Abstract
Autosomal dominant retinitis pigmentosa (adRP) is a heterogeneous set of progressive retinopathies caused by several distinct genes. One locus, the RP10 form of adRP, maps to human chromosome 7q31.1 and may account for 5–10% of adRP cases among Americans and Europeans. We identified two American families with the RP10 form of adRP by linkage mapping and used these families to reduce the linkage interval to 3.45 Mb between the flanking markers D7S686 and RP-STR8. Sequence and transcript analysis identified 54 independent genes within this region, at least 10 of which are retinal-expressed and thus candidates for the RP10 gene. A screen of retinal transcripts comparing retinas from normal mice to retinas from crx−/crx− knockout mice (with poorly differentiated photoreceptors) demonstrated a 6-fold reduction in one candidate, inosine monophosphate dehydrogenase 1 (IMPDH1; EC 1.1.1.205). Since many of the genes known to cause retinitis pigmentosa are under CRX control in photoreceptors, IMPDH1 became a high-priority candidate for mutation screening. DNA sequencing of affected individuals from the two American RP10 families revealed a GAC→AAC transition in codon 226 substituting an asparagine for an aspartic acid in both families. The identical mutation was also found in a British RP10 family. The Asp226Asn missense mutation is present in all affected individuals tested and absent from unaffected controls. The aspartic acid at codon 226 is conserved in all IMPDH genes, in all species examined, including bacteria, suggesting that this mutation is highly deleterious. Subsequent screening of probands from 60 other adRP families revealed an additional family with this mutation, confirming its association with retinitis pigmentosa and the relatively high frequency of this mutation. Another IMPDH1 substitution, Val268Ile, was also observed in this cohort of patients but not in controls. IMPDH1 is a ubiquitously expressed enzyme, functioning as a homotetramer, which catalyzed the rate-limiting step in de novo synthesis of guanine nucleotides. As such, it plays an important role in cyclic nucleoside metabolism within photoreceptors. Several classes of drugs are known to affect IMPDH isoezymes, including nucleotide and NAD analogs, suggesting that small-molecule therapy may be available, one day, for RP10 patients.
INTRODUCTION
Retinitis pigmentosa (RP) is a set of inherited retinopathies with an aggregate prevalence of approximately 1 in 3500 in the United States, Europe and elsewhere (1). Although symptoms vary considerably between individuals, even within families, the classical findings and symptoms are: (i) characteristic abnormalities in the electroretinogram (ERG) detectable at an early age, (ii) night blindness with onset in adolescence, (iii) subsequent appearance of ‘bone spicule’ deposits and other morphological abnormalities in the retina and (iv) progressive loss of vision in the mid-peripheral retina leading to ‘tunnel vision’ in adulthood. RP often culminates in severe visual impairment or blindness in mid-life. Thus RP accounts for a major fraction of inherited blindness worldwide.
The molecular causes of RP are strikingly heterogeneous. There are autosomal dominant, autosomal recessive and X-linked forms, and rare mitochondrial and digenic forms (1). Within these categories, many different genes can cause similar diseases. For example, 13 genes are known to cause dominant RP, 21 cause recessive RP, and five cause X-linked RP (RetNet, www.sph.uth.tmc.edu/RetNet/). Of the genes causing dominant RP, RP9, RP10 and RP17 have not yet been cloned. Of these, RP10 is likely to cause ~5–10% of cases of adRP in Americans of European origin and Europeans, based on linkage surveys and anecdotal evidence (2 and unpublished data).
The disease locus in a large Spanish family with dominant RP was mapped to human chromosome 7q in 1993 and was named the ‘RP10’ locus (3). Subsequently, the disease locus in a large American family with dominant RP, UTAD045, was mapped to the RP10 region (4). Linkage data from the American and Spanish families were then combined to map the RP10 locus to 7q31.1 and to develop a yeast artificial chromosome contig through the RP10 region (5). Two additional RP10 families have since been reported, one originating in the British Isles and a second of Spanish origin (6,7). Each of the published RP10 families have a LOD sore of 3.0 or greater for linkage, without recombination, to markers on 7q31.1, with a combined LOD score of over 16.
Earlier screening of potential candidates excluded blue cone pigment (BCP), ADP ribosylation factor 5 (ARF5), metabotrophic glutamate receptor 8 (GRM8), diacylglycerol kinase iota (DGKI), the human homolog of the Drosophila rdgA gene, a ubinuclein-like gene and a zinc finger-like gene as the RP10 gene (8,9 and unpublished data).
We report identification of the RP10 gene using positional candidate cloning, including refined linkage mapping, development of a dense transcript map on 7q31.1, and prioritization of retinal-expressed candidates based on differential expression in normal versus crx−/crx− knockout mice retinas (10). One mutation in inosine monophosphate dehydrogenase 1 (IMPDH1), at a highly conserved site, segregates with disease in the two large American families, in a British family and in an additional small American families with dominant RP. A second mutation was detected in one additional autosomal dominant RP (adRP) family (Table 1). Identification of IMPDH1 as the cause of RP10 will provide useful diagnostic and counseling benefits to affected families and suggests possible treatment opportunities based on small-molecule therapy.
Table 1.
AdRP families ascertained for this study with mutations in IMPDH1
| Family name | Number of affected individuals | Number of DNAs tested | IMPDH1 mutation |
|---|---|---|---|
| UTAD045 | 29 | 24 | Asp226Asn (GAC→AAC) |
| RFS015 | 23 | 13 | Asp226Asn (GAC→AAC) |
| UTAD278 | 7 | 4 | Asp226Asn (GAC→AAC) |
| UTAD177 | 8 | 1 | Asp226Asn (GAC→AAC) |
| UTAD389 | 3 | 1 | Val268Ile (GTC→ATC) |
RESULTS
Reducing the RP10-disease interval
As part of a positional candidate cloning approach, we sought additional RP10 families in the hope that the disease interval in any new RP10 family could be used to reduce the existing region. We identified one additional RP10 family, RFS015, through linkage analysis (Fig. 1). Linkage between the disease locus and RP10 markers was detected in this family with a maximum LOD score of 4.5. Linkage between the disease locus in this family and RP10 markers is further supported by LOD scores of <2.0 for all the remaining adRP loci tested (data not shown). Unfortunately, subsequent haplotype analysis of additional microsatellite markers determined that the minimal disease interval defined by individuals in RFS015 was larger than the existing minimal region. Identification of the RFS015 family, the 5th reported family whose disease has been linked to this region, further demonstrates the commonality of the RP10 form of adRP.
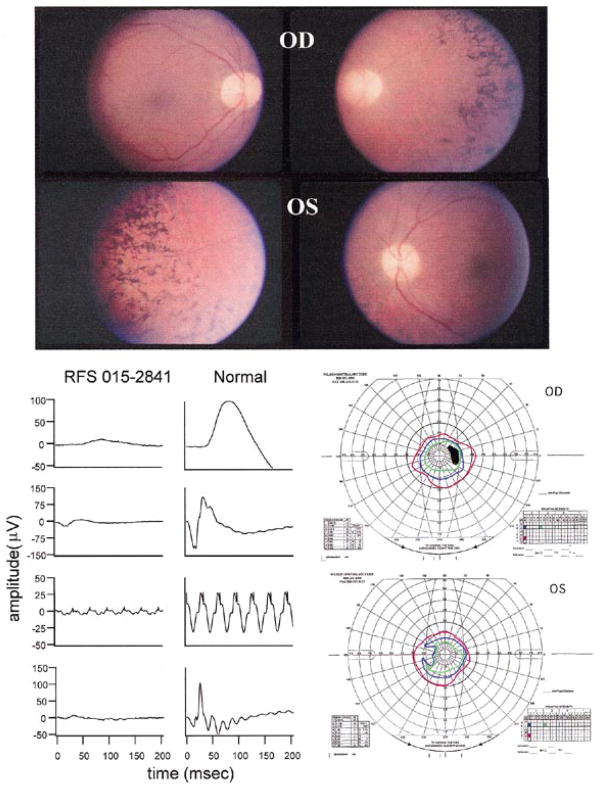
Figure 1.
Representative fundus photographs, ERGs and visual fields from the affected proband of family RFS015. The proband was aged 24 years when first seen at the Retina Foundation of the Southwest in 1988. Best-corrected vision in each eye was 20/50. The lenses of both eyes showed some posterior subcapsular cataract. He had been severely night blind for many years and voluntarily stopped driving at age 17. The fundus of each eye showed typical RP with attenuated retinal vessels and bone-spicule type pigmentary abnormalities throughout the midperiphery. ERG responses were significantly reduced in amplitude. However, rod responses were clearly detectable despite being reduced by ~90% below the lower limit of normal. Cone responses to 30 Hz flicker were reduced in amplitude by 90% and borderline delayed in b-wave implicit time, as reported previously for some patients with adRP (28). This pattern of ERG loss is unusual in that there is a similar reduction in rod and cone responses and could potentially be a phenotypic marker of IMPDH1 mutations. The more usual pattern in adRP is for greater loss of rod than cone responses. Consistent with the recordable rod ERG, final dark-adapted visual thresholds were elevated 0.6 log units above the upper limit of normal. This contrasts with the 3.0 log unit elevations typically seen in patients lacking rod ERG function. Visual fields were also consistent with the ERG in showing severe constriction to the IV-4-e test target. The IV-4-e isopter was within 20 degrees.
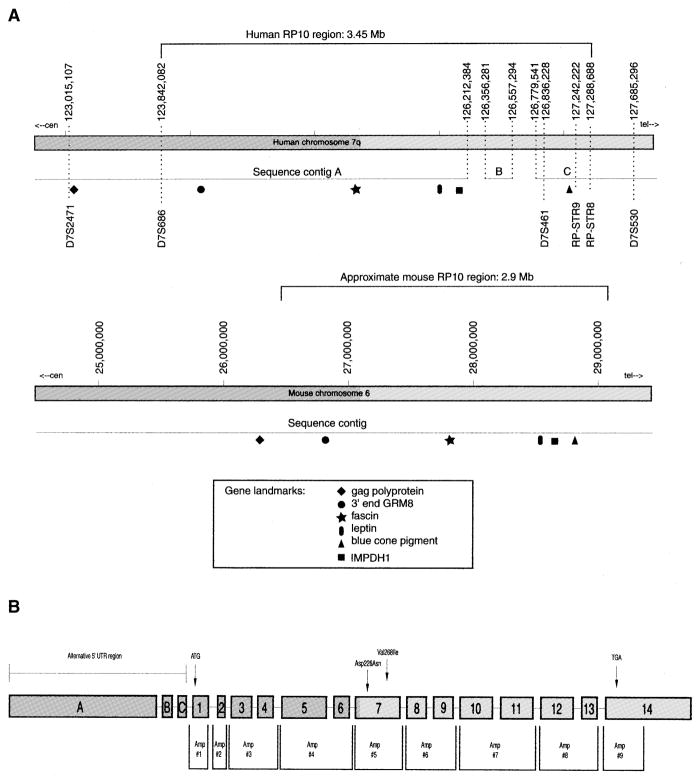
Another technique used to reduce the RP10 disease interval was fine-point haplotype analysis in select members of the UTAD045 family. Previous research demonstrated that affected members of a branch of this family were recombinant at marker D7S530, thereby setting it as the telomeric disease boundary (5). The distance between D7S530 and the closest non-recombinant marker, D7S461, was estimated to be ~1 cM, encompassing almost 20% of the disease interval. To reduce this region, we designed 14 short tandem repeat (STR) markers based on repetitive regions identified in the genomic sequence located between D7S461 and D7S530. Testing these markers placed the critical recombinant event between RP-STR8 and RP-STR9, thereby reducing the RP10 region by 300 kb (Fig. 2A).
Figure 2.
(A) Diagram of the RP10 candidate region on human chromosome 7q31.1 and the syntenic region on mouse chromosome 6. The recombinant event in UTAD045 that defines the RP10 critical region was localized to the region between RP-STR8 and RPSTR9. The approximate location of several known genes from this chromosomal region are shown for each species. Comparison of several genes and EST clusters from mouse and human led us to believe that contig B, as well as the majority of gap sequence located on either side of contig B, is erroneously placed. (B) IMPDH1 gene structure and location of screening amplimers used in this study. Exons are labeled accorded to nomenclature initially used by Gu et al. (12). Exons 1–14 contain the coding region of all three reported IMPDH1 transcripts. Exons A–C represent alternative 5′-UTRs used only in the 4.0 and 2.7 kb IMPDH1 transcripts.
RP10 candidate genes
Analysis of the publicly available Human Genome Working Draft, and the genomic sequence assembly generated by Celera, suggests that the genomic sequence through the RP10 region is essentially complete. Analysis of known genes from the region, and the previously determined marker order, indicates that the Celera database is more complete than the public data and, therefore, it is the one we chose to use for this study. As shown in Figure 2, the RP10 region is covered by three sequence contigs with two small gaps.
In addition, Celera has sequenced and assembled the syntenic region in mouse, which is located on mouse chromosome 6. This region is apparently 100% complete and can be aligned with the human sequence. Comparison of the mouse and human sequences suggests that the small human contig B is erroneously placed. This is supported by independent mapping of several of the genes in contig B to locations other than chromosome 7. Evaluation of the data suggests that contigs A and C are actually separated from each other by a very small gap and that the total length of the RP10 region is close to 3 Mb.
We used the Celera sequence annotation and independent bioinformatic analysis to create a transcription map of the RP10 region. This analysis identified 54 potential genes located in the RP10 critical disease region, at least 10 of which are retinal-expressed genes and are therefore believed to be good candidates for disease (data not shown).
Prioritization of candidate genes
The expression pattern of genes known to cause retinal degeneration strongly suggests that the RP10 disease-causing gene will be expressed in the retina, most likely in photoreceptors. Furthermore, the expression of several of these cloned disease genes is regulated by CRX, a photoreceptor and pineal gland-specific transcription factor (11).
As part of an independent research project, two co-authors (C.L.C. and S.B.) used serial analysis of gene expression (SAGE) methodology to identify retinal-specific or enriched genes by examining and comparing the expression levels of postnatal day 10.5 (P10.5) crx+/crx+ and crx−/crx− mice (10). Analysis of over 50 000 tags in the retinal libraries from these mice showed significant expression level changes in 12% (P < 0.0005) of the tags. This variation is believed to be due to the loss of crx and/or to the poorly differentiated photoreceptors that exist at P10.5. Initial mapping data suggested that three mouse genes determined to have reduced expression levels in the crx−/crx− mice have human homologs located near the RP10 region on chromosome 7q (Table 2) (10).
Table 2.
Candidate genes in the RP10 region on 7q31.1
Further analysis of these three genes determined that the genomic sequence corresponding to cDNA NM_014888 was located several megabases centromeric to the RP10 disease interval. A second gene, FLJ11350, mapped to the questionable gap sequence located between contigs A and B in Figure 2A. Analysis of the third gene identified using SAGE, IMPDH1, confirmed its location in the RP10 disease interval.
Mutations in IMPDH1
Since IMPDH1 is located in the refined RP10 disease interval, is expressed in the retina and its levels appear to be altered, either directly or indirectly, by CRX, we decided to test members of the UTAD045 and RFS015 families for disease-causing mutations.
Three different IMPDH1 transcripts have been reported in the literature, each of which consists of the same 14 coding exons (12). The exact locations of each exon was confirmed and the flanking intronic sequence determined by comparison of the cDNA (GenBank accession no. NM_000883) with available genomic sequence from both the public and private databases (Fig. 2B).
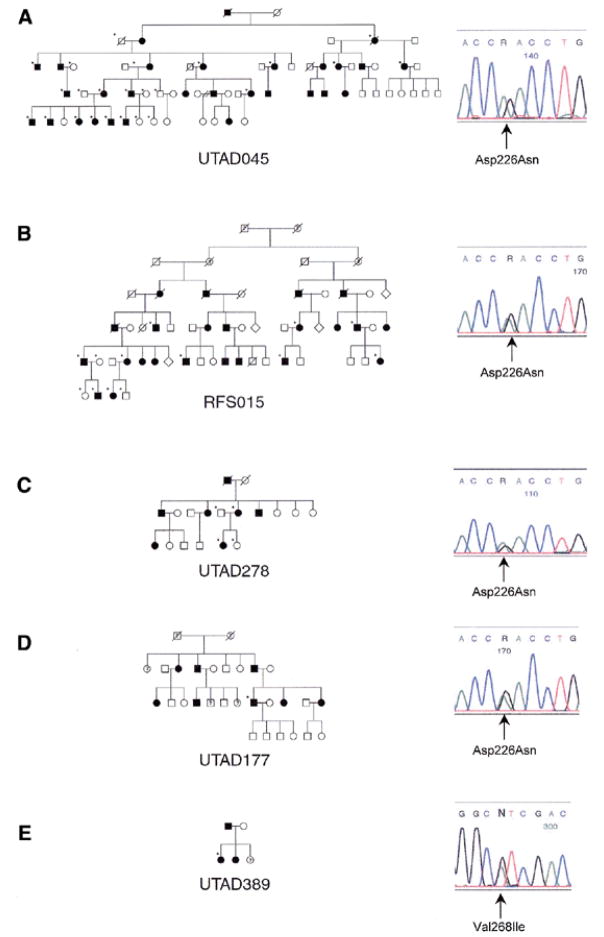
DNA samples from affected members of UTAD045 and RFS015 were tested by sequencing for the presence of disease-causing mutations in the entire IMPDH1 coding region and flanking intron/exon junctions. The same missense mutation in exon 7, Asp226Asn (GAC→AAC), was identified in affected members of both families tested. Sequence analysis of genomic DNA from all available family members demonstrates that this mutation segregates with disease in both families (Fig. 3A and B). Subsequent testing of four members of a British RP10 family, UTAD278, also identified the Asp226Asn mutation in affected individuals (Fig. 3C). Family history suggests that UTAD278 is a branch of the previously reported family from the British Isles (6 and J.Keen, personal communication).
Figure 3.
Pedigrees and electropherograms of five RP10 families with IMPDH1 mutations. All families are of American origin with the exception of UTAD278, which is a British family. (A) UTAD045, Asp226Asn; (B) RFS015, Asp226Asn; (C) UTAD278, Asp226Asn; (D) UTAD177, Asp226Asn; and (E) UTAD389, Val268Ile. Closed symbols represent affected individuals, question marks represent possibly affected individuals, and open symbols represent unaffected individuals. Tested individuals in each family are indicated by a star.
The Asp226Asn mutation was seen in three independently ascertained families and could therefore be a frequent cause of adRP. To test this hypothesis, we screened an additional 60 adRP probands for mutations in exon 7 of IMPDH1. SSCP analysis identified one proband, from family UTAD177, with a variant pattern similar to that of known Asp226Asn controls. Subsequent sequence analysis confirmed that this proband also had a G→A transition at nucleotide 676 resulting in the Asp226Asn mutation (Fig. 3D). SSCP analysis of exon 7 did not identify this substitution in 60 unrelated CEPH controls. Preliminary haplotype analysis of STRs and single nucleotide polymorphisms in these families suggests that the Asp226Asn mutation probably arose in a common ancestor, perhaps many generations ago (data not shown).
Because single strand conformation polymorphism (SSCP) analysis may not reveal all sequence variants, we analyzed the same 60 adRP probands by PCR product sequencing. This analysis revealed the presence of an A→G transition at base 802 in family UTAD389 that results in a Val268Ile substitution (Fig. 3E). This substitution was not seen in any of the other adRP probands tested nor in CEPH controls. Additional family members are currently being collected to determine whether this substitution segregates with disease. If the mutation does segregate with disease, further studies will be needed to determine if this substitution is truly pathogenic.
DISCUSSION
Mutations in IMPDH1 cause the RP10 form of adRP
We have identified an Asp226Asn IMPDH1 mutation in three adRP families, each with a disease locus that maps to the RP10 region with a LOD score of 3.0 or greater. In each family the mutation segregates with disease and the mutation has not been found in any unaffected family members or CEPH controls. We have identified the same mutation, Asp226Asn, in a proband from a fourth, unmapped, adRP family. As described below, this aspartic acid residue is conserved in all IMPDH proteins. In addition, a different substitution, Val268Ile, was seen in a proband from an adRP family not tested for linkage. Based on these data, we believe that mutations in IMPDH1 cause the RP10 form of adRP, and further confirm, based on the large number of families previously linked to the RP10 locus, that mutations in IMPDH1 may account for 5–10% of all adRP mutations. It is also likely that the Asp226Asn substitution will be frequent among IMPDH1 mutations, at least in individuals of American and western European origin.
In general, members of families with the Asp226Asn mutation, such as family RFS015 (Fig. 1), have early onset of symptoms, equal reduction in rod and cone responses, and rapid progression of retinopathy. This is consistent with symptoms described in other RP10 families (3,5).
Structure and conservation of IMPDH
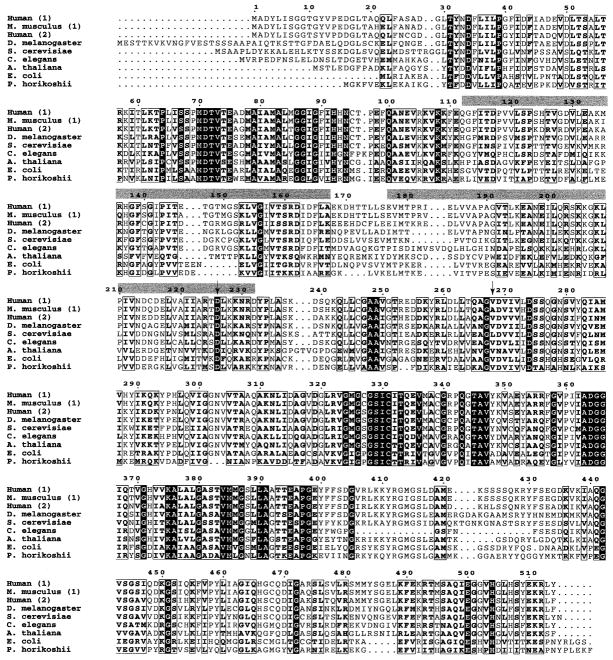
The amino acid sequence for 35 different IMPDH genes has been determined and there is extensive information about functional domains and structural motifs. An alignment of several IMPDH protein sequences (Fig. 4) reveals a high degree of sequence conservation in organisms as diverse as humans, fungi and archaebacteria. Four crystal structures of IMPDH are also available for comparison, including human (13), golden hamster (14), Streptococcus pyogenes (15) and Tritrichomonas foetus (16). These data indicate that the three-dimensional structure is highly conserved as well. In organisms that have more than one IMPDH gene, such as humans, the different isoforms are very similar in sequence and indistinguishable in terms of their catalytic activity, substrate affinities and interaction with inhibitors (17–19).
Figure 4.
IMPDH protein sequences from a range of organisms, aligned using ClustalW and formatted using ESPript 2.0 (prodes.toulouse.inra.fr/ESPript/cgi-bin/nph-ESPript_exe.cgi). In cases where multiple isoforms have been identified, isoform numbers are in parentheses. Sequence numbering is based on the human IMPDH1 sequence. Completely conserved residues are shown as white letters on a black background; highly conserved residues are boxed. The locations of the two CBS domains are indicated by gray bars and the location of the two mutations identified in this study are shown by arrows. SwissProtID and accession numbers: human-1 (imd1_human, P20829), Mus musculus-1 (imd1_mouse, P50096), human-2 (imd2_human, P12268), Drosophila melanogaster (imdh_drome, Q07152), Saccharomyces cerevisiae (imh1_yeast, P38697), Caenorhabditis elegans (PIR T3709), Arabidopsis thaliana (imh1_arath, P47996), E. coli (imdh_ecoli, P06981) and Pyrococcus horikoshii (impd_pyrho, O58045).
In all organisms, the active IMPDH enzymes are homotetramers and each monomer consists of two major functional domains—an eight-stranded α/β barrel that performs the catalytic function of the enzyme, and a smaller flanking domain. The flanking domain is inserted between the second α helix and third β strand of the barrel and is located approximately 35 Å away from the active site. The function of the flanking domain is unknown. It contains two regions of similarity to cystathionine β-synthase (CBS), which have also been found in a number of other proteins (20). In most cases, proteins contain either two or four copies of this CBS domain, which suggests that they interact in pairs (21). One possibility is that the CBS domain has a regulatory function (20), which is supported by the finding that the CBS domains in bacterial IMPDH form a structure that has a potential binding site for regulatory molecules (15).
Functional implications of the Asp226Asn and Val268Ile mutations
The Asp226Asn mutation occurs at a site that is conserved in all IMPDHs sequenced to date and is located within one of the two CBS domain sequences, which are components of the flanking domain. Experiments using Eschericia coli IMPDH indicate that the Asp226 residue (Asp200 in E. coli) is not involved in the enzymatic activity of the protein (22), although the high degree of sequence conservation at this site does imply the existence of another important function. This finding in E. coli is consistent with the observation that the entire flanking domain of human IMPDH2 can be deleted without significant effect on enzymatic activity in vitro (14). Mutations in the cystathionine β-synthase protein, for which the CBS domain was named, are associated with homocystinuria, further substantiating the functional importance of the CBS protein region (23). Further investigation into the function of CBS domains, both in IMPDH and in other proteins, should shed light on the role of this residue.
The Val268Ile change occurs at a less conserved site and is not within a CBS domain, although three-dimensional models show that this residue is located in the same region as Asp226Asn, away from the active site. Additional testing will be required to establish whether or not this is a disease-causing mutation, although this is likely given its occurrence in an adRP family and the lack of polymorphic variation in IMPDH1 in the individuals tested.
IMPDH function, expression and regulation
IMPDH catalyzes the rate-limiting step of de novo guanine nucleotide synthesis by oxidizing inosine monophosphate (IMP) to form xanthosine monophosphate (XMP). Guanine nucleotides play crucial roles in cellular growth, differentiation and apoptosis, and are important substrates for DNA and RNA synthesis and cell signaling (24). Human IMPDH activity comes from two isoenzymes, IMPDH1 and IMPDH2.
In humans, IMPDH1 and IMPDH2 are regulated differently. Expression of the IMPDH2 gene, located on human chromosome 3, is highly up-regulated in proliferating cells, especially in activated leukocytes and tumor cells. IMPDH1 activity is constitutively expressed, usually at lower levels than IMPDH2, and is not affected by proliferation (12,21,24,25). The presence of these two different isoforms has been explained by citing the vastly different guanine nucleotide level requirements of differentiated and proliferating cells (24).
Three different IMPDH1 transcripts have been identified to date. These transcripts differ in size (4.0, 2.7 and 2.5 kb) but contain identical coding sequences and 3′-untranslated regions (3′-UTRs), only varying in the length of their 5′-UTRs. In 1997, Gu et al. (12) performed studies to characterize the expression pattern and to determine the promoter regions of the three IMPDH1 transcripts. They determined that the 4.0 kb IMPDH1 transcript is expressed in activated peripheral blood lymphocytes and some tumor cell lines, while the 2.7 kb transcript is expressed only in tumor cell lines. The 2.5 kb transcript was detected in the majority of cell lines and tissues tested, indicating a more universal expression pattern. Furthermore, studies performed using chloramphenicol acetyltransferase (CAT) assays and different cell lines identified a region of the promoter, named P3, which regulates the expression of the 2.5 kb transcript. The P3 region identified is 700 bp in size and located immediately 5′ of the ATG in the genomic sequence. Additional data suggest that elements that regulate expression of this transcript in a cell type-specific manor are located in the genomic sequence 5′ of the P3 region.
Based on these observations and the reduced expression of IMPDH1 in crx−/crx− mice, we analyzed the human genomic sequence 5′ of the P3 promoter for CRX binding elements (CBEs). The promoter regions of CRX target genes contain a conserved motif comprised of a head-to-tail arrangement of one strong CBE and one weaker CBE (11). Our analysis of the available genomic sequence has identified an 11 bp sequence, TTAATGTGCTC, located at −745 bp, which matches the consensus CRX motif sequence. Further studies will determine if this sequence motif truly binds CRX and what role CRX may play in the regulation of IMPDH1 in photoreceptors.
IMPDH-directed therapy
Several IMPDH inhibitory drugs have been developed for use in antiviral, cancer and immunosuppression therapy (24). In general, these drugs, both nucleoside and non-nucleoside, inhibit both IMPDH1 and IMPDH2, although one drug, MPA, has a higher affinity for IMPDH2. The availability of compounds that bind IMPDH suggests that small-molecule therapy may be available, eventually, for RP10 patients. This discovery should heighten the urgency with which pharmaceutical companies develop IMPDH1-specific drugs.
MATERIALS AND METHODS
Subjects
Pedigrees of the UTAD045 and UTAD278 families and a description of their disease have been published previously, based on the expectation that UTAD278 is a branch of the published family (4,6). Newly identified subjects tested in this study were diagnosed at one of the following sites: (i) the Anderson Vision Research Center, Retina Foundation of the Southwest, Dallas, TX (26) or (ii) the Jules Stein Eye Institute, UCLA School of Medicine, Los Angeles, CA. All research was conducted under human subjects protocols approved by the respective academic institutions.
Peripheral blood or bucal swabs were obtained from each available family member. DNA was extracted from blood using previously reported methods (2). DNA was obtained from bucal swabs by soaking each swab overnight at 55°C in 1.0 ml cell lysis buffer (Gentra, Minneapolis, MN), 12.5 μl of 20 mg/ml Proteinase K (Qiagen, Chatsworth, CA), and 5 μl of 4 mg/ml RNase A (Gentra). The swab was placed in a Spin Ease extraction tube (Gibco BRL Life Technologies, Rockville, MD) and the digest buffer collected by centrifugation. The digest buffer was returned to the original tube and 335 μl of 10M NH4AC (Gentra) was added to precipitate protein. The supernatant was extracted twice with an equal volume of PCI (Sigma, St Louis, MO). DNA was precipitated using 2 vol ethanol and 1 μl of glycogen (Roche Molecular Biochemicals, Palo Alto, CA) at 4°C overnight. DNA was collected by centrifugation, washed with 70% EtOH, allowed to dry and resuspended in RNase/DNase-free H2O.
Linkage analysis
Four large adRP families from the Laboratory for Molecular Diagnosis of Inherited Eye Diseases were selected for linkage analysis (2). Selection was based on a negative mutation history for rhodopsin, peripherin/RDS, CRX and RP1, and the immediate availability of DNA from at least six affected family members. A minimum of two STR markers with tight linkage to the following nine known adRP loci were tested in each family: RP18 (D1S498, D1S2334), RP3 (D3S1587, D3S1589), RP9 (D7S795, D7S460), RP10 (D7S514, D7S504, D7S1875), RP1 (D8S591, D8S2607), RP13 (D17S1528, D17S1529), RP17 (D17S807, D17S787) and RP11 (D19S572, D19S927).
Primer pairs reported in GDB were used for each STR marker (www.gdb.org/gdb/gdbtop.html). One primer from each pair was end-labeled with [δ-P32]ATP at 37°C for 45 min using T4 polynucleotide kinase (Promega, Madison, WI). Genomic DNA was amplified using a labeled and unlabeled primer and standard-cycling conditions. Labeled PCR product was denatured and separated on 6% LongRanger gels (FMC Bioproducts, Rockland, MD) in 1× TBE for 90–180 min. Gels were dried and autoradiographed after electrophoresis. Autoradiographs were scored manually and the data for each family were recorded. Linkage and multipoint analysis was performed using the Vitesse program (27).
Haplotype analysis
Available genomic sequence located between D7S461 and D7S530 was analyzed for the presence of repeats using the RepeatMasker web server (ftp.genome.washington.edu/cgi-bin/RepeatMasker). Reports containing repeat location, length and type were analyzed manually to detect STR repeats with the highest probability of being polymorphic. Primer sequences were picked from the flanking genomic sequence of each STR using MacVector and the Primer 3 Web site (www-genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi).
Primers were end-labeled as described above. Genomic DNAs from the critical recombinant individuals of UTAD045, their immediate families, and two or three other affected members of the pedigree were amplified using the labeled primers and standard cycling conditions. Selected amplimers were digested with a restriction enzyme. PCR product was denatured and separated as described above. Gels were dried and autoradiographed after electrophoresis. Marker data were scored by hand and then assembled into individual haplotypes.
Identification of candidate genes
Celera’s gene annotation of the corresponding genomic sequence was the principal resource used to identify RP10 candidate genes (www.celera.com). Comparing the public genomic sequence with the expressed sequence tag (EST) division of GenBank identified additional candidate genes. Comparisons were made using the sequence from two large overlapping GenBank contigs, NT_000481 and NT_001521. These sequences were assembled manually and then broken into 25 000 bp pieces, which were compared against the human EST database using the Advanced BLAST server (www.ncbi.nlm.nih.gov/BLAST). ESTs were clustered using the UniGene database (www.ncbi.nlm.nih.gov/UniGene/), using the TIGR database (www.tigr.org/tdb/hgi/searching/reports.html), and manually.
Sequencing
One affected member of each of the RP10 families, UTAD045 and RFS015, was selected for screening. Nine PCR primer pairs were designed such that each of the 14 coding exons and any splice site sequences were amplified (Fig. 2B) (Table 3). These primers were used with standard cycling conditions to PCR amplify genomic DNA. PCR product was sequenced commercially by Seqwright (Houston, TX) using the ABI BigDye cycle sequencing dye terminator kit and an ABI 3700 Genetic Analyzer (Perkin Elmer, Branchburg, NJ). Alternatively, ~100–200 ng of PCR product was treated with a cocktail of shrimp alkaline phosphatase and Exonuclease I (United States Biochemical, Cleveland, OH) then sequenced in-house. Treated PCR product was sequenced according to the manufacturer’s protocols using the ABI BigDye cycle sequencing dye terminator kit (Applied Biosystems, Foster City, CA). Sequencing samples were purified using sephadex columns (Princeton Separations, Adelphia, NJ) and run on an ABI 310 Genetic Analyzer (Perkin Elmer).
Table 3.
Primers for sequencing and SSCP of IMPDH1
| Amplimer | Forward primer (5′→3′) | Reverse primer (5′→3′) | Annealing temperature (°C) |
|---|---|---|---|
| 1 | gcgtcagcagtagcagca | tgccacgtccgtctgctc | 62 |
| 2 | accccagtagacctttcgc | atgccctgcccctgagcaag | 62 |
| 3 | cttgttgccagtggtcg | gcagggagtgtagcagtgc | 54 |
| 4 | tctcagtggagccttggg | cagtctggttgctgggataac | 54 |
| 5 | cagtggaatctctggagtggtc | cctgggtcctcataaacctc | 56 |
| 6 | ttcatccactcaggctctcc | tggggaacaaaggcgagg | 56 |
| 7 | acactcatcctggtggtatttg | catctggggaagtcggtg | 56 |
| 8 | ttctggaaactgaggcacag | gggactaaaggacaaggaacag | 56 |
| 9 | gggaaagggttttgggaag | tggctggctgggctcggag | 56 |
SSCP analysis
Genomic DNA was amplified using the same primers used to sequence exon 7 (Table 3). PCR products were radiolabeled by incorporating 1 μCi of [32P]dCTP during amplification and then the resulting product was digested with StyI (Stratagene, La Jolla, CA). Digested PCR product was denatured and separated overnight on 0.6× MDE gels (FMC Bioproducts, Rockland, MD) at room temperature and 4°C. The gels were prepared in 0.6× TBE buffer and were dried and subjected to autoradiography after electrophoresis.
Acknowledgments
We thank the members of the several families involved in this study without whose enthusiasm and cooperation the project could not have been conducted. We also thank Dr Jeffery Keen and Prof. Chris Inglehearn, Leeds University, UK, for providing DNAs from family UTAD 278. This work was supported by grants from the Foundation Fighting Blindness and the George Gund Foundation, the Schissler Foundation, the William Stamps Farish Fund, the M.D. Anderson Foundation, the John S. Dunn Foundation, Alfred W. Lasher, III, and the Hermann Eye Fund; by the Presidents’ Research Scholarship from the University of Texas-Houston; and by grant EY07142 from the National Eye Institute—National Institutes of Health.
References
- 1.Heckenlively JR, Daiger SP. Hereditary retinal and choroidal degenerations. In: Rimoin DI, Connor JM, Pyeritz RE, editors. Principals and Practices of Medical Genetics. 4. Vol. 1. Churchill Livingston; New York: 2001. pp. 2255–2576. [Google Scholar]
- 2.Sohocki MM, Daiger SP, Bowne SJ, Rodriquez JA, Northrup H, Heckenlively JR, Birch DG, Mintz-Hittner H, Ruiz RS, Lewis RA, Saperstein DA, Sullivan LS. Prevalence of mutations causing retinitis pigmentosa and other inherited retinopathies. Hum Mutat. 2001;17:42–51. doi: 10.1002/1098-1004(2001)17:1<42::AID-HUMU5>3.0.CO;2-K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jordan SA, Farrar GJ, Kenna P, Humphries MM, Sheils DM, Kumar-Singh R, Sharp EM, Soriano N, Ayuso C, Benitez J, et al. Localization of an autosomal dominant retinitis pigmentosa gene to chromosome 7q. Nat Genet. 1993;4:54–58. doi: 10.1038/ng0593-54. [DOI] [PubMed] [Google Scholar]
- 4.McGuire RE, Gannon AM, Sadler-Sullivan LA, Rodriguez JA, Daiger SP. Evidence for a major gene (RP10) for autosomal dominant retinitis pigmentosa on chromosome 7q: linkage mapping in a second, unrelated family. Hum Genet. 1995;95:71–74. doi: 10.1007/BF00225078. [DOI] [PubMed] [Google Scholar]
- 5.McGuire RE, Jordan SA, Braden VV, Bouffard GG, Humphries P, Green ED, Daiger SP. Mapping the RP10 locus for autosomal dominant retinitis pigmentosa on 7q: refined genetic positioning and localization within a well-defined YAC contig. Genome Res. 1996;6:255–266. doi: 10.1101/gr.6.4.255. [DOI] [PubMed] [Google Scholar]
- 6.Mohamed Z, Bell C, Hammer HM, Converse CA, Esakowitz L, Haites NE. Linkage of a medium sized Scottish autosomal dominant retinitis pigmentosa family to chromosome 7q. J Med Genet. 1996;33:714–715. doi: 10.1136/jmg.33.8.714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Millán JM, Martínez F, Vilela C, Beneyto M, Prieto F, Nájera C. An autosomal dominant retinitis pigmentosa family with close linkage to D7S480 on 7q. Hum Genet. 1995;96:216–218. doi: 10.1007/BF00207382. [DOI] [PubMed] [Google Scholar]
- 8.Bowne SJ, Sullivan LS, Ding L, Traer E, Prescott SM, Birch DG, Kennan A, Humphries P, Daiger SP. Evaluation of human diacylglycerol kinase iota, a homolog of Drosophila rdgA, in inherited retinopathy mapping to 7q. Mol Vis. 2000;6:6–9. [PMC free article] [PubMed] [Google Scholar]
- 9.McGuire RE, Daiger SP, Green ED. Localization and characterization of the human ADP-ribosylation factor 5 (ARF5) gene. Genomics. 1997;41:481–484. doi: 10.1006/geno.1997.4689. [DOI] [PubMed] [Google Scholar]
- 10.Blackshaw S, Fraioli RE, Furukawa T, Cepko CL. Comprehensive analysis of photoreceptor gene expression and the identification of candidate retinal disease genes. Cell. 2001;107:1–20. doi: 10.1016/s0092-8674(01)00574-8. [DOI] [PubMed] [Google Scholar]
- 11.Livesey F, Furukawa T, Steffen M, Church G, Cepko C. Microarray analysis of the transcriptional network controlled by the photoreceptor homeobox gene CRX. Curr Biol. 2001;10:301–310. doi: 10.1016/s0960-9822(00)00379-1. [DOI] [PubMed] [Google Scholar]
- 12.Gu J, Spychala J, Mitchell B. Regulation of the human inosine monophosphate dehydrogenase type I gene. J Biol Chem. 1997;272:4458–4466. doi: 10.1074/jbc.272.7.4458. [DOI] [PubMed] [Google Scholar]
- 13.Colby TD, Vanderveen K, Strickler MD, Markham GD. Crystal structure of human type II inosine monophosphate dehydrogenase: implications for ligand binding and drug design. Proc Natl Acad Sci USA. 1999;96:3531–3536. doi: 10.1073/pnas.96.7.3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sintchak MD, Fleming MA, Futer O, Raybuck SA, Chambers SP, Caron PR, Murcko MA, Wilson KP. Structure and mechanism of inosine monophosphate dehydrogenase in complex with the immunosuppressant mycophenolic acid. Cell. 1996;85:921–930. doi: 10.1016/s0092-8674(00)81275-1. [DOI] [PubMed] [Google Scholar]
- 15.Zhang R, Evans G, Rotella FJ, Westbrook EM, Beno D, Huberman E, Joachimiak A, Collart FR. Characteristics and crystal structure of bacterial inosine-5-monophosphate dehydrogenase. Biochemistry. 1999;38:4691–4700. doi: 10.1021/bi982858v. [DOI] [PubMed] [Google Scholar]
- 16.Whitby FG, Luecke H, Kuhn P, Somoza JR, Huete-Perez JA, Phillips JD, Hill CP, Fletterick RJ, Wang CC. Crystal structure of Tritrichomonas foetus inosine-5′-monophosphate dehydrogenase and the enzyme-product complex. Biochemistry. 1997;36:10666–10674. doi: 10.1021/bi9708850. [DOI] [PubMed] [Google Scholar]
- 17.Konno Y, Natsumeda Y, Nagai M, Yamaji Y, Ohno S, Suzuki K, Weber G. Expression of human IMP dehydrogenase types I and II in Escherichia coli and distribution in human normal lymphocytes and leukemic cell lines. J Biol Chem. 1991;266:506–509. [PubMed] [Google Scholar]
- 18.Carr SF, Papp E, Wu JC, Natsumeda Y. Characterization of human type I and type II IMP dehydrogenases. J Biol Chem. 1993;268:27286–27290. [PubMed] [Google Scholar]
- 19.Hager PW, Collart FR, Huberman E, Mitchell BS. Recombinant human inosine monophosphate dehydrogenase type I and type II proteins. Purification and characterization of inhibitor binding. Biochem Pharmacol. 1995;49:1323–1329. doi: 10.1016/0006-2952(95)00026-v. [DOI] [PubMed] [Google Scholar]
- 20.Bateman A. The structure of a domain common to archaebacteria and the homocystinuria disease protein. Trends Biochem Sci. 1997;22:12–13. doi: 10.1016/s0968-0004(96)30046-7. [DOI] [PubMed] [Google Scholar]
- 21.Kery V, Poneleit L, Kraus JP. Trypsin cleavage of human cystathionine beta-synthase into an evolutionarily conserved active core: structural and functional consequences. Arch Biochem Biophys. 1998;355:222–232. doi: 10.1006/abbi.1998.0723. [DOI] [PubMed] [Google Scholar]
- 22.Kerr K, Hedstrom L. The roles of conserved carboxylate residue in IMP dehydrogenase and identification of a transition state analot. Biochemistry. 1997;36:13365–13373. doi: 10.1021/bi9714161. [DOI] [PubMed] [Google Scholar]
- 23.Kluijtmans LA, Bowers GH, Stevenes EM, Renier WO, Kraus JP, Trijbels FJ, van den Heuvel LP, Blom HJ. Defective cystathionine β-synthase regulation by S-adenosylmethioninie in a partially pyridoxine responsive homocystinuria patient. J Clin Invest. 1996;98:285–289. doi: 10.1172/JCI118791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yalowitz J, Hiremagalur J. Molecular targets of guanine nucleotides in differentiation proliferation and apoptosis. Anticancer Res. 2000;20:2329–2338. [PubMed] [Google Scholar]
- 25.Natsumeda Y, Ohno S, Kawasaki H, Konno Y, Weber G, Suzuki K. Two distinct cDNAs for human IMP dehyrogenase. J Biol Chem. 1990;265:5292–5295. [PubMed] [Google Scholar]
- 26.Wheaton DH, Daiger SP, Birch DG. The Southwest Eye Registry, distribution of disease types and mutations. In: Anderson RE, LaVail MM, Hollyfield JG, editors. New Insights Into Retinal Degenerative Diseases. Plenum Publishers; New York, NY: 2001. pp. 339–345. [Google Scholar]
- 27.O’Connell JR, Weeks DE. The VITESSE algorithm for rapid exact multilocus linkage analysis via genotype set-recoding and fuzzy inheritance. Nat Genet. 1995;11:402–408. doi: 10.1038/ng1295-402. [DOI] [PubMed] [Google Scholar]
- 28.Berson EL. Retinitis pigmentosa. Invest Ophthalmol Vis Sci. 1993;34:1659–1676. [PubMed] [Google Scholar]