Abstract
Plasma growth hormone (GH) profiles regulate the sexually dimorphic expression of cytochromes P450 and many other genes in rat and mouse liver, however, the proximal transcriptional regulators of these genes are unknown. Presently, we characterize three liver transcription factors that are expressed in adult female rat and mouse liver at levels up to 16-fold (Tox), 73-fold (Trim24/TIF1α), and 125-fold (Cutl2/Cux2) higher than in adult males, depending on the strain and species, with Tox expression only detected in mice. In rats, these sex differences first emerged at puberty, when the high prepubertal expression of Cutl2 and Trim24 was extinguished in males but was further increased in females. Rat hepatic expression of Cutl2 and Trim24 was abolished by hypophysectomy and, in the case of Cutl2, was restored to near-female levels by continuous GH replacement. Cutl2 and Trim24 were increased to female-like levels in livers of intact male rats and mice treated with GH continuously (female GH pattern), while Tox expression reached only about 40% of adult female levels. Expression of all three genes was also elevated to normal female levels or higher in male mice whose plasma GH profile was feminized secondary to somatostatin gene disruption. Cutl2 and Trim24 both responded to GH infusion in mice within 10–24 h and Tox within 4 d, as compared to at least 4–7 d required for the induced expression of several continuous GH-regulated cytochromes P450 and other female-specific hepatic genes. Cutl2, Trim24 and Tox were substantially up-regulated in livers of male mice deficient in either of two transcription factors implicated in GH regulation of liver sex specificity, namely, signal transducer and activator of transcription 5b (STAT5b) and hepatocyte nuclear factor 4α (HNF4α), with sex-specific expression being substantially reduced or lost in mice deficient in either nuclear factor. Cutl2 and Trim24 both display transcriptional repressor activity and could thus contribute to the loss of GH-regulated, male-specific liver gene expression seen in male mice deficient in STAT5b or HNF4α. Binding sites for Cutl1, whose DNA-binding specificity is very close to that of Cutl2, were statistically over-represented in STAT5b-dependent male-specific mouse genes, lending support to this hypothesis.
Keywords: growth hormone, STAT5b, HNF4α, liver gene expression, sex-specificity
Introduction
GH regulates the sexually dimorphic patterns of a large number of liver-expressed genes, including many receptors, signaling molecules and enzymes of steroid and drug metabolism, especially cytochrome P450s (CYPs) (for reviews, see (1, 2)). These sex differences are dictated by the sexual dimorphism of plasma GH profiles, which is especially prominent in rats and mice and is a major determinant of the sex-specificity of a large number of hepatic RNAs (3) and proteins (4). Plasma GH profiles are highly pulsatile in adult male rats, where high GH peaks occur every 3.5–4 hr and are interrupted by periods of no measurable hormone, while adult female rats are characterized by more frequent and overlapping plasma GH peaks, resulting in a nearly constant presence of GH in circulation (5, 6). The length of the GH-free interpulse interval is a key determinant of the sex-specific effect that GH has on liver gene expression (7, 8). Continuous infusion of GH in male rats and mice abolishes the male, pulsatile plasma GH profile and feminizes the expression of many liver genes (3, 9, 10).
GH signaling is initiated by GH binding to its membrane-bound receptor leading to activation/tyrosine phosphorylation of the GH receptor-associated protein tyrosine kinase Janus kinase 2 (JAK2) (11). JAK2, in turn, phosphorylates itself and the cytoplasmic domain of GH receptor, creating binding sites for signaling molecules implicated in downstream signaling, including STAT transcription factors. Following tyrosine phosphorylation, STAT proteins dimerize and translocate to the nucleus, where they bind specific DNA elements and activate gene transcription (12). One STAT family member, STAT5b, is repeatedly activated by each incoming male plasma GH pulse and is considered to be a key mediator of the sexually dimorphic response of liver CYPs to GH in rats and mice (13, 14). In contrast to males, which are characterized by high episodic bursts of liver STAT5b activity, the level of tyrosine phosphorylated, nuclear STAT5b is generally low in female liver (15–17). Additional transcription factors are likely to be required for establishing sexually dimorphic patterns of liver gene expression, however, as suggested by the absence of strong STAT5b response elements in the upstream sequences of several GH-regulated, male-specific liver CYP gene promoters (18, 19) and by the inability of STAT5b alone, when activated precociously in prepubertal rat liver, to induce male-specific liver gene expression (16). One such factor may be hepatocyte nuclear factor (HNF) 4α, which can acts in a synergistic manner with STAT5b to activate certain male-specific CYP promoters (20) and, like STAT5b, is required for the sex-dependent transcriptional control of several classes of GH-dependent liver genes (10, 21). Another liver-enriched transcription factor, HNF6, is expressed in a female-predominant, GH-regulated manner and may contribute to the female-specific expression of hepatic CYP gene 2C12 (22–24).
In the present study, we characterize three transcription factors, Cutl2, Trim24 and Tox, as novel, highly sexually dimorphic, liver expressed factors that are positively regulated by the female-characteristic plasma GH pattern. Cutl2, also known as Cux2, belongs to a family of CDP/Cut homeodomain transcription factors involved in the control of proliferation and differentiation (25) and was reported to be expressed primarily in nervous tissue (26). Cutl2 binds DNA in a sequence-specific manner and may act as a transcriptional repressor (26, 27). Trim24, also known as TIF-1α, is a transcriptional intermediary factor that interacts with ligand-bound nuclear receptors and is proposed to regulate transcriptional activity through chromatin remodeling (28, 29). The third female-specific factor, Tox, is a DNA sequence-independent high mobility group box-containing protein previously implicated in the regulation of T cell development (30).
Materials and Methods
Continuous GH treatment of ICR mice
Adult ICR mice, 7–8 wk of age, were purchased from Taconic, Inc. (Hudson, NY) and maintained under standardized conditions of light and temperature. Male ICR mice were given GH as a continuous infusion via Alzet osmotic mini-pumps (Durect Corp., Cupertino, CA) for up to 14 d as described earlier (10). Briefly, pumps were filled with recombinant rat GH (purchased from Dr. A.F. Parlow, Harbor-UCLA Medical Center, Torrance, CA) dissolved in 70 mM sodium bicarbonate, pH 9.5, containing 137 mM NaCl and 100 μg/ml rat albumin, or, in the case of vehicle-treated mice, with protein-free buffer. GH was delivered at a rate of 20 ng per g body weight per h for time periods ranging from 10 h to 14 d. At each time point, 6–7 GH-treated mice and 2–3 vehicle-treated mice were killed, their livers extracted, frozen in liquid nitrogen and stored at −80°C until use. Livers were also collected from untreated male (n=5) and female (n=5) ICR mice.
Knockout mouse models
Livers were obtained from male and female STAT5b-deficient mice (8–10 wk; 129 × BALB/c) (31), liver-specific HNF4α-deficient mice (7 wk; 129/SV x C57B6 × FVB) (32) and corresponding control mice (10, 21). Livers from adult male and female somatostatin-deficient mice (11–12 wk) (33) were kindly provided by Drs. R.M. Luque and R.D. Kineman (University of Illinois at Chicago, Chicago, IL) (34).
Rats
Livers were collected from untreated, sham-operated and hypophysectomized adult male and female adult Fischer 344 rats (9–13 wk of age) (35, 36). Where indicated, rats were subjected to GH treatment by continuous infusion via an Alzet osmotic mini-pump (20 ng of recombinant rat GH/g body weight/h for 7 d). Hypophysectomy was performed by the supplier of the rats (Taconic, Inc., Hudson, NY) at 8 wk of age. Livers from untreated Fischer 344 rats ranging in age from 4 d to 12 wk (16) were used for the post-natal developmental expression studies.
Western blot analysis of Cutl2
Liver nuclear extracts were prepared from freshly isolated livers from individual untreated adult male and female Fischer 344 rats, individual male rats given a continuous infusion of GH for 7 d, and pooled adult male and female ICR mice (n = 8 livers/group) using the method of Gorski et al (37) as described previously (35). Proteins were resolved on 6% SDS polyacrylamide gels (40 μg nuclear extract protein/lane) and subjected to Western blot analysis with anti-Cutl2 Ab 356, kindly provided by Dr. A. Nepveu, McGill University, as described previously (27), except that the Tris-buffered saline was 20 mM Tris-HCl, pH 7.6, 137 mM NaCl, and blocking was performed for 2 h at room temperature. For immunoprecipitation of Cutl2, male and female mouse liver nuclear extracts (1 mg protein/sample) were incubated overnight at 4°C with 1.2 μl of anti-Cutl2 Ab 356 in 0.6 ml of buffer A (100 mM sodium phosphate, pH 7.4, 0.9% NaCl, 1 mM EDTA, 0.1% Triton X-100, 0.5% IGEPAL CA-630 detergent, containing 1 mg/ml BSA and complete protease inhibitor cocktail (Roche Diagnostics GmbH, Mannheim, Germany). Antibody-antigen complexes were collected using Protein A-Sepharose CL-4B beads (GE Healthcare, Bio-Sciences Corp., Piscataway, NJ), washed 3 times with buffer A, once with 0.5 M NaCl and once with phosphate-buffered saline and analyzed by Western blotting. Whole cell extracts prepared from 293T cells transfected with mouse Cutl2 cDNA (plasmid pMX139/Myc/Cux2/HA, a gift from Dr. A. Nepveu (27)) served as a positive control.
Total RNA preparation and qPCR
Total RNA was purified from frozen liver using TRIzol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s protocol. Total RNA samples were treated with RQ1 RNase-free DNase (Promega, Madison, WI) for 1 h at 37°C followed by heating for 5 min at 75°C. Reverse transcription of 1 μg of RNA per sample was then performed as described earlier (10) or using a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA). SYBR Green I-based qPCR assays were performed as described (10) using either SYBR Green PCR Master Mix or Power SYBR Green PCR Master Mix (Applied Biosystems). The following primer pairs were designed using Primer Express software (Applied Biosystems) and used for qPCR amplifications (GeneBank accession numbers as indicated): mouse Cutl2 (NM_007804), 5′-CCTCAAGACGAACACCGTCAT-3′ (forward primer; exon 22) and 5′-GCGCATCCTGGACCTGTAGT-3′ (reverse primer; exon 23-exon 22 junction; Fig. 1A); rat Cutl2 (XM_222184), 5′-ATTGGCCAGCGTGTGTTTG-3′ and 5′-CATCCGACAGGAACTGCTTCA-3′; mouse Trim24 (NM_145076), 5′-GAGGCCTCCGTCAAACAGAAC-3′ and 5′-GAGCCAGAGCTTCCTCGACTT-3′; rat Trim24 (XM_575435), 5′-GAAGTATCTCCAGAGGCAGTTGGT-3′ and 5′-CAGCTTATCACACGTTTCACAGTAGAG-3′; mouse Tox (NM_145711), 5′-GTGAAGTGCTGCGGCTCTAGT-3′ and 5′-GGACCGTTTACCCCAGACATC-3′; rat CYP2C11 (NM_019184), 5′-AGAGGAGGCTCAGTGCCTTGT-3′ and 5′-CCCAGGATAAAGGTGGGATCA-3′; rat CYP2C12 (NM_031572), 5′-GCTCACCCTGTGATCCCAAA-3′ and 5′-TGACATTGCAGGGAGCACAT-3′. Rat Tox (XM_342800) was assayed using the following two primer sets: 5′-GCACACTGCTCTCCAATTCCA-3′ and 5′-CATGCTTGCCTGCTGTCTGA-3′ (primer set 1) and 5′-GAACATGGGAGGAACCAACGT-3′ and 5′-ACTTGCTCCCAGGTGGAGAAG-3′ (primer set 2). No amplification in rat samples was observed with either primer set. However, primer set 1, whose forward primer shows one mismatch to mouse Tox, successfully amplified Tox RNA from mouse liver. These findings suggest that Tox is not expressed in rat liver. The following qPCR primer pairs were used to determine the expression of different Cutl2 RNA variants (Fig. 1) in the livers of male and female ICR mice: for the exon 1A-contaning variant (CJ065459), 5′-CGGCTGCAGAAGGAGCTTAG-3′ (forward primer; exon 1A-exon 3 junction) and 5′-TTAAATTCCCGGCGGAGTT-3′ (reverse primer; exon 3); for the exon 1B-containing variant (U45665), 5′-CAGCCAGGACGCTCGGT-3′ (exon 1B) and 5′-GCTCCGAGGCGACAGAACT-3′ (exon 2); for the exon 1C-containing variant (NM_007804), 5′-CGTGCAAAGTCCAGGGTCTT-3′ (exon 1C) and 5′-GAAAGGGTCCCTGGCAAAG-3′ (exon 1C); and for exon 1B- and exon 1C-containing variants together, 5′-ACGGAGTACGGCGGTGTTC-3′ (exon 2) and 5′-TTAAATTCCCGGCGGAGTT-3′ (exon 3). Primers used to assay 18S rRNA assay were detailed previously (38).
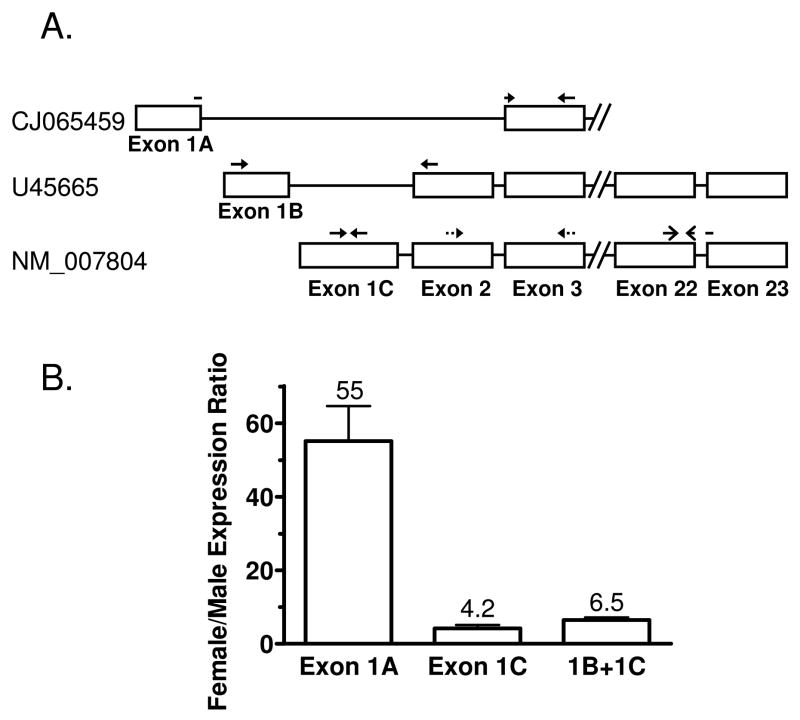
Figure 1. Expression of Cutl2 transcript variants in mouse liver.
Panel A: schematic representation of three different mouse Cutl2 RNAs and qPCR primer pairs used to determine their expression. Cutl2 transcript variants, based on the February 2006 build of the mouse genome, are identified by their GenBank accession numbers. Exon/intron structure shown is not drawn to scale; exons 1A and 1B are separated by 165 nt and exons 1B and 1C by 1951 nts. Exons 1–3 are non-coding. Exon 2 is absent from the exon 1A-containing RNA. The forward primer for this transcript (CJ065459) traverses the exon 1A/exon 3 junction, as indicated. The primer pair amplifying portions of exons 22 and 23 does not discriminate between the three Cutl2 RNA transcripts and was used to assay mouse Cutl2 in a majority of the experiments presented in this study. Panel B: female/male expression ratios of individual Cutl2 transcripts in ICR mouse liver. The ratios were determined for transcripts containing exon 1A, exon 1C and for the exon 1B + exon 1C transcripts together (‘1B + 1C’), as indicated. Relative RNA levels were determined by qPCR analysis of liver RNA from wild-type male and female mice (n=5 per group). The expression levels of each RNA transcript were normalized to the 18S rRNA content of each liver and then calculated relative to the average expression of the corresponding RNA transcript in males. Data shown are mean ± SE. The calculated mean ratios are indicated above each bar. Levels of each Cutl2 transcript showed statistically significant differences between females and males (Student’s t-test, p<0.01).
Analysis of sex-specific promoters for Cutl1/Cutl2 binding sites
Computational analysis of Cutl1/Cutl2 binding sites was carried out on a set of 21 group 1A male-specific genes, 18 group 2A male-specific genes and 20 group 1B female-specific genes (39), selected from the top 35 sex-specific genes in each group, as determined by microarray analysis (39). 207 liver-expressed genes that were identified by microarray analysis as non-sex-specific were selected as controls. The analysis was performed on an 8 kb region flanking the transcription start site (5 kb upstream + 3 kb downstream). The motif discovery program CLOVER (40) was used to search for significant Cutl1/Cutl2 motifs from the Transfac 7.0 public database using a p-value threshold of 0.005 and a motif score threshold of 6.5. The matrix comparison algorithm Possum (41) was used to scan for Cutl1/Cutl2 binding sites using a score threshold of 7.8.
Results
Female-specific expression of Cutl2, Trim24 and Tox RNAs
Microarray analysis has identified more than 750 liver-expressed genes, including several encoding transcription factors, that are more highly expressed in female compared to male mouse liver (39). qPCR was used to quantify the expression of three of these transcription factors, Cutl2, Trim24 and Tox, in livers of adult male and female mice and rats (Table I). All three factors were found to be expressed at higher levels in female compared to male liver in each of the four mouse strains examined. The female specificity was highest for Cutl2 (up to ≥ 100-fold higher expression than in males) in both mouse and rat liver. The female specificity of Trim24 RNA was substantially higher in rats (73-fold) than in mice (2.5 to 6.5-fold; Table I). Tox RNA could not be detected in male or female rat liver using two different qPCR primer pairs, one of which amplified mouse Tox RNA efficiently, suggesting the absence of Tox RNA in rat liver.
Table I.
Female-specific expression of Cutl2, Trim24 and Tox in mouse and rat liver
| Strain and species | Female/male expression ratio | ||
|---|---|---|---|
| Cutl2 | Trim24 | Tox | |
| ICR mice | 96 ± 18** | 6.5 ± 0.7** | 16 ± 2** |
| Black Swiss mice | 64 ± 6** | 3.9 ± 0.2** | 5.6 ± 0.3** |
| 129 × BALB/c mice | 107 ± 39* | 2.5 ± 0.6* | 9.5 ± 2.7** |
| 129/SV × C57B6 × FVB mice | 33 ± 13* | 2.8 ± 0.4** | 3.3 ± 0.2** |
| Fisher 344 rats | 126 ± 10** | 73 ± 11** | ND |
Relative expression of the each RNA was determined by qPCR analysis of wild-type male and female rat livers and mouse livers from the indicated strains. The expression levels in individual livers normalized to the 18S rRNA contents were calculated relative to the average expression in males. Data shown are mean ± SE based on the following numbers of animals per group (males/females): n=5/n=5 for ICR mice, n=3/n=5 for Black Swiss mice, n=6/n=5 for the mixed background strain of the STAT5-deficient mice (129 × BALB/c), n=8/n=8 for the mixed background strain of the HNF4α-deficient mice (129/SV × C57B6 × FVB), n=9/n=7 for Fischer 344 rats. The differences in expression between females and males were subjected to statistical analysis using Student’s t-test: * and **, p<0.05 and p<0.01, respectively. ND, not detected.
Mouse genomic and cDNA databases indicate the existence of at least three Cutl2 RNAs, which differ in the sequence of the first exon and in the presence or absence of exon 2 (Fig. 1, panel A), with exons 1 and 2 both being non-coding. qPCR primers that distinguish these three transcripts were designed and then used to determine relative expression of each Cutl2 transcript in male vs. female ICR mouse liver (Fig. 1B). Strong female-specific expression was observed for the exon 1A-containing cDNA (female/ male ratio = 55 ± 10), which was previously characterized as a spliced EST sequence (accession no. CJ065459). Reduced female specificity was apparent for the exon 1C-containing cDNA (NM_007804; female/male ratio = 4.2 ± 0.9). Amplification of the exon 1B-containing sequence produced very low signals, in both male and female samples (data not shown). When exon 1B- and exon 1C-containing transcripts were assayed together using a primer set targeting exons 2 and 3, a female/ male expression ratio of 6.5 ± 0.7 was determined. Liver Cutl2 RNA is thus represented by at least 2 transcripts preferentially expressed in females, with the transcripts containing exons 1A and 1C, respectively, being the major and minor sex-specific transcripts.
Female-specificity of liver nuclear Cutl2 protein
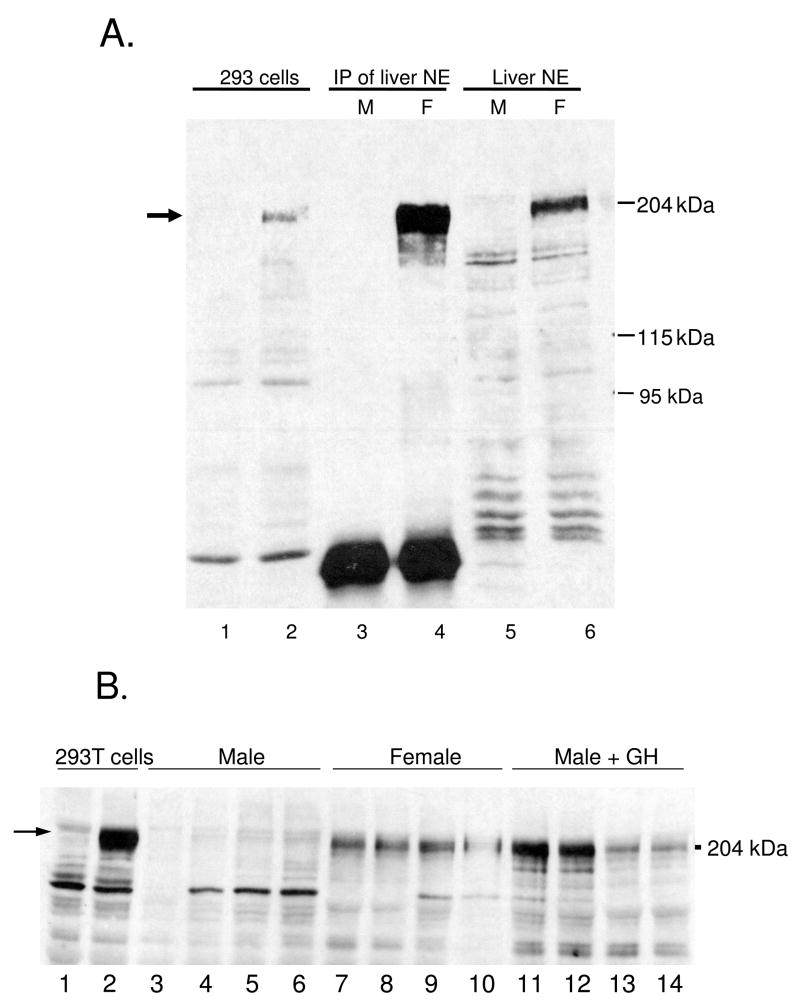
Next, we investigated whether the strong female specificity seen for Cutl2 RNA was reflected at the protein level. Liver nuclear extracts prepared from adult male and female mice were analyzed on Western blots probed with antibody to mouse Cutl2 (27). Fig. 2A shows that a protein of Mr ~ 200 kDa, corresponding in size to Cutl2 protein from 293T cells transfected with mouse Cutl2 plasmid, is highly enriched in female compared to male mouse liver. This female specificity is seen both in unfractionated liver nuclear extracts (lane 6 vs. lane 5) and after immunoprecipitation with anti-Cutl2 antibody (lane 4 vs. lane 3). Female-specific expression of Cutl2 protein was also seen in liver nuclear extracts from individual rats (Fig. 2B), consistent with the sex-specificity of rat Cutl2 RNA. Antibodies for Trim24 and Tox were not available.
Figure 2. Western blot analysis of liver Cutl2 protein.
Nuclear extracts prepared from pooled mouse livers (panel A) or from individual rat livers (panel B) were resolved on a 6% SDS polyacrylamide gel and subjected to Western blot analysis with anti-Cutl2 antibody 356 in comparison to cDNA-expressed mouse Cutl2. Panel A: whole cell extracts (40 μg) from untransfected 293T cells (lane 1) or from 293T cells transfected with Cutl2 cDNA (lane 2); Cutl2 protein immunoprecipitated with anti-Cutl2 antibody 356 from liver nuclear extract (1 mg) prepared from pools of adult male (lane 3) and female (lane 4) mouse liver (n=8 livers/group); and adult male (lane 5) and female (lane 6) mouse liver nuclear extract (40 μg/lane). Panel B, portion of Western blot showing, in lanes 1–2: cell extracts (40 μg) from untransfected or Cutl2 cDNA-transfected 293T cells, respectively; lanes 3–14: liver nuclear extracts prepared from individual adult male (lanes 3–6) and female (lanes 7–10) rats and from adult male rats given a continuous GH infusion for 7 d (lanes 11–14). Authentic, cDNA-expressed Cutl2 protein (lane 2, panels A and B; arrow) runs close to the 204 kDa protein marker shown on the right.
Developmental profiles of Cutl2 and Trim24
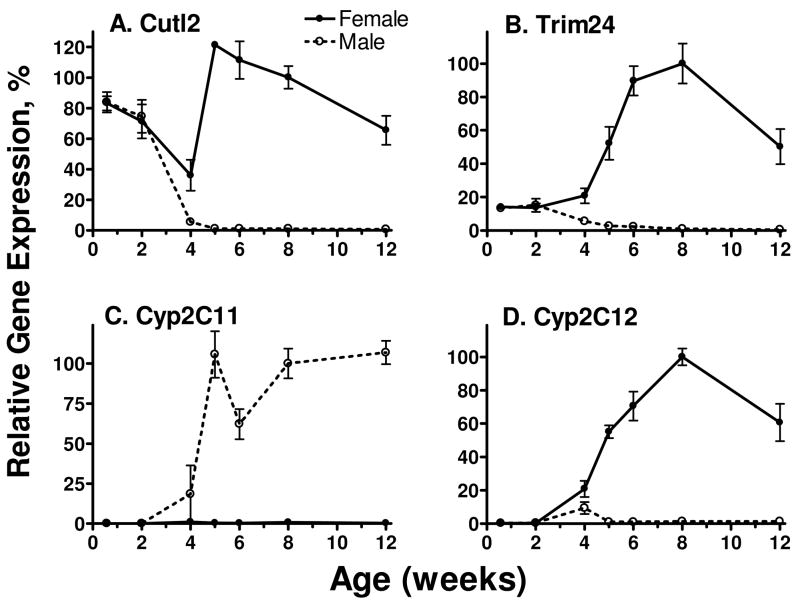
The sex-specificity of many liver RNAs and proteins emerges at the onset of puberty (~ 4 wk of age in the rat), coincident with a strong increase in pituitary GH release (1, 42). Investigation of the expression of Cutl2 and Trim24 during postnatal rat development revealed female-predominant expression of both RNAs beginning at 4 wk of age (Fig. 3A, 3B). Both RNAs were expressed in a sex-independent manner in prepubertal (4 d and 2 wk-old) rat liver at levels corresponding to ~ 15% (Trim24) and 70–80% (Cutl2) of 8 wk-old adult female liver. In males, Cutl2 and Trim24 both declined precipitously at 4 wk and were extinguished by 5 wk, whereas the expression in females increased after 4 wk and remained elevated through 12 wk of age. These developmental profiles contrast with those of the prototypical male-specific gene CYP2C11 (Fig. 3C) and the female-specific gene CYP2C12 (Fig. 3D). The GH pulse-regulated CYP2C11 and the continuous GH-regulated CYP2C12 were not expressed in neonatal and prepubertal rats; their expression first emerged at 4 wk of age in male and female liver, respectively.
Figure 3. Postnatal developmental profiles of rat liver Cutl2 and Trim24 RNAs in comparison with CYP2C11 and CYP2C12 RNAs.
Expression of the 4 indicated RNAs was determined by qPCR analysis of liver RNA prepared from individual male and female rats ranging from 4 d to 12 wk of age. RNA levels were normalized to the 18S rRNA content of each liver and expressed as a percentage of the average level in 8-wk-old females (panels A, B, D) or 8-wk-old males (panel C), which were set to 100%. The data shown are mean ± SE for each group (n=3–6 livers).
Responsiveness of rat liver Cutl2 and Trim24 to hypophysectomy and continuous GH treatment
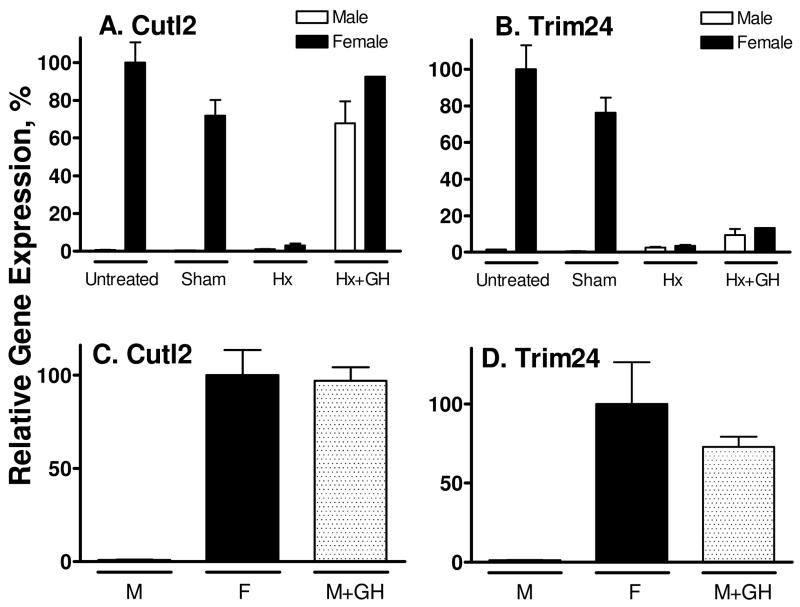
Given the dependence of many sex-specific hepatic RNAs and proteins on pituitary GH, we investigated the impact of hypophysectomy and GH replacement on the expression of Cutl2 and Trim24 (Fig. 4). Hypophysectomy of female rats ablated the expression of both liver transcription factors, indicating a strong dependence on pituitary or pituitary-derived hormone(s). Hypophysectomy of males did not lead to a substantial increase in Cutl2 or Trim24 RNA, indicating that neither transcription factor is subject to negative regulation by the male pituitary hormone pattern. Continuous GH treatment of hypophysectomized male and female rats using an osmotic mini-pump (20 ng GH/g BW/hr for 7 d) (7) restored normal female liver levels of Cutl2 (Fig. 4A). GH induced Trim24 ~ 3.7-fold in hypophysectomized males and females, however, this increase did not reach statistical significance (Fig. 4B).
Figure 4. Effect of hypophysectomy and continuous GH infusion on Cutl2 and Trim24 RNA in rat liver.
Cutl2 and Trim24 RNA levels were determined by qPCR analysis of livers isolated from individual untreated (n=3 livers/group), sham-operated (n=2/group) and hypophysectomized (Hx, n=4/group) male and female rats and from hypophysectomized rats given a continuous GH infusion via an osmotic pump (+GH) for 7 d (n=3 males and n=1 female) (panel A and panel B). RNA levels were also determined in individual livers from untreated male (M) and female (F) rats (n=4–5/group) and control male rats treated with continuous GH via an osmotic mini-pump for 7 d (n=5) (panel C and panel D). Data were analyzed as in Fig. 3, except that the RNA levels (mean ± SE for individual livers) are shown relative to the average level in untreated females, which was set to 100%.
The GH-responsiveness of Cutl2 and Trim24 was further established by the strong induction of both genes in livers of intact male rats given a 7 d continuous GH infusion (Fig. 4C, 4D). This treatment also increased Cutl2 protein in male liver nuclear extracts to normal female levels (Fig. 2B, lanes 11–14 vs. lanes 7–10). The substantial induction of Trim24 following continuous GH treatment of intact males but not hypophysectomized males (Fig. 4D vs. Fig. 4B) suggests a requirement for pituitary-dependent factor(s) other than GH, such as thyroid hormone and corticosteroids, for full female expression of Trim24. By contrast, Cutl2 RNA was fully feminized by continuous GH treatment of intact and hypophysectomized rats, i.e., even in the absence of other pituitary hormones. Neither gene was induced following hypophysectomy of male rats (Fig. 4A, 4B), indicating that Cutl2 and Trim24 are not subject to the strong negative regulation by the male GH profile seen with female-specific liver genes such as Cyp2b9 (10, 43).
Response to continuous GH in mouse liver
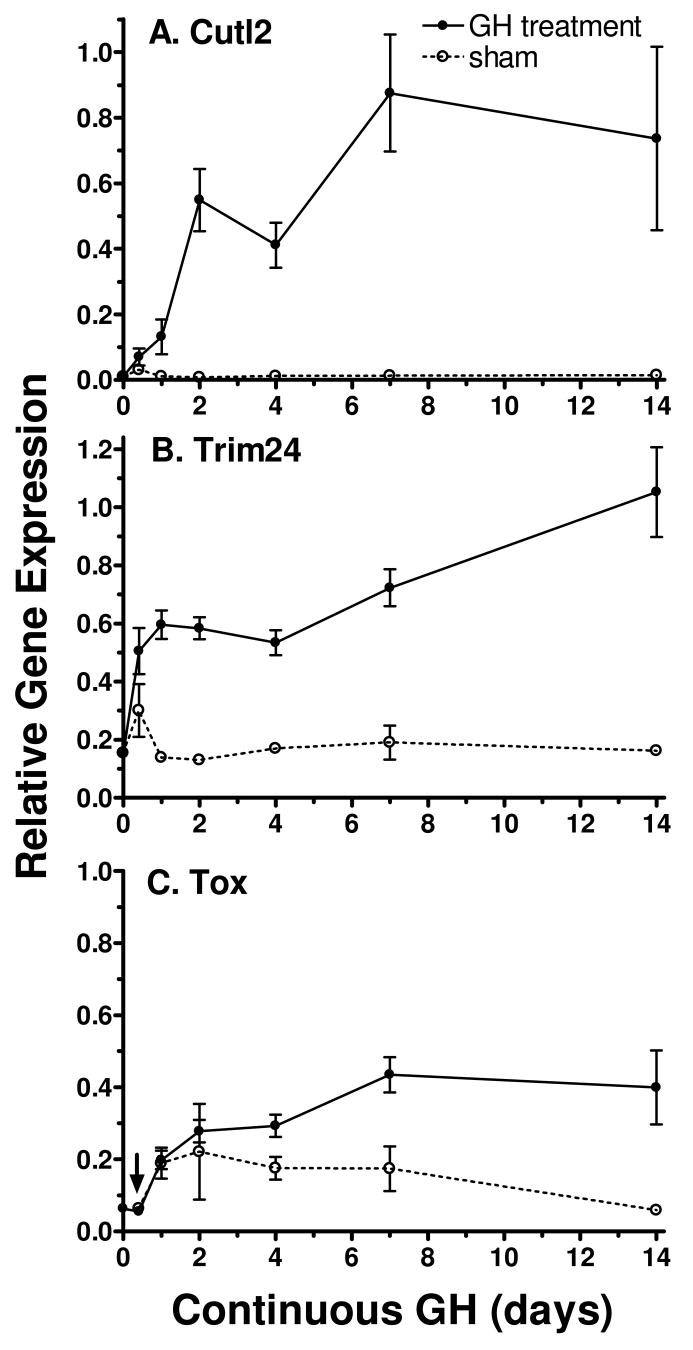
The impact of continuous GH treatment was investigated over a time course to determine the temporal relationship between changes in circulating GH profiles, induction of Cutl2 and Trim24, and the feminization of hepatic gene expression. This study was carried out in mice, where the time-dependent effects of continuous GH treatment on sex-specific Cyps and other liver-expressed genes are well established (10) and where the effects of continuous GH on the expression of Tox could be determined. Adult male mice were infused with GH continuously for time periods ranging from 10 h up to 14 d using osmotic mini-pumps. This treatment eliminates the GH-free interpulse period that is required for male liver gene expression and feminizes hepatic enzyme profiles (10). qPCR analysis revealed a time-dependent increase in all three RNAs. In the case of Cutl2 and Trim24, the induction of RNA levels was evident within the first day of GH infusion. Cutl2 RNA was up-regulated ~ 10-fold on day 1, 50-fold on day 2 and 80-fold on day 7, corresponding to ~ 85% of its adult female level (Fig. 5A). Trim24 increased about 4-fold, to 60% of its adult female level, within 1 d, and further increases were seen at later times (Fig. 5B). The early, and substantial increases in Cutl2 and Trim24 RNAs contrast with that of Tox RNA, whose induction was noticeably slower, reaching only ~ 40% of female levels after 7–14 d of continuous GH treatment (Fig. 5C).
Figure 5. Response of Cutl2, Trim24 and Tox RNAs to continuous GH infusion in mice.
Male ICR mice were implanted with osmotic mini-pumps delivering a continuous infusion of GH or vehicle control for time periods ranging from 10 h to 14 d. Individual livers were isolated and analyzed for Cutl2, Trim24 and Tox RNAs by qPCR. Data shown are based on the following number of individual mice per group: n=6–7 (each GH-treated time point) and n=2–3 (each vehicle-treated control group; sham). RNA levels (mean ± SE) were normalized to the 18S rRNA content of each liver and are presented relative to the average untreated female liver level, which was set to 1. All three RNAs showed stress-dependent responses to osmotic mini-pump implantation at the 10 h time point, as indicated by RNA increases in the vehicle-treated control group. The stress response of Tox RNA, evident at several of the early time points, was quite substantial and was indistinguishable for vehicle control and GH-treated livers at the 10h point (increase up to 80–95% of the untreated female control for both groups). Tox RNA data for the 10 h time point (arrow) was adjusted for the stress response by multiplying the relative expression levels in GH- and vehicle-receiving groups by the untreated male/vehicle control expression ratio.
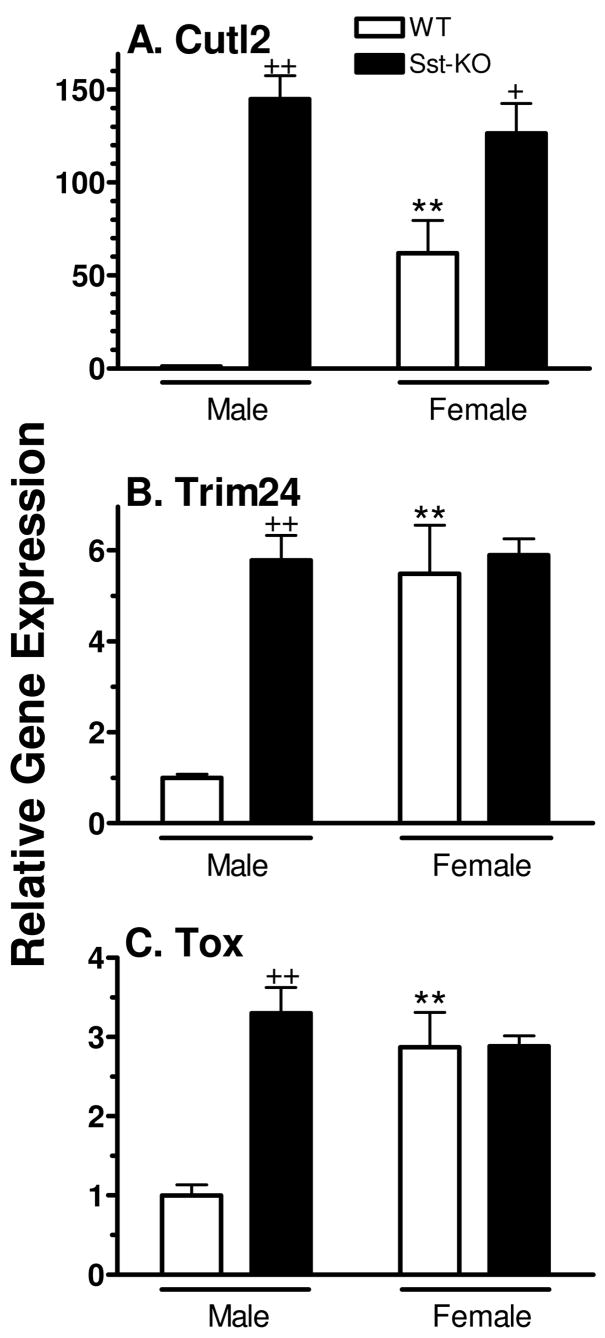
The responsiveness of Cutl2, Trim24 and Tox to changes in plasma GH profiles was further investigated in mice deficient in somatostatin, a hypothalamic suppressor of pituitary GH release whose absence leads to substantial increases in basal levels of GH between plasma pulses and increased expression of several female-specific hepatic genes (33). As show in Fig. 6, all three RNAs were increased to normal female levels or higher in somatostatin-deficient male mouse liver, resulting in a loss of sex-specificity in the somatostatin knockout strain. This increase reached ~150-fold in the case of Cutl2 RNA. In contrast, in somatostatin-deficient female liver, Cutl2 RNA but not Trim24 or Tox RNA was up-regulated by ~ 2-fold, consistent with the modest effect that somatostatin deficiency has on the overall plasma GH profile in females (33).
Figure 6. Effect of Sst gene disruption on mouse liver expression of Cutl2, Trim24 and Tox.
RNA levels were determined by qPCR analysis of individual livers from wild-type (WT) male (n=6) and female (n=3) mice and from Sst (somatostatin)-deficient (KO) male (n=5) and female (n=7) mice. RNA levels in individual livers were normalized to the 18S rRNA content and presented relative to the average RNA level in wild-type male mice, which was set at 1. Data shown are mean ± SE for each group. Data were analyzed using Student’s t-test: + and ++, p<0.05 and p<0.01, respectively, for somatostatin-deficient male (or female) vs. wild-type male (or female); * and **, p<0.05 and p<0.01, for wild-type (or somatostatin-deficient) female vs. wild-type (or somatostatin-deficient) male.
Dependence on STAT5b
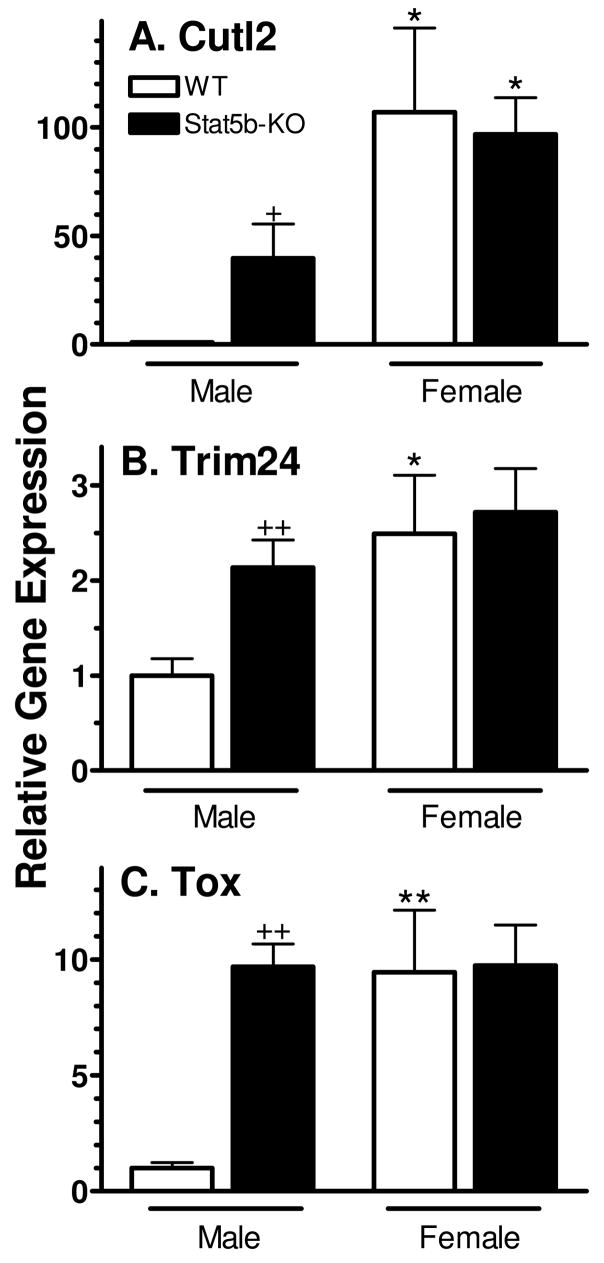
STAT5b is a GH pulse-responsive transcription factor whose expression is required for hepatic expression of many sex-dependent genes (39). The impact of STAT5b deficiency on Cutl2, Trim24 and Tox RNA levels was therefore investigated. In male mice deficient in STAT5b, Cutl2 RNA was up-regulated ~ 40-fold, to a level equal to 40% of wild-type female liver (Fig. 7A). Trim24 and Tox were increased by ~ 2-fold and 10-fold, respectively, and reached levels similar to wild-type females (Fig. 7B, 7C). In contrast, the loss of STAT5b in females had no effect on gene expression. Thus, sex-specificity is markedly decreased (Cutl2) or abolished (Trim24 and Tox) in the absence of STAT5b.
Figure 7. Effect of Stat5b gene disruption on mouse liver expression of Cutl2, Trim24 and Tox.
RNA levels were determined by qPCR analysis of individual livers from wild-type (WT) male (n=6) and female (n=5) mice and STAT5b-deficient (KO) male (n=6) and female (n=7) mice. Data were analyzed as described in Fig. 6, with the wild-type male RNA level set to 1.0 for each gene, and graphed as mean ± SE for each group. Data were analyzed using Student’s t-test: + and ++, p<0.05 and p<0.01, respectively, for STAT5b-deficient male (or female) vs. wild-type male (or female); * and **, p<0.05 and p<0.01, respectively, for wild-type (or STAT5b-deficient) female vs. wild-type (or STAT5b-deficient) male.
Co-dependence on HNF4α
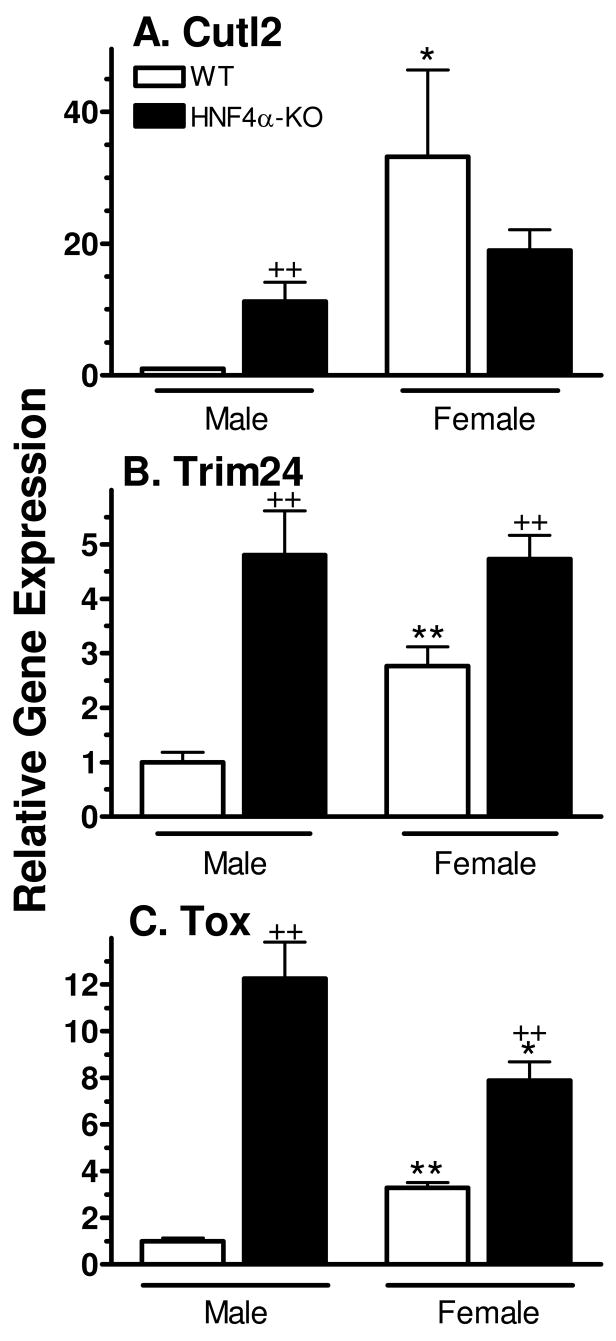
Many, but not all, sexually dimorphic mouse liver genes are co-dependent on HNF4α and STAT5b for sex-dependent expression (10). We therefore investigated the impact of HNF4α deficiency on the expression of Cutl2, Trim24 and Tox using a liver-specific HNF4α knockout mouse model (32). All three factors were strongly up-regulated in HNF4α-deficient male liver (Fig. 8). More modest increases in expression were seen for Trim24 and Tox, but not Cutl2, in HNF4α-deficient females. Overall, the loss of HNF4α led to loss of female-specificity for all three factors, as was also seen in the case of STAT5b deficiency (Fig. 7).
Figure 8. Effect of Hnf4α gene disruption on mouse liver Cutl2, Trim24 and Tox RNA.
RNA levels were assayed in livers of wild-type (WT) and HNF4α-deficient (KO) male and female mice by qPCR (n=8 livers/group). Data were analyzed as in Fig. 6, with the wild-type male level set to 1.0, and graphed as mean ± SE for each group. Data were analyzed using Student’s t-test: + and ++, p<0.05 and p<0.01, respectively, for HNF4α-deficient male (or female) vs. wild-type male (or female); * and **, p<0.05 and p<0.01, respectively, for wild-type (or HNF4α-deficient) female vs. wild-type (or HNF4α-deficient) male.
Cutl2 binding sites in sex-specific hepatic genes
Sex-specific genes have been grouped into sets of co-expressed genes based on their dependence on STAT5b for expression in males and in females (39). The two largest sets of co-expressed sex-specific genes, male-specific group 1A and female-specific group 1B, are regulated by STAT5b in a positive manner (group 1A) and in a negative manner (group 1B), respectively. Both gene sets were analyzed to determine whether DNA sequence motifs likely to be associated with Cutl2 binding are statistically over-represented in comparison to a control (background) set comprised of liver-expressed genes that are not sex-specific or subject to STAT5b-dependent regulation; the latter genes are expected to have a random distribution of Cutl2 binding sequences. Cutl2 has a DNA-binding specificity closely related to that of Cutl1 (27), whose binding specificity is well studied and, unlike Cutl2, is represented by binding site matrices in the Transfac database (44). These matrices were used by the motif analysis program Clover (40) to analyze 8 kb of DNA surrounding the transcription start sites of 21 group 1A male genes for the occurrence of Cutl1/Cutl2 binding sites. A set of 207 liver-expressed genes that is not sex-specific and does not display STAT5b-dependence served as a negative control. Binding sites represented by two Cutl1 Transfac matrices were found to be statistically over-represented, at p=0.002 and at p=0.004, in the group 1A male genes (Table II), suggesting that the repressor activity of Cutl2 (27) may contribute to the silencing of male gene expression in female liver. No such over-representation of binding sites was observed in a set of 20 group 1B female genes or in a second, distinctly regulated group of 18 male genes (group 2A), whose members require STAT5b for expression in both male and female liver (39). The prevalence of Cutl1/Cutl2 binding sites in group 1A male genes was confirmed using the matrix comparison algorithm Possum (Table II, legend).
Table II. Cutl1/Cutl2 binding sites identified in group 1A male-specific genes.
Shown are the Cutl1/Cutl2 binding sites, characterized by the two indicated Transfac matrices, identified in 16 of 21 group 1A male-specific genes by CLOVER analysis. The location of each binding site is shown relative to the transcription start site of each gene, along with the genomic DNA strand, binding sequence and associated motif score. Two motif scores are presented for binding sites that matched both Transfac matrices. In several cases, binding sites that overlapped with those shown below were found on the opposite strand with an offset of two nucleotides (not shown). The preponderance of Cutl1/Cutl2 sites in group 1A male-specific genes was confirmed using POSSUM to scan for binding sites, which were found in 11 group 1A genes but only 3 genes from female group 1B, 5 genes from male group 2A and 3–5 genes in each of three randomly selected sets of 21 background genes (not shown).
| Gene | TRANSFAC Motif ID | Location | Strand | Sequence | Motif Score |
|---|---|---|---|---|---|
| Cyp4a12 | M00106 | −3446 | − | ggcatcaacc | 7.04 |
| Moxd1 | M00106 | −4903 | − | ccgatcgacc | 7.45 |
| M00104, M00106 | −210 | + | catcgattcc | 6.94, 7.77 | |
| M00104, M00106 | 1966 | + | aatggatggg | 6.63, 6.84 | |
| M00106 | 2688 | + | catggatgcc | 7.24 | |
| Gstpi | M00104, M00106 | 1845 | − | cccatcaatg | 6.99, 8.26 |
| Aoh1 | M00104, M00106 | −3859 | + | catggatccc | 6.57, 7.94 |
| M00106 | −629 | − | cacatcaatc | 7.56 | |
| M00106 | −229 | + | catagatctg | 6.61 | |
| Dkk4 | M00106 | −4935 | − | gggatccatt | 7.29 |
| Ugt2b38 | M00104, M00106 | −4444 | + | gatcgatcca | 7.11, 7.28 |
| M00106 | −2683 | − | cacatcaatg | 7.46 | |
| M00106 | −778 | − | gggatggatc | 7.59 | |
| Sftpb | M00106 | −4198 | − | gggatccatg | 7.66 |
| M00106 | −2812 | − | gacatcaatg | 7.26 | |
| Il1r2 | M00104 | −4797 | − | gcgatggata | 6.66 |
| M00106 | −3468 | − | cagatcaatc | 8.01 | |
| M00106 | −2471 | + | catggatctg | 6.8 | |
| M00104 | −2169 | + | tattgattgc | 7.3 | |
| M00106 | −495 | − | cccatccatt | 6.66 | |
| Lamc3 | M00106 | −537 | − | gggatcattt | 6.62 |
| M00104, M00106 | 103 | + | catcgatggc | 8.16, 8.3 | |
| M00106 | 405 | − | cccatccatg | 6.7 | |
| Lck | M00106 | −4919 | + | gattgatttc | 6.6 |
| M00106 | −3263 | − | cggatggatg | 6.94 | |
| LOC382109 | M00104, M00106 | −4882 | − | gggatccatc | 6.65, 7.98 |
| M00106 | −3427 | − | cacatcaatc | 7.52 | |
| M00104, M00106 | −1513 | − | gggatccatc | 6.65, 7.98 | |
| Hba-x | M00106 | −4973 | + | catccatggc | 6.54 |
| M00106 | 2687 | − | cacatcaatt | 6.95 | |
| 4921504102Rik | M00104, M00106 | −4281 | − | gggatcaata | 6.73, 6.92 |
| M00106 | −2812 | − | cacatcaatc | 7.61 | |
| Irx2 | M00106 | −3684 | + | gatagatctg | 7.14 |
| M00106 | −2884 | − | gggatcattc | 6.89 | |
| M00104, M00106 | −1681 | + | aattgatccc | 7.56, 8.9 | |
| Ruvb12 | M00106 | −3498 | + | catggatctg | 6.7 |
| M00104, M00106 | −1057 | + | gattgatggc | 6.97, 7.97 | |
| Sox15 | M00104, M00106 | −4135 | + | aatcgatcgc | 9.79, 8.74 |
Discussion
STAT5b is a plasma GH profile-responsive transcription factor that plays a major role in establishing or maintaining sex-specific liver gene expression, with ~90% of male-specific genes down-regulated to wild-type female levels and ~60% of female-specific genes up-regulated in STAT5b-deficient male mice, as revealed by microarray analysis (39). Several observations suggest that some, perhaps many, of these gene regulatory effects of STAT5b are indirect, most notably the substantially delayed response of female-specific Cyps and other genes to changes in plasma GH profiles (10). These earlier findings prompted us to investigate novel signaling molecules and transcription factors that are expressed in liver in a sex-dependent manner and may potentially contribute to the STAT5b-dependent regulation of liver gene expression. Presently, three such liver transcription factors were characterized with respect to their sex-specificity, postnatal developmental expression, GH regulation and dependence on STAT5b and HNF4α for hepatic female specificity.
The three transcription factors investigated, Cutl2, Trim24 and Tox, were all expressed in liver in a female-specific manner, as determined by qPCR analysis of liver RNA. These results were confirmed at the protein level by analysis of liver nuclear extracts in the case of the CDP/Cut transcription factor family member Cutl2, which exhibited the highest sex-specificity (female:male ratio ~100) in both rats and mice. Female-predominant expression (female:male ~3) has previously been observed for another Cut domain protein, the liver-enriched transcription factor HNF6, which contributes to GH regulation of the female-specific rat gene CYP2C12 (23, 24). In the case of Trim24, a transcriptional intermediary factor 1 family member, the female/male specificity ratio in rats (~73) was substantially higher than in mice (2.5 to 6.5, depending on the strain).
Tox, an HMG box-containing transcription factor, exhibited up to 16-fold higher expression in female compared to male mouse liver. The delayed response of Tox to continuous GH treatment, as compared to Cutl2 and Trim24 (Fig. 5), rules out Tox as an early, upstream regulator of sex-dependent liver genes. Moreover, no expression of the predicted rat homolog of mouse Tox (95% nucleotide identity in their region of overlap) could be detected in either female or male rat liver. This absence indicates Tox does not play a fundamental, conserved regulatory role in the sex-dependent actions of GH in the liver.
High-level, female-like expression of Cutl2 and Trim24, and to a lesser extent Tox, could be induced in males given a continuous infusion of GH using osmotic mini-pumps. This treatment overrides the endogenous male, pulsatile plasma GH pattern and mimics the more continuous female GH pattern, leading to global feminization of liver gene expression (3). The dependence of these transcription factors on the female GH pattern was also demonstrated, in the case of Cutl2, in hypophysectomy and continuous GH replacement experiments and, in the case of all three factors, by their increased expression in mice deficient in somatostatin, an inhibitor of pituitary GH secretion. Somatostatin-deficient mice have a significantly elevated plasma GH baseline associated with feminization of several sex-dependent hepatic RNAs (33, 45). The mechanism whereby continuous GH induces hepatic expression of Cutl2, Trim24 and Tox is uncertain. However, it is not likely to be mediated by the low but sustained level of activated STAT5b found in intact adult female liver (15), insofar as STAT5b is dispensable for the high level expression of these genes seen in female mice (Fig. 7). This conclusion is further supported by the substantial expression of Cutl2, and to a lesser extent Trim24, in both male and female rats prior to puberty (Fig. 3), at which time hepatic STAT5b activity is low or undetectable (16).
While STAT5b may not be required for hepatic expression of Cutl2, Trim24 or Tox, it may, nevertheless, regulate these transcription factors, as indicated by the substantial increases in their expression in livers of male mice deficient in STAT5b. This apparent de-repression of all three factors in STAT5b-deficient males could indicate that the corresponding three genes are directly repressed by GH pulse-activated STAT5b, or perhaps by a STAT5b-dependent factor. This scenario seems unlikely for Cutl2 and Trim24, however, as their expression in males was not elevated following hypophysectomy (Fig. 4), where liver STAT5b is inactive due to the absence of stimulation by circulating GH. An alternative possibility is that the increased expression seen for the three genes in STAT5b-deficient liver reflects a loss of STAT5b-dependent feedback inhibition in the hypothalamus (46), which may lead to changes in plasma GH profiles that mimic an adult female pattern and thereby induce Cutl2, Trim24 and Tox expression.
Trim24, also known as TIF-1α, is a non-histone chromosomal protein kinase that forms complexes with TIF-1α/KAP-1 and KRAB motif-containing zinc finger repressors and exerts transcriptional repression activity, which is proposed to involve histone deacetylation and chromatin remodeling (28, 47, 48). Trim24 binds tightly to euchromatin, in particular at the borders between euchromatin and heterochromatin, and may positively modulate the interaction of liganded nuclear receptors, and perhaps other sequence-specific DNA-binding proteins, with their cognate binding sites (29, 48, 49). The female-specific expression of Trim24, which reached 73-fold in the case of rat liver, could play an important role in GH-dependent chromatin remodeling associated with expression of sex-dependent genes (1), potentially amplifying the sex-dependent effects of transcriptional regulators, such as HNF4α and HNF6, which exhibit more modest female-predominance (~3–6-fold) in expression and activity. Further study will be required to investigate these and other possible consequences of the sex-specificity of Trim24.
Cutl2, also known as Cux2, is a sequence-specific DNA-binding protein that was reported to be expressed exclusively in the central and peripheral nervous systems (26). The absence of Cutl2 RNA in adult mouse liver reported in earlier studies may readily be explained if the liver samples analyzed were from male mice. Presently, we found Cutl2 to be a female-specific, nuclear protein in both mouse and rat liver whose migration on SDS gels (Mr ~200 kDa) was somewhat slower than would be expected based on its predicted size of 1426 (mouse) or 1613 amino acids (rat), but is consistent with the apparent molecular weight of the similar length (1486 amino acids) human Cutl2 protein in a neuroblastoma cell line (27). Cutl2 contains three conserved Cut repeats and one COOH-terminal Cut homeodomain, which act in combination to dictate the sequence-specific DNA-binding activity of Cutl2. In contrast to Cutl1 (CCAAT-displacement protein), which exhibits both transcriptional repression and trans-activation functions, Cutl2 has, thus far, been exclusively associated with transcriptional repression of target genes (27). As in the case of Cutl1, repression by Cutl2 could involve displacement of the transcription factor C/EBP, which itself is subject to GH regulation (50, 51).
Conceivably, Cutl2 may contribute to sex-specific liver gene expression by repressing the expression of certain male-specific genes in female liver. This hypothesis is supported by our finding that binding sites for Cutl1, whose DNA-binding specificity is very close to that of Cutl2 (27), were statistically over-represented in a set of co-expressed male-specific mouse genes belonging to group 1A (39) but not in a corresponding set of female-specific mouse genes (group 1B) or in a distinctly regulated set of male-specific genes (group 2A). This hypothesis is also consistent with the dramatic induction of the male-specific rat gene CYP2C11 at the onset of puberty, i.e., at the same time that Cutl2 expression is silenced in male rat liver (Fig. 3). Similarly, the substantial up-regulation of Cutl2 seen in adult male mice deficient in HNF4α could contribute to the associated suppression of certain male-specific Cyps and other GH-regulated genes in this animal model. Further studies will be required to test these hypotheses and to evaluate the role, if any, that Cutl2 and the other factors characterized in this study play in the global expression patterns of sex-specific, GH-regulated genes in rat and mouse liver.
Acknowledgments
The authors thank Dr. Alain Nepveu, McGill University, for providing Cutl2 plasmid and antibody.
Supported in part by NIH Grant DK33765 (to D.J.W.)
Abbreviations
- CYP
cytochrome P450
- JAK
Janus kinase
- HNF
hepatocyte nuclear factor
- Cutl2
cut-like 2
- Trim 24
tripartite motif-containing 24
- Tox
thymus high mobility group box protein
Footnotes
Supported in part by NIH grant DK33765 (to D.J.W.)
Authors’ Disclosure Statement: The authors have nothing to disclose
This is an un-copyedited author manuscript copyrighted by The Endocrine Society. This may not be duplicated or reproduced, other than for personal use or within the rule of “Fair Use of Copyrighted Materials” (section 107, Title 17, U.S. Code) without permission of the copyright owner, The Endocrine Society. From the time of acceptance following peer review, the full text of this manuscript is made freely available by The Endocrine Society at http://www.endojournals.org/. The final copy edited article can be found at http://www.endojournals.org/. The Endocrine Society disclaims any responsibility or liability for errors or omissions in this version of the manuscript or in any version derived from it by the National Institutes of Health or other parties. The citation of this article must include the following information: author(s), article title, journal title, year of publication and DOI.
References
- 1.Waxman DJ, O’Connor C. Growth hormone regulation of sex-dependent liver gene expression. Molecular endocrinology (Baltimore, Md. 2006;20:2613–2629. doi: 10.1210/me.2006-0007. [DOI] [PubMed] [Google Scholar]
- 2.Mode A, Gustafsson JA. Sex and the liver - a journey through five decades. Drug Metab Rev. 2006;38:197–207. doi: 10.1080/03602530600570057. [DOI] [PubMed] [Google Scholar]
- 3.Ahluwalia A, Clodfelter KH, Waxman DJ. Sexual dimorphism of rat liver gene expression: regulatory role of growth hormone revealed by deoxyribonucleic Acid microarray analysis. Molecular endocrinology (Baltimore, Md. 2004;18:747–760. doi: 10.1210/me.2003-0138. [DOI] [PubMed] [Google Scholar]
- 4.Laz EV, Wiwi CA, Waxman DJ. Sexual dimorphism of rat liver nuclear proteins: regulatory role of growth hormone. Mol Cell Proteomics. 2004;3:1170–1180. doi: 10.1074/mcp.M400102-MCP200. [DOI] [PubMed] [Google Scholar]
- 5.Eden S. Age- and sex-related differences in episodic growth hormone secretion in the rat. Endocrinology. 1979;105:555–560. doi: 10.1210/endo-105-2-555. [DOI] [PubMed] [Google Scholar]
- 6.Shapiro BH, Agrawal AK, Pampori NA. Gender differences in drug metabolism regulated by growth hormone. Int J Biochem Cell Biol. 1995;27:9–20. doi: 10.1016/1357-2725(94)00056-5. [DOI] [PubMed] [Google Scholar]
- 7.Waxman DJ, Pampori NA, Ram PA, Agrawal AK, Shapiro BH. Interpulse interval in circulating growth hormone patterns regulates sexually dimorphic expression of hepatic cytochrome P450. Proc Natl Acad Sci U S A. 1991;88:6868–6872. doi: 10.1073/pnas.88.15.6868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.MacLeod JN, Pampori NA, Shapiro BH. Sex differences in the ultradian pattern of plasma growth hormone concentrations in mice. J Endocrinol. 1991;131:395–399. doi: 10.1677/joe.0.1310395. [DOI] [PubMed] [Google Scholar]
- 9.Pampori NA, Agrawal AK, Shapiro BH. Infusion of gender-dependent plasma growth hormone profiles into intact rats: effects of subcutaneous, intraperitoneal, and intravenous routes of rat and human growth hormone on endogenous circulating growth hormone profiles and expression of sexually dimorphic hepatic cyp isoforms. Drug Metab Dispos. 2001;29:8–16. [PubMed] [Google Scholar]
- 10.Holloway MG, Laz EV, Waxman DJ. Co-dependence of growth hormone-responsive, sexually dimorphic hepatic gene expression on signal transducer and activator of transcription 5b and hepatic nuclear factor 4-alpha Molecular endocrinology; Baltimore, Md. 2006. pp. 647–660. [DOI] [PubMed] [Google Scholar]
- 11.Herrington J, Carter-Su C. Signaling pathways activated by the growth hormone receptor. Trends Endocrinol Metab. 2001;12:252–257. doi: 10.1016/s1043-2760(01)00423-4. [DOI] [PubMed] [Google Scholar]
- 12.Darnell JE., Jr STATs and gene regulation. Science. 1997;277:1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 13.Waxman DJ, Ram PA, Park SH, Choi HK. Intermittent plasma growth hormone triggers tyrosine phosphorylation and nuclear translocation of a liver-expressed, Stat 5-related DNA binding protein. Proposed role as an intracellular regulator of male-specific liver gene transcription. The Journal of biological chemistry. 1995;270:13262–13270. doi: 10.1074/jbc.270.22.13262. [DOI] [PubMed] [Google Scholar]
- 14.Sueyoshi T, Yokomori N, Korach KS, Negishi M. Developmental action of estrogen receptor-alpha feminizes the growth hormone-Stat5b pathway and expression of Cyp2a4 and Cyp2d9 genes in mouse liver. Mol Pharmacol. 1999;56:473–477. doi: 10.1124/mol.56.3.473. [DOI] [PubMed] [Google Scholar]
- 15.Choi HK, Waxman DJ. Growth hormone, but not prolactin, maintains, low-level activation of STAT5a and STAT5b in female rat liver. Endocrinology. 1999;140:5126–5135. doi: 10.1210/endo.140.11.7106. [DOI] [PubMed] [Google Scholar]
- 16.Choi HK, Waxman DJ. Plasma growth hormone pulse activation of hepatic JAK-STAT5 signaling: developmental regulation and role in male-specific liver gene expression. Endocrinology. 2000;141:3245–3255. doi: 10.1210/endo.141.9.7638. [DOI] [PubMed] [Google Scholar]
- 17.Tannenbaum GS, Choi HK, Gurd W, Waxman DJ. Temporal relationship between the sexually dimorphic spontaneous GH secretory profiles and hepatic STAT5 activity. Endocrinology. 2001;142:4599–4606. doi: 10.1210/endo.142.11.8480. [DOI] [PubMed] [Google Scholar]
- 18.Park SH, Waxman DJ. Inhibitory cross-talk between STAT5b and liver nuclear factor HNF3beta: impact on the regulation of growth hormone pulse-stimulated, male-specific liver cytochrome P-450 gene expression. The Journal of biological chemistry. 2001;276:43031–43039. doi: 10.1074/jbc.M107597200. [DOI] [PubMed] [Google Scholar]
- 19.Subramanian A, Wang J, Gil G. STAT 5 and NF-Y are involved in expression and growth hormone-mediated sexually dimorphic regulation of cytochrome P450 3A10/lithocholic acid 6beta-hydroxylase. Nucleic acids research. 1998;26:2173–2178. doi: 10.1093/nar/26.9.2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wiwi CA, Waxman DJ. Role of hepatocyte nuclear factors in transcriptional regulation of male-specific CYP2A2. The Journal of biological chemistry. 2005;280:3259–3268. doi: 10.1074/jbc.M409294200. [DOI] [PubMed] [Google Scholar]
- 21.Wiwi CA, Gupte M, Waxman DJ. Sexually dimorphic P450 gene expression in liver-specific hepatocyte nuclear factor 4alpha-deficient mice. Molecular endocrinology (Baltimore, Md. 2004;18:1975–1987. doi: 10.1210/me.2004-0129. [DOI] [PubMed] [Google Scholar]
- 22.Lahuna O, Rastegar M, Maiter D, Thissen JP, Lemaigre FP, Rousseau GG. Involvement of STAT5 (signal transducer and activator of transcription 5) and HNF-4 (hepatocyte nuclear factor 4) in the transcriptional control of the hnf6 gene by growth hormone. Molecular endocrinology (Baltimore, Md. 2000;14:285–294. doi: 10.1210/mend.14.2.0423. [DOI] [PubMed] [Google Scholar]
- 23.Delesque-Touchard N, Park SH, Waxman DJ. Synergistic action of hepatocyte nuclear factors 3 and 6 on CYP2C12 gene expression and suppression by growth hormone-activated STAT5b. Proposed model for female specific expression of CYP2C12 in adult rat liver. The Journal of biological chemistry. 2000;275:34173–34182. doi: 10.1074/jbc.M004027200. [DOI] [PubMed] [Google Scholar]
- 24.Lahuna O, Fernandez L, Karlsson H, Maiter D, Lemaigre FP, Rousseau GG, Gustafsson J, Mode A. Expression of hepatocyte nuclear factor 6 in rat liver is sex-dependent and regulated by growth hormone. Proc Natl Acad Sci U S A. 1997;94:12309–12313. doi: 10.1073/pnas.94.23.12309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nepveu A. Role of the multifunctional CDP/Cut/Cux homeodomain transcription factor in regulating differentiation, cell growth and development. Gene. 2001;270:1–15. doi: 10.1016/s0378-1119(01)00485-1. [DOI] [PubMed] [Google Scholar]
- 26.Quaggin SE, Heuvel GB, Golden K, Bodmer R, Igarashi P. Primary structure, neural-specific expression, and chromosomal localization of Cux-2, a second murine homeobox gene related to Drosophila cut. The Journal of biological chemistry. 1996;271:22624–22634. doi: 10.1074/jbc.271.37.22624. [DOI] [PubMed] [Google Scholar]
- 27.Gingras H, Cases O, Krasilnikova M, Berube G, Nepveu A. Biochemical characterization of the mammalian Cux2 protein. Gene. 2005;344:273–285. doi: 10.1016/j.gene.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 28.Nielsen AL, Ortiz JA, You J, Oulad-Abdelghani M, Khechumian R, Gansmuller A, Chambon P, Losson R. Interaction with members of the heterochromatin protein 1 (HP1) family and histone deacetylation are differentially involved in transcriptional silencing by members of the TIF1 family. Embo J. 1999;18:6385–6395. doi: 10.1093/emboj/18.22.6385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thenot S, Henriquet C, Rochefort H, Cavailles V. Differential interaction of nuclear receptors with the putative human transcriptional coactivator hTIF1. The Journal of biological chemistry. 1997;272:12062–12068. doi: 10.1074/jbc.272.18.12062. [DOI] [PubMed] [Google Scholar]
- 30.Wilkinson B, Chen JY, Han P, Rufner KM, Goularte OD, Kaye J. TOX: an HMG box protein implicated in the regulation of thymocyte selection. Nat Immunol. 2002;3:272–280. doi: 10.1038/ni767. [DOI] [PubMed] [Google Scholar]
- 31.Udy GB, Towers RP, Snell RG, Wilkins RJ, Park SH, Ram PA, Waxman DJ, Davey HW. Requirement of STAT5b for sexual dimorphism of body growth rates and liver gene expression. Proc Natl Acad Sci U S A. 1997;94:7239–7244. doi: 10.1073/pnas.94.14.7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hayhurst GP, Lee YH, Lambert G, Ward JM, Gonzalez FJ. Hepatocyte nuclear factor 4alpha (nuclear receptor 2A1) is essential for maintenance of hepatic gene expression and lipid homeostasis. Mol Cell Biol. 2001;21:1393–1403. doi: 10.1128/MCB.21.4.1393-1403.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Low MJ, Otero-Corchon V, Parlow AF, Ramirez JL, Kumar U, Patel YC, Rubinstein M. Somatostatin is required for masculinization of growth hormone-regulated hepatic gene expression but not of somatic growth. J Clin Invest. 2001;107:1571–1580. doi: 10.1172/JCI11941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luque RM, Gahete MD, Hochgeschwender U, Kineman RD. Evidence that endogenous SST inhibits ACTH and ghrelin expression by independent pathways. Am J Physiol Endocrinol Metab. 2006;291:E395–403. doi: 10.1152/ajpendo.00038.2006. [DOI] [PubMed] [Google Scholar]
- 35.Sundseth SS, Alberta JA, Waxman DJ. Sex-specific, growth hormone-regulated transcription of the cytochrome P450 2C11 and 2C12 genes. The Journal of biological chemistry. 1992;267:3907–3914. [PubMed] [Google Scholar]
- 36.Ram PA, Waxman DJ. SOCS/CIS protein inhibition of growth hormone-stimulated STAT5 signaling by multiple mechanisms. The Journal of biological chemistry. 1999;274:35553–35561. doi: 10.1074/jbc.274.50.35553. [DOI] [PubMed] [Google Scholar]
- 37.Gorski K, Carneiro M, Schibler U. Tissue-specific in vitro transcription from the mouse albumin promoter. Cell. 1986;47:767–776. doi: 10.1016/0092-8674(86)90519-2. [DOI] [PubMed] [Google Scholar]
- 38.Apletalina EV, Li HC, Waxman DJ. Evaluation of thyroid hormone effects on liver P450 reductase translation. Arch Biochem Biophys. 2003;409:172–179. doi: 10.1016/s0003-9861(02)00417-4. [DOI] [PubMed] [Google Scholar]
- 39.Clodfelter K, Holloway MG, Hodor P, Park S-H, Ray WJ, Waxman DJ. Sex-dependent liver gene expression is extensive and largely dependent upon STAT5b: STAT5b-dependent activation of male genes and repression of female genes revealed by microarray analysis. Molecular endocrinology (Baltimore, Md. 2006;20:1333–1351. doi: 10.1210/me.2005-0489. [DOI] [PubMed] [Google Scholar]
- 40.Frith MC, Fu Y, Yu L, Chen JF, Hansen U, Weng Z. Detection of functional DNA motifs via statistical over-representation. Nucleic acids research. 2004;32:1372–1381. doi: 10.1093/nar/gkh299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fu Y, Frith MC, Haverty PM, Weng Z. MotifViz: an analysis and visualization tool for motif discovery. Nucleic acids research. 2004;32:W420–423. doi: 10.1093/nar/gkh426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rich KJ, Boobis AR. Expression and inducibility of P450 enzymes during liver ontogeny. Microsc Res Tech. 1997;39:424–435. doi: 10.1002/(SICI)1097-0029(19971201)39:5<424::AID-JEMT5>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 43.Sakuma T, Kitajima K, Nishiyama M, Mashino M, Hashita T, Nemoto N. Suppression of female-specific murine Cyp2b9 gene expression by growth or glucocorticoid hormones. Biochem Biophys Res Commun. 2004;323:776–781. doi: 10.1016/j.bbrc.2004.08.158. [DOI] [PubMed] [Google Scholar]
- 44.Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, Reuter I, Chekmenev D, Krull M, Hornischer K, Voss N, Stegmaier P, Lewicki-Potapov B, Saxel H, Kel AE, Wingender E. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic acids research. 2006;34:D108–110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zeyda T, Diehl N, Paylor R, Brennan MB, Hochgeschwender U. Impairment in motor learning of somatostatin null mutant mice. Brain Res. 2001;906:107–114. doi: 10.1016/s0006-8993(01)02563-x. [DOI] [PubMed] [Google Scholar]
- 46.Bennett E, McGuinness L, Gevers EF, Thomas GB, Robinson IC, Davey HW, Luckman SM. Hypothalamic STAT proteins: regulation of somatostatin neurones by growth hormone via STAT5b. J Neuroendocrinol. 2005;17:186–194. doi: 10.1111/j.1365-2826.2005.01296.x. [DOI] [PubMed] [Google Scholar]
- 47.Germain-Desprez D, Bazinet M, Bouvier M, Aubry M. Oligomerization of transcriptional intermediary factor 1 regulators and interaction with ZNF74 nuclear matrix protein revealed by bioluminescence resonance energy transfer in living cells. The Journal of biological chemistry. 2003;278:22367–22373. doi: 10.1074/jbc.M302234200. [DOI] [PubMed] [Google Scholar]
- 48.Remboutsika E, Lutz Y, Gansmuller A, Vonesch JL, Losson R, Chambon P. The putative nuclear receptor mediator TIF1alpha is tightly associated with euchromatin. J Cell Sci. 1999;112 ( Pt 11):1671–1683. doi: 10.1242/jcs.112.11.1671. [DOI] [PubMed] [Google Scholar]
- 49.Zhong S, Delva L, Rachez C, Cenciarelli C, Gandini D, Zhang H, Kalantry S, Freedman LP, Pandolfi PP. A RA-dependent, tumour-growth suppressive transcription complex is the target of the PML-RARalpha and T18 oncoproteins. Nat Genet. 1999;23:287–295. doi: 10.1038/15463. [DOI] [PubMed] [Google Scholar]
- 50.Cui TX, Piwien-Pilipuk G, Huo JS, Kaplani J, Kwok R, Schwartz J. Endogenous CCAAT/enhancer binding protein beta and p300 are both regulated by growth hormone to mediate transcriptional activation. Molecular endocrinology (Baltimore, Md. 2005;19:2175–2186. doi: 10.1210/me.2004-0502. [DOI] [PubMed] [Google Scholar]
- 51.Huo JS, McEachin RC, Cui TX, Duggal NK, Hai T, States DJ, Schwartz J. Profiles of growth hormone (GH)-regulated genes reveal time-dependent responses and identify a mechanism for regulation of activating transcription factor 3 by GH. The Journal of biological chemistry. 2006;281:4132–4141. doi: 10.1074/jbc.M508492200. [DOI] [PubMed] [Google Scholar]