti-nuclear antibodies, and increased memory T cells. These observations are consistent with an important role for Ifi200-family genes in lupus susceptibility in NZB and (NZB × NZW)F1 mice.
In our study [44], a comparison of gene expression analysis in splenic cells between C57BL/6 and B6.Nba2 congenic female mice revealed that: (i) Ifi202 (probably both Ifi202a and Ifi202b genes; see below) expression is up-regulated in B6.Nba2 mice; (ii) Ifi203 expression is down-regulated in B6.Nba2 mice; and (iii) Ifi204 expression remains unchanged between C57BL/6 and B6.Nba2 mice. Interestingly, a SNP in the promoter region of the Ifi202 gene correlated well with levels of expression: high in NZB and Balb/C mice that share the 95 C-allele, low in NZW, C57BL/6, and B10 mice that are characterized by a 95 T-allele. Although, these observations provided evidence that a particular SNP (the 95-C allele) in the promoter region of the Ifi202 gene is associated with increased steady-state levels of Ifi202 mRNA in congenic mice and the development of lupus-like disease, these observations did not provide a molecular basis for allelic variation contributing to alterations in the expression of Ifi202 gene. Our further analysis (see below) of the promoter region of the Ifi202 gene in C57BL/6 and NZB mice has identified additional polymorphisms in the promoter region that could account for differential expression of Ifi202 gene between the C57BL/6 and NZB mice. Additionally, we have identified polymorphisms in the coding region of the Ifi202 gene in the NZB mice (see below), which could account for increased levels of p202 protein in the B6.Nba2 mice through post-translational mechanisms.
Notably, steady-state levels of Ifi202 mRNA are not detectable in splenic cells from C57BL/6 mice and cells derived from these mice [6, 44, 46]. However, the steady-state levels of Ifi203 mRNA are readily detectable in C57BL/6 mice [44]. Interestingly, a recent study [47] has revealed that the Ifi203 mRNA is multiply spliced and encodes for at least two p203 proteins. Intriguingly, the study also revealed that p203 proteins are exclusively detected in the liver of adult C129/SvJ and C57BL/6 mice. Because p200-family proteins are predicted to homo- and heterodimerize [35, 36, 48], and levels of p200-family proteins are also regulated by post-transcriptional mechanisms [36], further studies are needed to examine potential role for other p200-family proteins, such as p203 and p204 proteins, in lupus susceptibility.
4. The Ifi202 genes in mouse models of SLE
The Ifi202 gene sub-family includes Ifi202a, Ifi202b and Ifi202c genes [35, 36, 49]. The Ifi202a gene encodes p202a protein and the Ifi202b gene encodes p202b protein. Co-expression of Ifi202a and Ifi202b genes has been reported [49, 50] in mouse tissues. Interestingly, ratios of Ifi202a and Ifi202b mRNAs differ among various adult mouse organs and tissues and the ratio is 1:1.2 in the spleen [49]. Of note, Ifi202a knockout (Ifi202a−/−) mice do not have any phenotype [49]. This is in part because in organs (including in spleen) and in mouse embryonic fibroblasts (MEFs) from the knockout mice the Ifi202b mRNA and protein levels are detectable. Moreover, there is a post-transcriptional compensatory increase in p202b protein levels in the Ifi202a−/− MEFs [49], raising the possibility that the compensatory increases in p202b protein levels in Ifi202a knockout mice could account for the lack of a phenotype. Importantly, our studies [50] have revealed that B6.Nba2 splenic cells express detectable levels of both Ifi202a and Ifi202b mRNAs.
In addition to our studies involving generation of B6.Nba2 congenic mice [36, 44], several recent studies (see below) have provided additional evidences for the idea that increased expression of Ifi202 (probably both Ifi202a and Ifi202b) gene in certain strains of mice is strongly associated with increased susceptibility to develop lupus disease.
4.1. Increased expression of Ifi202 in Apcs knockout mice
Mice with targeted disruption of the Apcs gene (encodes serum amyloid P-component) on a mixed genetic background (129/Sv × C57BL/6; F2) were reported [51] to develop high titers of anti-nuclear antibodies (ANA) and severe glomerulonephritis, a phenotype resembling SLE patients. However, in a subsequent study [52], which assessed the effect of genetic background and linked genes on autoimmunity in Apcs-null mice, revealed that genetic disruption of the Apcs gene is not sufficient for autoimmunity. Interestingly, the study found that production of ANA in Apcs−/− mice as compared to Apcs+/+ (129/Sv//Ev × C57BL6 × C3H/He) mice was associated with increased expression of Ifi202 mRNA in splenic cells. Because the study [52] ruled out the possibility that Sap protein expression suppresses the expression of the Ifi202 gene on a C57BL/6 background, the study suggested that autoimmunity in the Apcs−/− 129/Sv//Ev × C57BL/6 mice is due to the expression of 129/Sv//Ev-type genes, such as Ifi202. Furthermore, because an interval (87.9–105 cM) from 129-derived strain of mice on C57BL/6 genetic background is known to develop lupus-like disease in hybrid mice [53], the above observations raise the possibility that increased expression of 129 allele of Ifi202 gene in the hybrid (129 × C57BL/6) mice contributes to lupus-like disease.
4.2. Functional interactions between Yaa locus and Nba2 interval in congenic mice
Y-linked autoimmune accelerator (Yaa) locus, which is present on the Y chromosome of the BXSB lupus-prone mice, is responsible for the accelerated development of lupus-like autoimmune syndrome in BXSB mice and in their F1 hybrids with NZB or NZW mice [24, 25]. Interestingly, the Yaa locus has been identified to overexpress the toll-like receptor 7 (TLR7) [54, 55], activation of which by single-stranded RNA is likely to increase expression of interferon-α. Notably, Yaa locus itself is not sufficient to induce significant autoimmune responses in non-lupus-prone mice. However, it can induce and accelerate the development of SLE in combinations with autosomal susceptibility alleles present in lupus-prone mice [25]. It has been shown that the Yaa defect is functionally expressed in B cells, but not in T cells [25]. Thus, it has been postulated that the Yaa defect may decrease the threshold of BCR-mediated signaling thereby triggering and excessively stimulating autoreactive B cells. Interestingly, comparative assessment of three different B6.Yaa mice congenic for each three different NZB susceptibility interval (Nba2, Nba5, or Sgp3) [56], together with analysis of backcross mice, has revealed that the Nba2 interval regulates all three traits (anti-nuclear autoantibody production, gp70-anti-gp70 immune complex (gp70 IC) formation, and glomerulonephritis) that are associated with SLE disease. Importantly, the study [56] suggested that the presence of both Yaa and Nba2 is sufficient to induce a lethal form of lupus-like nephritis in C57BL/6 mice. Because our study [36, 44] suggested that interferon-inducible Ifi202 gene is a major contribution from the Nba2 interval, further studies are desirable to assess the relative contribution of the Ifi202 gene in this mouse model of lupus disease.
4.3. Increased expression of Ifi202 gene in Balb/c.C1 (77–105 cM) congenic mice
A recent study [57] utilized a murine model of lupus that is congenic for the NZB chromosome 1 on Balb/c genetic background to refine the pattern of differential transcriptional activation of Ifi200 genes between the two strains of mice. Comparison of the expression of Ifi200-family genes among splenic CD19+ B cells and CD4+ T-cells from NZB, Balb/C, and Balb/c.C1(77–105 cM) congenic mice revealed a significant over-expression of Ifi202 gene in congenic mice and reduced expression of Ifi203, Ifi204, and Ifi205 genes [57]. Of note, reduced expression of Ifi203 gene in Balb/c.C1(77–105 cM) congenic mice in this study is consistent with our previous studies [44] indicating that increased expression of Ifi202 gene (or increased levels of p202 protein) in B6.Nba2 mice is associated with reduced expression of Ifi203 mRNA. Together, these observations raise the possibility that increased expression of p202 protein in the B6.Nba2 congenic mice regulates the expression of Ifi203 gene. Therefore, further work will be needed to determine how increased expression of p202 (p202a and p202b) protein regulates the expression of Ifi203gene in lupus-prone mice.
4.4. Identification of Ifi202 gene as a candidate for lupus susceptibility in B10.Yaa.Bxs2/3 congenic mice
Generation of B10.Yaa.Bxs2/3 congenic mice, coupled with gene expression analyses, has identified the IFN-inducible Ifi202 gene as a lupus susceptibility gene [25]. This independent study further suggests a role for p202 protein in lupus susceptibility in B10.Yaa.Bxs2/3 mouse model.
4.5. Autoimmunity in MRLlpr/lpr mice and p202 expression
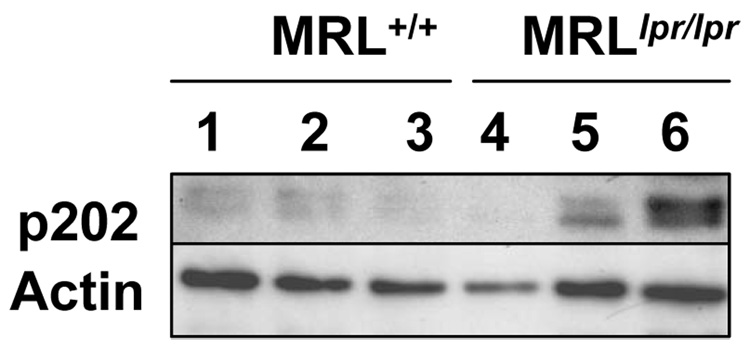
To determine whether increases in levels of p202 (p202a and p202b) protein in cells of the immune system contribute to lupus susceptibility in other lupus-prone mice, we have compared p202 protein levels in splenocytes isolated from age-matched MRLlpr/lpr mice and wild type (MRL+/+) mice [22, 24]. As shown in Fig. 1, levels of p202 protein were much higher in MRL lpr/lpr female mice than age-matched (~12 months) wild type female mice. These observations indicated that the development of autoimmunity in MRLlpr/lpr female mice is also associated with increases in p202 protein levels. Consistent with our above observations, a study [58] has revealed that increased expression of Ifi202 in spleen and kidney cells of MRL/lpr female mice is associated with development of lupus disease.
Figure 1.
Total cell extracts from splenocytes isolated from age-matched MRL+/+ (lanes 1–3) or MRLlpr/lpr (lanes 4–6) mice were analyzed by immunoblotting using antibodies specific to the indicated proteins.
Although, the above studies [25, 55, 56, 57, and Fig. 1] have provided evidences that increased expression of Ifi202 gene in certain strains of mice is associated with the development of lupus disease, signaling pathways that regulate the expression of the Ifi202 gene in immune cells remain to be elucidated. Moreover, it remains unclear how increased expression levels of the p202 protein in immune cells of lupus-prone mice contribute to the development of lupus disease.
5. Regulation of Ifi202 expression
Most cells produce basal low constitutive levels of type I IFNs (IFN-α and IFN-β) [59]. Therefore, it seems likely that the IFN-induced signaling pathways contribute to the regulation of Ifi202 gene expression in IFN-responsive cells. However, there are studies (see below) that suggest that IFN-independent signaling pathways also regulate expression of Ifi202 gene in immune and non-immune cells.
5.1. IFN-mediated Ifi202 regulation
Previous studies [6, 46] using Northern hybridization technique to detect Ifi202 mRNA levels had indicated that most cultured mouse fibroblasts and certain adult mouse tissues express low basal levels of Ifi202 mRNA. Moreover, IFN (α or β) treatment of cultured fibroblasts increases the steady-state levels of Ifi202 mRNA [6]. Furthermore, the increase in Ifi202 mRNA (and protein) is, in part, due to increases in the transcriptional rate of the Ifi202 gene [6]. Of note, studies using Northern hybridization indicated that splenic cells from non-lupus-prone C57BL/6 mice and immortalized embryonic fibroblasts from these mice do not express detectable levels of Ifi202 mRNA [6, 46]. Although, these studies provided very useful information concerning steady-state levels of Ifi202 mRNA, the technique failed to detect low basal steady-state levels of Ifi202 mRNA and did not distinguish between the expression levels of Ifi202a and Ifi202b mRNAs.
Using primers specific to Ifi202a or Ifi202b gene and semi-quantitative RT-PCR, a study [49] indicated that most cells and cell type that were tested express mRNAs encoded by both Ifi202a and Ifi202b genes. Therefore, using the gene-specific primers in RT-PCR, we tested whether splenic cells from B6.Nba2, C57BL/6, and NZB mice express detectable levels of Ifi202a, Ifi202b, or both mRNAs. Our experiments [50] revealed that NZB, B6.Nba2, and C57BL/6 cells express detectable levels of both mRNAs. Interestingly, levels of Ifi202a mRNA are relatively higher than the Ifi202b in NZB and B6.Nba2 mice. In contrast, levels of Ifi202b mRNA are higher than Ifi202a in C57BL/6 mice. Furthermore, using antibodies to p202 protein, which allow detection of both p202a and p202b proteins in immunoblotting [49], we could detect p202 protein in NZB and B6.Nba2, but not in C57BL/6, mice. These observations raise the possibility that post-transcriptional and post-translational mechanisms contribute to the lack of detection of p202 protein in the C57BL/6 splenic cells.
A recent study [60] reported that sera derived from lupus prone NZB mice had higher levels of IFN-α as compared to age-matched Balb/C mice after injection of the TLR9 ligand, indicating that the NZB mice mount faster and stronger response to microbial material than the Balb/C mice. Importantly, consistent with IFN-mediated up-regulation of Ifi202 expression [35], the study [60] also revealed that dendritic CD4+ and CD4− cells derived from bone marrow of NZB mice expressed much higher levels of Ifi202 mRNA than Balb/C mice.
It has been reported [27–29] that the effects of double-stranded RNA or IFN-α depend on the genetic background of mice. A recent study [29] showed that treatment of (B6.Nba2 × NZW)F1 mice, but not (B6 × NZW)F1 or parental strains (B6.Nba2, C57BL/6, or NZW), with polyI:C (or double-stranded RNA), which increases the expression of IFN-α/β, resulted in increased levels of proteinurea and lupus-like nephritis in mice. Because increased expression of Ifi202 gene in B6.Nba2 mice in part depends on IFN-signaling [30], the above observations raise the possibility that IFN-induced increased levels of p202 protein in (B6.Nba2 × NZW)F1 mice contribute to increased levels of proteinurea and lupus-like nephritis. Further work will be needed to test this hypothesis.
5.2. IFN-independent Ifi202 regulation
There are studies that IFN-independent signaling pathways also regulate expression of the Ifi202 gene in immune and non-immune cells: (i) during the differentiation of C2C12 myoblasts to myotubes in vitro, steady-state levels of the Ifi202 mRNA and protein are increased [61]; (ii) under reduced serum culture conditions, levels of the Ifi202 mRNA and protein increase in mouse fibroblasts [62]; (iii) Notch signaling positively regulates the expression of the Ifi202 mRNA in CD4+ and CD8+ double positive thymocytes [63]; (iv) steady-state levels of Ifi202 mRNA and protein decrease after increases in steady-state levels of wild type p53 protein and activation of p53 by DNA-damaging agents in cultured fibroblasts [64]; (v) Ifi202 mRNA and protein levels decrease after serum (or growth factor) stimulation of serum-starved growth-arrested fibroblasts and the decrease is associated with activation of certain E2F-family of transcription factors [65], and (vi) transformation of NIH 3T3 cells with activated H-Ras results in up-regulation of Ifi202 mRNA and protein [66].
Consistent with the above observations that expression of Ifi202 is also regulated by IFN-independent mechanisms; our studies revealed that in B6.Nba2 splenocytes expression of Ifi202 gene is up-regulated by IL-6 treatment [67]. Likewise, we recently noted [68] that stimulation of splenic T cells in vitro with anti-CD3 and anti-CD28 up-regulates the expression of the Ifi202 gene. Furthermore, generation of B6.Nba2 mice that were defective in type I IFN-signaling revealed that the defect resulted in only two fold reduction in Ifi202 mRNA levels in splenocytes as compared to the wild type mice [30]. Together, these observations suggest that both IFN-dependent and independent signaling pathways contribute to Ifi202 expression in immune cells of the B6.Nba2 lupus-prone mice.
5.3. Promoter polymorphisms and differential expression of Ifi202 gene
The 5’-regulatory region of Ifi202 gene is polymorphic [44]. Therefore, it seems likely that these polymorphisms could account for differential regulation of the Ifi202 gene expression in certain strains of mice. Because an earlier study [69] had indicated that ~800-bp 5’-regulatory region of the Ifi202 gene contributes to transcription regulation, we sequenced the 5’-regulatory region of Ifi202 gene from C57BL/6 and NZB mice and compared the sequence. The sequence comparison revealed numerous polymorphisms between C57BL/6 and NZB mice (Fig. 2), including a 3 bp (TTT) insertion in the NZB allele. These sequence polymorphisms between the C57BL/6 and NZB mice are predicted to result in differences in binding of transcription factors to the Ifi202 gene in vivo (Table 1).
Figure 2.
A comparison of the 5’-regulatory sequences of the Ifi202 gene between C57BL/6 (B6) and NZB mice. A complete nucleotide sequence for the B6 mice is shown and a sequence for the NZB mice is shown only when it is either polymorphic or there is an insertion of nucleotides.
Table 1.
Locations of polymorphic sites in the 5’-regulatory region of the Ifi202 gene in C57BL/6 and NZB mice. The polymorphic sites that are predicted to result in alterations potential DNA-binding sites for transcription factors are indicated.
| Relative location of polymorphisms | C57BL/6 allele | NZB allele | Predicted change in binding of a transcription factor(s) to NZB allele |
|---|---|---|---|
| −696 | T | G | A new site for NF-GMB |
| −600 | A | G | A new site for TCF-1α |
| −511 | G | A | A new site for GATA-1 |
| −487 | G | A | A new site for glucocorticoid receptor (GR) |
| −414 | T | C | A site for CP2 is lost |
| −394 | C | T | A new TATA-binding site |
| −311 | - | Insertion of TTT | A new site for Krupple-like factor |
| −231 | T | G | A new site for KLF6 and AP-1 |
| −164 | C | T | A site for Sry-β lost |
| −139 | T | G | A new site for GATA-1 and GR |
Of note, a polymorphism at nucleotide −394 in the 5’-regulatory region of Ifi202 gene in NZB mice results in a typical TATA box sequence (TATAAAA). The TATA-binding protein (TBP) recognizes the TATA box [70] that is often found in genes that are highly transcribed. Importantly, the polymorphism at −394 position results in a weak TATA box (TACAAAA) in the C57BL/6 allele (Fig. 3). Additionally, a strong TATA box is also found in the promoter region of the Ifi202gene in AKR mice [71]. Consistent with the presence of a strong TATA box in the AKR and NZB alleles of the Ifi202 gene, cells from these mice contain relatively higher steady-state levels of Ifi202 mRNA. Furthermore, consistent with the presence of a strong TATA box in the NZB, but not in C57BL/6 (B6), allele of Ifi202 gene, we noted appreciable (~2.5-fold) differences between the activity of B6-202-luc and NZB-202-luc reporter in B6 MEFs under normal (10%) and reduced (1%) serum conditions (data not shown). Interestingly, consistent with regulation of Ifi202 gene by serum growth factors [62], the difference between the two reporter activities was more (~3.5-fold) under reduced serum conditions (data not shown). Together, these observations suggest that the promoter region polymorphisms contribute to differential expression of the Ifi202 gene between C57BL/6 and NZB mice.
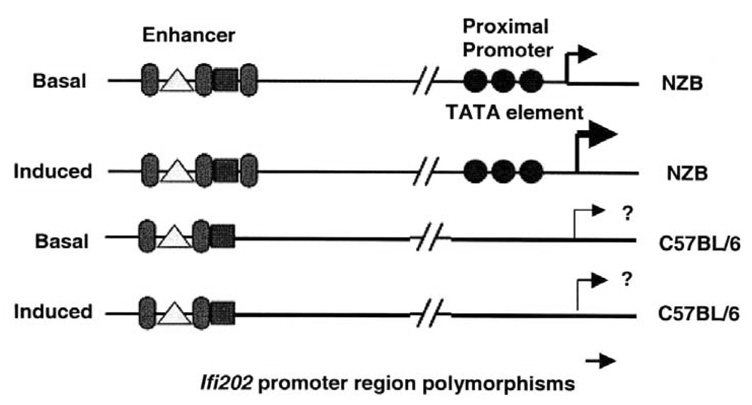
Figure 3.
The presence of a typical TATA sequence in the NZB allele is predicted to result in higher basal and induced (induced by IFN, or IL-6) levels of transcription of the Ifi202 gene in the B6.Nba2 congenic mice.
5.4. Polymorphisms in the coding region of the Ifi202 gene
We have also sequenced (NCBI sequence accession # DQ222946.1) the coding region of the Ifi202 gene in the NZB mice and compared (Fig. 4) the coding sequence with our previous Ifi202 sequence [71]. The comparison revealed that the coding region of the Ifi202 gene in NZB mice is polymorphic, resulting in changes in six amino acids in the p202 protein (Fig. 4). Although, the significance of these amino acids changes in NZB-derived p202 protein remains under investigation, these polymorphic changes in the amino acid sequence of the p202 protein have potential to affect its functions through altering the protein stability, sub-cellular localization, and/or protein-protein interactions. Further work will be needed to test this hypothesis.
Figure 4.
A comparison of the amino acid sequences of the p202 protein, which was previously published (ref. 71; the sequence was derived from mouse EAT cell line) by us, with that of the NZB mice derived p202 protein sequence. A complete amino acid sequence of the p202 protein from EAT cell line is shown and the sequence for the NZB-derived p202 protein is shown only when it is polymorphic.
5.5. Gender bias in the development of SLE and Ifi202 expression
Gender bias is observed in the development of human SLE [72, 73]. Similarly, it is known that female (NZB × NZW)F1 mice are more prone to lupus disease than male mice [74–76]. Consistent with these observations, a recent study [75] noted that female B6.Nba2 mice produce higher levels of anti-dsDNA, anti-ssDNA, anti-chromatin, and anti-histone autoantibodies than age-matched male mice. Moreover enhancement of autoantibody production in female mice is also apparent in (B6.Nba2 × NZW)F1 mice, which develop glomerulonephritis. Female (B6.Nba2 × NZW)F1 mice produce more anti-ssDNA, anti-chromatin, anti-dsDNA, and anti-histone autoantibodies than male (B6.Nba2 × NZW)F1 mice. Furthermore, total IgG levels are also 2-fold higher in female mice than male mice and female mice also develop more severe renal disease. These observations are consistent with the possibility that gender bias in lupus susceptibility that is observed in B6.Nba2 and (B6.Nba2 × NZW)F1 mice is associated with differential expression of lupus susceptibility genes, such as Ifi202 gene, between female and male mice. Therefore, further work will be needed to determine whether gender-dependent factors contribute to the regulation of Ifi202 expression.
It has been reported [77] that activation of TLR7 in cells derived from female SLE patients induces production of IFN-α at much higher levels than male patients. This could account for gender bias in the development of human SLE. However, it remains unknown whether gender-specific differences in the expression of TLR7 (or other TLRs) could account for gender bias in the development of lupus in mice.
6. The p202 proteins
The protein p202a (encoded by Ifi202a gene) is the first and the best-characterized member of the p200-protein family [6, 35, 36]. The protein, p202b (encoded by Ifi202b gene) differs from the p202a protein only in 7 N-terminal amino acids out of 445 [49]. Based on detection of steady-state levels of the Ifi202a or Ifi202b mRNA levels by semi-quantitative RT-PCR, both the mRNAs are co-expressed in several mouse tissues [49] and splenic cells from NZB and B6.Nba2 mice [50]. Interestingly, the steady-state levels of Ifi202a mRNA are relatively higher than the Ifi202b in B6.Nba2 mice [50]. However, because polyclonal antibodies that were raised against the full-length p202a protein [78] detect p202a and p202b proteins (Xin et al., unpublished data), the relative contributions of the p202a and p202b proteins in total cellular levels of p202 protein remain unknown. Therefore, it remains to be seen whether in B6.Nba2 mice (and in other lupus-prone mice) levels of p202 proteins (p202a and p202b) are differentially regulated in immune cells in cell type-specific manner.
The protein p202 (p202a) is detected as a phosphoprotein (52-kDa) in mouse fibroblasts in immunoblotting [78]. Moreover, the IFN-induced levels of p202 protein in cultured mouse AKR-2B fibroblasts are detected both in the cytoplasm and in the nucleus [78]. The cytoplasmic localization of p202 protein in B6.Nba2 fibroblasts is consistent with the lack of a classical nuclear localization signal (NLS) in the p202 protein [35, 78]. Therefore, it is likely that the nuclear localization of p202 protein is regulated (see below) in immune cells and may depend on cell type-specific factors. Consistent with this idea, it has been demonstrated that the large increases in the levels of p202 protein during differentiation of C2C12 myoblasts in vitro are associated with translocation of p202 protein into the cytoplasm [36].
6.1. Regulation of sub-cellular localization of p202 proteins by IFNs
In IFN-treated cultured fibroblasts, p202 protein (possibly both p202a and p202b) is detected both in the cytoplasm and in the nucleus [78]. Consistent with the presence of a mitochondrial targeting sequence in the p202a protein in the N-terminus [78, 79], in the cytoplasm, a significant fraction of p202 protein (possibly both p202a and p202b) associates with the mitochondria [79]. Additionally, we found that the constitutive levels of p202 protein are primarily detected in the cytoplasm of B6.Nba2 MEFs and IFN-α treatment of B6.Nba2 MEFs potentiated nuclear accumulation of p202 protein [79]. Because the p202a and the p202b proteins differ in 7 amino acids in the N-terminus [49], it is conceivable that these differences account for differential association of these two proteins with the mitochondrial membranes. Furthermore, because p202 (p202a) protein has the ability to forms homo- and heterodimers [35, 48], further work will be needed to determine what factors regulate sub-cellular localization of p202 proteins in immune cells.
Interestingly, a recent study [30] has indicated that B6.Nba2 and (B6.Nba2 × NZW)F1 mice, which are deficient in IFN-α/β-receptor signaling, fail to develop anti-nuclear antibodies and renal disease. Because nuclear localization of p202 protein in B6.Nba2 mouse embryonic fibroblasts (MEFs) is stimulated by IFN-α treatment [79], it seems likely that in B6.Nba2 mice the lack of signaling through the Type I IFN receptor results in nuclear exclusion of the p202 proteins, resulting in defects in nuclear functions of p202 proteins. Thus, functional inactivation of p202 proteins in the IFNAR−/− B6.Nba2 mice could account for the lack of anti-nuclear antibodies and renal disease. Obviously, further studies are needed to test the above hypothesis.
6.2. Inhibition of cell growth and modulation of apoptosis by p202 proteins
The ability of p202 (possibly both p202a and p202b) protein to retard cell proliferation and modulate cell survival is thought to depend on its ability to bind with certain transcription factors and modulate their transcriptional activities [7, 35, 36]. These transcription factors include: c-Fos, c-Jun, c-Myc, NF-κB (p50 and p65), E2Fs (E2F1 and E2F4), and p53 [35, 36]. Binding of p202 protein to most (excluding the p53) of these transcription factors results in inhibition of the sequence-specific DNA-binding activity [35].
Of note, increased expression of Ifi202 in dendritic cells from the NZB mice is associated with retardation of cell proliferation [60]. This observation is consistent with our previous observations [35, 36] that increased expression of p202 in a variety of cultured cells retards cell proliferation.
Because p53 [80–83] and E2F-family [83–87] of transcription factors are thought to contribute to the development of autoimmunity through defects in apoptosis of immune cells, we have further investigated the functional interactions between p202 protein and these two transcription factors in the B6.Nba2 mice to understand the molecular mechanisms by which increased expression of p202 protein in these lupus-prone mice contributes to lupus susceptibility.
6.2.1. Disruption of mutually-negative regulatory feedback loop between interferon-inducible p202 protein and p53 in lupus-prone mice
p53 protein is a transcription factor, which is activated in cells in response to several stimuli, including DNA damage, hypoxia, and oxidative stress [88, 89]. The activation of p53 in cells results in binding of p53 to its DNA-binding consensus sequence that is present in its target genes. The binding of p53 to its target genes results in either transcriptional activation or repression of genes [89]. Importantly, proteins that are encoded by the p53 target genes contribute to either cell growth arrest or apoptosis. For example, transcriptional activation of p21 gene results in increases in p21CIP1 protein levels and cell growth arrest. Likewise, transcriptional repression of the Ifi202 gene by p53 results in increased susceptibility to apoptosis [64]. Moreover, the lack of expression of p53 target genes, such as Gadd45a [81] or p21 [90] in mice has been reported to result in lupus-like disease.
Studies have provided evidence that p53 is required for spontaneous autoantibody production in B6/lpr lupus mice [82]. Generation of double mutant mice (p53−/− lpr) having defects in both p53- and Fas-dependent pathways revealed that these mice have lower autoantibody levels than the single mutant lpr mice. These studies suggested a role for p53 in the progression of autoimmunity and the production of autoantibodies. Consistent with the above studies, a recent study [83] revealed that the p53-deficient mice are more susceptible to streptozotocin-induced diabetes than control mice. Moreover, these mice produced higher levels of pro-inflammatory cytokines, such as interleukin-1, -6, and -12. The innate immune response of p53−/− macrophages to lipopolysaccharides and γ-interferon was significantly enhanced compared with p53+/+ cells. Additionally, p53−/− macrophages produced more pro-inflammatory cytokines and higher levels of total and phosphorylated signal transducer and activator of transcription (STAT)-1 [91]. These results indicate that p53 inhibits autoimmune diabetes and innate immune responses through down-regulating STAT-1 transcription factor and pro-inflammatory cytokines.
Because p53 represses transcription of Ifi202 gene through binding to a p53 DNA-binding site [64] and increased expression levels of p202 protein inhibit the p53-mediated transcription [92], the above observations prompted us to examine whether increased expression of p202 protein in immune cells of B6.Nba2 mice has any effect on p53 functions. We found [93] that increased expression of p202 protein in the B6.Nba2 splenocytes, as compared with cells derived from the parental C57BL/6 mice, was associated with increased levels of p53 protein and inhibition of p53-mediated transcription of its target genes that encode pro-apoptotic proteins, such as Gadd45a and Puma. Conversely, knockdown of p202 expression in B6.Nba2 cells resulted in stimulation of p53-mediated transcription. Furthermore, we also noted that: (i) p202 protein bound to p53 in the N-terminal region (amino acids 44–83) comprising the proline-rich region that is important for p53-mediated apoptosis; and (ii) increased expression of p202 protein in B6.Nba2 mouse embryonic fibroblasts inhibited p53-mediated apoptosis. Taken together, our observations support the idea that increased levels of p202 protein (possibly both p202a and p202b) in B6.Nba2 mice increases the susceptibility to develop lupus, in part, by inhibiting p53-mediated apoptosis. Further work will be needed to determine whether the functional interactions between p53 and p202 protein are regulated in cell-type specific manner.
6.2.2. Disruption of mutually-negative regulatory feedback loop between interferon-inducible p202 protein and the E2F-family of transcription factors in lupus-prone mice
The E2F family of transcription factors consists of six E2Fs which heterodimerize with one of the two different DP proteins to create 12 different DNA-binding transcriptional regulators [94–96]. The formation of heterodimeric protein complexes is shown to be essential for the production of high affinity E2F protein-DNA complexes since E2F homodimers have minimal DNA-binding activity and DP homodimers have little or no affinity for DNA-binding [94, 96].
The E2F factors can be divided into three subgroups: (i) E2F1, E2F2, and E2F3, which are highly related and display overall maximal expression in G1 to early S phase; (ii) E2F4 and E2F5, which are less responsive to changes in proliferation and lack an N-terminal domain present in E2Fs 1 to 3; and (iii) E2F6 that lacks both the N-terminal region common to E2Fs 1 to 3 and the C-terminal trans-activation domain common to E2Fs 1 to 5. The E2F trans-activation domain is encoded within an acidic carboxy-terminal region that also carries a site for binding by pocket proteins, such as pRb, p107, and p130 [96].
Known E2F target genes are numerous and include critical cell cycle regulators (e.g., cyclins, Cdks, and Cdk inhibitors), as well as important mediators of DNA synthesis (e.g., DNA polymerase α, DHFR, and thymidine kinase) [94, 96, 97], and pro-apoptosis genes (e.g., p73, Bim, Puma). Genes controlled by E2F family of transcription factors show low promoter activity in quiescent and early G1 phase cells and high promoter activity in late G1 and S phase cells.
The activity of E2F2 is required for suppression of T cell proliferation and immunologic self-tolerance [85]. Consequently, mice null for E2F2 develop late-onset autoimmune features, characterized by widespread inflammatory infiltrates, glomerular immunocomplex deposition, and anti-nuclear antibodies. Importantly, E2F2 appears to repress the transcription of the E2F1 whose activity is required for normal S phase entry [85]. On the contrary, E2F1 positively regulates the expression of E2F2 [85]. Because p202 inhibits the transcriptional activity of a subset of E2F family members [98, 99], it is likely that alterations in the levels of p202 in a subset of T cells affect E2F2 levels and/or activity, resulting in defects in immunologic self-tolerance.
Consistent with the above observations, we noted [100] that increased expression of Ifi202 in the B6.Nba2 congenic mice was associated with inhibition of E2F1-mediated transcription and decreased expression of E2F1 and its target genes that encode pro-apoptotic proteins. These observations support the idea that increased levels of p202 in certain strain of mice contribute to lupus susceptibility in part by inhibiting E2F1-mediated pro-apoptotic functions.
7. The human p200-family proteins in autoimmune diseases
Increased serum levels of interferon-α are detected in the majority of SLE patients [17–19]. Moreover, PBMCs from these patients exhibit interferon gene expression signature: expression levels of mRNAs encoded by the ISGs are up-regulated. Interestingly, knockdown of FcγRIIB in human PBMCs, which results in up-regulation ISGs, also results in up-regulation of steady-state levels of IFI16 mRNA [101]. Consistent with a potential role for the IFI16 protein in immune regulation, a study has suggested a role for IFI16 protein in T-cell development [102]. Furthermore, the naïve CD8+ T cells express relatively low levels of IFI16 mRNA. However, memory CD8+ cells express intermediate levels of IFI16 mRNA whereas the effector CD8+ cells express the highest levels of IFI16 mRNA [102]. These observations provide support for the idea that IFI16 protein may have role in differentiation of naïve CD8+ cells.
The IFI16 proteins (IFI16A, B, and C proteins) share the structure similarities with the murine p202 proteins (p202a and p202b) and appear to have a role in autoimmune diseases [36, 39]. Moreover, increased expression of IFI16 protein in endothelial cells is shown to induce expression of pro-inflammatory cytokines [103]. Importantly, basal and IFN-induced expression of IFI16 gene varies among individuals and may depend on the race [39]. Moreover, we found that the coding region of the IFI16 gene has polymorphisms, which could account for its differential regulation and functions in immune cells among individuals [39], raising the possibility that increased expression levels of IFI16 protein in immune cells contributes to lupus susceptibility. Consistent with this idea, it has been reported that the protein may be involved in the pathophysiological mechanisms of connective tissue disorders, such as systemic sclerosis [104, 105]. Moreover, up to 29% SLE [104] and 70% Sjogren’s syndrome [106] patients develop autoantibodies to the IFI16 protein. Although, the significance of these observations remains unclear in the development of SLE and other autoimmune disease, it is conceivable that increased levels of IFI16 proteins in immune cells contribute to defects in cell survival and apoptosis, resulting in increased susceptibility to develop SLE and other autoimmune diseases.
A study has revealed that increased steady-state levels of MNDA mRNA are associated with IFN signature in glomeruli isolated from a group of SLE patients [107]. However, the significance of the increased levels of MNDA mRNA in SLE-associated pathophysiology of glomeruli remains unclear.
In a recent study [57], authors fine mapped 317 kb of genome region encompassing the human Ifi200 gene cluster using single nucleotide polymorphism (SNP)-based genotyping in 350 nuclear United Kingdom (UK) SLE patients. This study revealed an association signal in the PYHIN1-IFI16 intergenic region. Moreover, the strongest association signal was noted within the 3’-untranslated region of PYHIN1. These observations raise the possibility that polymorphisms in the human Ifi200-family genes contribute to alterations in expression of certain IFI200-family genes. Therefore, further work will be needed to examine this possibility.
8. Conclusions
Several independent studies have provided evidence that the p200-family proteins participate in the regulation of cell growth and survival. Moreover, these studies have suggested that increased expression of Ifi202 gene in certain strains of mice is associated with defects in apoptosis of immune cells.
Based on recent studies using various congenic strains of mice, it is evident that increased levels of Ifi202 (probably both Ifi202a and Ifi202b) mRNA in immune cells are associated with development of lupus-like disease. Because cellular levels of p202 protein and its nuclear localization are regulated, it will be important to examine the regulation and role of p202 (probably both p202a and p202b) proteins in lupus-prone strains of mice.
The demonstrated ability of IFN-inducible p202 protein (possibly both p202a and p202b proteins) to inhibit cell proliferation and modulate cell survival in a variety of cell systems provides support for the idea that the p202 protein mediates the biological activities of IFNs. However, several IFN-independent signaling pathways appear to regulate the expression levels of p202 (probably both p202a and p202b) protein. Therefore, a complete understanding of the molecular mechanisms is needed to understand how p202 protein levels are regulated in immune cells.
The p202 protein has the ability to form homo- and heterodimers. Therefore, it is likely that physical interactions between the p202 protein and other family members, such as p203 and p204 (and may be other proteins), contribute to the regulation of nuclear localization p202 protein and its ability to modulate the transcriptional activities of factors that regulate cell survival, such as NF-κB, AP-1, E2Fs, and p53. Consistent with this idea, we have found that increased expression levels of p202 protein in female B6.Nba2 mice are associated with inhibition of p53 and E2F-mediated transcription of pro-apoptotic genes. Therefore, further work will be needed to determine whether increased levels of p202 protein in other lupus-prone mice modulate the transcriptional activities of pro- and/or anti-apoptotic factors.
Our understanding of the regulation and role of IFN-inducible p202 proteins in immune cells will provide novel insights into the signaling pathways and the molecular mechanisms that contribute to the development of lupus disease. These studies are likely to advance our knowledge of IFN-inducible proteins in the development of SLE. Increased understanding of the role of IFN-inducible genes and encoded proteins in SLE has potential to develop new approaches to diagnose and treat SLE patients.
Acknowledgments
This work is supported by a grant (AI066261) from the National Institutes of Health to D. C. The authors would like to thank Drs. Peter Lengyel, Brian Kotzin, Steven Rozzo, and Argyrios Theofilopoulos for their helpful suggestions. The authors would also like to thank Ms. Amruta Desai for her help in typing of references.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD. How cells respond to interferons? Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 2.Borden EC. Gene regulation and clinical roles for interferons in neoplastic diseases. Oncologist. 1998;3:198–203. [PubMed] [Google Scholar]
- 3.van Boxel-Dezaire AH, Rani MR, Stark GR. Complex modulation of cell type-specific signaling in response to type I interferons. Immunity. 2006;25:361–372. doi: 10.1016/j.immuni.2006.08.014. [DOI] [PubMed] [Google Scholar]
- 4.Garcia-Sastre A, Biron CA. Type I interferons and the virus-host relationship: a lesson in détente. Science. 2006;312:879–882. doi: 10.1126/science.1125676. [DOI] [PubMed] [Google Scholar]
- 5.Sen GC. Novel functions of interferon-induced proteins. Semin Cancer Biol. 2000;10:93–101. doi: 10.1006/scbi.2000.0312. [DOI] [PubMed] [Google Scholar]
- 6.Lengyel P, Choubey D, Li S-J, Datta B. The interferon-activatable gene 200-cluster: From structure toward function. Semin Virol. 1995;6:203–213. [Google Scholar]
- 7.Johnstone RW, Trapani JA. Transcription and growth regulatory functions of the HIN-200 family proteins. Mol Cell Biol. 1999;19:5833–5838. doi: 10.1128/mcb.19.9.5833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kotzin BL. Systemic lupus erythematosus. Cell. 1996;85:303–306. doi: 10.1016/s0092-8674(00)81108-3. [DOI] [PubMed] [Google Scholar]
- 9.Wakeland EK, Liu K, Graham RR, Behrens TW. Delineating the genetic basis of systemic lupus erythematosus. Immunity. 2001;15:397–408. doi: 10.1016/s1074-7613(01)00201-1. [DOI] [PubMed] [Google Scholar]
- 10.Vyse TJ, Kotzin BL. Genetic susceptibility to systemic lupus erythematosus. Ann Rev Immunol. 1998;16:261–292. doi: 10.1146/annurev.immunol.16.1.261. [DOI] [PubMed] [Google Scholar]
- 11.Tsokos GC, Kammer GM. Molecular aberrations in human systemic lupus erythematosus. Mol Med Today. 2000;6:418–424. doi: 10.1016/s1357-4310(00)01798-6. [DOI] [PubMed] [Google Scholar]
- 12.Tsao BP. An update on genetic studies of systemic lupus erythematosus. Curr Rheumatol Rep. 2002;4:359–367. doi: 10.1007/s11926-002-0046-5. [DOI] [PubMed] [Google Scholar]
- 13.Baechler EC, Batliwalla FM, Karypis G, Gaffney PM, Ortmann WA, Espe KJ, Shark KB, Grande WJ, Hughes KM, Kapur V, Gregersen PK, Behrens TW. Interferon-inducible gene expression signature in peripheral blood cells of patients with severe lupus. Proc Natl Acad Sci USA. 2003;100:2610–2615. doi: 10.1073/pnas.0337679100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bennett L, Palucka AK, Arce E, Cantrell V, Borvak J, Banchereau J, Pascual V. Interferon and granulopoiesis signatures in systemic lupus erythematosus blood. J Exp Med. 2003;197:711–723. doi: 10.1084/jem.20021553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feng X, Wu H, Grossman JM, Hanvivadhanakul P, Fitzgerald JD, Park GS, Dong X, Chen W, Kim MH, Weng HH. Association of increased interferon-inducible gene expression with disease activity and lupus nephritis in patients with systemic lupus erythematosus. Arthritis Rheum. 2006;54:2951–2962. doi: 10.1002/art.22044. [DOI] [PubMed] [Google Scholar]
- 16.Nikpour M, Dempsey AA, Urowitz MB, Gladman DD, Barnes DA. Association of a gene expression profile from whole blood with disease activity in systemic lupus erythematosus. Ann Rheum Dis. 2007 Dec 6; doi: 10.1136/ard.2007.074765. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 17.Crow MK. Type I interferon in systemic lupus erythematosus. Curr Top Microbiol Immunol. 2007;316:359–386. doi: 10.1007/978-3-540-71329-6_17. [DOI] [PubMed] [Google Scholar]
- 18.Niewold TB, Hua J, Lehman TJA, Harley JB, Crow MK. High serum IFN-α activity is a heritable risk factor for systemic lupus erythematosus. Genes Immun. 2007;8:492–502. doi: 10.1038/sj.gene.6364408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Niewold TB, Harley J, Hua J, Crow M, Lehman T. Over-expression of interferon-α in lupus families: evidence for a complex heritable trait. Clin Immunol. 2007;123:89. [Google Scholar]
- 20.Banchereau J, Pascual V. Type I interferon in systemic lupus erythematosus and other autoimmune diseases. Immunity. 2006;25:383–392. doi: 10.1016/j.immuni.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 21.Theofilopoulos AN, Dixon FJ. Murine models of systemic lupus erythematosus. Adv Immunol. 1985;37:269–290. doi: 10.1016/s0065-2776(08)60342-9. [DOI] [PubMed] [Google Scholar]
- 22.Zhu J, Mohan C. SLE 1, 2, 3‥genetic dissection of lupus. Adv Exp Med Biol. 2007;601:85–95. doi: 10.1007/978-0-387-72005-0_9. [DOI] [PubMed] [Google Scholar]
- 23.Jorgensen TN, Gubbels MR, Kotzin BL. New insights into disease pathogenesis from mouse lupus genetics. Curr Opin Immunol. 2004;16:787–793. doi: 10.1016/j.coi.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 24.Santiago-Raber ML, Laporte C, Reininger L, Izui S. Genetic basis of murine lupus. Autoimmune Rev. 2004;3:33–39. doi: 10.1016/S1568-9972(03)00062-4. [DOI] [PubMed] [Google Scholar]
- 25.Haywood MEK, Rose SJ, Horswell S, Lees MJ, Fu G, Walport MJ, Morley BJ. Overlapping BXSB congenic intervals, in combination with microarray gene expression, reveal novel lupus candidate genes. Genes Immun. 2006;7:250–263. doi: 10.1038/sj.gene.6364294. [DOI] [PubMed] [Google Scholar]
- 26.Santiago-Raber ML, Baccala R, Haraldsson KM, Choubey D, Stewart TA, Kono DH, Theofilopoulos AN. Type-I interferon receptor deficiency reduces lupus-like disease in NZB mice. J Exp Med. 2003;197:777–788. doi: 10.1084/jem.20021996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Theofilopoulos AN, Baccala R, Beutler B, Kono DH. Type I interferons (α/β) in immunity and autoimmunity. Annu Rev. Immunol. 2005;23:307–336. doi: 10.1146/annurev.immunol.23.021704.115843. [DOI] [PubMed] [Google Scholar]
- 28.Mathian A, Weinberg A, Gallegos M, Banchereau J, Koutouzov S. IFN-α Induces early lethal lupus in pre-autoimmune (New Zealand Black x New Zealand White)F1 but not in BALB/c mice. J Immunol. 2005;174:2499–2506. doi: 10.4049/jimmunol.174.5.2499. [DOI] [PubMed] [Google Scholar]
- 29.Jφrgensen TN, Thurman J, Izui S, Falta MT, Metzger TE, Flannery SA, Kappler J, Marrack P, Kotzin BL. Genetic susceptibility to PolyI:C-induced IFN-α/β -dependent accelerated disease in lupus-prone mice. Genes Immun. 2006;7:555–567. doi: 10.1038/sj.gene.6364329. [DOI] [PubMed] [Google Scholar]
- 30.Jφrgensen TN, Roper E, Thurman JM, Marrack P, Kotzin BL. Type I interferon signaling is involved in the spontaneous development of lupus-like disease in B6. Nba2 and (B6. Nba2 X NZW)F mice. Genes Immun. 2007;8:653–662. doi: 10.1038/sj.gene.6364430. [DOI] [PubMed] [Google Scholar]
- 31.Nacionales DC, Kelly-Scumpia KM, Lee PY, Weinstein JS, Lyons R, Sobel E, Satoh M, Reeves WH. Deficiency of the type I interferon receptor protects mice from experimental lupus. Arthritis Rheum. 2007;56:3770–3783. doi: 10.1002/art.23023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu Q, Shen N, Li XM, Chen SL. Genomic view of IFN-alpha response in pre-autoimmune NZB/W and MRL/lpr mice. Genes Immun. 2007;8:590–603. doi: 10.1038/sj.gene.6364421. [DOI] [PubMed] [Google Scholar]
- 33.Hron JD, Peng SL. Type I IFN protects against murine lupus 1. J Immunol. 2004;173:2134–2142. doi: 10.4049/jimmunol.173.3.2134. [DOI] [PubMed] [Google Scholar]
- 34.Li J, Liu Y, Xie C, Zhu J, Kreska D, Morel L, Mohan C. Deficiency of type I interferon contributes to Sle2-associated component lupus phenotypes. Arthritis Rheum. 2005;52:3063–3072. doi: 10.1002/art.21307. [DOI] [PubMed] [Google Scholar]
- 35.Choubey D. p202: an interferon-inducible negative regulator of cell growth. J Biol Regul Homeost Agents. 2000;14:187–192. [PubMed] [Google Scholar]
- 36.Choubey D, Kotzin BL. Interferon-inducible p202 in the susceptibility to systemic lupus. Front Biosci. 2002;7:e252–e262. doi: 10.2741/A921. [DOI] [PubMed] [Google Scholar]
- 37.Asefa B, Klarmann KD, Copeland NG, Gilbert DJ, Jenkins NA, Keller JR. The interferon-inducible p200 family of proteins: a perspective on their role in cell cycle regulation and differentiation. Blood Cells Mol Dis. 2004;32:155–167. doi: 10.1016/j.bcmd.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 38.Ludlow LE, Johnstone RW, Clarke CJ. The HIN-200 family: more than interferon-inducible genes? Exp Cell Res. 2005;308:1–17. doi: 10.1016/j.yexcr.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 39.Choubey D, Deka R, Ho SM. Interferon-inducible IFI16 protein in human cancers and autoimmune diseases. Front Biosci. 2008;13:598–608. doi: 10.2741/2705. [DOI] [PubMed] [Google Scholar]
- 40.Ouchi M, Ouchi T. Role of IFI16 in DNA damage and checkpoint. Front Biosci. 2008;13:236–239. doi: 10.2741/2673. [DOI] [PubMed] [Google Scholar]
- 41.Ahmad YA, Bruce IN. Genetic epidemiology: systemic lupus erythematosus. Arthritis Res. 2001;3:331–336. doi: 10.1186/ar324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Olson JM, Song Y, Dudke DM, Moser KL, Kelly JA, Bruner GR, Downing KJ, Berry CK, James JA, Harley JB. A genome screen of systemic lupus erythematosus using affected-relative-pair linkage with covariates demonstrates genetic heterogeneity. Genes Immun. 2002;3 Suppl 1:S5–S12. doi: 10.1038/sj.gene.6363860. [DOI] [PubMed] [Google Scholar]
- 43.Kotzin BL. Susceptibility loci for lupus: a guiding light from murine models? J Clin Invest. 1997;99:557–558. doi: 10.1172/JCI119194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rozzo SJ, Allard JD, Choubey D, Vyse TJ, Izui S, Peltz G, Kotzin BL. Evidence for an interferon-inducible gene, Ifi202, in the susceptibility to systemic lupus. Immunity. 2001;15:435–443. doi: 10.1016/s1074-7613(01)00196-0. [DOI] [PubMed] [Google Scholar]
- 45.Wither JE, Lajoie G, Heinrichs S, Cai YC, Chang N, Ciofani A, Cheung YH, MacLeod R. Functional dissection of lupus susceptibility loci on the New Zealand Black mouse chromosome 1: Evidence for independent genetic loci affecting T and B cell activation. J Immunol. 2003;171:1697–1706. doi: 10.4049/jimmunol.171.4.1697. [DOI] [PubMed] [Google Scholar]
- 46.Gariglio M, Panico S, Cavallo G, Divaker C, Lengyel P, Landolfo S. Impaired transcription of the poly rI:rC- and interferon-activatable 202 gene in mice and cell lines from the C57BL/6 strain. Virology. 1992;187:115–123. doi: 10.1016/0042-6822(92)90300-e. [DOI] [PubMed] [Google Scholar]
- 47.Zhang Y, Tian Q, Du Y, Cao H, Lengyel P, Kong W. Multiple splicing results in at least two p203 proteins that are expressed in the liver and down-regulated during liver generation. Front Biosci. 2008;13:2444–2451. doi: 10.2741/2857. [DOI] [PubMed] [Google Scholar]
- 48.Koul D, Obeyesekere NU, Gutterman JU, Choubey D. p202 self-associates through a sequence conserved among the members of the p200-family proteins. FEBS Lett. 1998;438:21–24. doi: 10.1016/s0014-5793(98)01263-0. [DOI] [PubMed] [Google Scholar]
- 49.Wang C, Chatterjee G, Meyer JJ, Liu CJ, Manunath NA, Bray-Ward P, Lengyel P. Characteristics of three homologous 202 genes (Ifi202a Ifi202b, and Ifi202c) from the murine interferon-activatable gene 200 cluster. Genomics. 1999;60:281–294. doi: 10.1006/geno.1999.5923. [DOI] [PubMed] [Google Scholar]
- 50.Choubey D. Comment on: The candidate lupus susceptibility gene Ifi202a is largely dispensable for B-cell function. Rheumatology. 2008;47:558–559. doi: 10.1093/rheumatology/ken018. [DOI] [PubMed] [Google Scholar]
- 51.Bickerstaff MC, Botto M, Hutchinson WL, Herbert J, Tennent GA, Bybee A, Mitchell DA, Cook HT, Butler PJ, Walport MJ, Pepys MB. Serum amyloid P component controls chromatin degradation and prevents antinuclear autoimmunity. Nat Med. 1999;5:694–697. doi: 10.1038/9544. [DOI] [PubMed] [Google Scholar]
- 52.Tamaoki T, Tezuka H, Okada Y, Ito S, Shimura H, Sakamoto M, Endo T, Ozaki Y, Kanba S, Maeda S. Avoiding the effect of linked genes is crucial to elucidate the role of Apcs in autoimmunity. Nature Medicine. 2005;11:11–12. doi: 10.1038/nm0105-11. [DOI] [PubMed] [Google Scholar]
- 53.Bygrave AE, Rose KL, Cortes-Hernandez J, Warren J, Rigby RJ, Cook HT, Walport MJ, Vyse TJ, Botto M. Spontaneous autoimmunity in 129 and C57BL/6 mice. Implications for autoimmunity described in gene-targeted mice. PLoS Biol. 2004;2:1081–1090. doi: 10.1371/journal.pbio.0020243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Subramanian S, Tus K, Li QZ, Tian XH, Zhou J, Liang C, Bartov G, McDaniel LD, Zhou XJ, Schultz RA, Wakeland EK. A Tlr7 translocation accelerates systemic autoimmunity in murine lupus. Proc Natl Acad Sci USA. 2006;103:9970–9975. doi: 10.1073/pnas.0603912103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pisitkun P, Deane JA, Difilippantonio MJ, Tarasenko T, Satterthwaite AB, Bolland S. Autoreactive B cell response to RNA-related antigens due to TLR7 gene duplication. Science. 2006;312:1669–1672. doi: 10.1126/science.1124978. [DOI] [PubMed] [Google Scholar]
- 56.Kikuchi S, Fossati-Jimack L, Moll T, Amano H, Amano E, Ida A, Ibnou-Zekri N, Laporte C, Santiago-Raber ML, Rozzo SJ. Differential role of three major New Zealand Black-derived loci linked with Yaa-induced murine lupus nephritis. J Immunol. 2005;174:1111–1117. doi: 10.4049/jimmunol.174.2.1111. [DOI] [PubMed] [Google Scholar]
- 57.Fernando MM, Rigby RJ, Roberton CA, Rioux JD, Vyse TJ. Interferon-inducible genes on chromosome 1 contribute to lupus susceptibility. Rheumatology. 2006;45:i15–i17. [Google Scholar]
- 58.Teramoto K, Negoro N, Kitamoto K, Iwai T, Iwao H, Okamura M, Miura K. Microarray analysis of glomerular gene expression in murine lupus nephritis. J Pharmaco Sci. 2008;106:56–67. doi: 10.1254/jphs.fp0071337. [DOI] [PubMed] [Google Scholar]
- 59.Taniguchi T, Takaoka AA. A weak signal for strong responses: interferon-α/β revisited. Nat Rev Mol Cell Biol. 2001;2:378–386. doi: 10.1038/35073080. [DOI] [PubMed] [Google Scholar]
- 60.Lian ZX, Kikuchi K, Yang GX, Ansari AA, Ikehara S, Gershwin ME. Expansion of bone marrow IFN-α-producing dendritic cells in New Zealand Black (NZB) mice: high level expression of TLR9 and secretion of IFN-α in NZB bone marrow. J Immunol. 2004;173:5283–5289. doi: 10.4049/jimmunol.173.8.5283. [DOI] [PubMed] [Google Scholar]
- 61.Datta B, Min W, Burma S, Lengyel P. Increase in p202 expression during skeletal muscle differentiation: inhibition of MyoD protein expression and activity by p202. Mol Cell Biol. 1998;18:1074–1083. doi: 10.1128/mcb.18.2.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Geng Y, D’Souza S, Xin H, Walter S, Choubey D. p202 levels are negatively regulated by serum growth factors. Cell Growth Deffer. 2000;11:475–483. [PubMed] [Google Scholar]
- 63.Deftos ML, Huang E, Ojala EW, Forbush KA, Bevan MJ. Notch1 signaling promotes the maturation of CD4+ and CD8+ SP thymocytes. Immunity. 2000;13:73–84. doi: 10.1016/s1074-7613(00)00009-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.D'Souza S, Xin H, Walter S, Choubey D. The gene encoding p202, an interferon-inducible negative regulator of the p53 tumor suppressor, is a target of p53-mediated transcriptional repression. J Biol Chem. 2001;276:298–305. doi: 10.1074/jbc.M007155200. [DOI] [PubMed] [Google Scholar]
- 65.Xin H, Pramanik R, Choubey D. Retinoblastoma (Rb) protein up-regulates expression of the Ifi202 gene encoding an interferon-inducible negative regulator of cell growth. Oncogene. 2003;22:4775–4785. doi: 10.1038/sj.onc.1206780. [DOI] [PubMed] [Google Scholar]
- 66.Xin H, Geng Y, Pramanik R, Choubey D. Induction of p202, a modulator of apoptosis, during oncogenic transformation of NIH 3T3 cells by activated H-Ras (Q61L) contributes to cell survival. J Cell Biochem. 2003;88:191–204. doi: 10.1002/jcb.10372. [DOI] [PubMed] [Google Scholar]
- 67.Pramanik R, Jorgensen TN, Xin H, Kotzin BL, Choubey D. Interleukin-6 induces expression of Ifi202, an interferon-inducible candidate gene for lupus susceptibility. J Biol Chem. 2004;279:16121–16127. doi: 10.1074/jbc.M313140200. [DOI] [PubMed] [Google Scholar]
- 68.Chen J, Panchanathan R, Choubey D. Stimulation of T cells up-regulates expression of Ifi202, an interferon-inducible lupus susceptibility gene, through activation of JNK/c-Jun pathway. Immunol Lett. 2008;118:13–20. doi: 10.1016/j.imlet.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gribaudo G, Toniato E, Engel DA, Lengyel P. Interferons as gene activators. Characteristic of an interferon-activatable enhancer. J Biol Chem. 1987;262:11878–11883. [PubMed] [Google Scholar]
- 70.White RJ, Jackson SP. The TATA-binding protein: a central role in transcription by RNA polymerase I, II, and III. Trends Genet. 1992;8:284–288. doi: 10.1016/0168-9525(92)90255-3. [DOI] [PubMed] [Google Scholar]
- 71.Choubey D, Snoddy J, Chaturvedi V, Toniato E, Opdenakker G, Thakur A, Samanta H, Engel DA, Lengyel P. Interferons as gene activators. Indications for repeated gene duplication during the evolution of a cluster of interferon-activatable genes on murine chromosome 1. J Biol Chem. 1989;264:17182–17189. [PubMed] [Google Scholar]
- 72.Yacoub-Wasef SZ. Gender differences in systemic lupus erythematosus. Gend Med. 2004;1:12–17. doi: 10.1016/s1550-8579(04)80006-8. [DOI] [PubMed] [Google Scholar]
- 73.Zandman-Goddaed G, Peeva E, Shoenfeld Y. Gender and autoimmunity. Autoimmun Rev. 2007;6:366–372. doi: 10.1016/j.autrev.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 74.Cohen-Solal JF, Jeganathan V, Grimaldi CM, Peeva E, Diamond B. Sex hormones and SLE: influencing the fate of autoreactive B cells. Curr Top Microbiol Immunol. 2006;305:67–88. doi: 10.1007/3-540-29714-6_4. [DOI] [PubMed] [Google Scholar]
- 75.Gubbels MR, Jorgensen TN, Metzger TE, Menze K, Steele H, Flannery SA, Rozzo SJ, Kotzin BL. Effects of MHC and Gender on Lupus-Like Autoimmunity in Nba2 Congenic Mice. J Immunol. 2005;175:6190–6196. doi: 10.4049/jimmunol.175.9.6190. [DOI] [PubMed] [Google Scholar]
- 76.Gubbels Bupp MR, Jorgensen TN, Kotzin BL. Indetification of candidate genes that influence sex hormone-dependent disease phenotypes in mouse lupus. Genes Immun. 2008;9:47–56. doi: 10.1038/sj.gene.6364447. [DOI] [PubMed] [Google Scholar]
- 77.Berghofer B, Frommer T, Haley G, Fink L, Bein G, Hackstein H. TLR7 ligands induce higher IFN-α production in females. J Immunol. 2006;177:2088–2096. doi: 10.4049/jimmunol.177.4.2088. [DOI] [PubMed] [Google Scholar]
- 78.Choubey D, Lengyel P. Interferon action: cytoplasmic and nuclear localization of the interferon-inducible 52-kD protein that is encoded by the Ifi 200 gene from the gene 200 cluster. J Interferon Res. 1993;13:43–52. doi: 10.1089/jir.1993.13.43. [DOI] [PubMed] [Google Scholar]
- 79.Choubey DR, Pramanik R, Xin H. Sub-cellular localization and mechanisms of nucleocytoplasmic distribution of p202, an interferon-inducible candidate for lupus susceptibility. FEBS Lett. 2003;553:245–249. doi: 10.1016/s0014-5793(03)01006-8. [DOI] [PubMed] [Google Scholar]
- 80.Herkel J, Mimran A, Erez N, Kam N, Lohse AW, Marker-Hermann E, Rotter V, Cohen IR. Autoimmunity to the p53 protein is a feature of systemic lupus erythematosus (SLE) related to anti-DNA antibodies. J Autoimmun. 2001;17:63–69. doi: 10.1006/jaut.2001.0518. [DOI] [PubMed] [Google Scholar]
- 81.Salvador JM, Hollander MC, Nguyen AT, Kopp JB, Barisoni L, Moore JK, Ashwell JD, Fornace AJ., Jr Mice lacking the p53-effector gene Gadd45a develop a lupus-like syndrome. Immunity. 2002;16:499–508. doi: 10.1016/s1074-7613(02)00302-3. [DOI] [PubMed] [Google Scholar]
- 82.Kuan AP, Cohen PL. p53 is required for spontaneous autoantibody production in B6/lpr lupus mice. Eur J Immunol. 2005;35(5):1653–1660. doi: 10.1002/eji.200525982. [DOI] [PubMed] [Google Scholar]
- 83.Zheng SJ, Lamhamedi-Cherradi SE, Wang P, Xu L, Chen YH. Tumor suppressor p53 inhibits autoimmune inflammation and macrophage function. Diabetes. 2005;54:1423–1428. doi: 10.2337/diabetes.54.5.1423. [DOI] [PubMed] [Google Scholar]
- 84.Lissy NA, Davis PK, Irwin M, Kaelin WG, Dowdy SF. A common E2F-1 and p73 pathway mediates cell death induced by TCR activation. Nature. 2000;407:642–645. doi: 10.1038/35036608. [DOI] [PubMed] [Google Scholar]
- 85.Murga M, Fernandez,-Capetillo O, Field SJ, Moreno B, Borlado LR, Fujiwara Y, Balomenos D, Vicario A, Carrera AC, Orkin SH, Greenberg ME, Zubiaga AM. Mutation of E2F2 in mice causes enhanced T lymphocyte proliferation, leading to the development of autoimmunity. Immunity. 2001;15:959–970. doi: 10.1016/s1074-7613(01)00254-0. [DOI] [PubMed] [Google Scholar]
- 86.Zhu JW, Field SJ, Gore L, Thomson M, Yang H, Fujiwara Y, Cardiff RD, Greenberg M, Orkin SH, DeGregori J. E2F1 and E2F2 determine thresholds for antigen-induced T-cell proliferation and suppress tumorigenesis. Mol Cell Biol. 2001;21:8547–8564. doi: 10.1128/MCB.21.24.8547-8564.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.DeRyckere D, DeGregori J. E2F1 and E2F2 are differentially required for homeostasis-driven and antigen-induced T cell proliferation in vivo. J Immunol. 2005;175:647–655. doi: 10.4049/jimmunol.175.2.647. [DOI] [PubMed] [Google Scholar]
- 88.Vousden KH, Lu X. Live or let die: the cell's response to p53. Nat Rev Cancer. 2002;2:594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- 89.Sionov RV, Haput Y. The cellular response to p53: the decision between life and death. Oncogene. 1999;18:6145–6157. doi: 10.1038/sj.onc.1203130. [DOI] [PubMed] [Google Scholar]
- 90.Balomenos D, Martin-Caballero J, Garcia MI, Prieto I, Flores JM, Serrano M, Martinez AC. The cell cycle inhibitor p21 controls T-cell proliferation and sex-linked lupus development. Nat Med. 2000;6:171–176. doi: 10.1038/72272. [DOI] [PubMed] [Google Scholar]
- 91.Youlyouz-Marfak I, Gachard N, Le Clorennec C, Najjar I, Baran-Marszak F, Reminieras L, May E, Bornkamm GW, Fagard R, Feuillard J. Identification of a novel p53-dependent activation pathway of STAT1 by antitumour genotoxic agents. Cell Death Differ. 2008;15:376–385. doi: 10.1038/sj.cdd.4402270. [DOI] [PubMed] [Google Scholar]
- 92.Datta B, Li B, Choubey D, Nallur G, Lengyel P. p202, an interferon-inducible modulator of transcription, inhibits transcriptional activation by the p53 tumor suppressor protein, and a segment from the p53-binding protein 1 that binds to p202 overcomes this inhibition. J Biol Chem. 1996;271:27544–27557. doi: 10.1074/jbc.271.44.27544. [DOI] [PubMed] [Google Scholar]
- 93.Xin H, D'Souza S, Jorgensen TN, Vaughan AT, Lengyel P, Kotzin BL, Choubey D. Increased expression of Ifi202, an IFN-activatable gene, in B6. Nba2 lupus susceptible mice inhibits p53-mediated apoptosis. J Immunol. 2006;176:5863–5870. doi: 10.4049/jimmunol.176.10.5863. [DOI] [PubMed] [Google Scholar]
- 94.Slansky JE, Farnham PJ. Introduction to the E2F family: protein structure and gene regulation. Curr Top Microbiol Immunol. 1996;208:1–30. doi: 10.1007/978-3-642-79910-5_1. [DOI] [PubMed] [Google Scholar]
- 95.Nevins JR. Toward an understanding of the functional complexity of the E2F and retinoblastoma families. Cell Growth Differ. 1998;9:585–593. [PubMed] [Google Scholar]
- 96.DeGregori J. The genetics of the E2F family of transcription factors: shared functions and unique roles. Biochem Biophys Acta. 2002;1602:131–150. doi: 10.1016/s0304-419x(02)00051-3. [DOI] [PubMed] [Google Scholar]
- 97.Johnson DG, Ohtani K, Nevins JR. Auto-regulatory control of E2F1 expression in response to positive and negative regulators of cell cycle progression. Genes Dev. 1994;8:1514–1525. doi: 10.1101/gad.8.13.1514. [DOI] [PubMed] [Google Scholar]
- 98.Choubey D, Li S-J, Datta B, Gutterman JU, Lengyel P. Inhibition of E2F-mediated transcription by p202. EMBO J. 1996;15:5668–5678. [PMC free article] [PubMed] [Google Scholar]
- 99.Choubey D, Gutterman JU. Inhibition of E2F-4/DP-1-stimulated transcription by p202. Oncogene. 1997;15:291–301. doi: 10.1038/sj.onc.1201184. [DOI] [PubMed] [Google Scholar]
- 100.Panchanathan R, Xin H, Choubey D. Disruption of mutually negative regulatory feedback loop between interferon-inducible p202 protein and the E2F family of transcription factors in lupus-prone mice. J Immunol. 2008;180:5927–5934. doi: 10.4049/jimmunol.180.9.5927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Dhodapkar KM, Banerjee D, Connolly J, Kukreja A, Matayeva E, Veri MC, Ravetch JV, Steinman RM, Dhodapkar MV. Selective blockade of the inhibitory Fcγ receptor (Fcγ RIIB) in human dendritic cells and monocytes induces a type I interferon response program. J Exp Med. 2007;204:1359–1369. doi: 10.1084/jem.20062545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhuang H, Narain S, Sobel E, Lee PY, Nacionales DC, Kelly KM, Richards HB, Segal M, Stewart C, Satoh M, Reeves WH. Association of anti-nucleoprotein autoantibodies with up-regulation of Type I interferon-inducible gene transcripts and dendritic cell maturation in systemic lupus erythematosus. Clin Immunol. 2005;117:238–250. doi: 10.1016/j.clim.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 103.Caposio P, Gugliesi F, Zannetti C, Sponza S, Mondini M, Medico E, Hiscott J, Young HA, Gribaudo G, Gariglio M, Landolfo S. A novel role of the interferon-inducible protein IFI16 as inducer of proinflammatory molecules in endothelial cells. J Biol Chem. 2007;282:33515–33529. doi: 10.1074/jbc.M701846200. [DOI] [PubMed] [Google Scholar]
- 104.Mondini M, Vidali M, De Andrea M, Azzimonti B, Airo P, D’Ambrosio R, Riboldi P, Meroni PL, Albano E, Shoenfeld Y, Garglio M, Landolfo S. A novel autoantigen to differentiate limited cutaneous systemic sclerosis from diffuse cutaneous systemic sclerosis. Arthritis & Rheumatism. 2006;54:3939–3944. doi: 10.1002/art.22266. [DOI] [PubMed] [Google Scholar]
- 105.Mondini M, Vidali M, Airó P, De Andrea M, Riboldi P, Meroni PL, Gariglio M, Landolfo S. Role of the interferon-inducible gene IFI16 in the etiopathogenesis of systemic autoimmune disorders. Ann New York Acad Sci. 2007;1110:47–56. doi: 10.1196/annals.1423.006. [DOI] [PubMed] [Google Scholar]
- 106.Uchida K, Akita Y, Matsuo K, Fujiwara S, Nakagawa A, Kazoka Y, Hachiya H, Naganawa Y, Oh-iwa I, Ohura K, Saga S, Kawai T, Matsumoto Y, Shimozato K, Kozaki K. Identification of specific autoantigens in Sjogren’s syndrome by SEREX. Immunol. 2005;116:53–63. doi: 10.1111/j.1365-2567.2005.02197.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Peterson KS, Huang J-F, Zhu J, Agati VD, Liu X, Miller N, Erlander MG, Jackson MR, Winchester RJ. Characterization of heterogeneity in the molecular pathogenesis of lupus nephritis from transcriptional profiles of laser-captured glomeruli. J Clin Invest. 2004;113:1722–1733. doi: 10.1172/JCI19139. [DOI] [PMC free article] [PubMed] [Google Scholar]