Abstract
Bcr-Abl acquires its transforming abilitythro ugh its upregulated Abl tyrosine kinase activity. Bcr is a phosphoprotein with a novel serine/threonine kinase activitye ncoded by its first exon. In chronic myelogenous leukemia (CML) cells, Bcr-Abl phosphorylates Bcr on tyrosine residues reducing its kinase activity. Overexpression of BCR in BCR-ABL+ cells produces a phosphoserine form of Bcr, which inhibits the oncogenic effects of BCR-ABL. To investigate the inhibitory effects of Bcr on Bcr-Abl, we expressed BCR/GFP in TonB210 cells, which contain a tetracycline-inducible BCR-ABL. In nude mice injected with cell clones of TonB210/BCR/GFP, tumor formation was delayed, and tumors were 50% smaller compared with the TonB210/GFP. In addition, TonB210/BCR/GFP cells had little colony-forming ability in soft agar compared with TonB210/GFP cells. In contrast, a point mutant of BCR (Y360F), which disrupts its kinase activity, not only blocked Bcr’s inhibitory effects but also enhanced the oncogenic effects of Bcr-Abl in a solid tumor model and in soft agar colony assays. Similar effects were observed with a second BCR kinase domain mutant, S354A. These results indicate that the inhibitory function of Bcr directed toward Bcr-Abl requires its kinase function
Keywords: BCR-ABL, CML, BCR kinase-defective BCR
Introduction
The Bcr-Abl oncoprotein, responsible for causing chronic myeloid leukemia (CML), is derived from the fusion of the ABL and BCR sequences, which results in the formation of the Philadelphia chromosome. The Bcr-Abl oncoprotein acquires its transforming ability through its upregulated Abl tyrosine kinase activity. Normal Bcr is a 160 000-Da phosphoprotein, with a novel serine/threonine kinase activity localized in its first exon (Stam et al., 1987; Li et al., 1989; Maru and Witte, 1991). The first exon also contains two serine-rich domains that bind specifically to the SH2 domain of Abl in a non-phosphotyrosine-dependent manner (Pendergast et al., 1991). These serine-rich sequences in the phosphoserine form are involved in down regulating Bcr-Abl oncogenic activity (Wu et al., 1999; Lin et al., 2001; Hawk et al., 2002).
Perturbation of normal Bcr function is important in the generation of CML. Oncogenic Bcr-Abl phosphorylates endogenous Bcr protein on various tyrosine residues localized in Bcr’s first exon (Liu et al., 1993, 1996; Lu et al., 1993). In this tyrosine-phosphorylated form, Bcr serine/threonine kinase activity is reduced (Liu et al., 1996). However, overexpression of Bcr can interfere with Bcr-Abl oncogenic effects (Wu et al., 1999). In fact, Bcr becomes resistant to tyrosine phosphorylation in Bcr-Abl-positive cells when its expression is in molar excess. In this condition, the excess Bcr protein is mostly in the phosphoserine form and reduces the phosphotyrosine content of Bcr-Abl, strongly inhibiting its oncogenic activity (Wu et al., 1999; Lin et al., 2001). This phosphoserine form of Bcr is also predominant after overexpression of Bcr in soft agar clones of the CML K562 line containing an inducible BCR gene. As a consequence, these clones have reduced ability to induce extramedullary leukemia (Lin et al., 2001). On the basis of these results, we have proposed that the Bcr protein plays two roles in CML (Arlinghaus, 2002). In the tyrosine-phosphorylated form, Bcr would be neutralized as an inhibitor of Bcr-Abl effects and would serve as an important facilitator of Bcr-Abl-induced leukemia, possibly in the form of a heterotetramer structure with Bcr-Abl (McWhirter et al., 1993). On the other hand, in the serine/threonine-phosphorylated form, Bcr would function as an inhibitor of Bcr-Abl oncogenic ability. Here, we studied the effects of Bcr and two kinase domain mutants of Bcr on Bcr-Abl oncogenicity. We show for the first time in a nude mouse model and in soft agar colony assays, that kinase domain mutants of Bcr not only reverse Bcr inhibitory activity but also enhance Bcr-Abl oncogenic effects.
Results
Overexpression of BCR in TonB210/Bcr-Abl has no effect in cell culture
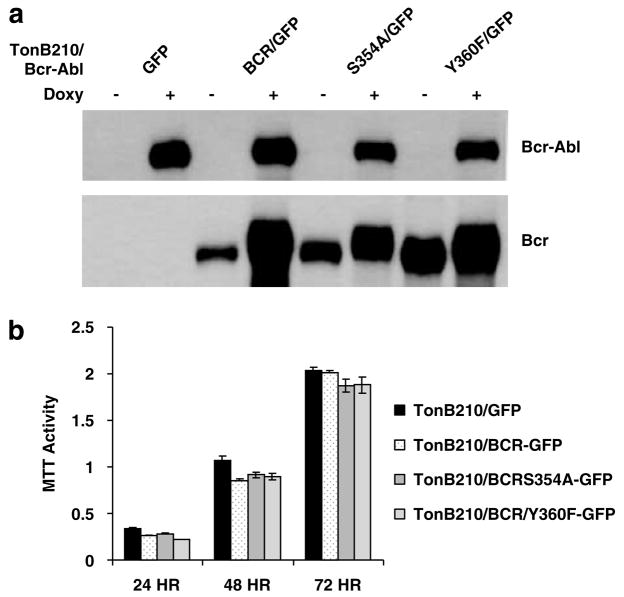
We used a lentiviral vector system to transduce TonB210 cells, originated from hematopoietic BaF3 cells, with green fluorescent protein (GFP) alone, HA-tagged BCRIRES-GFP and two independent HA-tagged BCR mutant-IRES-GFP (S354A and Y360) constructs. In TonB210, Bcr-Abl expression is tightly regulated by a tetracycline-dependent expression system (Klucher et al., 1998). Western blotting showed that Bcr-Abl expression is induced when the cells are grown in the presence of doxycycline, but with no significant expression without induction. Bcr is constitutively expressed in the lentivirus tranduced cells, but for reasons that are not understood higher levels of the Bcr protein are detected in Bcr-Ablinduced cells (Figure 1a). It remains to be determined whether forced Bcr expression affects the Bcr-Abl/Bcr heterotetramer ratio, which might affect Bcr-Abl oncogenicity. In cell culture, overexpression of BCR did not alter any of the normal cell functions. Cell proliferation and viability measured by using the 3-(4,5-dimethylthiazolyl-2)-2,5-diphenyltetrazolium bromide (MTT) assay showed no differences in the different cell lines even after 72 h of BCR-ABL induction (Figure 1b). Bcr expression did not affect Bcr-Abl’s ability to maintain viable growth in the absence of added interleukin-3. This is consistent with data collected for another over-expressing Bcr cell line, the Tet-off BCR-ABL 32D (X Ling and R Arlinghaus, unpublished results). These results indicate that overexpression of Bcr does not affect the normal physiology of the cell per se, but it has a specific function in the regulation of Bcr-Abl oncogenic process as shown by Lin et al. (2001).
Figure 1.
Western blot analysis of Bcr-Abl and exogenous Bcr in TonB210 cells. (a) The indicated cells were cultured with or without doxycycline (1 μg ml−1) for 24h. Bcr-Abl was detected with anti-Abl 8E9 antibody at 1:8000 dilution. Anti-HA antibody was used to detect retroviral expressed Bcr to distinguish it from endogenous Bcr protein. (b) Cell proliferation and viability were measured by the MTT assay over a 3-day period as indicated following the induction of Bcr-Abl with 1 μg/ml of doxycycline for 72 h.
Bcr inhibits Bcr-Abl-dependent tumor formation in a nude mouse model
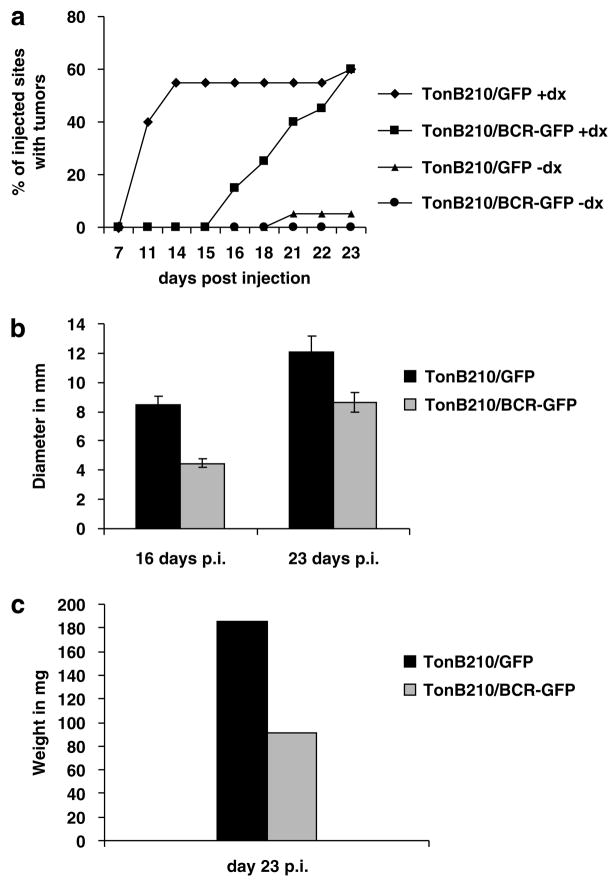
Since Bcr-Abl-expressing cells are able to induce tumors in nude mice (Daley and Baltimore, 1988), we performed several independent mouse experiments. The initial experiments were performed with cell-sorted (for GFP expression) but uncloned populations of TonB210/BCR-ABL/BCR/GFP cells. We observed a slight delay in tumor formation compared with control TonB210/BCR-ABL/GFP (data not shown). Subsequently, before injection into the mice, cells overexpressing wild-type BCR/GFP were isolated from single clones after serial dilution in liquid culture and selected by western blotting analysis for their high levels of Bcr expression. We then analysed the ability of these clones to form tumors. All groups contained 90–95% of GFP-positive cells before injection. TonB210/GFP-injected mice showed tumors in 40% of the injected sites at day 11 post injection, while at the same time no tumors formed in TonB210/BCR/GFP mice (Figure 2a). Tumors started to appear in the BCR/GFP group only at day 16 post injection, with a 5-day delay compared with the GFP group. By the end of the experiment, at day 23 post injection, both groups had developed the same number of tumors. However, the tumors in the TonB210/BCR/GFP injected mice were significantly smaller in diameter and weight than those in the TonB210/GFP group (Figures 2b and c). We hypothesize that these late-emerging tumors originated from ‘escaped’ cells that did not contained the Bcr protein. In order to test this idea, we isolated genomic DNA from a sample of tumors from the two groups and screened them by polymerase chain reaction for the presence of BCR-ABL and of BCR retroviral DNA. Even though we were always able to amplify BCR-ABL, we could not detect exogenous BCR sequences in any of the tumors tested (Table 1). Western blotting analysis of total protein isolated from all the tumors and performed using two different antibodies also lacked Bcr expression (data not shown). These results indicate that although 90–95% of injected TonB210/BCR-GFP cells were GFP+ and consequently should have had Bcr protein expression, a relative small number of ‘escaped’ cells that lacked Bcr in this population actually allowed tumors to form after a significant delay. Therefore, we conclude that in mice injected with TonB210/BCR/GFP cells, Bcr protein completely blocked Bcr-Abl’s ability to form tumors.
Figure 2.
BCR overexpression blocks the formation of Bcr-Ablinduced solid tumors in nude mice. (a) A total of 107 cells of each cell line were injected subcutaneously into the flanks of nude mice that were given water with of without 800 μg ml−1 doxycycline 1 week before injections, and during the whole duration of the experiment. We injected 10 mice (20 injection sites) for the TonB210/GFP+doxy and for TonB210/BCR/GFP+doxy groups, and five mice (10 injection sites) for the same cell lines, but in the −doxy conditions. Number of tumors was counted and expressed as a percentage of the total number of injected sites. An insignificant number of Bcr-Abl-independent tumors were also observed in the control group TonB210/GFP−doxy. (b) The diameter of tumors was measured by using a caliper at day 16 post injection and at day 23 when the animals were killed. Comparison analysis of tumor diameters between the two groups showed significant differences with P-values of 2.26449E-05 and 0.01513 at days 16 and 23, respectively. (c) After the animals were euthanized, tumors were isolated and immediately weighed. Comparison analysis of tumor weights for the two groups showed a significant difference between TonB210/GFP+doxy and TonB210/BCR/GFP+doxy (P-value= 0.008525). doxy, doxycycline; GFP, green fluorescent protein.
Table 1.
Tumor genomic DNA analysis by PCR for the presence of BCR-ABL and retroviral BCR
| BCR-ABL | BCR | BCR-ABL | BCR | ||
|---|---|---|---|---|---|
| GFP DNA | S354A DNA | ||||
| Mouse 1 | + | − | Mouse 1 | + | + |
| Mouse 2 | + | − | Mouse 2 | + | + |
| Mouse 3 | + | − | Mouse 3 | + | + |
| Mouse 4 | + | − | Mouse 4 | + | + |
| Mouse 5 | + | − | |||
| BCR DNA | Y360F DNA | ||||
| Mouse 1 Exp 1 | + | − | Mouse 1 | + | − |
| Mouse 2 Exp 1 | + | − | Mouse 2 | + | − |
| Mouse 3 Exp 1 | + | − | Mouse 3 | + | − |
| Mouse 4 Exp 1 | + | − | Mouse 4 | + | + |
| Mouse 5 Exp 1 | + | − | Mouse 5 | + | − |
| Mouse 6 Exp 1 | + | − | Mouse 6 | + | − |
| Mouse 7 Exp 1 | + | − | Mouse 7 | + | − |
| Mouse 1 Exp 2 | + | − | Mouse 10 | + | − |
| Mouse 2 Exp 2 | + | + | |||
| Mouse 3 Exp 2 | + | + | |||
| Mouse 4 Exp 2 | + | − | |||
| Mouse 5 Exp 2 | + | − | |||
| Mouse 6 Exp 2 | + | − | |||
| Mouse 7 Exp 2 | + | − | |||
| Mouse 8 Exp 2 | + | − |
Abbreviations: Exp, experiment; GFP, green fluorescent protein; PCR, polymerase chain reaction.
Kinase domain mutants of BCR enhanced the tumor forming ability of TonB210 cells
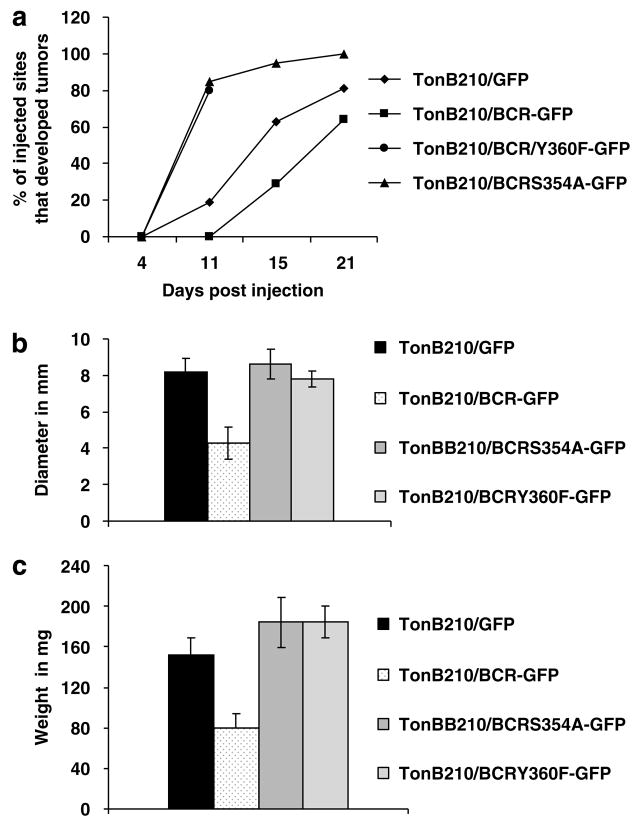
Amino-acid residue serine 354has been shown to be essential for the inhibition of Bcr-Abl oncogenic activity in in vitro experiments performed with a truncated form of Bcr (64–413) lacking the oligomerization domain (Hawk et al., 2002). Similarly, phosphorylation of tyrosine residue 360 by Bcr-Abl was shown to cause downregulation of Bcr serine/threonine kinase activity (Liu et al., 1996). Consequently, we predicted that injections of mice with Bcr-Abl-positive TonB210/BCR-ABL cells expressing Bcr with mutations either at S354A or at Y360F would restore the full oncogenic potential of Bcr-Abl. Thus, we conducted other mouse experiments to test the two BCR mutants S354A and Y360F, in addition to wild-type BCR and the BCRABL/GFP only cells. As above, overexpressing BCR wild type and BCR-mutant single cell clones were selected for high levels of BCR. The rate of tumor formation in mice injected with wild type BCR cells followed the same trend seen in the previous mouse experiments (see Figures 2a and 3a), with delayed tumors that were 50% smaller than those isolated from mice injected with BCR-ABL/GFP-only containing cells. Tumors in the two groups injected with BCR kinase domain-mutant expressing cells started to appear as early as 8 days after injection. As a result, BCR kinase mutants not only did not inhibit Bcr-Abl-induced oncogenicity, but actually, they dramatically increased the rate of tumor formation (Figure 3a), yielding larger tumors compared with the wild-type BCR group (Figures 3b and c), and even larger than the ones found in theTonB210/GFP-injected mice. Tumors in the Bcrmutant- injected mice contained high levels of green fluorescent cells, indicating that the tumor cells contained the Y360F and the S354A BCR-mutant proteins. Polymerase chain reaction analysis of genomic DNA isolated from TonB210/BCR-ABL/BCRS354A/GFP tumors showed the presence of BCR in all the samples tested (Table 1). However, contrary to our expectations, we detected Bcr only in one of the TonB210/BCR-ABL/BCRY360F/GFP tumors. We detected exogenous Bcr sequences in only two of eight tumors screened from the TonB210/BCR-ABL/BCR/GFP group. These results indicate that BCR expression in TonB210 cells inhibits the formation of Bcr-Abl-dependent tumors in mice and that Bcr serine kinase activity is required for this inhibitory function. Interestingly, the S354A mutant also enhanced Bcr-Abl’s oncogenic activity. This mutant reduces the level of the serine hyperphosphorylated form of BCR (64–413) (Hawk et al., 2002). Finally, these results show for the first time in a mouse model system, that the integrity of the serine kinase function of Bcr is indispensable for its inhibitory activity toward Bcr-Abl.
Figure 3.
Kinase domain BCR mutants S354A and Y360F enhance the formation of Bcr-Abl-induced solid tumors in nude mice. (a) Conditions are the same as described in Figure 2. Ten animals were used for TonB210/BCRS354A/GFP and TonB210/BCRY360F/GFP cells. Seven and eight animals were used for TonB210/GFP and TonB210/BCR/GFP, respectively. Mice injected with TonB210/BCRY360F/GFP cells were euthanized at day 12 post injection; the other groups were killed between days 21 and 22 post injection. (b) Tumor diameters were measured as in Figure 2. Significant differences were observed between TonB210/GFP and TonB210/BCR/GFP (P-value = 0.00214), TonB210/BCR/GFP and TonB210/BCRS354A (P-value = 0.00029), and TonB210/BCR/GFP and TonB210/BCRY360F (P-value = 0.00199). (c) Tumor weights were measured as in Figure 2. Significant differences are observed between TonB210/GFP and TonB210/BCR/GFP (P-value = 0.00429), TonB210/BCR/GFP and TonB210/BCRS354A/GFP (P-value = 0.00321), and TonB210/BCR/GFP and TonB210/BCRY360F/GFP (P-value = 0.00022). GFP, green fluorescent protein.
Wild-type BCR, but not kinase domain mutants, interferes with the growth of TonB210/BCR-ABL in soft agar
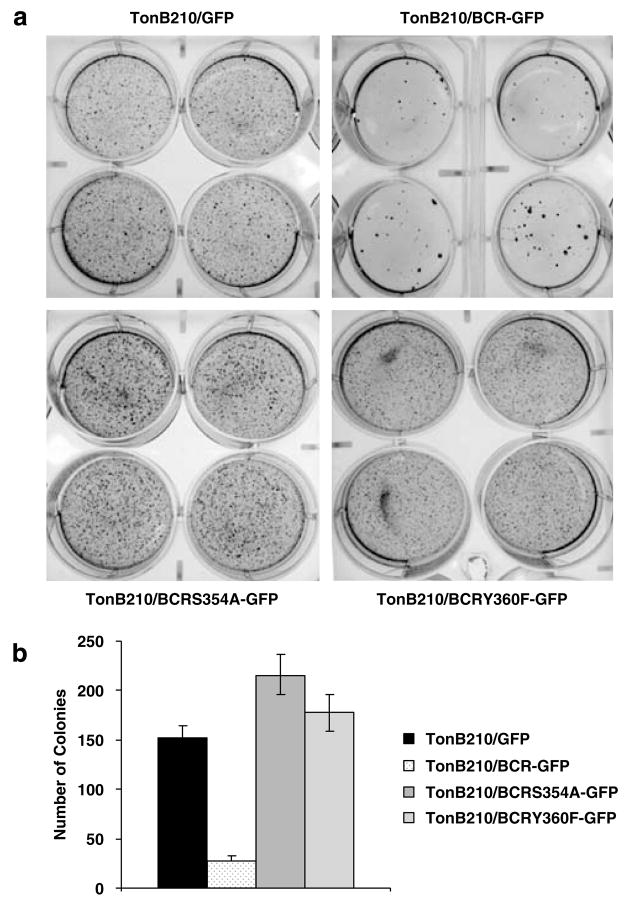
We assessed the effects of Bcr expression on Bcr-Abl oncogenic effects in soft agar colony assays, which is another way to measure oncogenic potential. Overexpression of wild-type BCR blocked colony formation almost completely as shown in Figure 4. In contrast, either the Y360F- or the S354A-mutant BCR generated a higher number of colonies than the TonB210/BCRABL/GFP cells. These in vitro results paralleled the results obtained from the mouse model studies and reenforce the concept that although TonB210/BCR-ABL/BCR/GFP cells do not show any apparent deficit in cell culture, they lack the ability to form colonies in vitro agar assay. However, this phenotype is reversed in TonB210 cells expressing kinase domain mutants of BCR.
Figure 4.
Wild-type Bcr but not kinase domain-mutant BCR proteins blocks the formation of colonies in a soft agar. (a) Thirty thousand cells were seeded in four different plates for each cell line. The assay was repeated four times for TonB210/GFP and TonB210/BCR/GFP and two times for TonB210/BCRS354A/GFP and TonB210/BCRY360F/GFP. (b) Colonies were counted after staining with p-iodonitrotetrazolium violet 14 days post inoculation. GFP, green fluorescent protein.
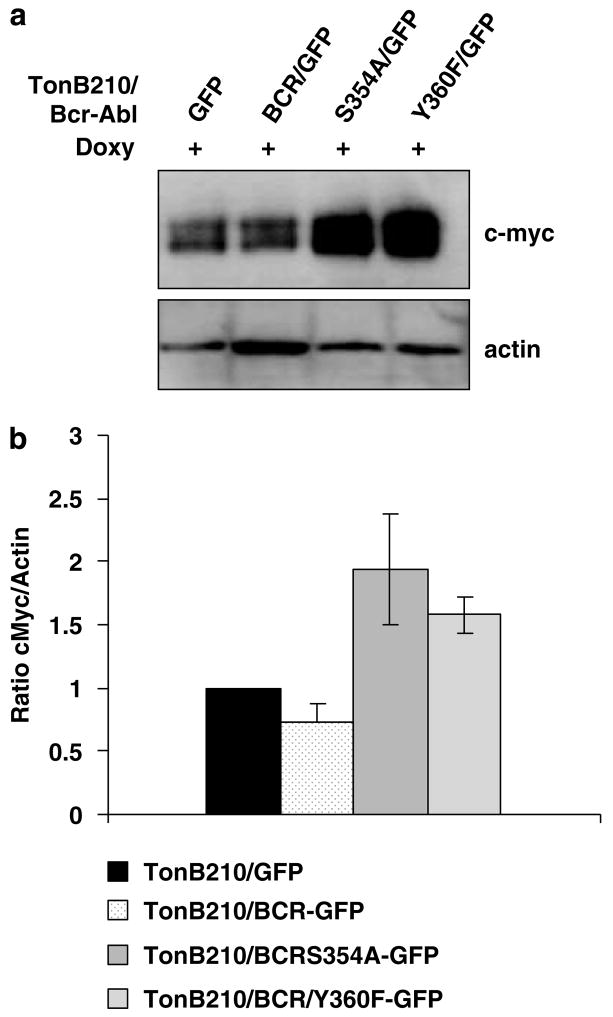
BCR expression reduces c-Myc expression in Bcr-Abl+ cells, but kinase domain mutants of Bcr enhance c-Myc expression
Bcr-Abl transformation requires enhanced expression of c-Myc (Sawyers et al., 1992). Importantly, the requirement of c-Myc for Bcr-Abl transformation is not observed in cell culture, but instead positively affects tumor forming ability of Bcr-Abl + cells (Xie et al., 2002). Therefore, we determined the level of c-Myc expression in TonB210/BCR-ABL expressing wild-type and mutant BCRs (Figure 5). In cells expressing wild-type BCR, c-Myc levels were reduced compared with TonB210/BCR-ABL/GFP cells, correlating with the reduction observed in tumor and colony formation results. In contrast, c-Myc protein levels were enhanced in the kinase-defective BCR and in the S354A mutant, which also showed increased tumor forming ability and enhanced oncogenicity in the soft agar colony assay.
Figure 5.
Kinase domain mutants of Bcr enhance c-Myc expression. (a) The upper panel shows a representative western blot of TonB210 cells with various Bcr proteins and control GFP grown as described in Figure 1. c-Myc was detected by using an rabbit anticMyc (N-262) antibody from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Quantification of protein levels was by volume densitometry using the Scion Image program, and intensity values were normalized for actin content. Five different determinations were averaged for the GFP control and the wild-type BCR groups, and two different determinations were averaged for the S354A and the Y360F groups. (b) Comparison analysis of the intensities of the c-Myc bands showed a significant difference between TonB210/GFP and TonB210/BCR/GFP (P-value = 0.0617), TonB210/BCR/GFP and TonB210/BCRS354A/GFP (P-value = 0.0068), and TonB210/BCR/GFP and TonB210/BCRY360F/GFP (P-value = 0.0004). GFP, green fluorescent protein.
Discussion
In this report, we show that overexpression of Bcr to levels that exceed the amount of endogenous Bcr and are in molar excess compared with the Bcr-Abl oncoprotein, inhibited tumor formation in a nude mouse solid tumor model (Figures 2 and 3) and blocked colony formation in soft agar assays (Figure 4). Cell cultures of overexpressing Bcr cells had no effects on the proliferation and survival of these cells (Figure 1b), indicating that the inhibitory effect is specific to the interaction of Bcr and Bcr-Abl in the oncogenic process. However, Bcr-Abl induction increased levels of Bcr expression (Figure 1a), which could change the ratio of Bcr/Bcr-Abl and impact the effects of Bcr on Bcr-Abl oncogenicity through a change in the heterotetramer composition. The antitumor effects of Bcr were completely blocked with either a kinase-defective BCR mutant (Y360F) or a BCR kinase domain mutant (S354A) (Figure 3). Importantly, the oncogenic effects of Bcr-Abl were significantly enhanced by coexpression of kinase domain-defective mutants of BCR.
The Y360F mutant of Bcr is defective in phosphorylating added substrates like casein, although maintaining its autophosphorylation activity on serine residues (Liu et al., 1996).
How a Bcr protein that is defective in trans-phosphorylating activity (Y360F Bcr) would enhance the oncogenic effects of Bcr-Abl is unknown. It is possible that Bcr down regulates Bcr-Abl tyrosine kinase activity by phosphorylating Bcr-Abl on serine/threonine residues. Crystallography results (Nagar et al., 2006) support the idea that phosphoserine residue 69 in Abl is involved in the down regulation of Abl kinase activity. The same residue in Bcr-Abl might be the target of wild-type Bcr phosphorylation, thus contributing to the down regulation of the Abl kinase activity within the Bcr-Abl oncoprotein. We hypothesize that this inhibitory mechanism would be defective in the Y360F Bcr mutant. In support of the role of Bcr in regulating Bcr-Abl tyrosine kinase activity, our preliminary studies indicate that when we introduced the Y360F mutation in BCR-ABL expressed in Rat1 cells, the mutant Bcr-Abl showed increased intrinsic kinase activity compared with wild-type Bcr-Abl. Moreover, agar colony formation of Bcr-Abl + Rat1 cells transduced with the Y360F Bcr-Abl mutant was also increased (data not shown). These results suggest that cis Bcr sequences within Bcr-Abl regulate the tyrosine kinase of Abl kinase domain and that the tyrosine 360 residue is involved in this regulation.
The mechanism for the lack of inhibition of Bcr-Abl by the S354A mutant of BCR is less clear. In our previous findings with the truncated form of Bcr, (Bcr 64–413), lacking the oligomerization domain, we found that, the S354A mutation of Bcr (64–413) maintains its kinase function but is unable to form a hyperphosphorylated form. This S354A mutant form of Bcr (64–413) is also unable to bind to the SH2 domain of c-Abl. Loss of binding activity correlates with the decrease of Bcr (64–413) inhibition of BCR-ABL oncogenicity (Hawk et al., 2002). We are currently investigating the biochemical mechanisms involved in the enhancement of Bcr-Abl oncogenicity by this S354A Bcr mutant.
We have previously showed that overexpression of Bcr relative to Bcr-Abl is required for Bcr’s inhibitory effects (Wu et al., 1999; Lin et al., 2001). In addition, it has been reported that BCR-ABL/BCR transcript ratios can be used as an important diagnostic parameter for aggressive behavior of CML patients, suggesting a possible role of normal Bcr in CML pathogenesis (Moravcová et al., 2005). Therefore it would be important to define the level of Bcr expression relative to Bcr-Abl necessary to achieve the inhibitory effect. Surprisingly, we found that induction of BCR-ABL expression also increased Bcr protein levels (Figure 1a), making it difficult to control the ratio of these two proteins in cell culture. Of interest, it is possible that replacing Bcr in the heterotetramer Bcr-Abl/Bcr structure with a kinase-defective mutant of Bcr increases oncogenic potential of these cells, suggesting another mechanism for increased oncogenicity in CML patients.
The antioncogenic effects of Bcr are likely to be explained by the effects of Bcr on c-Myc expression (Mahon et al., 2003). Mahon et al. (2003) showed that BCR expression downregulates the activity of c-Myc and competes with Max for c-Myc binding. In the CML cell line K562 when excess BCR is induced, the c-Myc protein is downregulated. Furthermore, in studies of Jak2 inhibition effects in the CML cell line K562 (Xie et al., 2002), c-Myc protein is also downregulated. Although the effects of c-Myc reduction are not observed in cell culture, lower levels of c-Myc correlate with lower tumor induction in our mouse studies (Xie et al., 2002). The c-Myc protein is known to be required for the leukemic effects of Bcr-Abl (Sawyers et al., 1992; Xie et al., 2002). In this report, we were able to show that TonB210/BCR-ABL/BCR/GFP has decreased levels of c-Myc (Figure 5), and this decrease correlated with a decrease in tumor formation. Importantly, expression of Bcr that is defective in kinase activity enhanced the tumorigenic effects of Bcr-Abl and strongly increased the level of c-Myc (Figure 5). Similar results were obtained with the S354A mutant form of BCR.
Finally, another possible mechanism by which Bcr exerts its inhibitory function is through the β-catenin pathway. Ress and Moelling (2005) showed that BCR expression reduces levels of β-catenin. Importantly, Bcr-Abl reverses these effects by tyrosine phosphorylation of Bcr first exon sequences (Ress and Moelling, 2005). We hypothesize that overexpression of wild-type Bcr inhibits tumorigenicity of Bcr-Abl + cells by downregulation of β-catenin. Further studies are underway to investigate the involvement of c-Myc and β-catenin pathways in Bcr inhibitory effects on Bcr-Abl oncogenicity.
Materials and methods
Cell lines
TonB210 cells were provided by Dr George Daley (Whitehead Institute) and grown as described (Klucher et al., 1998). Bcr- Abl expression was induced with 0.3–1 μgml−1 of doxicycline (Sigma Aldrich, St Louis, MO, USA). Mutations in the BCR sequence were introduced by using QuickChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA, USA). The mutations were confirmed by direct sequencing.
The TonB210/GFP, TonB210/BCR/GFP, TonB210/BCR Y360F/GFP and TonB210/BCR S354A/GFP cell lines were created by infection of non-induced TonB210 cells with recombinant lentiviruses encoding BCR-HA tagged (wild type or mutants) or GFP prepared as described in Ling et al. (2003). Infected cells were enriched for Bcr expression by sorting of GFP+ cells by fluorescence-activated cell sorting, and high Bcr expressing clones were selected by single-cell cloning of GFP+ cells.
Cell proliferation assay
Cell proliferation was measured by using the MTT assay (Sigma Aldrich).
Protein analysis
For western blot analysis of Bcr-Abl and Bcr, total proteins were isolated as described (Abe et al., 1996). Blots were probed with either anti-HA (Invitrogen, Carlsbad, CA, USA), anti-Bcr (1–16) or anti-Abl 8E9 monoclonal antibody (Guo et al., 1994).
Tumorigenicity studies
Nude mice were from Jackson Laboratories (Bar Harbor, ME, USA). Feeding regimen with doxycycline was as described (Klucher et al., 1998). Ten million cells were injected subcutaneously into each flank of the nude mice in 250 μl of phosphate-buffered saline containing 2 μgml−1 doxycycline.
Soft agar assay for colony formation
Approximately 30 000 cells were seeded into a six-well plate on 3.5% agarose. Colonies were allowed to grow for 2 weeks and were stained with p-iodonitrotetrazolium violet (Sigma Aldrich).
Polymerase chain reaction analysis
Genomic DNA was isolated from solid tumors by using Trizol reagent (Invitrogen). Exogenous BCR was detected by using the following set of primers: 5′TGATGATGAGCGAGATG GACGTGA3′ in 3′Bcr and 5′AACAGACCTTGCATTCCTT TGGCG3′ in the pHR vector. Endogenous BCR and BCRABL were amplified by using 5′AGAACCTGAGAGCCAGA AGCAAC3′ and 5′TCAGGAACGTGTAACTCTTGCCG3′.
Acknowledgments
This research was supported in part by Grant CA49639 and by funds from the Hendrick Marrow Foundation.
References
- Abe J, Kusuhara M, Ulevitch RJ, Berk BC, Lee JD. Big mitogenactivated protein kinase 1 (BMK1) is a redox-sensitive kinase. J Biol Chem. 1996;271:16586–16590. doi: 10.1074/jbc.271.28.16586. [DOI] [PubMed] [Google Scholar]
- Arlinghaus RB. BCR: a negative regulator of the Bcr-Abl oncoprotein in leukemia. Oncogene. 2002;21:8560–8567. doi: 10.1038/sj.onc.1206083. [DOI] [PubMed] [Google Scholar]
- Daley GQ, Baltimore D. Transformation of an interleukin 3-dependent hematopoietic cell line by the chronic myelogenous leukemia-specific P210bcr/abl protein. Proc Natl Acad Sci USA. 1988;85:9312–9316. doi: 10.1073/pnas.85.23.9312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JQ, Lian JY, Xian YM, Lee M-S, Deisseroth AB, Stass SA, et al. BCR-ABL protein expression in peripheral blood cells of chronic myelogenous leukemia patients undergoing therapy. Blood. 1994;83:3629–3637. [PubMed] [Google Scholar]
- Hawk N, Sun T, Xie S, Wang Y, Wu Y, Liu J, et al. Inhibition of the BCR-ABL oncoprotein by BCR requires phosphotyrosine 354. Cancer Res. 2002;62:386–390. [PubMed] [Google Scholar]
- Klucher KM, Lopez DV, Daley GQ. Secondary mutation maintains the trasnformation state in BaF3 cells with inducible BCR-ABL expression. Blood. 1998;91:3927–3934. [PubMed] [Google Scholar]
- Li WJ, Dreazan O, Kloetzer W, Gale RP, Arlinghaus RB. Characterization of the bcr gene products in hematopoietic cells. Oncogene. 1989;4:127–138. [PubMed] [Google Scholar]
- Lin F, Monaco G, Sun T, Liu J, Lin H, Stephens C, et al. BCR gene expression blocks Bcr-Abl induced pathogenicity in a mouse model. Oncogene. 2001;20:1873–1881. doi: 10.1038/sj.onc.1204409. [DOI] [PubMed] [Google Scholar]
- Ling X, Ma G, Sun T, Liu J, Arlinghaus RB. Bcr and Abl interaction. Oncogenic activation of c-Abl by sequestering Bcr. Cancer Res. 2003;63:298–303. [PubMed] [Google Scholar]
- Liu J, Campbell M, Guo JQ, Lu D, Xian YM, Anderson BS, et al. BCR-ABL tyrosine kinase is autophosphorylated or transphosphorylates P160 BCR on tyrosine predominantly within the first BCR exon. Oncogene. 1993;1:101–109. [PubMed] [Google Scholar]
- Liu J, Wu Y, Ma GZ, Lu D, Haataja L, Heisterkamp N, et al. Inhibition of BCR serine kinase by tyrosine phosphorylation. Mol Cell Biol. 1996;3:998–1005. doi: 10.1128/mcb.16.3.998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D, Liu J, Campbell M, Guo JQ, Heisterkamp N, Groffen J, et al. Tyrosine phosphorylation of P160 BCR by P210 BCR-ABL. Blood. 1993;82:1257–1263. [PubMed] [Google Scholar]
- Mahon GM, Wang Y, Korus M, Kostenko E, Cheng L, Sun T, et al. The c-Myc oncoprotein interacts with Bcr. Curr Biol. 2003;5:437–441. doi: 10.1016/s0960-9822(03)00090-3. [DOI] [PubMed] [Google Scholar]
- Maru Y, Witte ON. The BCR gene encodes a novel serine/threonine kinase activity within a single exon. Cell. 1991;67:459–468. doi: 10.1016/0092-8674(91)90521-y. [DOI] [PubMed] [Google Scholar]
- McWhirter JR, Galasso DL, Wang JY. A coiled-coil oligomerization domain of BCR is essential for the transforming function of BCR-ABL oncoproteins. Mol Cell Biol. 1993;13:7587–7595. doi: 10.1128/mcb.13.12.7587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moravcová J, Regner J, Mouèková D, Fišer K, Zmeková V, Maláèová R, et al. Disease status in patients with chronic myeloid leukemia is better characterized by BCR-ABL/BCR transcript ratio than by BCRABL transcript level, which may suggest a role for normal BCR gene in the disease pathogenesis. Neoplasma. 2005;52:119–125. [PubMed] [Google Scholar]
- Nagar B, Hantschel O, Seeliger M, Davies JM, Weis WI, Superti-Furga G, et al. Organization of the SH3-SH2 unit in active and inactive forms of the c-Abl tyrosine kinase. Mol Cell. 2006;21:787–798. doi: 10.1016/j.molcel.2006.01.035. [DOI] [PubMed] [Google Scholar]
- Pendergast AM, Muller AJ, Havlik MH, Maru Y, Witte ON. BCR sequences essential for transformation by the BCR-ABL oncogene bind to the ABL SH2 regulatory domain in a nonphosphotyrosine- dependent manner. Cell. 1991;66:161–171. doi: 10.1016/0092-8674(91)90148-r. [DOI] [PubMed] [Google Scholar]
- Ress A, Moelling K. Bcr is a negative regulator of the Wnt signaling pathway. EMBO Rep. 2005;11:1095–1100. doi: 10.1038/sj.embor.7400536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyers CL, Callahan W, Witte ON. Dominant negative MYC blocks transformation of ABL oncogenes. Cell. 1992;70:901–910. doi: 10.1016/0092-8674(92)90241-4. [DOI] [PubMed] [Google Scholar]
- Stam K, Heisterkamp N, Reynolds FH, Jr, Groffen J. Evidence that the phl gene encodes a 160 000-dalton phosphoprotein with associated kinase activity. Mol Cell Biol. 1987;5:1955–1960. doi: 10.1128/mcb.7.5.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Ma G, Lu D, Lin F, Xu HJ, Liu J, et al. BCR: a negative regulator of the Bcr-Abl oncoprotein. Oncogene. 1999;18:4416–4424. doi: 10.1038/sj.onc.1202828. [DOI] [PubMed] [Google Scholar]
- Xie S, Lin H, Sun T, Arlinghaus RB. Jak2 is involved in c-Myc induction of Bcr-Abl. Oncogene. 2002;21:7137–7146. doi: 10.1038/sj.onc.1205942. [DOI] [PubMed] [Google Scholar]