Abstract
In most eukaryotic cells, the nucleus is localized to a specific location. This highlight article focuses on recent advances describing the mechanisms of nuclear migration and anchorage. Central to nuclear positioning mechanisms is the communication between the nuclear envelope and the cytoskeleton. All three components of the cytoskeleton—microtubules, actin filaments and intermediate filaments—are involved in nuclear positioning to varying degrees in different cell types. KASH proteins on the outer nuclear membrane connect to SUN proteins on the inner nuclear membrane. Together they transfer forces between the cytoskeleton and the nuclear lamina. Once at the outer nuclear membrane, KASH proteins can interact with the cytoskeleton. Nuclear migrations are a component of many cellular migration events and defects in nuclear positioning lead to human diseases, most notably lissencephaly.
Introduction
The human body contains a wide variety of amazingly specialized cells. One striking example is a skeletal muscle cell, a myotube, which extends along the entire length of a muscle (up to tens of centimeters) and contains hundreds to thousands of nuclei. The nuclei are evenly spaced along the entire myotube and are apposed to the plasma membrane.1 Exceptions include the neuromuscular junction where a cluster of myonuclei form immediately below the synapse, the myotendinous junction, and in developing, injured, or pathological myotubes where nuclei are mispositioned to the interior of the myotube. A nuclear positioning phenotype in a myotube, from certain mutants described below, where large aggregates of nuclei form, is striking (see Fig. 1). The challenges of positioning nuclei in most eukaryotic cells are not as severe as those for a skeletal muscle cell. Nonetheless, the textbook model where the nucleus randomly floats around in the middle of the cytoplasm is overly simplistic. Many, if not most, cell and developmental processes, including fertilization, cell division, cell migration, and establishment of polarity, depend on positioning the nucleus to a specific location within the cell. In this review, while discussing some of the better-understood mechanisms of nuclear positioning, I will touch on selected examples spread across all eukaryotes.
Fig. 1.
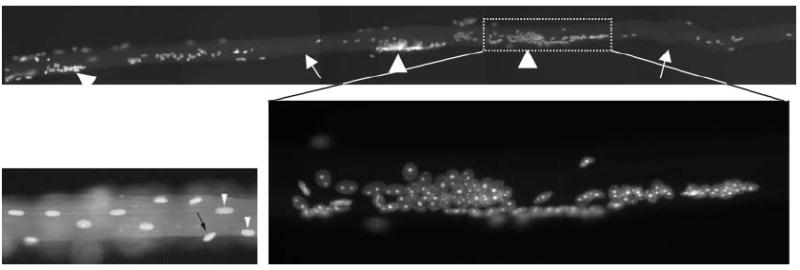
Nuclear anchorage is aberrant in desmin knockout mouse myotubes. A skeletal muscle fiber from a desmin knockout mouse is stained with Hoechst to detect nuclei. Arrows indicate areas lacking nuclei and arrowheads mark large nuclear aggregates. One cluster is magnified in the inset. A portion of a wildtype myotube is shown on the left.56 Copyright © 2006, John Wiley & Sons, Inc. Reprinted with premission of Wiley-Liss, Inc., a subsidiary of John Wiley & Sons, inc
Two related processes are required to control the specific positioning of nuclei. First, nuclei migrate through the cytoplasm to the appropriate location within a cell, and then they are anchored at that position to prevent drift. Different mechanisms exist for nuclear migration and anchorage and different mechanisms function in different cell types and organisms. In addition, the switch between migration and anchorage is predicted to be a carefully coordinated event, for which no molecular mechanisms have been identified. Nonetheless, common to all nuclear positioning events, the nucleus must communicate with the cytoskeleton. This is accomplished by two general mechanisms; either the nuclear envelope actively engages and manipulates the cytoskeleton or an active cytoskeletal network passively positions the nucleus. All three components of the cytoskeleton, microtubules, actin filaments, and intermediate filaments, function together to position nuclei, although the role of each type of filament varies in different cell types. This review will first summarize mechanisms that bridge the nuclear envelope and target proteins to the outer nuclear membrane, and then focus on the role of the three types of cytoskeletal filaments that function during nuclear positioning. Finally, I will discuss how defects in nuclear positioning could block neuronal migration events, leading to disease.
KASH and SUN proteins bridge the nuclear envelope
Central to any nuclear positioning mechanism, is communication between the nuclear envelope and the cytoskeleton. The nuclear envelope is a specialized extension of the endoplasmic reticulum (ER) with two membranes and a perinuclear space that together separate the nucleoplasm from the cytoplasm (Fig. 2; for review see ref. 2). The outer nuclear membrane (ONM) is contiguous with the ER. The ONM is also contiguous with the inner nuclear membrane (INM) at nuclear pores. Finally, a nuclear lamina, which provides support to the nuclear membrane in higher eukaryotes, lies immediately below the INM. INM proteins are targeted to the nucleoplasmic face of the nuclear envelope by karyopherins and retained in the INM by interactions with the lamina.3,4 The structure of the nuclear envelope presents two problems for nuclear positioning. First, a mechanism must exist to transfer forces across both membranes and the intermembrane space to physically link the cytoskeleton to the nuclear matrix. Second, the cell needs to target proteins specifically to the ONM to distinguish it from the ER.
Fig. 2.
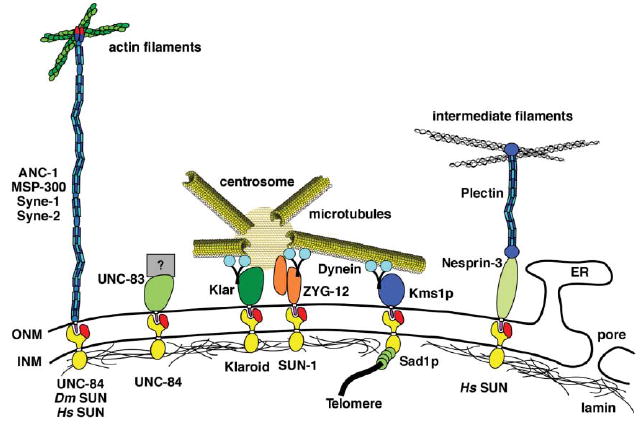
KASH and SUN proteins bridge the nuclear envelope and connect the cytoskeleton to the nucleus. Proteins with SUN domains (red) localize to the inner nuclear membrane (INM) and proteins with KASH domains (purple) localize to the outer nuclear membrane (ONM). Various KASH/SUN interactions from different organisms are depicted here. From left to right: the ANC-1/MSP-300/Syne family connects actin to the ONM, C. elegans UNC-83 and UNC-84 function during nuclear migration by unknown mechanisms, Drosophila Klar mediates dynein and is necessary to link the centrosome to the ONM, unrelated C. elegans ZYG-12 dimerizes to connect the centrosome to the ONM, S. pombe Sad1p and Kms1p transfer the forces from cytoplasmic dynein to move telomeres at the INM, and human Nesprin-3 connects intermediate filaments to the ONM. For more details, see text.
A genetic approach in C. elegans identified three genes required for nuclear positioning (for review see ref. 5,6). Mutations in unc-83 or unc-84 disrupt nuclear migration events in at least three tissues7-9 while mutations in anc-1 or unc-84 disrupt nuclear anchorage in a wide variety of cells.8,10 The UNC-83, UNC-84, and ANC-1 proteins were all shown to localize to the nuclear envelope; localization of either UNC-83 or ANC-1 requires UNC-84 at the nuclear envelope.8,9,11 UNC-84 contains a conserved SUN domain (for Sad1, UNC-84 homology; see below).8 UNC-83 and ANC-1 both have KASH domains at their C-termini (for Klarsicht, ANC-1, Syne homology; see below).11,12 Genetic and cell biological studies on UNC-83, UNC-84, and ANC-1 implicate for the first time an essential role for integral nuclear envelope proteins in the mechanisms of nuclear positioning.
KASH and SUN proteins have since been identified across eukaryotes (Fig. 2; for review see6,13-15). In general, SUN proteins contain a conserved SUN domain at their C-termini and at least one transmembrane domain. The rest of the protein is not obviously conserved at the sequence level, although it is likely conserved at a functional level.15-17 KASH proteins play multiple functions. All KASH proteins contain a C-terminal KASH domain consisting of a predicted trans-membrane domain followed by 10–30 conserved residues before the stop codon.13 The KASH domain is necessary and sufficient for nuclear envelope localization.18 The large N-terminal domains of KASH proteins play multiple roles, suggesting that the KASH domain functions as a targeting module for different proteins.13 Adding more complexity to the role of KASH proteins is that in examined cases, multiple transcripts exist, some without KASH domains.19,20
A model has emerged where KASH and SUN proteins function together to bridge the nuclear envelope, effectively connecting the nuclear lamina to the cytoskeleton (see Fig. 2; for review see ref. 6,13-15). The nuclear envelope bridging model has been refined by four groups who have each concluded that SUN proteins reside in the INM where they function to target and retain KASH proteins to the ONM through a direct interaction.12,21-23 The nuclear envelope bridging model predicts that the N-termini of SUN proteins face the nucleoplasm. Consistent with the model, SUN proteins have been shown to interact with lamins and to serve as linkers to telomeres during meiosis.22,24,25 The large N-terminal domains of KASH proteins are predicted to extend into the cytoplasm. In support, KASH proteins have been shown to interact with actin filaments, intermediate filaments and centrosomes (see below for examples). Finally, the nuclear envelope bridging model addresses two major cell biological problems. First, KASH and SUN proteins likely transfer forces from the cytoskeleton to the nucleoskeleton through a direct interaction in the perinuclear space. Second, SUN proteins recruit KASH proteins specifically to the ONM, marking this unique membrane domain apart from the contiguous rough endoplasmic reticulum.
The role of microtubules, centrosomes and dynein in nuclear migration
The role of microtubules in nuclear positioning is relatively well understood and thoroughly reviewed.26-30 Most of the data implicating microtubules and the dynein motor in nuclear positioning come from genetic studies in fungi. The growing hyphae of filamentous fungi undergo multiple divisions of the nucleus without cytokinesis (for review see ref. 26,29,30). The nuclei migrate throughout hyphal growth by a microtubule dependent mechanism, which allows them to remain evenly spaced.31 Molecular characterization of a series of nud mutations in Aspergillus (for nuclear distribution) have demonstrated a role for dynein and associated proteins in nuclear migration and distribution (for review see ref. 30). Microtubules and dynein are also essential for nuclear migration events in budding and fission yeast. During mitosis, the nucleus must migrate to the proper position to ensure accurate chromosomal segregation to daughter cells of appropriate size. Microtubules and dynein function to position fission yeast nuclei to the middle of the cells and to control migration of budding yeast nuclei to the bud neck prior to mitosis.30,32,33 During meiosis in S. pombe the nucleus rapidly migrates back and forth along the long axis of the cell in a motion termed the horsetail movement, which is thought to induce recombination between homologous chromosomes. Horsetail movement is led by the spindle pole body (the yeast version of a centrosome) and requires microtubules and dynein.34
Many advances in understanding the contributions of microtubules in nuclear positioning in animal cells come from studies in C. elegans and Drosophila. Dynein and microtubules play essential roles in pronuclear migration, nuclear rotation, and asymmetric spindle positioning in the early C. elegans embryo.28,35-37 In addition, dynein and microtubules clearly function during nuclear migration in the developing Drosophila eye and nuclear positioning in the syncytial embryo.38,39
How does the microtubule network interact with the nucleus? KASH and SUN proteins function at the nuclear envelope to communicate with microtubules and centrosomes. A striking example is in the C. elegans zygote. ZYG-12 (a KASH protein) and SUN-1 (a SUN protein also known as MTF-1) were identified through genetic screens for mutants defective in centrosome attachment to the nuclear envelope (Fig. 2).40 ZYG-12 exists as both a KASH-less isoform that binds to the centrosome and a full-length KASH isoform that localizes to the outer nuclear membrane. Localization of ZYG-12 to the nuclear envelope requires SUN-1, which functions analogously to UNC-84 to specify the outer nuclear membrane. The centrosome and nuclear envelope isoforms of ZYG-12 bind to one another to physically connect the centrosome to the nuclear envelope.40 ZYG-12 also recruits dynein to the nuclear envelope where it likely plays multiple roles in nuclear positioning.40 A second example of the role of KASH proteins and microtubules in nuclear positioning is in the Drosophila eye disk. The KASH protein Klarsicht functions to connect the centrosome to the migrating nucleus (Fig. 2).13,41 Klarsicht function requires lamin and the SUN protein Klaroid; Klarsicht is thought to coordinate opposing forces from dynein and kinesins.17,41 One final example is the role of the SUN protein Sad1p in S. pombe meiosis (Fig. 2). Sad1p interacts with telomeres at the nuclear envelope and transports them to the spindle pole body.24 Telomere localization requires dynein and microtubules in the cytoplasm. The forces of the microtubule motors are thought to be transmitted into the nucleus through Kms1p, a KASH protein at the outer nuclear membrane and Sad1p.24 Recently two Drosophila nuclear envelope proteins have been shown to play an important role in nuclear positioning during blastoderm cellularization. Mutations in charleston/kugelkern or kurzkern disrupt nuclear shape and nuclear anchoring to the apical side of the cell. The Char/Kuk and Kur proteins are thought to act in the nuclear matrix.42,43 Microtubules also function in this process,43 although the mechanism and interactions with any KASH or SUN proteins remain to be elucidated.
Actin filaments and the Syne/ANC-1/MSP-300 family in nuclear positioning
A family of huge KASH proteins, Syne-1 and -2 in mammals (also known as nesprin-1 and -2), MSP-300 in Drosophila, and ANC-1 in C. elegans, functions to anchor nuclei to the actin network (Fig. 2). Dictyostelium interaptin may also be a member of this family, although it is significantly smaller and its role in nuclear positioning has not been described.44 Full-length isoforms of ANC-1/MSP-300/Syne each contain a conserved N-terminal actin-binding domain of the calponin family and a C-terminal KASH domain that targets the protein to the outer nuclear membrane. The KASH and calponin domains are connected by a huge (over 8000 residues) spectrin-like domain. The ANC-1/MSP-300/Syne proteins are related to Dystrophin, which functions to connect actin filaments to the plasma membrane. The major difference is that the C-terminus of Dystrophin does not contain a KASH domain and instead interacts with the plasma membrane.45,46 The ANC-1/MSP-300/Syne proteins are thought to act as a rope to connect actin to the nuclear membrane (for review see ref. 6,13).
Null mutations that affect the KASH isoforms of Syne-1 in mice, MSP-300 in Drosophila or ANC-1 in C. elegans disrupt nuclear anchorage. Mutations in C. elegans anc-1 were originally isolated in screens for nuclear positioning mutants in the syncytial hypodermis. Normally nuclei of the hypodermis are evenly spaced apart, but in an anc-1 mutant animal, the nuclei are moved back and forth by underlying muscles and aggregate in large clusters10 (Fig. 3A–B). Mutations in Drosophila msp-300 lead to embryonic lethality because of muscle morphology defects.47 To investigate the role of msp-300 in nuclear positioning, germ-line clones were examined. Developing oocyte cysts in female germline clones of homozygous msp-300 null mutations have a strong dumping phenotype caused by a lack of nurse cell nuclear anchorage.48 Normally the nuclei of the 15 nurse cells remain anchored by an actin-dependent process while they contract and “dump” their cytoplasm into the oocyte through narrow ring canals; nurse cell nuclei must remain anchored to prevent their movement into ring canals.49 msp-300 mutant nurse cells have a dumping phenotype resulting in small eggs. Nurse cell nuclei are misplaced in oocytes, and others block ring canals. The oocyte nucleus is also unanchored and floats freely in the oocyte cytoplasm (Fig. 3C–D).48 To determine if the roles of ANC-1 and MSP-300 are conserved in the human homologues Syne-1 and Syne-2, Zhang et al.50 created mice with the KASH domains of Syne-1 and/or Syne-2 knocked out. Disruption of both Syne-1 and Syne-2 was lethal shortly after birth, as the pups never breathed.50 Syne-1 KASH knockout mice were viable, but had severe nuclear positioning defects in the muscle (Fig. 2E–F); the Syne-2 KASH knockout mice had no obvious phenotype.50 The Syne-1 knockout was more severe than the previously described dominant negative Syne mice where overexpression of the KASH domain in skeletal muscles displaced endogenous Syne proteins and disrupted the positioning of nuclei at the neuromuscular junction.50,51 The Syne-1 KASH knockout mice had no nuclei clustering under the neuromuscular junction. The non-synaptic nuclei in the myotube were also displaced, often aggregating in large clusters.50 Additionally, non-synaptic nuclei were occasionally observed in the center of a cross-section of a myotube, a phenotype associated with regeneration and disease pathology.50 Together these genetic experiments show that the ANC-1/MSP-300/Syne family of proteins function to anchor nuclei and that the mechanism is conserved from nematodes to insects and mammals.
Fig. 3.
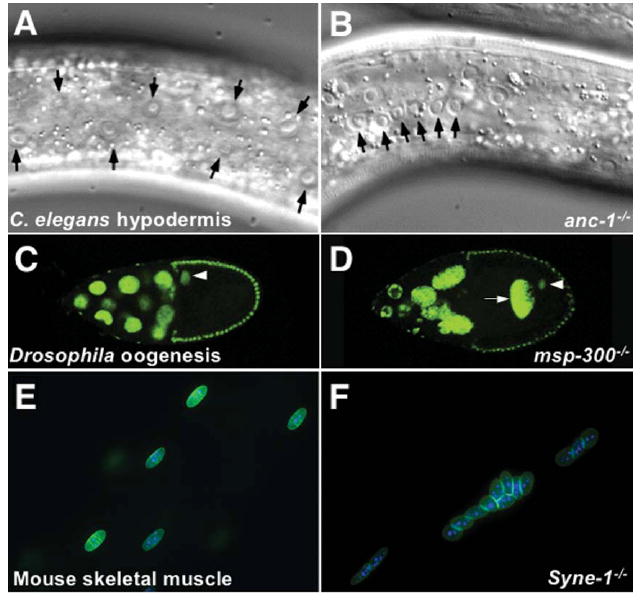
The ANC-1/MSP-300/Syne-1 family of proteins function to anchor nuclei. (A–B) Normarski microscopy images of C. elegans syncytial hypodermal cells from wildtype (A) and anc-1 mutant (B) adults. Normally, the nuclei (black arrowheads) are evenly spaced apart whereas in the anc-1 mutant animals nuclei float freely through the syncytium. (Reproduced with permission from ref.11.) (C–D) Fluorescent images showing GFP-histone marked nuclei in stage 10A developing Drosophila oocytes from wildtype (C) and msp-300 mutant (D) ovaries. In the msp-300 mutant ovary, nurse cells are misplaced; one (arrow) is in the oocyte cytoplasm. The oocyte nucleus (arrowhead) is also mispositioned. Reproduced with permission from ref. 48, copyright Elsevier (2006). (E–F) Fluorescent images showing nuclei in a single mouse muscle fiber from heterozygous control siblings (E) or homozygous Syne-1KASH knockout mice (F). In control fibers the nuclei are evenly spaced apart while in the mutant fiber the nuclei cluster. (Reproduced with permission of the Company of Biologists from ref. 50.)
The importance of actin networks in nuclear migration has recently been described for an unexpected situation. It had been known that NIH 3T3 fibroblasts in tissue culture polarize in response to wounding, which results in the centrosomes facing the wound.52 It was believed that this was due to an active movement of the centrosome towards the wound edge via microtubules. However, video microscopy has demonstrated that the centrosomes remain anchored in the middle of the cell while the nucleus actively migrates behind the centrosome.53 This rearward nuclear movement requires actin, myosin, and the small G-protein cdc42 and is coupled with retrograde actin flow.53 It is not known how the nuclear envelope interacts with the actin network during rearward nuclear migration, but Syne-1 and Syne-2 are prime candidates to anchor the nucleus to the retrograde actin flow.53
There are also some less well understood examples of roles for actin filaments in nuclear positioning.46 In Drosophila early embryonic nuclear migrations towards the periphery of the syncytial blastoderm, the actin gel-like network around migrating nuclei depolymerizes, allowing the nucleus to passively move forward along the depolymerizing front.54 Additionally, actin depolymerizing drugs completely abolished the rapid intracellular nuclear migration events in Arabidopsis root hairs through unknown mechanisms, whereas drugs that disrupted microtubules had no effect.55
Intermediate filaments and nuclear anchorage
The role of intermediate filaments in nuclear positioning is less well understood. Since there are no motors that move along intermediate filaments and because they do not have the polar dynamic growth properties of microtubules or actin filaments, intermediate filaments are relegated to a more static scaffolding role to anchor nuclei in place or to recruit components that can interact with other members of the cytoskeleton. Desmin is the major component of intermediate filaments in mammalian muscle fibers and the desmin knockout mouse has a striking nuclear anchorage phenotype56 (Fig. 1). Normally the hundreds of nuclei in a mouse skeletal muscle are evenly spaced apart and at the periphery of the cell. In desmin knockout muscles, nuclei cluster together in dense aggregates reminiscent of the mouse Syne-1 or C. elegans anc-1 mutant phenotypes. Additionally, the oval nuclei fail to align normally with the long axis of the muscle fiber.56 The mispositioned nuclei in the desmin null muscles do, however, retain their preferential association with blood vessels, suggesting that desmin and blood vessels play independent roles in anchoring nuclei.56 It remains to be determined whether Syne-1 functions in conjunction with either desmin or blood vessels. In other cells, intermediate filaments might function in part to connect the plasma membrane to the nucleus. For example, vimentin is found closely associated with the nucleus and mutant forms of vimentin exhibit nuclear morphology defects.57
A newly identified KASH protein, nesprin-3, likely functions to connect the outer nuclear membrane to intermediate filaments. Nesprin-3 localizes to the nuclear envelope where it interacts with plectin in a yeast two-hybrid assay (Fig. 2).58 Plectin, a plakin family member, consists of an actin-binding domain, an extended coiled-coil domain, and an intermediate filament-binding domain and can crosslink actin filaments to intermediate filaments.59 The actin-binding domain can alternatively interact with other proteins including integrins.60 Nesprin-3 was found to bind to the actin-binding domain of plectin in a two-hybrid screen.58 It is hypothesized that nesprin-3 and plectin together could extend from the outer nuclear membrane into the cytoplasm to interact with intermediate filaments; this would be analogous to how the ANC-1/MSP-300/Syne proteins connect the outer nuclear membrane with actin filaments.58
Lamin, a type V intermediate filament, is the major structural component of the nuclear matrix immediately underlying the inner nuclear membrane (Fig. 2). It plays both structural and scaffolding roles during nuclear positioning (reviewed in ref. 2). Some, but not all, SUN proteins require lamins to localize to the inner nuclear membrane.22,61 Since SUN proteins recruit KASH proteins to the outer nuclear membrane, lamins function in a variety of the mechanisms for nuclear positioning discussed above.
The role of nuclear migration in neuronal migration and disease
Nuclear migration and anchorage events have been described for a wide variety of cellular processes. Thus, it is not surprising that defects in nuclear positioning would lead to disease. Characterization of migrating neurons has implicated both microtubules and actin–myosin contraction in nuclear migration. The growth cone, the leading edge of a migrating neuron, extends at a constant rate while the nucleus moves through the cytoplasm in jumps and pauses. Each nuclear migration translocation of about 10 microns is preceded by a movement of the centrosome away from the nucleus and into a swelling of the cellular process at the site the nucleus will jump to.62-64 Nuclear translocation requires myosin II contraction at the rearward side to propel the nucleus forward.64 Thus neuronal migration is an example of how the microtubule and actin networks can function together to control nuclear positioning.
Lissencephaly (Greek for smooth brain) is a neuro-developmental disease where neurons fail to properly migrate to the cortex. Mutations in any of three proteins, LIS1, Doublecortin or α-tubulin, have been linked to lissencephaly.65-67 LIS1 regulates dynein and is a homologue of Aspergillus NudF, which is required for nuclear positioning (for review see ref. 68). Doublecortin is a microtubule-associated protein68 and the mutation in α-tubulin blocks GTP binding, slowing microtubule dynamics.67 Recently developed techniques to study neuronal migration in siRNA-treated living brain and mouse knockout systems have implicated the lissencephaly mutations in nuclear migration.63,67,69 Thus microtubule and dynein driven nuclear migration play an essential role in human brain development.
One KASH protein has been directly implicated in a disease. Mutations in Syne-1 lead to autosomal cerebellar recessive ataxia type 1 (ARCA1), also known as recessive ataxia of Beauce, a late-onset ataxia with a slow progression.70 Further studies are required to understand if and how nuclear positioning contributes to this neurodegenerative disease or if the ataxia represents a defect independent of KASH function. It also remains to be seen if other human KASH or SUN proteins are also involved in the pathology of disease.
Conclusions and directions for the field
The position of the nucleus within the cell is essential to a wide variety of cellular and developmental events across eukaryotes. In the past five years, great progress has been made in understanding some of the mechanisms of nuclear positioning. The main focus of these studies was on the role KASH and SUN proteins play to position the nucleus. KASH proteins specify the outer nuclear membrane and together with SUN proteins, transfer forces across the nuclear envelope. KASH and SUN proteins act as the central link to connect all three components of the cytoskeleton to the nuclear matrix. There are certainly more players and mechanisms to be identified in nuclear positioning. Proteins remain to be identified to explain how nuclear migration is controlled, and many unanswered questions remain.
Microtubules, actin filaments, and intermediate filaments all play important roles in positioning nuclei. However, little is known about how they function together or why some components of the cytoskeleton play more important roles than others in different tissue types. For example, in the early C. elegans embryo, the microtubule system plays the major role in positioning the nucleus through the KASH protein ZYG-12.40 However, other systems function to migrate nuclei later in development through the KASH protein UNC-83.9,61 The identity of the cytoskeleton components that UNC-83 interacts with is being actively pursued. Most likely, multiple cytoskeletal components function together to position nuclei. This is best described in budding yeast where the actin and microtubule networks function together.71
Some of the questions to be addressed include: How are polar cues incorporated to move the nucleus directionally? What happens to the ER during migration, is it dragged behind or does it actively rearrange during nuclear migration? And, how do nuclei switch from an anchored position to a migrating one? The switch must be carefully coordinated; this could be accomplished by controlling the specificity of KASH SUN interactions. UNC-84 interacts with one KASH protein, UNC-83, during migration and a different KASH protein, ANC-1, during anchorage. Nothing is known about how this switch is made. Furthermore, a sub-molecular in vitro characterization of the KASH/SUN interaction should help determine how these proteins transfer force across the nuclear envelope. During the next few years the field will likely identify additional nuclear envelope and cytoskeletal interactions that function to position the nucleus. Both genetic and biochemical approaches are likely to clarify the mechanisms of nuclear migration.
Finally, the link between nuclear positioning and disease needs further examination. There is an exciting possibility that proteins involved in nuclear positioning are also involved in disease. The list of top candidates today include Syne-1, implicated in an ataxia,70 Lis1, one cause of lissencephaly,65 and lamins, which may act through SUN proteins to cause a variety of diseases termed laminopothies (reviewed in ref. 2). Furthermore, since nuclear migration is an important component of many cellular migration events, it is exciting to speculate that nuclear migration might play a role in all cellular migration events including metastasis. It is likely that defects in KASH and SUN proteins lead to disease, although further studies are required to determine if they play causal or correlative roles in cancer and other diseases. Therefore, KASH and SUN proteins may be good targets for chemical genetics to both better understand the processes of nuclear and cellular migration and ultimately as potential medical treatments.
Acknowledgments
I am grateful to Lesilee Rose and the members of my laboratory for comments on this manuscript and thoughtful discussions. I apologize to those whose work was not included because of space. The studies in my lab are supported by a grant from the NIH (GM073874) and a March of Dimes Basil O’Conner award.
Biography

Daniel Starr grew up in Minneapolis, MN and earned a BA in Biology in 1992 from Colby College in Waterville, ME. He earned a PhD in Genetics from Cornell University in 1998. His thesis, under the guidance of Michael Goldberg, focused on the role of the kinetochore protein ZW10 and the recruitment of dynein in Drosophila and human tissue culture. Dr Starr completed a postdoctoral fellowship with Min Han at the University of Colorado-Boulder from 1998–2003 where he focused on the molecular characterization of unc-83 and anc-1 and their roles in nuclear positioning in Caenorhabditis elegans. Dr Starr joined the Section of Molecular and Cellular Biology and the Center for Genetics and Development at the University of California, Davis in 2003 as an assistant professor where he runs a laboratory focusing on the role of KASH and SUN proteins in nuclear positioning in C. elegans.
References
- 1.Bruusgaard JC, Liestol K, Ekmark M, Kollstad K, Gundersen K. J Physiol (Oxford, UK) 2003;551:467–478. doi: 10.1113/jphysiol.2003.045328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gruenbaum Y, Margalit A, Goldman RD, Shumaker DK, Wilson KL. Nat Rev. 2005;6:21–31. doi: 10.1038/nrm1550. [DOI] [PubMed] [Google Scholar]
- 3.King MC, Lusk CP, Blobel G. Nature. 2006;442:1003–1007. doi: 10.1038/nature05075. [DOI] [PubMed] [Google Scholar]
- 4.Ohba T, Schirmer EC, Nishimoto T, Gerace L. J Cell Biol. 2004;167:1051–1062. doi: 10.1083/jcb.200409149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Starr DA, Han M. Novartis Found Symp. 2005;264:208–219. discussion 219–230. [PubMed] [Google Scholar]
- 6.Wilhelmsen K, Ketema M, Truong H, Sonnenberg A. J Cell Sci. 2006;119:5021–5029. doi: 10.1242/jcs.03295. [DOI] [PubMed] [Google Scholar]
- 7.Horvitz HR, Sulston JE. Genetics. 1980;96:435–454. doi: 10.1093/genetics/96.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Malone CJ, Fixsen WD, Horvitz HR, Han M. Development (Cambridge, UK) 1999;126:3171–3181. doi: 10.1242/dev.126.14.3171. [DOI] [PubMed] [Google Scholar]
- 9.Starr DA, Hermann GJ, Malone CJ, Fixsen W, Priess JR, Horvitz HR, Han M. Development (Cambridge, UK) 2001;128:5039–5050. doi: 10.1242/dev.128.24.5039. [DOI] [PubMed] [Google Scholar]
- 10.Hedgecock EM, Thomson JN. Cell. 1982;30:321–330. doi: 10.1016/0092-8674(82)90038-1. [DOI] [PubMed] [Google Scholar]
- 11.Starr DA, Han M. Science. 2002;298:406–409. doi: 10.1126/science.1075119. [DOI] [PubMed] [Google Scholar]
- 12.McGee MD, Rillo R, Anderson AS, Starr DA. Mol Biol Cell. 2006;17:1790–1801. doi: 10.1091/mbc.E05-09-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Starr DA, Fischer JA. Bioessays. 2005;27:1136–1146. doi: 10.1002/bies.20312. [DOI] [PubMed] [Google Scholar]
- 14.Tzur YB, Wilson KL, Gruenbaum Y. Nat Rev. 2006;7:782–788. doi: 10.1038/nrm2003. [DOI] [PubMed] [Google Scholar]
- 15.Worman HJ, Gundersen GG. Trends Cell Biol. 2006;16:67–69. doi: 10.1016/j.tcb.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 16.Jaspersen SL, Martin AE, Glazko G, Giddings TH, Jr, Morgan G, Mushegian A, Winey M. J Cell Biol. 2006;174:665–675. doi: 10.1083/jcb.200601062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kracklauer MP, Banks SML, Xie X, Wu Y, Fischer JA. Fly. 2007;1:75–85. doi: 10.4161/fly.4254. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Q, Skepper JN, Yang F, Davies JD, Hegyi L, Roberts RG, Weissberg PL, Ellis JA, Shanahan CM. J Cell Sci. 2001;114:4485–4498. doi: 10.1242/jcs.114.24.4485. [DOI] [PubMed] [Google Scholar]
- 19.Guo Y, Jangi S, Welte MA. Mol Biol Cell. 2005;16:1406–1416. doi: 10.1091/mbc.E04-10-0920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Warren DT, Zhang Q, Weissberg PL, Shanahan CM. Expert Rev Mol Med. 2005;7:1–15. doi: 10.1017/S1462399405009294. [DOI] [PubMed] [Google Scholar]
- 21.Crisp M, Liu Q, Roux K, Rattner JB, Shanahan C, Burke B, Stahl PD, Hodzic D. J Cell Biol. 2006;172:41–53. doi: 10.1083/jcb.200509124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haque F, Lloyd DJ, Smallwood DT, Dent CL, Shanahan CM, Fry AM, Trembath RC, Shackleton S. Mol Cell Biol. 2006;26:3738–3751. doi: 10.1128/MCB.26.10.3738-3751.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Padmakumar VC, Libotte T, Lu W, Zaim H, Abraham S, Noegel AA, Gotzmann J, Foisner R, Karakesisoglou I. J Cell Sci. 2005;118:3419–3430. doi: 10.1242/jcs.02471. [DOI] [PubMed] [Google Scholar]
- 24.Chikashige Y, Tsutsumi C, Yamane M, Okamasa K, Haraguchi T, Hiraoka Y. Cell. 2006;125:59–69. doi: 10.1016/j.cell.2006.01.048. [DOI] [PubMed] [Google Scholar]
- 25.Schmitt J, Benavente R, Hodzic D, Hoog C, Stewart CL, Alsheimer M. Proc Natl Acad Sci U S A. 2007 doi: 10.1073/pnas.0609198104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morris NR. J Cell Biol. 2000;148:1097–1101. doi: 10.1083/jcb.148.6.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morris NR. Curr Opin Cell Biol. 2003;15:54–59. doi: 10.1016/s0955-0674(02)00004-2. [DOI] [PubMed] [Google Scholar]
- 28.Reinsch S, Gonczy P. J Cell Sci. 1998;111:2283–2295. doi: 10.1242/jcs.111.16.2283. [DOI] [PubMed] [Google Scholar]
- 29.Xiang X, Fischer R. Fungal Genet Biol. 2004;41:411–419. doi: 10.1016/j.fgb.2003.11.010. [DOI] [PubMed] [Google Scholar]
- 30.Yamamoto A, Hiraoka Y. J Cell Sci. 2003;116:4501–4512. doi: 10.1242/jcs.00835. [DOI] [PubMed] [Google Scholar]
- 31.Oakley BR, Morris NR. Cell. 1980;19:255–262. doi: 10.1016/0092-8674(80)90407-9. [DOI] [PubMed] [Google Scholar]
- 32.Palmer RE, Sullivan DS, Huffaker T, Koshland D. J Cell Biol. 1992;119:583–593. doi: 10.1083/jcb.119.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tran PT, Marsh L, Doye V, Inoue S, Chang F. J Cell Biol. 2001;153:397–411. doi: 10.1083/jcb.153.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ding DQ, Chikashige Y, Haraguchi T, Hiraoka Y. J Cell Sci. 1998;111(Pt 6):701–712. doi: 10.1242/jcs.111.6.701. [DOI] [PubMed] [Google Scholar]
- 35.Gonczy P, Pichler S, Kirkham M, Hyman AA. J Cell Biol. 1999;147:135–150. doi: 10.1083/jcb.147.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Skop AR, White JG. Curr Biol. 1998;8:1110–1116. doi: 10.1016/s0960-9822(98)70465-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sonnichsen B, Koski LB, Walsh A, Marschall P, Neumann B, Brehm M, Alleaume AM, Artelt J, Bettencourt P, Cassin E, Hewitson M, Holz C, Khan M, Lazik S, Martin C, Nitzsche B, Ruer M, Stamford J, Winzi M, Heinkel R, Roder M, Finell J, Hantsch H, Jones SJ, Jones M, Piano F, Gunsalus KC, Oegema K, Gonczy P, Coulson A, Hyman AA, Echeverri CJ. Nature. 2005;434:462–469. doi: 10.1038/nature03353. [DOI] [PubMed] [Google Scholar]
- 38.Fan SS, Ready DF. Development (Cambridge, UK) 1997;124:1497–1507. doi: 10.1242/dev.124.8.1497. [DOI] [PubMed] [Google Scholar]
- 39.Whited JL, Cassell A, Brouillette M, Garrity PA. Development (Cambridge, UK) 2004;131:4677–4686. doi: 10.1242/dev.01366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Malone CJ, Misner L, Le Bot N, Tsai MC, Campbell JM, Ahringer J, White JG. Cell. 2003;115:825–836. doi: 10.1016/s0092-8674(03)00985-1. [DOI] [PubMed] [Google Scholar]
- 41.Patterson K, Molofsky AB, Robinson C, Acosta S, Cater C, Fischer JA. Mol Biol Cell. 2004;15:600–610. doi: 10.1091/mbc.E03-06-0374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brandt A, Papagiannouli F, Wagner N, Wilsch-Brauninger M, Braun M, Furlong EE, Loserth S, Wenzl C, Pilot F, Vogt N, Lecuit T, Krohne G, Grosshans J. Curr Biol. 2006;16:543–552. doi: 10.1016/j.cub.2006.01.051. [DOI] [PubMed] [Google Scholar]
- 43.Pilot F, Philippe JM, Lemmers C, Chauvin JP, Lecuit T. Development (Cambridge, UK) 2006;133:711–723. doi: 10.1242/dev.02251. [DOI] [PubMed] [Google Scholar]
- 44.Rivero F, Kuspa A, Brokamp R, Matzner M, Noegel AA. J Cell Biol. 1998;142:735–750. doi: 10.1083/jcb.142.3.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rando TA. Muscle Nerve. 2001;24:1575–1594. doi: 10.1002/mus.1192. [DOI] [PubMed] [Google Scholar]
- 46.Starr DA, Han M. J Cell Sci. 2003;116:211–216. doi: 10.1242/jcs.00248. [DOI] [PubMed] [Google Scholar]
- 47.Rosenberg-Hasson Y, Renert-Pasca M, Volk T. Mech Dev. 1996;60:83–94. doi: 10.1016/s0925-4773(96)00602-8. [DOI] [PubMed] [Google Scholar]
- 48.Yu J, Starr DA, Wu X, Parkhurst SM, Zhuang Y, Xu T, Xu R, Han M. Dev Biol. 2006;289:336–345. doi: 10.1016/j.ydbio.2005.10.027. [DOI] [PubMed] [Google Scholar]
- 49.Robinson DN, Cooley L. Annu Rev Cell Dev Biol. 1997;13:147–170. doi: 10.1146/annurev.cellbio.13.1.147. [DOI] [PubMed] [Google Scholar]
- 50.Zhang X, Xu R, Zhu B, Yang X, Ding X, Duan S, Xu T, Zhuang Y, Han M. Development (Cambridge, UK) 2007;134:901–908. doi: 10.1242/dev.02783. [DOI] [PubMed] [Google Scholar]
- 51.Grady RM, Starr DA, Ackerman GL, Sanes JR, Han M. Proc Natl Acad Sci U S A. 2005 [Google Scholar]
- 52.Kupfer A, Louvard D, Singer SJ. Proc Natl Acad Sci U S A. 1982;79:2603–2607. doi: 10.1073/pnas.79.8.2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gomes ER, Jani S, Gundersen GG. Cell. 2005;121:451–463. doi: 10.1016/j.cell.2005.02.022. [DOI] [PubMed] [Google Scholar]
- 54.von Dassow G, Schubiger G. J Cell Biol. 1994;127:1637–1653. doi: 10.1083/jcb.127.6.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chytilova E, Macas J, Sliwinska E, Rafelski SM, Lambert GM, Galbraith DW. Mol Biol Cell. 2000;11:2733–2741. doi: 10.1091/mbc.11.8.2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ralston E, Lu Z, Biscocho N, Soumaka E, Mavroidis M, Prats C, Lomo T, Capetanaki Y, Ploug T. J Cell Physiol. 2006;209:874–882. doi: 10.1002/jcp.20780. [DOI] [PubMed] [Google Scholar]
- 57.Toivola DM, Tao GZ, Habtezion A, Liao J, Omary MB. Trends Cell Biol. 2005;15:608–617. doi: 10.1016/j.tcb.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 58.Wilhelmsen K, Litjens SH, Kuikman I, Tshimbalanga N, Janssen H, van den Bout I, Raymond K, Sonnenberg A. J Cell Biol. 2005;171:799–810. doi: 10.1083/jcb.200506083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fuchs E, Karakesisoglou I. Genes Dev. 2001;15:1–14. doi: 10.1101/gad.861501. [DOI] [PubMed] [Google Scholar]
- 60.Geerts D, Fontao L, Nievers MG, Schaapveld RQ, Purkis PE, Wheeler GN, Lane EB, Leigh IM, Sonnenberg A. J Cell Biol. 1999;147:417–434. doi: 10.1083/jcb.147.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee KK, Starr DA, Cohen M, Liu J, Han M, Wilson KL, Gruenbaum Y. Mol Biol Cell. 2002;13:892–901. doi: 10.1091/mbc.01-06-0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bellion A, Baudoin JP, Alvarez C, Bornens M, Metin C. J Neurosci. 2005;25:5691–5699. doi: 10.1523/JNEUROSCI.1030-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kappeler C, Saillour Y, Baudoin JP, Tuy FP, Alvarez C, Houbron C, Gaspar P, Hamard G, Chelly J, Metin C, Francis F. Hum Mol Genet. 2006;15:1387–1400. doi: 10.1093/hmg/ddl062. [DOI] [PubMed] [Google Scholar]
- 64.Schaar BT, McConnell SK. Proc Natl Acad Sci U S A. 2005;102:13652–13657. doi: 10.1073/pnas.0506008102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Reiner O, Carrozzo R, Shen Y, Wehnert M, Faustinella F, Dobyns WB, Caskey CT, Ledbetter DH. Nature. 1993;364:717–721. doi: 10.1038/364717a0. [DOI] [PubMed] [Google Scholar]
- 66.Gleeson JG, Allen KM, Fox JW, Lamperti ED, Berkovic S, Scheffer I, Cooper EC, Dobyns WB, Minnerath SR, Ross ME, Walsh CA. Cell. 1998;92:63–72. doi: 10.1016/s0092-8674(00)80899-5. [DOI] [PubMed] [Google Scholar]
- 67.Keays DA, Tian G, Poirier K, Huang GJ, Siebold C, Cleak J, Oliver PL, Fray M, Harvey RJ, Molnar Z, Pinon MC, Dear N, Valdar W, Brown SD, Davies KE, Rawlins JN, Cowan NJ, Nolan P, Chelly J, Flint J. Cell. 2007;128:45–57. doi: 10.1016/j.cell.2006.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vallee RB, Tsai JW. Genes Dev. 2006;20:1384–1393. doi: 10.1101/gad.1417206. [DOI] [PubMed] [Google Scholar]
- 69.Tsai JW, Chen Y, Kriegstein AR, Vallee RB. J Cell Biol. 2005;170:935–945. doi: 10.1083/jcb.200505166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gros-Louis F, Dupre N, Dion P, Fox MA, Laurent S, Verreault S, Sanes JR, Bouchard JP, Rouleau GA. Nat Genet. 2007;39:80–85. doi: 10.1038/ng1927. [DOI] [PubMed] [Google Scholar]
- 71.Pearson CG, Bloom K. Nat Rev. 2004;5:481–492. doi: 10.1038/nrm1402. [DOI] [PubMed] [Google Scholar]


