Abstract
Chronic lymphocytic leukemia (CLL) B cells characteristically exhibit low or undetectable surface B cell receptor (BCR) and diminished responses to BCR-mediated signaling. These features suggest that CLL cells may have sustained mutations affecting one or more of the BCR proteins required for receptor surface assembly and signal transduction. Loss of expression and mutations in the critical BCR protein B29 (Igβ, CD79b), are prevalent in CLL and could produce the hallmark features of these leukemic B cells. Because patient CLL cells are intractable to manipulation, we developed a model system to analyze B29 mutations. Jurkat T cells stably expressing μ, κ, and mb1 efficiently assembled a functional BCR when infected with recombinant vaccinia virus bearing wild-type B29. In contrast, a B29 CLL mutant protein truncated in the transmembrane domain did not associate with μ or mb1 at the cell surface. Another B29 CLL mutant lacking the C-terminal immunoreceptor tyrosine activation motif tyrosine and distal residues brought the receptor to the surface as well as wild-type B29 but showed significant impairment in anti-IgM-stimulated signaling events including mitogen-activated protein kinase activation. These findings demonstrate that B29 mutations previously identified in CLL patients can affect BCR-dependent signaling and may contribute to the unresponsive B cell phenotype in CLL. Finally, the features of the B29 mutations in CLL predict that they may be generated by somatic hypermutation.
Chronic lymphocytic leukemia (CLL) is the most common leukemia in the Western world, but the genetic events leading to this disease remain unknown. CLL patients typically accumulate CD5+, nonresponsive, growth-arrested B cells that express little or no surface Ig. CLL is unique among B cell malignancies: although multiple chromosomal abnormalities exist in ≈50% of all patients (1), no single translocation predominates. Trisomy 12 and deletions of chromosome 13 (13q14) each occur in 20–30% of CLL patients, but these lesions are seen in only a portion of a patient's B cells, indicating that they likely are secondary events (2–4). Deletions in p53 occur late in CLL in a minority of patients and correlate with aggressive transformation (5). Bcl2 expression is elevated in CLL B cells in the absence of Bcl2 gene rearrangements (5) and is correlated with promoter hypomethylation (6). Increased Bcl2 expression has been shown to protect B cells against apoptosis in a transgenic mouse (7, 8) and could prolong B cell survival, leading to the peripheral accumulation of noncycling B cells in CLL. Impaired B-cell receptor (BCR) signaling has been documented in CLL B cells (9–11). Altered Src family (11) or Syk (10) tyrosine kinase function may contribute to the decreased protein tyrosine phosphorylation and calcium flux exhibited by selected CLL B cell populations (9). However, none of these defects are likely to account for the reduction in surface BCR that is a hallmark of CLL.
B29 and the associated transmembrane protein mb1 are crucial for the assembly and membrane display of the BCR (12–14). Mice deficient for B29 are blocked in B cell development at the earliest stage of membrane μ expression, demonstrating the central role of B29 in B cell development (15). Initiation and amplification of BCR signaling is coordinated by the immunoreceptor tyrosine activation motifs (ITAMs) of the B29/mb1 heterodimers (16). Substitutions of both conserved tyrosine residues within the ITAM abrogate B29 signal transduction (17–20). Continuous signaling through the BCR is required for B cell survival in the periphery (21).
Two studies have documented mutations in the B29 genes of B cells from CLL patients (22, 23). Mutations in the B29-coding region were seen in 65% of 26 CLL patients in these two studies. The functional consequences of most of these B29 mutations is not known, although several mutants were predicted to produce truncated B29 proteins that would likely be impaired in BCR assembly and surface expression. Another study (24) of six CLL patients did not detect B29 mutations and instead attributed the primary CLL lesion to alternative splicing of B29 and mb1 mRNA, resulting in deletion of all (B29) or part (mb1) of the extracellular domain in B-CLL cells. However, the significance of these latter findings remains unclear as these splice variants were found previously in a range of human B cell lines and normal peripheral B cells (14, 25).
The critical role of B29 in B cell development and activation led us to directly test the functional consequences of selected CLL B29 mutations on BCR assembly, membrane transport, and signal transduction. Because CLL cells cannot be cultured long term and the cells are resistant to gene transfer, we adapted a model lymphoid cell system to evaluate the effects of selected mutations. Recombinant vaccinia viruses were used to efficiently deliver wild-type or mutant B29 genes into Jurkat model cells that require B29 for surface BCR display and signaling. The findings with this model indicate that B29 mutations from CLL patients are sufficient to replicate the reduced surface BCR and impaired signaling responses characteristic of CLL B cells.
Materials and Methods
Viral Vectors.
Mutations identified in the coding region of B29 from CLL patients (22) were introduced by site-directed mutagenesis (Stratagene QuikChange) in a Bluescript plasmid carrying the AspI–EcoRI fragment of B29 cDNA, pBSKS-hB29. pBSKS-hB29.44 deletes A1352, pBSKS-hB29Δcyto inserts T1291, and pBSKS-hB29.18 inserts G1222 and G1233 (Fig. 1). BamHI–EcoRV fragments carrying wild-type B29 and B29.44 and B29Δcyto mutations were cloned into the BglII–SmaI sites of pSC65 (26) to create pSC65-B29, pSC65-B29.44, and pSC65-B29Δcyto; a NotI–EcoRV fragment of B29.18 cloned into NotI–SmaI of pSC66 yielded pSC66-B29.18. These plasmids then were used to generate recombinant vaccinia viral vectors as described (refs. 26 and 27 and Fig. 1).
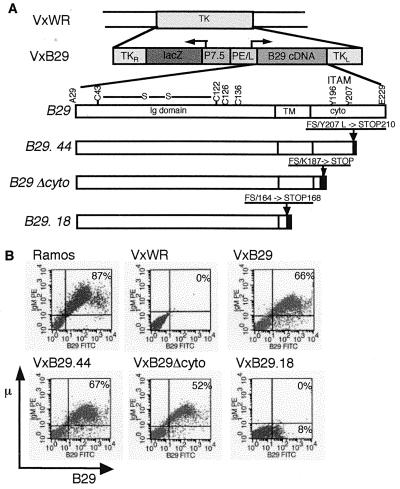
Figure 1.
Vaccinia vector for B29 expression in Jurkat model cells. (A) Mutations identified in the coding region of B29 from CLL patients were generated by site-directed mutagenesis and used to construct recombinant vaccinia viral vectors. B29.44 expresses B29 with a leucine substitution in the second ITAM tyrsosine. B29Δcyto deletes the cytoplasmic domain. B29.18 truncates the protein in the transmembrane domain. (B) Jurkat μ+κ+mb1+ were infected with empty virus (VxWR) or recombinant vaccinia carrying wild-type (VxB29) or mutated (VxB29.44, VxB29Δcyto, or VxB29.18) B29 at an multiplicity of infection of 5 for 15 h. Infected cells and Ramos B cells (positive control) were analyzed for μ and B29 surface protein by flow cytometry.
Jurkat Model Cells.
The Ramos B cell line (American Type Culture Collection) served as a positive control. Linearized p468 expressing human μ and κ (a kind gift of M. Nussenzweig, The Rockefeller University, New York; ref. 13) was introduced into Jurkat T cells (American Type Culture Collection) by electroporation (Cell-Porator, GIBCO/BRL). Stable transfectants were selected in 1 mg/ml geneticin (GIBCO/BRL), cloned by limiting dilution and screened for expression of the transgene by Western blot and flow cytometry of permeabilized cells. Stable expression of mb1 was added to Jurkat μ+κ+ cells by infection with a murine stem cell retroviral vector carrying human mb1 cDNA under the control of a viral long terminal repeat promoter and the puromycin resistance gene (28). Infected cells were selected in 0.8 μg/ml puromycin (Sigma), cloned by limiting dilution, and screened as described above. The 2 × 106/ml Jurkat model cells were infected at multiplicity of infection of 5 for 15 h with empty (VxWR) or recombinant vaccinia (VxB29, VxB29.44, VxB29Δcyto, and VxB29.18).
Flow Cytometry.
Jurkat μ+κ+ and Jurkat μ+κ+mb1+ stable transfectants were analyzed for cytoplasmic expression of the transgenes after permeabilization with Fix&Perm (CalTag, South San Francisco, CA, the manufacturer's directions) and double staining with goat anti-IgM-phycoerythin (1:5,000, Southern Biotechnology Associates) and mouse monoclonal anti-mb1 (clone 1839 1:50, Chemicon) followed by goat anti-mouse fluorescein isothiocyanate (1:1,000, CalTag) or the appropriate isotype controls. To analyze IgM and B29 surface expression, 5 × 105 infected cells were stained with anti-IgM-phycoerythin and mouse monoclonal anti-B29 (clone SN8 1:2, Dako) followed by goat anti-mouse- fluorescein isothiocyanate in 50 μl of PBS with 2% FBS for 30 min in the dark at 4°C. Stained cells were analyzed on a FACSCalibur flow cytometer by using cellquest software (Becton-Dickinson).
Anti-IgM Stimulation, Protein Extraction, Immunoprecipitation, and Western Blots.
Infected cells were stimulated with RPMI medium 1640 alone (resting) or with (stimulated) 40 μg/ml goat anti-IgM F(ab′)2 fragment (Jackson Laboratories) for 5 min at 37°C. Stimulation was quenched with excess cold PBS, and cells were lysed in cold 1% NP-40 lysis buffer (50 mM Tris, pH 8.0/150 mM NaCl/1 mM EDTA/10 mM PMSF/1 mM Na3VO4/1 mM NaF/1 μg/ml each aprotinin, leupeptin, and pepstatin) on ice for 30 min. Tyrosine-phosphorylated proteins were immunoprecipitated at 4°C overnight with 10 μg/5 × 105 cells 4G10 (mouse monoclonal antiphosphotyrosine antibody, Upstate Biotechnology, Lake Placid, NY) and 15 μl of Protein G-Sepharose beads (Amersham Pharmacia). Whole-cell lysates (106 cells/lane) and immunoprecipitates (2 × 106 cells/lane) were separated by electrophoresis on 12% ProSieve gels (FMC, manufacturer's directions), blotted to nitrocellulose, and blocked overnight with TBST (40 mM Tris, pH 7.4/300 mM NaCl/0.1% Tween-20) containing 5% fatty acid-free BSA (Boehringer Mannheim). Western blots were performed with 1 μg/ml 4G10, 1:10,000 rabbit anti-B29, 1:200 mouse anti-mb1, 1:200 rabbit anti-PLCγ1, 1:1,000 rabbit anti-Erk, and 1:600 mouse anti-phospho-Erk (all Santa Cruz Biotechnology) followed by 1:10,000 donkey anti-mouse (Jackson Laboratories) or 1:3,000 goat anti-rabbit (Promega) conjugated to horseradish peroxidase. Rabbit anti-B29 antiserum was generated against the extracellular domain of B29 fused to glutathione S-transferase. Glutathione S-transferase-B29 fusion proteins were purified over a glutathione Sepharose column and eluted with excess reduced glutathione (Amersham Pharmacia, manufacturer's directions) and used to inject animals (Antibodies Inc.). Blotted proteins were detected by Renaissance Enhanced Chemiluminescence (NEN) and exposed to Kodak XOMATBlue film. Proteins were quantitated by densitometry using bioimage (Ann Arbor, MI) software. Data shown are representative of at least three experiments.
Analysis of Intracellular Free Calcium Concentrations.
The 5 × 106 infected cells were washed and resuspended to 107/ml in RPMI medium 1640 with 2% FBS and 5 μg/ml Indo-1 (Molecular Probes). Cells were stimulated with anti-IgM as described above or with 75 nM ionomycin (Chemicon). Intracellular calcium was measured at 350-nm excitation and 400 nm (monochrometer)/488 nm (filter) emission by spectrofluorimetry and felix software (Photon Technology International, Princeton). Data are representative of four experiments.
Results
Surface BCR Expression by Wild-Type and CLL Mutant B29.
Because CLL cells proliferate poorly in culture (29) and many patient samples are limited or no longer available, we have used a Jurkat T cell model (13) to test the B29 protein mutations found in leukemic B cells from CLL patients. Surface BCR in these cells is capable of transducing the signals required for tyrosine phosphorylation, inositol triphosphate (IP3) production, calcium flux, and IL-2 secretion at levels comparable to those produced by endogenous T cell antigen receptor (13). Two independent B29 mutations identified in CLL patients with normal levels of B29 mRNA were analyzed in this model system (22). The frameshift mutation found in patient 18 (B29.18) is predicted to truncate B29 in the transmembrane domain (Fig. 1A). This mutation removes the region expected to interact with the distal polar patch of the μ transmembrane (tm) domain (30) but retains the cysteine residues for linking B29 to mb1. The frameshift mutation in B29 mutant, B29.44, (22) substitutes leucine for the second ITAM tyrosine (i.e., Y207L) and truncates the protein at residue 210 (Fig. 1A). Replacement of this ITAM tyrosine with phenylalanine inhibited tyrosine phosphorylation and calcium flux by chimeric B29 complexes without mb1 (20). B29.44 expressed in the context of the BCR is predicted to exhibit reduced signaling capacity. B29Δcyto is an engineered mutation removing all ITAM residues and the cytoplasmic tail (Fig. 1A). This construct was based on a murine mb1 transgene with the same cytoplasmic tail deletion. The reduction in perpherial B cells in these animals is predicted to result from impaired BCR signaling (31).
Jurkat model cells stably transfected with μ heavy chain, κ light chain, and mb1 expressed these cytoplasmic proteins at levels equivalent to those of control Ramos B cells but failed to transport Ig to the membrane in the absence of B29 (data not shown). When assayed by flow cytometry, 40–66% of the Jurkat cells infected with a vaccinia vector carrying wild-type B29 (VxB29), but not with empty virus (VxWR), expressed μ and B29 at the surface (Fig. 1B). Similarly, 45–67% of Jurkat cells infected with the mutant B29 protein from CLL patient 44 (VxB29.44) and 34–52% of cells infected with the engineered B29 cytoplasmic deletion mutant (VxB29Δcyto) also expressed surface IgM, indicating that both these B29 mutants functioned as well as wild-type B29 in promoting BCR assembly and surface expression (Fig. 1B).
In contrast to VxB29, VxB29.44, and VxB29Δcyto, the CLL mutant protein from patient 18 (VxB29.18) failed to promote surface μ expression in infected Jurkat cells (Fig. 1B). A minor population of VxB29.18-infected Jurkat cells (8–12% in independent experiments) transported B29 to the membrane. However, flow cytometry showed that it was displayed in the absence of surface mb1, and coimmunoprecipitation of VxB29.18-infected cell extracts with anti-mb1 antibody confirmed that this B29 mutation abrogated B29/mb1 dimerization (data not shown). B29 and mb1 previously have been found to independently associate with μ in pre-B and mature B cell lines, suggesting that previous association with μ is necessary for formation of the B29/mb1 heterodimer (32, 33). Thus in B29.18, the removal of transmembrane residues required for μ interaction also may prevent its binding to mb1 (data not shown).
The second B29 allele of patient 18 is essentially normal containing only two conservative mutations, which are not predicted to interfere with BCR assembly or function (22). Analysis of B-CLL cells from this patient revealed normal amounts of B29 mRNA but significantly decreased surface IgM and B29 (22). The B29.18 mutation could have a dominant negative effect on protein expressed from the second allele. To test this hypothesis and to simulate the B-CLL cells of patient 18, Jurkat model cells were infected with equal titres of VxB29 and VxB29.18. Coinfected cells exhibited a 40% decrease in surface IgM (data not shown). These data suggest that this phenotype may not result from a dominant negative effect by the B29.18 mutation, but this issue remains to be fully resolved.
Decreased Tyrosine Phosphorylation in Response to Anti-IgM Stimulation in Lymphoid Cells Expressing B29 CLL Mutant Proteins.
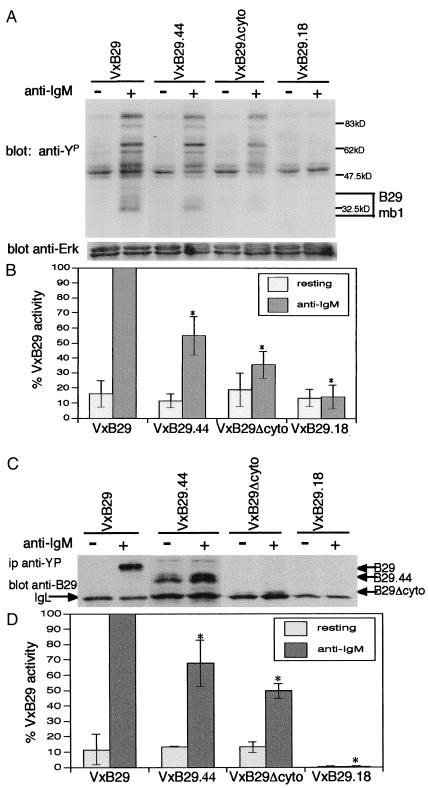
Some surface IgM+ B-CLL cells exhibit reduced signaling in repsonse to anti-IgM stimulation (9–11, 34). The protein tyrosine kinase, Syk, has been found to be decreased in only a limited subset of these CLL cells (10, 11). Reduced tyrosine phosphorylation of B29 and mb1 also has been reported in leukemic B cells of CLL patients (10). Because wild-type B29, B29.44, and B29Δcyto proteins promoted equivalent surface BCR expression in infected Jurkat cells, we were able to conduct direct comparisons of their signaling responses to BCR stimulation. Jurkat model cells were infected with VxB29, VxB29.44, VxB29Δcyto, or VxB29.18 (the latter was used as a negative control). Infected cells then were stimulated with anti-IgM and cell lysates were assayed for tyrosine phosphorylation by Western blot with antiphosphotyrosine antibody. As expected from previous studies, tyrosine phosphorylation of multiple proteins was strongly induced by anti-IgM crosslinking of model Jurkat cells infected with wild-type VxB29 (Fig. 2A). Model cells infected with the CLL mutation, VxB29.44, exhibited a significant reduction in total tyrosine phosphorylation after anti-IgM stimulation (Fig. 2A). This reduction was evident within 30 sec of anti-IgM stimulation and persisted throughout an extended kinetic analysis of activated cells (data not shown). Jurkat cells infected with the engineered mutant, VxB29Δcyto, showed even greater reduction in tyrosine phosphorylation in response to anti-IgM stimulation (Fig. 2A). Western blots of total tyrosine phosphorylation proteins were quantified by densitometry and normalized to both a loading control (total Erk protein) and an infection efficiency control (% mIgM+ cells). This quantitation showed that deletion of the B29 C-terminal ITAM tyrosine and distal residues in B29.44 decreased total tyrosine phosphorylation an average of 45% compared to wild-type B29 (Fig. 2B). Removal of the majority of the B29 cytoplasmic tail and complete ITAM in B29Δcyto resulted in a 64% average reduction of phosphotyrosine (Fig. 2B). These results show statistically significant decreases in total phosphotyrosine responses that are proportional to the stepwise deletion of critical B29 ITAM tyrosine residues.
Figure 2.
Total tyrosine phosphorylation and phosphorylation of the B29/mb1 heterodimer are decreased in lymphoid cells infected with VxB29 mutants. Model cells infected with wild-type B29 (VxB29), vectors carrying signaling mutations (VxB29.44 or VxB29Δcyto), or a negative control (VxB29.18) were stimulated with anti-μ antibody and cell lysates assayed for phosphotyrosine by Western blot. *, Statistically significant differences relative to VxB29 (P < 0.05). (A) Total tyrosine phosphorylated proteins. (B) Densitometric quantitation of phosphotyrosine normalized to total Erk protein and infection efficiency (mIgM). (C) Lysates of infected and stimulated cells were immunoprecipitated with antiphosphotyrosine antibody, and B29/mb1 was detected by Western blot with anti-B29 antibody. (D) Phosphorylation of the BCR was quantitated as described above.
Phosphorylation of B29/mb1 Is Reduced in Receptors Containing the CLL B29.44 Mutant Protein.
BCR aggregation triggers immediate tyrosine phosphorylation of the B29/mb1 heterodimer in the BCR (35). B29 and mb1 phosphotyrosines recruit the protein tyrosine kinase, Syk, to the BCR contributing to its activation. Binding of phospholipase Cγ (PLCγ) and the nucleotide exchange complexes Shc-Grb2-Sos and Vav-SLP-65 to activated BCR-Syk, translocates these complexes to their membrane substrates leading to IP3 production, calcium flux, and activation of Ras, Rac, and the mitogen-activated protein (MAP) kinase cascade (36–38). To quantitate the effects of CLL B29 mutations on BCR signaling, infected model Jurkat cells were stimulated with anti-IgM, immunoprecipitated with antiphosphotyrosine, and assayed for the presence of the B29/mb1 heterodimer among tyrosine-phosphorylated proteins by Western blot with anti-B29. Cells infected with wild-type VxB29 exhibited robust phosphorylation of B29/mb1 in response to anti-IgM antibody (Fig. 2C). Deletion of the entire cytoplasmic domain in B29Δcyto resulted in an average reduction in tyrosine phosphorylation of B29/mb1 of 50% in comparison to wild-type B29 (Fig. 2 C and D). This reduction is consistent with a BCR complex containing only one-half of the ITAM tyrosines (those contributed by mb1) as a B29/mb1 complex with wild-type B29. VxB29.44 showed an intermediate reduction (averaging 32%, Fig. 2 C and D) in tyrosine phosphorylation compared to wild-type B29. This intermediate phenotype is consistent with the removal of one-fourth of the BCR ITAM tyrosine residues.
PLCγ Activation Is Reduced but Calcium Signaling Remains Intact in Model Cells Expressing a BCR Containing the CLL B29.44 Mutant Protein.
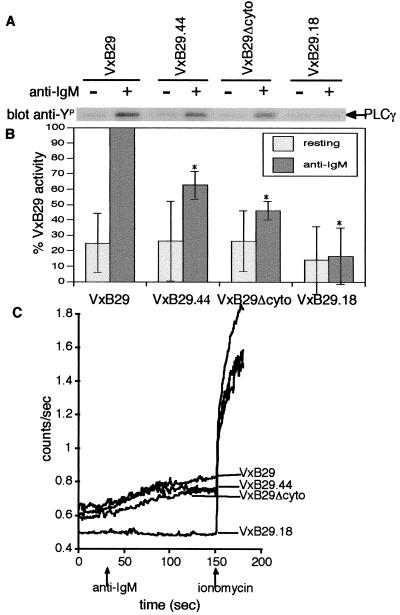
Tyrosine phosphorylation of PLCγ after BCR stimulation enhances its enzymatic activity, leading to increased IP3 production and release of calcium from IP3-gated stores (39). Removal of B29 ITAM tyrosine residues again produced a stepwise reduction in PLCγ activation in response to anti-IgM stimulation (Fig. 3A). PLCγ tyrosine phosphorylation in Jurkat cells infected with VxB29.44 was reduced 37% on average compared to cells infected with wild-type VxB29 (Fig. 3B). Cells infected with VxB29Δcyto showed an average 54% reduction in PLCγ phosphorylation compared to wild-type VxB29 (Fig. 3B).
Figure 3.
PLCγ activation, but not calcium mobilization, is reduced by VxB29 mutants. (A) Infected lymphoid cells were stimulated as before and analyzed by Western blot with antiphosphotyrosine antibody. PLCγ phosphorylation was determined by scanning the region of the gel corresponding to 110 kDa. The identity of PLCγ1 was confirmed by stripping and reprobing with specific antibody against PLCγ1. (B) Densitometric quantitation normalized to total PLCγ and mIgM. (C) Cells were infected as before, loaded with Indo-1, and analyzed for calcium mobilization in response to anti-IgM and ionomycin by bulk spectrofluorimetry.
These deficits in PLCγ activation led us to test the ability of these B29 mutants to mobilize calcium in response to BCR crosslinking. Model cells were infected as before with recombinant vaccinia, stimulated with anti-IgM, and assayed for calcium flux by spectrofluorimetry. Uninfected Jurkat cells or cells infected with VxB29.18 did not flux calcium in response to anti-IgM (Fig. 3C). Interestingly, despite the significant reductions in PLCγ phosphorylation consistently seen with B29 mutants, no marked difference in calcium fluxes was evident in Jurkat cells infected with the VxB29.44 and VxB29Δcyto mutants as compared to cells infected with wild-type VxB29 in our assay and may reflect the limited sensitivity of spectrofluorimetry and/or differences in adapter or PLCγ1 activation in this T cell system (Fig. 3C).
MAP Kinase Activation Is Significantly Reduced in Lymphoid Cells Expressing CLL B29 Mutant Proteins.
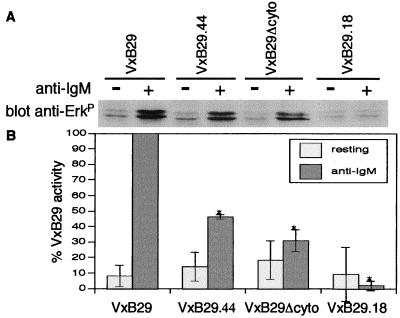
Activation of MAP kinases relays cytoplasmic signaling events to the nucleus. Full activation of these effectors requires stimulatory input from both PLCγ and the small G proteins, Ras and Rac (40, 41). We next determined the activation of the MAP kinases, Erk 1 and 2, in Western blots of anti-IgM stimulated cell lysates using mAbs specific to the dually phosphorylated proteins. Strikingly, the BCR signaling defects of the CLL mutant protein, B29.44, were amplified at the level of Erk activation (Fig. 4A). Jurkat cells infected with VxB29.44 exhibited Erk 1 and 2 phosphorylation that was reduced an average of 55% compared to cells infected with wild-type VxB29 (Fig. 4B). Jurkat cells infected with VxB29Δcyto again showed a greater reduction in Erk 1 and Erk 2 phosphorylation (average of 70%, Fig. 4B), which also was amplified relative to the signaling deficits seen in earlier steps in BCR signaling.
Figure 4.
MAP kinase activation is markedly impaired by VxB29 mutants. (A) Model cells were infected and stimulated as before and analyzed for activation of Erk 1 and 2 by Western blot using antibody specific for dually phosphorylated Erk1/2. (B) Densitometric quantitation normalized to total Erk protein and mIgM.
Discussion
The data presented here establish a model lymphoid cell/vaccinia virus gene delivery system for testing B29 mutations identified in the B cells of CLL patients. This model system has several important advantages: (i) efficient B29 gene delivery and expression by vaccinia vectors; (ii) B29 mutations are analyzed in the context of a complete BCR; and (iii) BCR signaling in Jurkat T cells closely parallels that in B cells. We have focused on two CLL B29 mutations predicted to impair BCR assembly and membrane display, or signal transduction: a frameshift that truncates B29 in the transmembrane domain (i.e., CLL patient 18, B29.18) and a mutation in the second critical ITAM tyrosine (Y207L) that truncates the remainder of the cytoplasmic tail (i.e., CLL patient 44, B29.44). These CLL mutants were analyzed in comparison to an engineered B29 mutant in which the cytoplasmic tail was deleted (i.e., B29Δcyto). This latter B29 mutant construct is equivalent to the mb1Δcyto mouse (31, 42, 43).
CLL mutant protein B29.18 failed to transport the BCR to the cell membrane and was not associated with mb1. Both the B29.44 and B29Δcyto mutants promoted surface BCR display in infected cells equivalent to wild-type VxB29. Model cells infected with these mutants showed significant quantitative reductions in signaling responses that were directly related to the number of remaining B29 and mb1 ITAM tyrosine residues. Relative to cells infected with wild-type VxB29 (eight ITAM tyrosines/BCR), total cellular phosphotyrosine was reduced by 45% in VxB29.44-infected cells (six ITAM tyrosines/BCR) and by 65% in VxB29Δcyto-infected cells (four ITAM tyrosines/BCR). The effects of these mutations were further amplified in late signaling events. Most notably MAP kinase activation was reduced by 55% in cells expressing B29.44 and by 70% in cells expressing B29Δcyto compared to wild-type B29. This reduction in MAP kinase activation (which directly induces gene transcription and resultant cell proliferation) strongly suggests that the mutations tested here could account for the unresponsiveness characteristic of B-CLL cells. This system provides a model for testing the signaling capacity of other B29 CLL mutant proteins.
Earlier studies performed primarily with chimeric constructs have not resolved the debate regarding redundant (17, 19, 44, 45) or unique (46–49) roles for B29 and mb1 in the BCR. The data obtained from this model system in conjunction with recent results from mb1Δcyto mice (42, 43) suggest differential signaling by B29 and mb1. Mutation (B29.44) or deletion (B29Δcyto) of key B29-signaling residues diminished total tyrosine phosphorylation in response to anti-IgM stimulation. In contrast, total tyrosine phosphorylation is slightly increased and calcium mobilization exaggerated and prolonged in immature B cells from mb1Δcyto mice (42, 43). Together these observations suggest that B29 contributes to activating signals by the BCR, which are then modulated by mb1.
The frequent association of B29 mutations in CLL suggests that these changes may be involved in the disease phenotype. Limited BCR expression and tonic signaling in B-CLL cells may be sufficient to maintain survival of either naïve or germinal center B cells. Elevated Bcl2 expression, characteristic of B-CLL cells, also may contribute to their survival. After initial splenic or germinal center selection events, deficient BCR signaling may protect B-CLL cells against additional BCR signals, which normally limit the lifespan of peripheral B cells. This deficiency may permit these cells to persist and acquire additional genetic alterations leading to a transformed phenotype. It will be critical to determine whether restoration of functional B29 can alter the phenotype in primary B-CLL cells, and whether such restoration is sufficient to alter cell survival in response to BCR and costimulatory signals. Recent adaptation by our laboratories of the CD40L culture system for in vitro culture of B-CLL cells (ref. 50 and unpublished results) in association with the viral vectors described here can be used to directly address these questions.
The spectrum of B29 gene mutations in B-CLL cells is strikingly similar to those features of somatic hypermutations in Ig V-regions (51, 52) and suggests an origin for these changes. First, the predominant B29 mutations in CLL involve amino acid replacements and silent mutations (i.e., 23/29) caused by single base changes in which transitions are more frequent than transversions (22, 23). Second, deletions and insertions in somatically hypermutated V-regions often involve single nucleotides, which, for insertions, are likely to be duplications of adjacent nucleotides (53–56). Single nucleotide deletions/insertions account for 4/29 B29 mutations documented in CLL (22, 23). Three of these are insertions of a single G after a G in the wild-type B29 sequence (22). Third, larger insertions/deletions as well as multiple insertions/deletions also occur in hypermutated V-regions (53–56). One mutant B29 allele with two in-frame internal deletions of 228 and 5 nts was reported (22). Finally, hypermutated V-regions frequently have multiple mutations [i.e., ≈2–5% nucleotide changes relative to the germ-line V region sequences (51, 52)]. The frequency of B29 mutations in CLL is not this high, although 6/17 CLL patients with mutated B29 alleles had two or more mutations (22, 23). Interestingly, replacement of the natural promoter of a VH transgene with the B29 promoter lowered the frequency, but not the distribution, of VH region somatic hypermutations (57). The frequencies and other features of B29 gene mutations in CLL are essentially identical to the somatic hypermutations in BCL-6 genes in memory B cells and B-cell lymphomas (57, 58). Taken together, these findings predict that B29 mutations in CLL arise through somatic hypermutation and that these mutations are likely to occur only in postgerminal center populations. More than one-half of circulating B-CLL cells contain Ig VH- and VL-regions bearing somatic hypermutations indicative of memory B cells and postgerminal center derivation (59–62). Recent studies have correlated significantly improved clinical prognosis for patients whose B-CLL cells have somatic hypermutated Ig V-regions versus those whose B-CLL cells have unmutated Ig V-regions (59–62). Demonstration of somatic hypermutation in B29 genes in CLL would provide a readily ascertained clinical test for predicting outcome and selecting an appropriate course of therapy in CLL patients.
Acknowledgments
We thank the members of the Wall, Rawlings, and Teitell laboratories for their discussions. This work was supported by National Institutes of Health Grants CA12800, GM40185, CA85841, HD37091, and CA81140 and by the American Cancer Society and the facilities of the UCLA Johnson Comprehensive Cancer Center. D.J.R. is a recipient of a McDonnell Scholar Award, a Leukemia and Lymphoma Society Scholar Award, UCLA Child Health Award, Howard Hughes Medical Institute and Pennington Research Award, and the Joan J. Drake Grant for Excellence in Cancer Research.
Abbreviations
- CLL
chronic lymphocytic leukemia
- BCR
B cell receptor
- ITAM
immunoreceptor tyrosine activation motif
- IP3
inositol triphosphate
- PLCγ
phospholipase Cγ
- MAP
mitogen-activated protein
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.090087097.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.090087097
References
- 1.Jurlander J. Crit Rev Oncol Hematol. 1998;27:29–52. doi: 10.1016/s1040-8428(97)10008-7. [DOI] [PubMed] [Google Scholar]
- 2.Garcia-Marco J, Matutes E, Morilla R, Ellis J, Oscier D, Fantes J, Catovsky D, Price C M. Br J Haematol. 1994;87:44–50. doi: 10.1111/j.1365-2141.1994.tb04868.x. [DOI] [PubMed] [Google Scholar]
- 3.Jabbar S A, Ganeshaguru K, Wickremasinghe R G, Hoffbrand A V, Foroni L. Br J Haematol. 1995;90:476–478. doi: 10.1111/j.1365-2141.1995.tb05180.x. [DOI] [PubMed] [Google Scholar]
- 4.Crossen P E. Cancer Genet Cytogenet. 1997;94:44–51. doi: 10.1016/s0165-4608(96)00253-1. [DOI] [PubMed] [Google Scholar]
- 5.Reed J C. Semin Oncol. 1998;25:11–18. [PubMed] [Google Scholar]
- 6.Hanada M, Delia D, Aiello A, Stadtmauer E, Reed J C. Blood. 1993;82:1820–1828. [PubMed] [Google Scholar]
- 7.McDonnell T J, Deane N, Platt F M, Nunez G, Jaeger U, McKearn J P, Korsmeyer S J. Cell. 1989;57:79–88. doi: 10.1016/0092-8674(89)90174-8. [DOI] [PubMed] [Google Scholar]
- 8.Strasser A, Whittingham S, Vaux D L, Bath M L, Adams J M, Cory S, Harris A W. Proc Natl Acad Sci USA. 1991;88:8661–8665. doi: 10.1073/pnas.88.19.8661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Michel F, Merle-Beral H, Legac E, Michel A, Debre P, Bismuth G. J Immunol. 1993;150:3624–3633. [PubMed] [Google Scholar]
- 10.Lankester A C, van Schijndel G M, van der Schoot C E, van Oers M H, van Noesel C J, van Lier R A. Blood. 1995;86:1090–1097. [PubMed] [Google Scholar]
- 11.Semichon M, Merle-Beral H, Lang V, Bismuth G. Leukemia. 1997;11:1921–1928. doi: 10.1038/sj.leu.2400832. [DOI] [PubMed] [Google Scholar]
- 12.Hombach J, Tsubata T, Leclercq L, Stappert H, Reth M. Nature (London) 1990;343:760–762. doi: 10.1038/343760a0. [DOI] [PubMed] [Google Scholar]
- 13.Costa T E, Franke R R, Sanchez M, Misulovin Z, Nussenzweig M C. J Exp Med. 1992;175:1669–1676. doi: 10.1084/jem.175.6.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koyama M, Nakamura T, Higashihara M, Herren B, Kuwata S, Shibata Y, Okumura K, Kurokawa K. Immunol Lett. 1995;47:151–156. doi: 10.1016/0165-2478(95)00071-x. [DOI] [PubMed] [Google Scholar]
- 15.Gong S, Nussenzweig M C. Science. 1996;272:411–414. doi: 10.1126/science.272.5260.411. [DOI] [PubMed] [Google Scholar]
- 16.Cambier J C, Jensen W A. Curr Opin Genet Dev. 1994;4:55–63. doi: 10.1016/0959-437x(94)90091-4. [DOI] [PubMed] [Google Scholar]
- 17.Sanchez M, Misulovin Z, Burkhardt A L, Mahajan S, Costa T, Franke R, Bolen J B, Nussenzweig M. J Exp Med. 1993;178:1049–1055. doi: 10.1084/jem.178.3.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Papavasiliou F, Jankovic M, Suh H, Nussenzweig M C. J Exp Med. 1995;182:1389–1394. doi: 10.1084/jem.182.5.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Teh Y M, Neuberger M S. J Exp Med. 1997;185:1753–1758. doi: 10.1084/jem.185.10.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pao L I, Famiglietti S J, Cambier J C. J Immunol. 1998;160:3305–3314. [PubMed] [Google Scholar]
- 21.Lam K P, Kuhn R, Rajewsky K. Cell. 1997;90:1073–1083. doi: 10.1016/s0092-8674(00)80373-6. [DOI] [PubMed] [Google Scholar]
- 22.Thompson A A, Talley J A, Do H N, Kagan H L, Kunkel L, Berenson J, Cooper M D, Saxon A, Wall R. Blood. 1997;90:1387–1394. [PubMed] [Google Scholar]
- 23.Payelle-Brogard B, Magnac C, Mauro F R, Mandelli F, Dighiero G. Blood. 1999;94:3516–3522. [PubMed] [Google Scholar]
- 24.Alfarano A, Indraccolo S, Circosta P, Minuzzo S, Vallario A, Zamarchi R, Fregonese A, Calderazzo F, Faldella A, Aragno M, et al. Blood. 1999;93:2327–2335. [PubMed] [Google Scholar]
- 25.Hashimoto S, Chiorazzi N, Gregersen P K. Mol Immunol. 1995;32:651–659. doi: 10.1016/0161-5890(95)00023-8. [DOI] [PubMed] [Google Scholar]
- 26.Scharenberg A M, Lin S, Cuenod B, Yamamura H, Kinet J P. EMBO J. 1995;14:3385–3394. doi: 10.1002/j.1460-2075.1995.tb07344.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rawlings D J, Scharenberg A M, Park H, Wahl M I, Lin S, Kato R M, Fluckiger A C, Witte O N, Kinet J P. Science. 1996;271:822–825. doi: 10.1126/science.271.5250.822. [DOI] [PubMed] [Google Scholar]
- 28.Hawley R G. Ann NY Acad Sci. 1994;716:327–330. doi: 10.1111/j.1749-6632.1994.tb21724.x. [DOI] [PubMed] [Google Scholar]
- 29.Planken E V, Dijkstra N H, Willemze R, Kluin-Nelemans J C. Leukemia. 1996;10:488–493. [PubMed] [Google Scholar]
- 30.Reth M. Immunol Today. 1995;16:310–313. doi: 10.1016/0167-5699(95)80141-3. [DOI] [PubMed] [Google Scholar]
- 31.Torres R M, Flaswinkel H, Reth M, Rajewsky K. Science. 1996;272:1804–1808. doi: 10.1126/science.272.5269.1804. [DOI] [PubMed] [Google Scholar]
- 32.Brouns G S, de Vries E, Borst J. Int Immunol. 1995;7:359–368. doi: 10.1093/intimm/7.3.359. [DOI] [PubMed] [Google Scholar]
- 33.Lassoued K, Illges H, Benlagha K, Cooper M D. J Exp Med. 1996;183:421–429. doi: 10.1084/jem.183.2.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hivroz C, Grillot-Courvalin C, Labaume S, Miglierina R, Brouet J C. Eur J Immunol. 1988;18:1811–1817. doi: 10.1002/eji.1830181124. [DOI] [PubMed] [Google Scholar]
- 35.DeFranco A L. Curr Opin Immunol. 1997;9:296–308. doi: 10.1016/s0952-7915(97)80074-x. [DOI] [PubMed] [Google Scholar]
- 36.Baumann G, Maier D, Freuler F, Tschopp C, Baudisch K, Wienands J. Eur J Immunol. 1994;24:1799–1807. doi: 10.1002/eji.1830240812. [DOI] [PubMed] [Google Scholar]
- 37.Saxton T M, van Oostveen I, Bowtell D, Aebersold R, Gold M R. J Immunol. 1994;153:623–636. [PubMed] [Google Scholar]
- 38.Fu C, Turck C W, Kurosaki T, Chan A C. Immunity. 1998;9:93–103. doi: 10.1016/s1074-7613(00)80591-9. [DOI] [PubMed] [Google Scholar]
- 39.Berridge M J. Crit Rev Immunol. 1997;17:155–178. doi: 10.1615/critrevimmunol.v17.i2.30. [DOI] [PubMed] [Google Scholar]
- 40.Taylor-Fishwick D A, Siegel J N. Eur J Immunol. 1995;25:3215–3221. doi: 10.1002/eji.1830251203. [DOI] [PubMed] [Google Scholar]
- 41.Hashimoto A, Okada H, Jiang A, Kurosaki M, Greenberg S, Clark E A, Kurosaki T. J Exp Med. 1998;188:1287–1295. doi: 10.1084/jem.188.7.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kraus M, Saijo K, Torres R M, Rajewsky K. Immunity. 1999;11:537–545. doi: 10.1016/s1074-7613(00)80129-6. [DOI] [PubMed] [Google Scholar]
- 43.Torres R M, Hafen K. Immunity. 1999;11:527–536. doi: 10.1016/s1074-7613(00)80128-4. [DOI] [PubMed] [Google Scholar]
- 44.Mitchell R N, Barnes K A, Grupp S A, Sanchez M, Misulovin Z, Nussenzweig M C, Abbas A K. J Exp Med. 1995;181:1705–1714. doi: 10.1084/jem.181.5.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yao X R, Flaswinkel H, Reth M, Scott D W. J Immunol. 1995;155:652–661. [PubMed] [Google Scholar]
- 46.Clark M R, Johnson S A, Cambier J C. EMBO J. 1994;13:1911–1919. doi: 10.1002/j.1460-2075.1994.tb06460.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bonnerot C, Lankar D, Hanau D, Spehner D, Davoust J, Salamero J, Fridman W H. Immunity. 1995;3:335–347. doi: 10.1016/1074-7613(95)90118-3. [DOI] [PubMed] [Google Scholar]
- 48.D'Ambrosio D, Hippen K L, Cambier J C. Eur J Immunol. 1996;26:1960–1965. doi: 10.1002/eji.1830260842. [DOI] [PubMed] [Google Scholar]
- 49.Siemasko K, Eisfelder B J, Stebbins C, Kabak S, Sant A J, Song W, Clark M R. J Immunol. 1999;162:6518–6525. [PubMed] [Google Scholar]
- 50.Fluckiger A C, Rossi J F, Bussel A, Bryon P, Banchereau J, Defrance T. Blood. 1992;80:3173–3181. [PubMed] [Google Scholar]
- 51.Klein U, Goossens T, Fischer M, Kanzler H, Braeuninger A, Rajewsky K, Kuppers R. Immunol Rev. 1998;162:261–280. doi: 10.1111/j.1600-065x.1998.tb01447.x. [DOI] [PubMed] [Google Scholar]
- 52.Neuberger M S, Milstein C. Curr Opin Immunol. 1995;7:248–254. doi: 10.1016/0952-7915(95)80010-7. [DOI] [PubMed] [Google Scholar]
- 53.Goossens T, Klein U, Kuppers R. Proc Natl Acad Sci USA. 1998;95:2463–2468. doi: 10.1073/pnas.95.5.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sale J E, Neuberger M S. Immunity. 1998;9:859–869. doi: 10.1016/s1074-7613(00)80651-2. [DOI] [PubMed] [Google Scholar]
- 55.Wilson P C, de Bouteiller O, Liu Y J, Potter K, Banchereau J, Capra J D, Pascual V. J Exp Med. 1998;187:59–70. doi: 10.1084/jem.187.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tumas-Brundage K, Manser T. J Exp Med. 1997;185:239–250. doi: 10.1084/jem.185.2.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shen H M, Peters A, Baron B, Zhu X, Storb U. Science. 1998;280:1750–1752. doi: 10.1126/science.280.5370.1750. [DOI] [PubMed] [Google Scholar]
- 58.Migliazza A, Martinotti S, Chen W, Fusco C, Ye B H, Knowles D M, Offit K, Chaganti R S, Dalla-Favera R. Proc Natl Acad Sci USA. 1995;92:12520–12524. doi: 10.1073/pnas.92.26.12520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hamblin T J, Davis Z, Gardiner A, Oscier D G, Stevenson F K. Blood. 1999;94:1848–1854. [PubMed] [Google Scholar]
- 60.Damle R N, Wasil T, Fais F, Ghiotto F, Valetto A, Allen S L, Buchbinder A, Budman D, Dittmar K, Kolitz J, et al. Blood. 1999;94:1840–1847. [PubMed] [Google Scholar]
- 61.Fais F, Ghiotto F, Hashimoto S, Sellars B, Valetto A, Allen S L, Schulman P, Vinciguerra V P, Rai K, Rassenti L Z, et al. J Clin Invest. 1998;102:1515–1525. doi: 10.1172/JCI3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Naylor M, Capra J D. Blood. 1999;94:1837–1839. [PubMed] [Google Scholar]