Abstract
Mutations resulting in overexpression of intracellular Notch1 (ICN1) are frequently observed in human T cell acute lymphoblastic leukemia (T-ALL). We have determined the consequences of ICN1 overexpression from retroviral vectors introduced into bone marrow cells. Early consequences are the generation of polyclonal nontumorigenic CD4+8+ T cell receptor (TCR)-αβ+ cells that do not qualify as tumor precursors despite the observation that they overexpress Notch 1 and c-Myc and degrade the tumor suppressor E2A by posttranslational modification. The first tumorigenic cells are detected among more immature CD4−8+TCR-αβ− cells that give rise to monoclonal tumors with a single, unique TCR-β chain and diverse TCR-α chains, pinpointing malignant transformation to a stage after pre-TCR signaling and before completion of TCR-α rearrangement. In T-ALL, E2A deficiency is accompanied by further transcriptional up-regulation of c-Myc and concomitant dysregulation of the c-Myc-p53 axis at the transcriptional level. Even though the tumors consist of phenotypically heterogeneous cells, no evidence for tumor stem cells was found. As judged by array-based comparative genomic hybridization (array CGH) and spectral karyotype (SKY) analysis, none of the tumors arise because of genomic instability.
Notch receptors play an important role in cell growth and differentiation. When binding to their ligands, these transmembrane receptors undergo a series of proteolytic cleavages that result in the generation of intracellular Notch (ICN), which regulates transcription in a transcriptional complex with several other proteins after nuclear translocation (1). Mutations that result in overexpression of ICN1 can be found in more than half of human T cell acute lymphoblastic lymphoma (T-ALL) samples (2–4). These findings have sparked renewed interest in ICN1-initiated lymphomagenesis. Recent results have indicated that c-Myc is a target of ICN1 and contributes in a major way to the growth of Notch-dependent T-ALL cell lines (5, 6). However, overexpression of c-Myc by itself is insufficient to cause T-ALL, and additional events are required to initiate clonal growth of tumor cells (7, 8). Girard et al. observed that frequent proviral insertions into the Notch1 locus enhanced T-ALL in c-Myc transgenic mice, suggesting a collaboration of c-Myc and Notch1 in oncogenesis that may result from the observation that ICN1 and c-Myc target overlapping genes (5), or that ICN1 targets distinct genes that facilitate c-Myc–dependent oncogenesis. One of the genes that is affected by Notch1 is the tumor suppressor E2A, which has an important role in the differentiation of T and B lymphocytes (9).
Crucial events that eventually lead to malignant transformation are difficult to study retrospectively in cell lines with unknown etiology that may have accumulated additional mutations permitting growth in vitro. Therefore, we have systematically studied the consequences of overexpression of intracellular Notch 1 introduced into BM cells by retroviral vectors (10). The results identify a series of cellular and molecular changes that accompany the generation of tumor cells (T-ALL) with extensively dysregulated gene expression in spite of a normal karyotype and genomic stability.
RESULTS
Abnormal lymphoid development from ICN1-overexpressing hematopoietic precursors
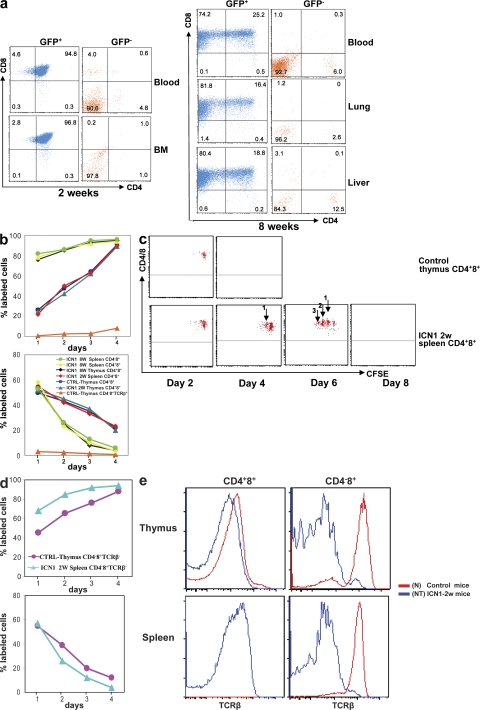
ICN1 overexpression in murine hematopoietic precursor cells from a retroviral IRES vector containing ICN1 and EGFP results in a shutdown of B cell development and ectopic T cell development (Fig. 1) (10). Early after BM transfer (BMT) of retrovirally transduced cells into sublethally irradiated recipients, most EGFP+ cells resemble CD4+8+ thymocytes, with the exception that they can be found in lymphoid organs outside of the thymus (Fig. 1 a). Such cells become apparent 2 wk after BMT and do not cause tumors when transplanted into nu/nu recipient mice (Table S1, available at http://www.jem.org/cgi/content/full/jem.20081561/DC1). Consistent with their nonmalignant and nontumorigenic phenotype, these ICN1-overexpressing cells are polyclonal in regard to TCR-β rearrangement that is completed before the CD4+8+ stage of T cell development (11, 12). As determined by BrdU labeling, the nontumorigenic but abnormal cells have a rapid turnover, much like CD4+8+ thymocytes, which contain ∼15% of dividing cells that go through one or two divisions before becoming small cells that subsequently die by apoptosis (13). Accordingly, normal CD4+8+ thymocytes and the abnormal ICN1-overexpressing CD4+8+ lymphoid cells incorporate BrdU quickly. After pulse labeling, the label of normal CD4+8+ thymocytes and the abnormal ICN1-overexpressing CD4+8+ cells is also quickly lost, indicating that both cell types do not have a long intermitotic lifespan (Fig. 1 b). However, CFSE labeling of these cell types followed by injection into nu/nu mice (in this case, the CFSE-labeled ICN1-overexpressing cells express a retroviral vector containing DsRed) revealed a clear difference in that control CD4+8+ thymocytes did not divide measurably and disappeared 2 d after transplantation, whereas some of the abnormal, nontumorigenic cells went through three cell divisions before their disappearance by day six. Thus, abnormal ICN1-overexpressing CD4+8+ cells early after BMT transfer can be distinguished from normal CD4+8+ thymocytes by their greater, but still limited, proliferation/survival potential (Fig. 1 c). Thus, these early appearing polyclonal cells represent neither tumor cells nor their precursors because they do not generate tumors upon transfer into nude mice.
Figure 1.
Phenotypic analysis of GFP+ cells in various tissues after transplantation of ICN1-overexpressing BM cells. (a) Single-cell suspensions were prepared from blood and BM at 2 wk and from blood, lung, and liver at 8 wk after reconstitution with retrovirally transduced BM. GFP+ and GFP− cells were stained with CD4 and CD8 antibodies. Infiltration by GFP+CD4+8+ and GFP+CD4−8+ malignant cells was analyzed in nonlymphoid organs such as lung and liver. Numbers indicate the percentage of cells within each quadrant. Data are representative of three independent experiments. (b) BrdU–labeling kinetics: continuous (top) and pulse (bottom) BrdU labeling of lymphocytes subsets. BrdU was administered to control mice and mice at 2 or 8 wk after BM transplantation with ICN1-overexpressing cells. The percentage of BrdU-labeled cells was analyzed by flow cytometry as previously described (13). (c) CFSE labeling: normal CD4+8+ thymocytes and ICN1-overexpressing CD4+8+ splenocytes were labeled with CFSE and injected into nu/nu mice. Dilution of CFSE label was analyzed by FACS analysis. (d) BrdU–labeling kinetics: continuous (top) and pulse (bottom) BrdU labeling of ICN1-overexpressing TCR-αβ− CD4−8+ splenocytes versus control normal CD4−8+ thymocytes. (e) TCR-β expression by various lymphocyte subsets in ICN1-transplanted mice at 2 wk after BMT, as well as control mice. TCR-β staining was performed with the pan–TCR-β antibody H57.
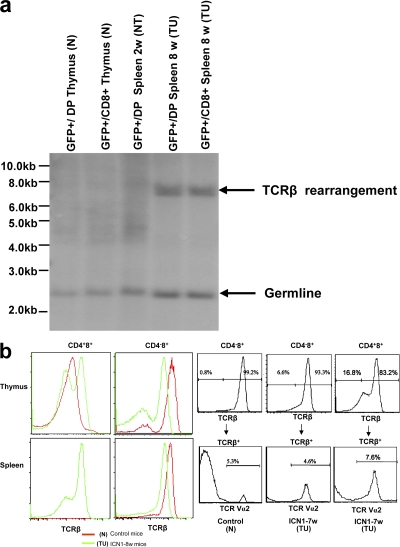
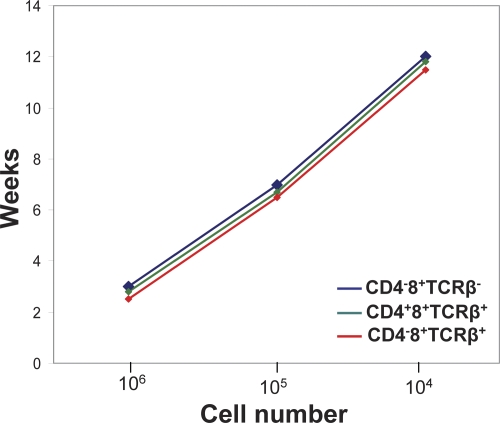
At 2 wk, a minor subset of CD4−8+ EGFP+ cells could be observed in peripheral lymphoid tissue, which, unlike the abnormal CD4+8+ cells, did not express an αβ TCR, and thus corresponded in surface phenotype to CD4−8+ immature thymocytes that represent precursors of CD4+8+ DP cells (Fig. 1, d and e). When these cells were transplanted into nu/nu mice, they generated tumors comprised of CD4+8+ and CD4−8+ cells with αβ TCRs on the cell surface. These data suggested that cells with malignant potential are generated after pre-TCR signaling–dependent TCR-β selection (14) and before TCR-α rearrangement. This was supported by the notion that the resulting monoclonal T-ALL carry a single TCR-β rearrangement (Fig. 2 a) but express diverse TCR-α chains, as indicated by the partial staining of αβ TCRs with antibodies specific for one particular Vα subgroup only (Fig. 2 b). Thus, malignant transformation of ICN1-overexpressing cells is an event that occurs after pre-TCR signaling, but before the acquisition of the CD4+8+ TCR-αβ+ phenotype. In fact, some of the monoclonal tumor cells do not express an αβ TCR on the cell surface, which is likely caused by the observation that they did not succeed in generating productive TCR-α rearrangements. The malignant potential of the phenotypically distinct tumor cells is reflected in their DNA labeling kinetics that are faster than those of their normal counterparts with the same surface phenotype. This is true for the “early” ICN1-overexpressing CD4−8+ TCR-αβ− cells that cause malignancy after transfer into nu/nu mice and also for the CD4+8+ and CD4−8+ TCR-αβ+ tumor cells that each can cause malignant tumors on their own after transfer into nu/nu recipients even when injected in low cell numbers. The analysis showing that each phenotypically distinct subset causes tumors with identical kinetics when injected in different doses refutes the possible notion that the subset with the most immature phenotype, i.e., the CD4−8+ TCR-αβ− subset features tumor stem cells, whereas the more mature subsets feature tumor mass but not necessarily highly tumorigenic cells (Fig. 1 d and Fig. 3). On the basis of these data, it appears useful to distinguish between normal CD4+8+ TCR-αβ+ thymocytes, abnormal nonmalignant and nontumorigenic ICN1-overexpressing CD4+8+ TCR-αβ+ cells, and tumorigenic CD4−8+ TCR-αβ−, as well as CD4+8+ TCR-αβ+ and CD4−8+ TCR-αβ+ tumor cells when further analyzing tumor development at the subcellular level. We assume that the nontumorigenic but abnormal CD4+8+ cells are derived from ICN1-overexpressing CD4−8+ TCR-αβ− precursors that have failed to undergo malignant transformation because of a genetic status not suited to cooperate with Notch1 overexpression to generate malignant transformation (see Discussion). Because this sequence of events after ICN1 overexpression has not been elaborated previously, it seems important to remember the contribution of the different subsets to malignancy when further analyzing gene expression.
Figure 2.
TCR expression by malignant and nonmalignant cells. (a) TCR-β rearrangement in various cell types. Analysis of TCR-β rearrangement by Southern blot analysis in GFP+CD4+8+(GFP+/DP) and GFP+CD4−8+(GFP+/CD8+) thymocytes derived from retroviral GFP-empty vector–transplanted mice, as well as GFP+CD4+8+(GFP+/DP) and GFP+CD4−8+(GFP+/CD8+) splenocytes derived from retroviral GFP-ICN1–transplanted mice at 2 and 8 wk after BMT. Molecular markers are shown on the left. (b) TCR-β and TCRV-α2 expression by various lymphocyte subsets. (left) Staining with the pan TCR-β antibody H57. (right) Staining with Vα2 TCR antibody.
Figure 3.
Kinetic analysis of tumor development. Various cell numbers of ICN1-overexpressing cells with a distinct surface phenotype (as indicated) were injected into nu/nu mice and the kinetics of tumor development were monitored.
Posttranslational elimination of E2A-encoded tumor suppressor proteins
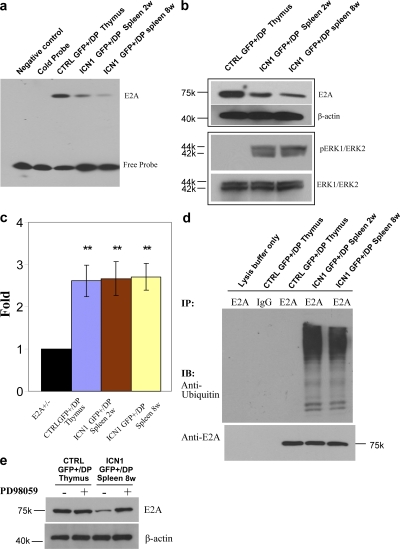
Because ICN1 overexpression results in ablation of B cell development (Fig. 1 a), we investigated its impact on the basic helix-loop-helix proteins encoded by the E2A gene. These proteins bind as homo- or heterodimers with other basic helix-loop-helix proteins as transcription factors to ubiquitous E-box motifs and have an essential function in B cell development (15). In fact, E2A-encoded proteins have an important role in T cell development as well (14), and E2A-deficient mice regularly develop T-ALL (16). When analyzing binding of E2A protein to E-box motifs by EMSA, it became clear that nuclear extracts of both ICN1-overexpressing polyclonal, nontumorigenic, and tumor cells exhibited strongly reduced activity (Fig. 4 a). According to the literature, the results could have been caused by distinct mechanisms; the reduced activity of E2A in EMSA could have been caused by either up-regulation of the pre-TCR by overexpressed ICN1 or by increased pre-TCR signaling resulting in increased Id1 or Id3 levels that prevent binding of E2A-encoded proteins to E-box motifs (17, 18). Alternatively, overexpressed ICN1 could have resulted in E2A protein phosphorylation and degradation (19). Apparently, the second mechanism was responsible because there were reduced levels of E2A protein as determined by Western blotting (Fig. 4 b), but not reduced RNA levels as analyzed by quantitative RT-PCR analysis (Fig. 4 c). The reduced levels of E2A-encoded proteins were caused by ubiquitination-dependent degradation of E2A depending on phosphorylation by activated ERK kinase (Fig. 4 d). In fact, inhibition of ERK phosphorylation (19) by the specific MEK1 inhibitor PD98059 restored the levels of E2A-encoded proteins (Fig. 4 e). Similarly, reduced levels of E2A proteins were observed in both nontumorigenic and tumor cells. Thus, as observed in ICN1-overexpressing fibroblasts (19), the E2A-encoded tumor suppressors were eliminated in ICN1-overexpressing lymphoid cells by posttranslational modifications. The ICN1-dependent reduction of E2A-encoded tumor suppressors by itself was insufficient to cause tumors within the first few months of observation.
Figure 4.
Overexpressed ICN1 inhibits E2A activity by promoting protein degradation at different stages of disease development. (a) E2A E-box motif binding activities were analyzed by electrophoretic mobility shift assay (EMSA) using nuclear extracts of GFP+CD4+8+ (GFP+/DP) cells derived from various mice. (b) E2A and phosphorylated ERK1/2 protein expression levels were analyzed by Western blot in whole-cell lysates from various cells. (c) Quantitative analysis of E47 mRNA expression in various cell types. GAPDH mRNA was used as control. **, P < 0.01, compared with E2A+/− group. Data are reported as mean ± SEM for triplicate experiments. (d) Ubiquitination assay: whole-cell lysates were immunoprecipitated by E2A antibody. Immunoprecipitates were blotted with antiubiquitin antibody. (e) ICN1-induced degradation and phosphorylation of E2A by ERK1/2 MAP kinases. Whole-cell lysates were from various cells that have incubated in the absence or presence of 50 μM PD98059 for 90 min; PD98059 is a specific inhibitor of MEK1 and MEK2. Western blot analysis was performed to evaluate E2A protein expression levels. β-Actin is shown as loading control.
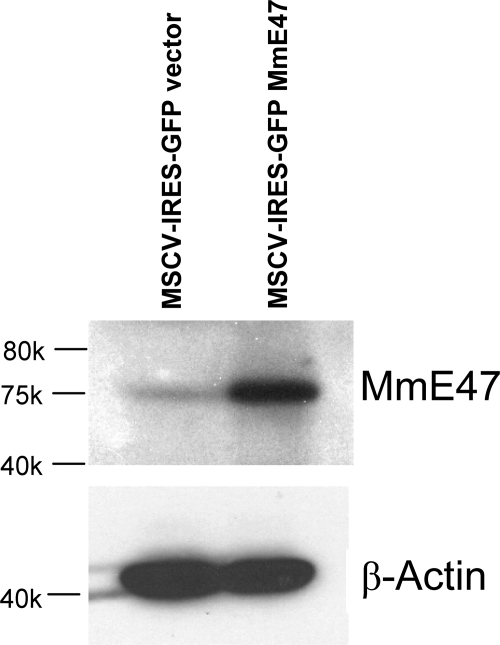
To evaluate the role of E2A-encoded E47 protein as a potential tumor suppressor in T-ALL, nondegradable mutant E47 (19) was introduced into ICN1-overexpressing tumors. Double fluorescence–labeled tumor cells were sorted and injected into nu/nu recipient mice, whereas control mice received tumor cells transduced with an empty EGFP vector. Cells overexpressing E47 (Fig. 5) had a clear growth disadvantage because they generated tumors more slowly than ICN1-overexpressing cells transduced with an empty vector. Also, after injection of 98% pure double-labeled tumor cells expressing the two retroviral vectors, there was some selection favoring cells no longer expressing mutant E47. This selection resulted in tumors in which half of the cells no longer expressed mutant E47, indicating that in ICN1-overexpressing tumor cells, E47 can act as a tumor-suppressor, albeit not one that is sufficiently strong to completely prevent the outgrowth of E47+ tumors (Fig. 6). Thus, the posttranslational elimination of the E2A-encoded tumor suppressor by overexpressed ICN1 contributes to Notch1-induced malignant growth, which is consistent with the observation that this tumor suppressor has a role in preventing proliferation and facilitating differentiation in normal T cell development. On the other hand, it appears that E2A reduction may not be essential for tumor formation after ICNI overexpression because E47-overexpressing tumors could still be detected, albeit at a later stage.
Figure 5.
Nondegradable mutant E47 is overexpressed in ICN1-DsRed tumor cells. Western blot analysis of mutant E47 protein expression in whole-cell lysates from ICN1 tumor cells transduced with the empty retroviral vector MSCV-IRES-GFP or retroviral construct MSCV-IRES-GFP-MmE47. β-Actin is shown as loading sample.
Figure 6.
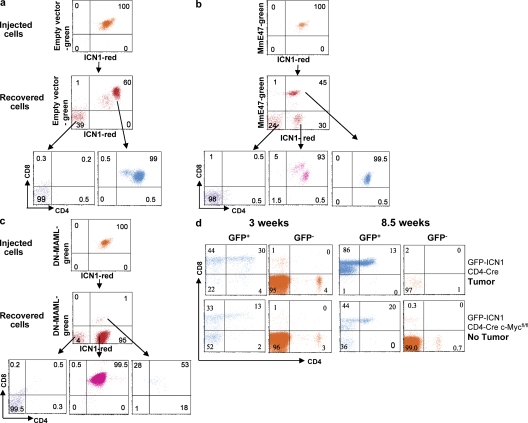
Essential role of ICN1 and c-Myc, but not E47, down-regulation in tumor growth. Tumor development and growth dependent on c-Myc and ICN1, respectively. The empty GFP-IRES retroviral vector (a), the GFP-IRES retroviral vector containing mutant E47 (b), or the GFP-IRES retroviral vector containing DNMAML1 (c) were introduced into tumors expressing a DsRed-ICN1 retrovirus. Double-fluorescent tumor cells were sorted and 106 cells were injected into nu/nu recipient mice. Tumor cells were analyzed when mice had developed palpable tumors. Nude mice injected with tumors containing empty retroviral vectors died of lymphoma 3 wk of after transfer. Mice injected with cells containing MmE47 or DN-MAML1 developed tumors 7 wk after injection. Double-fluorescent cells did not form tumors in c. (d) Analysis of GFP+ and GFP− cells in blood of Rag2−/−, γc−/− mice that were γ irradiated and reconstituted with retrovirally transduced BM cells from either floxed c-Myc, CD4-cre, or CD4-cre mice. CD4-cre mice were killed at 8.5 wk when they exhibited large lymphoblastic tumors. In contrast, CD4-cre c-MycFL/FL mice developed no tumors up to 15 wk.
Tumor-specific gene expression
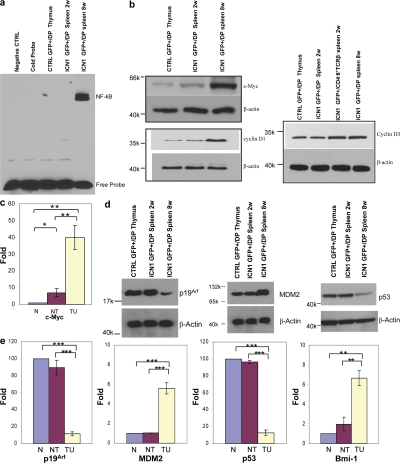
When analyzing the expression of a few specific candidate genes such as c-Myc, NF-κB, cyclin D1, and cyclin D3 by Western blot, it became apparent that tumor cells exhibited higher levels of these proteins compared with their nontumorigenic but abnormal or normal counterparts (Fig. 7, a and b). ICN1 has been described to bind specifically in a transcriptional complex to c-Myc (5, 6), and we found c-Myc to be up-regulated in nontumorigenic ICN1-overexpressing cells, but further up-regulated in tumor cells at the level of both RNA and protein (Fig. 7 c). It is interesting to note that in studies looking at cyclin D3 deficiency, it was concluded that cyclin D3 was essential for growth of T-ALL (20). These data, however, only addressed human T-ALL and murine nontumorigenic cells. Our results showing that murine T-ALL overexpress cyclin D1 are, in fact, not easily compatible with the notion that cyclin D3 is essential for growth of murine T-ALL.
Figure 7.
Specific gene expression. (a) NF-κB DNA binding activity was analyzed by electrophoretic mobility shift assay (EMSA) using nuclear extracts of GFP+/CD4+8+ DP cells derived from various mice. (b) c-Myc, cyclin D1, and cyclin D3 protein levels in various cells were analyzed by Western blot in whole-cell lysates. β-Actin is shown as loading control. (c) Total RNA isolated from various cells was analyzed by quantitative real-time PCR using specific primers for c-Myc. GAPDH mRNA was used as control. *, P < 0.05; **, P < 0.01. Data are shown as mean ± SEM for triplicate experiments. (d) The c-Myc-p53 axis: Western blot analysis in whole-cell lysates from various cells was performed. β-Actin is shown as loading sample. (e) Quantitative analysis of p19Arf, Mdm2, p53, and Bmi-1 mRNA expression in various cells. GAPDH mRNA was used as control. **, P < 0.01; ***, P < 0.001. Data are shown as mean ± SEM for triplicate experiments. N, CTRL GFP+/DP thymus; NT, ICN1, GFP+/DP spleen 2 wk; TU, ICN1 GFP+/DP spleen 8 wk.
To obtain further insight into differential gene expression, comparative analysis of global gene expression was conducted after isolation of RNA from normal CD4+8+ DP thymocytes, nontumorigenic ICN1-overexpressing CD4+8+ cells from peripheral lymphoid tissue, and CD4+8+ tumor cells by Affymetrix chip analysis. Direct targets of Notch, such as Hes1, Hes5, and pTα were elevated in nontumorigenic versus normal cells and either maintained or down-regulated in malignant (tumor) cells, in contrast to c-Myc that was further up-regulated in tumor cells (Fig. S1, a and b, available at http://www.jem.org/cgi/content/full/jem.20081561/DC1), consistent with quantitative RT-PCR data and Western blot analysis (Fig. 7, b and c). In regard to cell cycle control and death pathways, both nontumorigenic and tumor cells exhibited reduced transcription of the death-associated protein kinase 1 (DAPK1). Reduced levels of DAPK1 are often found in tumors because of the hypermethylation of its promoter (21). Fas was down-regulated in both nontumorigenic and tumor cells versus normal controls. Casting a wider net, the data showed that tumor cells versus nontumorigenic cells were especially deficient in tumor suppressors such as the large tumor suppressor 2 (LATS2), often found down-regulated in human lymphoma, and inhibitors of cyclin-dependent kinases, such as p19 and p18 (Fig. S1 a). On the other hand, several oncogenes, including several Ras family oncogenes, the Fyn-proto-oncogene or general transcripts of genes enhancing proliferation (CDK4) and antagonizing cell death were specifically up-regulated in tumor cells, as were genes enhancing transcription and translation and representing c-Myc target genes (Fig. S1 b) (5, 22). These data suggest a scenario in which high levels of ICN1 alone result in elimination or down-regulation of several tumor suppressors, such as E2A-encoded proteins and DAPK1, and in up-regulation of several Notch targets and survival genes in nontumorigenic cells, but only moderately up-regulated c-Myc levels. These changes, however, are insufficient to cause malignant transformation and lead only to moderately increased survival/proliferation of cells with a CD4+8+ phenotype. The nontumorigenic cells do, in fact, express higher Annexin V levels than tumor cells, indicating their higher susceptibility to apoptosis. Also, nontumorigenic cells still express appreciable levels of tumor suppressors that are down-regulated in malignant tumor cells (Fig. S1, a and b). Of special interest was the observation that the p53 inducible nuclear protein 1 gene was up in nontumorigenic versus tumor cells, indicating that p53 activity was increased in nontumorigenic versus tumor cells. These observations may explain the nonmalignant phenotype of the ICN1-overexpressing nontumorigenic cells.
C-Myc and ICN1 overexpression are essential for tumor development and growth
The gene expression data suggest that up-regulation of c-Myc may play an essential role in tumorigenesis, as has been suggested by analysis of select T-ALL cell lines (5). The essential role of c-Myc for tumor growth was confirmed in experiments where floxed c-Mycfl/fl (23) mice were crossed with CD4-cre (24) mice and BM cells from c-Mycfl/fl CD4-cre+/− mice were infected with the ICN1 and EGFP bicistronic retroviral vector. As shown in Fig. 6 d, the deletion of c-Myc at the CD4+8+ stage of T cell development prevented tumor formation, but did not prevent the generation of EGFP+ extrathymic nontumorigenic cells, whereas in CD4-cre mice, tumors developed normally. Thus, c-Myc plays an essential role in early tumor growth because it is deleted in CD4-expressing tumor cells, but not their CD4−8+ TCR-αβ− precursors. This result is in line with reports on T-ALL cell lines (23).
The essential role of ICN1 for tumor growth in vivo was shown by introducing dominant-negative mastermind-like 1 (DNMAML1) (25) through retroviral transduction into primary ICN1-overexpressing tumor cells using an approach similar to that used to test the effect of mutant E47 on tumor development (Fig. 6), i.e., the retroviral vector containing ICN1 and DsRed was used to generate tumors and the retroviral vector containing DNMAML1 and EGFP was introduced into tumor cells before transplantation into nude mice. Even though 99% pure double-fluorescent tumor cells were sorted and injected into nu/nu recipients, double-fluorescent cells did not form tumors, whereas DsRed, ICN1-expressing cells survived selectively in the injected hosts and generated tumors at a much later stage compared with ICN1-expressing controls (Fig. 6 c). These results indicate that c-Myc and ICN1 are both required for the maintenance of tumors as previously established for cell lines (5, 25).
The c-Myc—p53 pathway in malignant transformation
The tumor suppressor p53 can counteract the oncogenic effect of elevated c-Myc by inducing cell death. This can occur through c-Myc–dependent increased production of p19Arf that sequesters the p53-degrading enzyme Mdm2 in the nucleolus, preventing it from reaching p53, which is thereby stabilized and induces apoptosis in c-Myc–overexpressing cells (26). Thus, this scenario depicts the regulation of the c-myc-p53 axis mostly by posttranscriptional mechanism. Several mechanisms can interfere with this protective pathway, in particular mutations in either c-Myc (27) or p53 (28), or the activation of the c-Myc–dependent transcriptional repressor Bmi-1 that can down-regulate p19Arf (29). To investigate whether the c-Myc-p53 axis (28) is intact in ICN1-dependent tumor cells with elevated levels of c-Myc, we determined c-Myc and p53 levels by Western blotting. As shown in Fig. 7 d, in spite of the highly elevated levels of the c-Myc protein in tumor cells, the level of the p53 protein was down compared with nontumorigenic ICN1-overexpressing cells, indicating that the c-Myc–dependent pathway of p53 stabilization was not operative in tumor cells. Furthermore, p19Arf protein was down-regulated and Mdm2 protein was up-regulated in tumor cells compared with nontumorigenic ICN1-overexpressing cells. Curiously, all changes in protein expression were mirrored by changes at the level of RNA, indicating the existence of regulatory mechanisms in addition to posttranscriptional modifications. Also, the transcriptional repressor Bmi-1 was up-regulated in tumor cells at the level of RNA, providing an explanation for the low-level expression of p19Arf at the level of RNA.
Normal karyotype and genomic stability of tumors
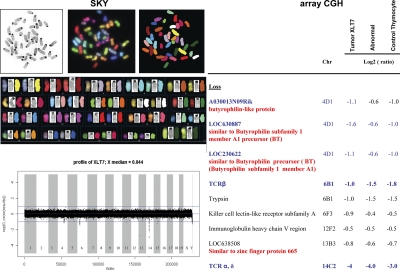
Recent analysis of human T-ALL lines has indicated considerable genomic instability, whereas genomic instability of murine T-ALL was artificially introduced (30). Also, overexpression of c-Myc was reported to result in genomic instability (31). When several independent primary T-ALL samples from ICN1-overexpressing mice samples were subjected to SKY analysis, it became clear that the independent T-ALL samples exhibited a normal karyotype (Fig. 8) as has also been observed in subsets of primary human T-ALL samples (32, 33). Furthermore, high-resolution oligo array–based comparative genomic hybridization of four independent primary T-ALL samples revealed expected deletions at the TCR-β, TCR-α, and TCR-γ loci, as well as the lgH locus; these deletions are also found among the majority of normal CD4+8+ thymocytes. Importantly, however, there were no deletions or amplifications that occurred only in tumor cells, but not in normal cells or nontumorigenic cells, excluding genomic instability as the cause of malignant transformation in this mouse model. These results therefore show that dysregulated gene expression in murine T-ALL initiated by ICN1 overexpression does not result from genomic instability.
Figure 8.
Genomic analysis. (left) SKY analysis of ICN1-induced tumor cells. SKY table of stained ICN1-induced tumor chromosomes using SKY paint shows the spectral, classified, and banded-inverted chromosomes. (right) Array CGH. Genomic DNAs extracted from ICN1-induced malignant tumor GFP+CD4+8+ (Tumor XLT7), abnormal nonmalignant ICN1-overexpressing cells GFP+CD4+8+, and control GFP+CD4+8+ thymocytes were subjected to high-resolution oligo array–based comparative genomic hybridization. Tail DNA from the same mouse was used as reference for the hybridization. Malignant tumor GFP+CD4+8+ cells from four independent tumor mice were analyzed by Array CGH.
DISCUSSION
Cellular and molecular pathways involved in tumorigenesis initiated by a single aberrantly expressed gene are difficult to study retrospectively in human tumor cell lines because of the unknown impact of unrelated genetic changes that may have independently contributed to tumor formation, as well as further genetic changes that are required for in vitro growth (30). For this reason, disease progression was explored in a murine model of T-ALL initiated exclusively by retrovirus-mediated overexpression of ICN1 in lin− BM cells. In fact, mutations resulting in overexpression of ICN1 are frequently observed in human T-ALL samples (2–4). However, in all cases, it is unknown whether such mutations initiated tumor development or just contributed later to the progression of disease. In a mouse model of ICN1-initiated T-ALL, it was shown by Aster et al. that retrovirally induced overexpression of ICN1 in lineage-negative BM cells resulted in extrathymic T cell development and T-ALL (10). In this study, we have characterized various stages during tumor development with the aim to define cellular and molecular pathways in ICN1-dependent tumorigenesis.
BMT with cells retrovirally transduced with a bicistronic retroviral vector containing ICN1 and a fluorescence marker resulted after 2 wk in accumulation of CD4+8+ cells outside of the thymus. Such ICN1-overexpressing cells were not only abnormal in regard to their anatomical location but also exhibited an extended proliferation/survival potential compared with their intrathymic counterparts. These cells, however, were nontumorigenic after transplantation into nude mice, and hence do not qualify as tumor progenitors. Molecular analysis of these early appearing nontumorigenic CD4+8+ cells revealed the ICN1-dependent, degradation of E2A tumor suppressor that normally binds to ubiquitous E-box motifs and in the thymus reduces proliferation while promoting differentiation of pre-TCR–expressing thymocytes (16).The ubiquitin-dependent degradation of E2A-encoded proteins in ICN1-overexpressing cells explains the lack of B cell development of ICN1-overexpressing BM cells (15).
Nontumorigenic CD4+8+ cells exhibit multiple TCR-β rearrangements consistent with their polyclonal nonmalignant and nontumorigenic phenotype. Global gene expression analysis confirmed that these cells differ extensively from normal CD4+8+ thymocytes; expression of several Notch target genes is increased, as is the expression of genes involved in proliferation/survival, whereas RNAs coding for cell cycle inhibitors and death-promoting proteins are reduced. These data fit well with the functional studies showing increased survival and proliferation of ICN1 overexpression among nontumorigenic CD4+8+ cells.
The proliferative capacity, TCR-β rearrangement status, and gene expression profile of these abnormal but nontumorigenic cells are distinct from those of monoclonal, malignant CD4+8+ tumor cells that express a single TCR-β chain but diverse TCR-α chains, implying that pre-TCR signaling (the pre-TCR consisting of a TCR-β chain that is disulfide linked to the invariable pre-TCR-α chain) (34) makes a major contribution to malignancy, being absolutely essential for T-ALL generation in some (10) but not all studies (35, 36). In fact, Notch1 signaling and pre-TCR signaling do cooperate in normal T cell development to induce survival and proliferation (37), as they do cooperate in the initiation of ICN1-dependent lymphoma. Therefore, malignant transformation causing independent tumors in different mice must have taken place before the completion of TCR-α rearrangement because of the expression of a single TCR-β chain paired with diverse TCR-α chains in different cells from the same tumor. This fits well with the finding that TCR-αβ− CD4−8+ cells, i.e., immature cells that have undergone pre-TCR signaling are tumorigenic and exhibit a significantly higher proliferation than their normal counterparts (Fig. 1 d). Because the ICN1-overexpressing CD 4+8+ cells that appear early in blood are not tumorigenic, the data indicate that malignant transformation must occur in precursors of CD4+8+ cells and does take place within a few weeks after BMT with ICN1-overexpressing lineage-negative cells.
In contrast to the nonmalignant (nontumorigenic) CD4+8+ TCR-αβ+ cells, the malignant CD4+8+ TCR-αβ+ or CD4−8+ TCR-αβ+ cells derived from the immature CD4−8+ TCR-αβ− cells continued to grow, and each phenotypically distinct subset was able to independently cause tumors in nu/nu recipient mice with very similar kinetics; this is incompatible with the notion that the “early” CD4−8+ cells represent tumor stem cells that generate nonmalignant TCR-αβ+ progeny that constitutes the tumor mass. The tumorigenic potential was mirrored by the up-regulation of several oncogenes, most prominently the c-Myc oncogene that represents an ICN1 target, but also others, such as Akt and the oncogenes of the Ras family, the latter being considered targets of c-Myc. Furthermore, transformed cells exhibited reduced levels of cyclin-dependent kinase inhibitors p18 and p19, as well as elevated expression of cyclin-dependent kinase 4 (another c-Myc target) compared with their nonmalignant CD4+8+ counterparts. Increased c-Myc RNA levels were also reflected in increased levels of RNAs transcribed from genes involved in transcription and translation such as RNA polymerases, as well as initiation and elongation factors, which are likewise considered to represent targets of c-Myc. However, c-Myc overexpression by itself is not sufficient to cause monoclonal tumors (7, 8), and needs to be accompanied by further genetic changes to result in outgrowth of monoclonal tumors.
Apart from being an ICN1 target, c-Myc expression can be dramatically increased by chromosomal translocation (38). However, SKY analysis of independent tumors did not reveal any translocations, indicating that overexpression was dependent on transcriptional regulation of c-Myc by ICN1 (5, 6). Furthermore, analysis by high-resolution oligo array–based comparative genomic hybridization (array CGH) did not exhibit any tumor-specific amplifications or deletions, suggesting that genomic instability does not contribute to the generation of tumors initiated by ICN1 overexpression, in spite of the observation that c-Myc overexpression can be associated with genomic instability. Thus, mechanisms other than genomic instability must account for the observation that highly elevated levels of c-Myc in tumors did not result in stabilization of apoptosis-inducing p53. Normally this occurs via up-regulation of p19Arf, which then sequesters Mdm2 that can then no longer degrade p53. In fact, p53 levels in tumor cells were much reduced compared with those in the abnormal nonmalignant or normal CD4+8+ cells, as were levels of p19Arf both at the level of RNA and protein. Down-regulation of the p19Arf protein explains the lack of compensatory p53 up-regulation that could have prevented tumor outgrowth and probably did so in the nonmalignant CD4+8+ cells. However, it does not explain the up-regulation of Mdm2 and down-regulation of p53 at the transcriptional level. The reduced expression of p19Arf is at least in part dependent on its transcriptional repressor Bmi-1, which can be up-regulated by c-Myc (29) and was specifically up-regulated in T-ALL. Thus, the reduced levels of p19Arf protein in the absence of any mutation are best explained by Bmi-1 up-regulation. It is important to stress however, that the dysregulation of the c-myc-p53 axis in T-ALL does not only involve the usually suspected posttranscriptional mechanisms, but also results from the unexplained dysregulation at the transcriptional level of Mdm2 as well as p53.
Because of the monoclonal nature of the tumors that eventually develop, it is likely that another thus far unknown genetic event is required for malignant transformation that does not depend on genomic instability, as evident from the CGH data. The cooperating mutation(s) require further scrutiny, and could conceivably involve mutations caused by genetic instability. However, because the tumors develop relatively early, and we did not find mutation by sequencing of genes in the c-Myc-p53 axis, we consider it more likely that the cooperating event is caused by insertional mutagenesis of the retroviral vector itself. This hypothesis will be examined by determining the retroviral integration sites in tumors versus nontumorigenic ICN1-overexpressing cells.
MATERIALS AND METHODS
Mice.
BALB/c WT and BALB/c nu/nu mice were purchased from Taconic. C57BL/6 and Rag-2−/− γc−/− mice were purchased from The Jackson Laboratory. All mice were kept in specific pathogen–free animal facilities at the Dana-Farber Cancer Institute. All animal procedures were performed in compliance with the guidelines of the Dana-Farber Cancer Institute Animal Resources Facility, which operates under regulatory requirements of the U.S. Department of Agriculture and Association for Assessment and Accreditation of Laboratory Animal Care, and experimental protocols were approved by the Animal Care and Use Committee of the Dana-Farber Cancer Institute.
Tumor transplantation.
Nude mice were injected with various numbers of tumor cells (1 × 104 to 2 × 106) and tumor development was subsequently monitored.
Recombinant DNA constructs.
The intracellular domain of the murine Notch1 (ICN1) retroviral vector was created by cloning a PCR-amplified ICN1 full-length sequence into a MSCV-IRES-DsRed vector, Mutant E47 cDNA was cloned into a retroviral vector MSCV-IRES-GFP. The MSCV-IRES-DsRed vector and MSCV-IRES-GFP vectors were provided by D. Vignali (St. Jude Children's Research Hospital, Memphis, TN). Mutant E47 cDNA was provided by X.H. Sun (Oklahoma Medical Research Center, Oklahoma City, OK) and has been previously described (19). The murine ICN1 retroviral plasmid CMMP-ICN1-IRES-EGFP and its parent vector CMMP-IRES-EGFP were used in most infection experiments with ICN1. The retroviral construct encoding a truncated N-terminal fragment of mastermind-like 1 fused to EGFP (DN-MAML), and the control vector MigR1 was provided by J.C. Aster (Brigham & Women's Hospital, Boston, MA) and has been previously described (25, 39).
Retrovirus production and BM transplantation.
Retroviral supernatants were produced by transient transfection of 293T cells with the retroviral construct and appropriate packaging plasmids (provided by H. Cantor, Dana-Farber Cancer Institute, Boston, MA). Retroviral infection and BM reconstitution experiments were performed as previously described (20, 36, 40). In brief, lineage-negative (CD3e, TCR-β, NK1.1, DX5, CD19, Ter-119, Mac-1, and Gr-1) BM cells were sorted from BALB/c or CD4-Cre c-Mycflox/flox and C57BL/6 mice and cultured in the presence of Flt3L, SCF, IL-6, and IL-7 (all from R&D Systems). On day 2 and 4 after sorting, cells were subjected to retroviral transduction by centrifugation at 2,300 rpm for 1.5 h at room temperature with 8 μg/ml polybrene. On day 5, 1–2 × 106 cells were injected intravenously into irradiated (550 rad) 5–8-wk-old syngeneic BALB/c or RAG 2−/− γc−/− recipients.
Flow cytometry and cell sorting.
Monoclonal antibodies specific for CD4 (RM4-5, GK1.5), CD8 (53–6.7), TCR-β (H57-597), CD3e(145-2C11) Gr-1 (RB6-8C5), erythroid cell marker (Ter-119), CD19 (1D3), CD11b (M1/70), pan-NK (DX5), and NK1.1 (PK136) were purchased from BD and were used as biotin, FITC, PE, peridinin chlorophyll protein (PerCP), PerCP-Cy5.5, PE-Cy7, allophycocyanin (APC), or APC-Cy7 conjugates. Single-cell suspensions were prepared from peripheral blood, BM, spleen, lymph node, thymus, lung, and liver of ICN1-transplanted animals and related controls at the indicated time points. Cells were analyzed by FACSCalibur cytometer (BD) equipped with CellQuest Software (BD Biosciences) and FlowJo (Tree Star, Inc.) and were sorted by a FACSAria (BD) or a MoFlo cell sorter (Cytomation). Sorted cells were of >98% purity, as determined by postsort analysis.
BrdU labeling.
BrdU incorporation procedure was performed as previously described (13). In brief, mice were injected intraperitoneally with 1.8 μg freshly resuspended BrdU in water every day. Sorted cells from BrdU-treated mice were fixed, permeabilized, and stained intracellularly using the BrdU labeling kit from BD.
CFSE labeling.
Sorted CD4+8+ cells were incubated with 10 μM CFSE (Invitrogen) at 5 × 106 cells/ml PBS, 0.1% BSA for 10 min at 37°C. Cells were washed two times with RPMI 1640 medium to eliminate the remaining CFSE and injected intravenously into nude mice. Cell division of CFSE-labeled cells was analyzed by flow cytometry on day 2, 4, 6, and day 8 after injection.
SKY.
Metaphase chromosomes were prepared from tumor CD4+8+ cells. In brief, cells were treated with colcemid for 2 h, and then incubated in 0.075 M KCl for 20 min at 37°C. Chromosomes were subsequently fixed in 3:1 methanol/acetic acid. Metaphases were dropped on slides, dried overnight, and stained with DAPI before microscopic examination. SKY was done using the SkyPaint DNA kit for mouse samples (Applied Spectral Imaging) according to the manufacturer's instructions. Spectral images were captured using a microscope (E800; Nikon) equipped with ASI spectral cube, 60× objective, and analyzed using SKYView software from Applied Spectral Imaging. At last 20 metaphases were analyzed per sample.
Array CGH Profiling.
Genomic DNA from sorted cells was extracted using DNeasy Tissue kit (QIAGEN) and processed for hybridization by the Belfer Cancer Genomics Center, Dana-Farber Cancer Institute. The DNA was hybridized to 244 K Whole Mouse Genome Chip (G4122A; Agilent Technologies) according to the manufacturer's instructions. Tail DNA from the same mouse as reference. The median spatial resolution for this chip is 8.05 Kb; probes cover all known genes. Data were assembled and analyzed using CGH Analytics software (Agilent Technologies).
Southern blot analysis.
10 μg of genomic DNA was digested with EcoRI overnight at 37°C, separated on a 1.0% agarose gel, transferred onto nitrocellulose membrane. After UV cross-linking, the blot was hybridized with a p32-labeled probe from the Jβ2 region of the TCR-β gene.
Electrophoretic Mobility Shift Assays (EMSA).
Nuclear extracts were prepared from cells using nuclear and cytoplasmic extraction reagents (PIERCE). Protein concentration of each extract was measured by OD 595 nm. 5 μg of each nuclear extract was used in each EMSA. To generate probes for EMSAs, 5′ end biotin-labeled double-stranded oligonucleotides containing the binding sites for E2A and NF-κB were synthesized from Integrated DNA Technologies. The following oligonucleotides were used for EMSA: E2A E-box consensus sequences, CCCCAACACCTGCTGCCTGAG; NF-κB binding motif, AGTTGAGGGGACTTTCCCAGGC. The probes were purified with PAGE. EMSA was performed using LightShift chemiluminescent EMSA kit (Thermo Fisher Scientific), following the manufacturer's instructions.
Immunoprecipitation and Western blot.
Cells were lysed in radioimmunoprecipitation assay (RIPA) buffer supplemented with Protease Inhibitor Cocktail (Sigma-Aldrich). A 1,000-μg aliquot of total cell lysates was incubated with 10 μg of specific antibodies or control IgG for 2 h or overnight at 4°C. A 40-μl aliquot of 50% protein A–agarose slurry was added and incubated for another hour, followed by several washes with 0.1% NP-40 in PBS. The precipitates bound to the beads were analyzed by immunoblotting with Ubiquitin antibody (Santa Cruz Biotechnology, Inc.).
For Western blot analysis, cells were lysed in RIPA buffer supplemented with Protease Inhibitor Cocktail (Sigma-Aldrich). Lysates (40 μg) were separated on 4–20% SDS-PAGE and transferred onto PVDF membrane (Millipore). Nonspecific binding was blocked by incubation in blocking buffer (5% nonfat milk in TBST), followed by incubation with the primary antibodies and the appropriate horseradish peroxidase–conjugated secondary antibodies diluted in blocking buffer. Immunoreactive proteins were detected using enhanced chemiluminescence (ECL) reagents (GE Healthcare). The E2A, Mdm2 antibodies were purchased from BD. ERK1/2, phosph-ERK1/2 antibodies, and MEK1/2-specific inhibitor PD98059 were purchased from Cell Signaling Technology. Cyclin D3 antibody was obtained from Santa Cruz Biotechnology. Cyclin D1 antibody was purchased from NeoMarker, and the p19Arf and c-Myc antibodies were purchased from Abcam. p53 antibody was provided by R. Depinho (Dana-Farber Cancer Institute, Boston, MA).
Quantitative RT-PCR.
cDNA was prepared by oligo-dT primer using the Superscript II Reverse Transcription kit (Invitrogen). Quantitative RT-PCR was performed using an ABI7300 machine (Applied Biosystems). c-Myc, E47, Bmi-1, p19Arf, Mdm2, and p53 RNA was determined relative to GAPDH expression using the TaqMan Gene Expression Assay (Applied Biosystems). All quantitative RT-PCR reactions were done in triplicate in 20 μl volume.
Microarray analysis.
Sorted cells were homogenized for RNA isolation using the TRIZOL reagent (Invitrogen) and purified using QIAGEN RNeasy minicolumns. RNA samples were processed for hybridization on the Mouse Expression-Array-430.2.0 Genechips (Affymetrix) by the Microarray Core Facility of the Dana-Farber Cancer Institute. Data were loaded onto the DNA-Chip Analyzer (dChip; http://www.dchip.org) program for normalization and quantification. Normalization was performed using the default settings of the dChip software, genes were selected for expression differences higher than 1.5-fold by Student's t test (P < 0.05) or for significant changes (P < 0.05). The microarray data have been deposited in the GEO database under accession no. GSE12948.
Online supplemental material.
Fig. S1 shows global gene expression profiling in control, abnormal nonmalignant cells, and tumor cells. Table S1 provides tumorigenicity assays in nu/nu homozygous mice. The online version of this article is available at http://www.jem.org/cgi/content/full/jem.20081561/DC1.
Supplementary Material
Acknowledgments
The authors wish to thank Dr. Scott J. Rodig (Brigham & Women's Hospital) for histopathological analysis, Xiaoyan Li for technical assistance with BMT, and Rui Chang and Marei Dose for their help with the work of CD4-Cre c-MycFL/FL mice. We would also like to thank Frances Lundblad and Linnea Benson for editorial help.
This work was supported by National Institutes of Health (NIH) grants P01 CA109901 and R01 AI45846. X. Li is supported by the Lymphoma Research Foundation, Lee Fellowship, and NIH training grant T32-CA70083.
The authors have no conflicting financial interests.
Abbreviations used: array CGH, array-based comparative genomic hybridization; BMT, BM transfer; DAPK, death-associated protein kinase; DNMAML1, dominant-negative mastermind-like 1; ICN, intracellular Notch; SKY, spectral karyotype; T-ALL, T cell acute lymphoblastic leukemia.
References
- 1.Artavanis-Tsakonas, S., M.D. Rand, and R.J. Lake. 1999. Notch Signaling: Cell fate control and signal integration in development. Science. 284:770–776. [DOI] [PubMed] [Google Scholar]
- 2.O'Neil, J., J. Grim, P. Strack, S. Rao, D. Tibbitts, C. Winter, J. Hardwick, M. Welcker, J.P. Meijerink, R. Pieters, et al. 2007. FBW7 mutations in leukemic cells mediate NOTCH pathway activation and resistance to gamma-secretase inhibitors. J. Exp. Med. 204:1813–1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thompson, B.J., S. Buonamici, M.L. Sulis, T. Palomero, T. Vilimas, G. Basso, A. Ferrando, and I. Aifantis. 2007. The SCFFBW7 ubiquitin ligase complex as a tumor suppressor in T cell leukemia. J. Exp. Med. 204:1825–1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weng, A.P., A.A. Ferrando, W. Lee, J.P.T. Morris, L.B. Silverman, C. Sanchez-Irizarry, S.C. Blacklow, A.T. Look, and J.C. Aster. 2004. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 306:269–271. [DOI] [PubMed] [Google Scholar]
- 5.Palomero, T., W.K. Lim, D.T. Odom, M.L. Sulis, P.J. Real, A. Margolin, K.C. Barnes, J. O'Neil, D. Neuberg, A.P. Weng, et al. 2006. NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc. Natl. Acad. Sci. USA. 103:18261–18266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weng, A.P., J.M. Millholland, Y. Yashiro-Ohtani, M.L. Arcangeli, A. Lau, C. Wai, C. del Bianco, C.G. Rodriguez, H. Sai, J. Tobias, et al. 2006. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev. 20:2096–2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Felsher, D.W., and J.M. Bishop. 1999. Reversible tumorigenesis by MYC in hematopoietic lineages. Mol. Cell. 4:199–207. [DOI] [PubMed] [Google Scholar]
- 8.Girard, L., Z. Hanna, N. Beaulieu, C.D. Hoemann, C. Simard, C.A. Kozak, and P. Jolicoeur. 1996. Frequent provirus insertional mutagenesis of Notch1 in thymomas of MMTVD/myc transgenic mice suggests a collaboration of c-myc and Notch1 for oncogenesis. Genes Dev. 10:1930–1944. [DOI] [PubMed] [Google Scholar]
- 9.Ikawa, T., H. Kawamoto, A.W. Goldrath, and C. Murre. 2006. E proteins and Notch signaling cooperate to promote T cell lineage specification and commitment. J. Exp. Med. 203:1329–1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pear, W.S., and J.C. Aster. 2004. T cell acute lymphoblastic leukemia/lymphoma: a human cancer commonly associated with aberrant NOTCH1 signaling. Curr. Opin. Hematol. 11:426–433. [DOI] [PubMed] [Google Scholar]
- 11.Raulet, D.H., R.D. Garman, H. Saito, and S. Tonegawa. 1985. Developmental regulation of T-cell receptor gene expression. Nature. 314:103–107. [DOI] [PubMed] [Google Scholar]
- 12.Snodgrass, H.R., Z. Dembic, M. Steinmetz, and H. von Boehmer. 1985. Expression of T-cell antigen receptor genes during fetal development in the thymus. Nature. 315:232–233. [DOI] [PubMed] [Google Scholar]
- 13.Huesmann, M., B. Scott, P. Kisielow, and H. von Boehmer. 1991. Kinetics and efficacy of positive selection in the thymus of normal and T cell receptor transgenic mice. Cell. 66:533–540. [DOI] [PubMed] [Google Scholar]
- 14.Fehling, H.J., A. Krotkova, C. Saint-Ruf, and H. von Boehmer. 1995. Crucial Role of the pre-T-cell receptor alpha gene in development of alpha beta but not gamma delta T cells. Nature. 375:795–798. [DOI] [PubMed] [Google Scholar]
- 15.Matthias, P., and A.G. Rolink. 2005. Transcriptional networks in developing and mature B cells. Nat. Rev. Immunol. 5:497–508. [DOI] [PubMed] [Google Scholar]
- 16.Bain, G., I. Engel, E.C. Robanus Maandag, H.P. te Riele, J.R. Voland, L.L. Sharp, J. Chun, B. Huey, D. Pinkel, and C. Murre. 1997. E2A deficiency leads to abnormalities in alphabeta T-cell development and to rapid development of T-cell lymphomas. Mol. Cell. Biol. 17:4782–4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Engel, I., and C. Murre. 2002. Disruption of pre-TCR expression accelerates lymphomagenesis in E2A-deficient mice. Proc. Natl. Acad. Sci. USA. 99:11322–11327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Talora, C., A.F. Campese, D. Bellavia, M. Pascucci, S. Checquolo, M. Groppioni, L. Frati, H. von Boehmer, A. Gulino, and I. Screpanti. 2003. Pre-TCR triggered ERK signaling-dependent downregulation of E2A activity in Notch3-induced T-cell lymphoma. EMBO Rep. 4:1067–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nie, L., M. Xu, A. Vladimirova, and X.H. Sun. 2003. Notch-induced E2A ubiquitination and degradation are controlled by MAP kinase activities. EMBO J. 22:5780–5792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sicinska, E., I. Aifantis, L. Le Cam, W. Swat, C. Borowski, Q. Yu, A.A. Ferrando, S.D. Levin, Y. Geng, H. von Boehmer, and P. Sicinski. 2003. Requirement for cyclin D3 in lymphocyte development and T-cell leukemias. Cancer Cell. 4:451–461. [DOI] [PubMed] [Google Scholar]
- 21.Raval, A., S.M. Tanner, J.C. Byrd, E.B. Angerman, J.D. Perko, S.S. Chen, B. Hackanson, M.R. Grever, D.M. Lucas, J.J. Matkovic, et al. 2007. Downregulation of death-associated protein kinase 1 (DAPK1) in chronic lymphocytic leukemia. Cell. 129:879–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Menssen, A., and H. Hermeking. 2002. Characterization of the c-MYC-regulated transcriptome by SAGE: identification and analysis of c-MYC target genes. Proc. Natl. Acad. Sci. USA. 99:6274–6279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Alboran, I.M., R.C. O'Hagan, F. Gartner, B. Malynn, L. Davidson, R. Rickert, K. Rajewsky, R.A. DePinho, and F.W. Alt. 2001. Analysis of C-MYC function in normal cells via conditional gene-targeted mutation. Immunity. 14:45–55. [DOI] [PubMed] [Google Scholar]
- 24.Wolfer, A., T. Bakker, A. Wilson, M. Nicolas, V. Ioannidis, D.R. Littman, P.P. Lee, C.B. Wilson, W. Held, H.R. MacDonald, and F. Radtke. 2001. Inactivation of Notch 1 in immature thymocytes does not perturb CD4 or CD8T cell development. Nat. Immunol. 2:235–241. [DOI] [PubMed] [Google Scholar]
- 25.Weng, A.P., Y. Nam, M.S. Wolfe, W.S. Pear, J.D. Griffin, S.C. Blacklow, and J.C. Aster. 2003. Growth suppression of pre-T acute lymphoblastic leukemia cells by inhibition of notch signaling. Mol. Cell. Biol. 23:655–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adhikary, S., and M. Eilers. 2005. Transcriptional regulation and transformation by Myc proteins. Nat. Rev. Mol. Cell Biol. 6:635–645. [DOI] [PubMed] [Google Scholar]
- 27.Bahram, F., N. von der Lehr, C. Cetinkaya, and L.G. Larsson. 2000. c-Myc hot spot mutations in lymphomas result in inefficient ubiquitination and decreased proteasome-mediated turnover. Blood. 95:2104–2110. [PubMed] [Google Scholar]
- 28.Eischen, C.M., J.D. Weber, M.F. Roussel, C.J. Sherr, and J.L. Cleveland. 1999. Disruption of the ARF-Mdm2-p53 tumor suppressor pathway in Myc-induced lymphomagenesis. Genes Dev. 13:2658–2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jacobs, J.J., B. Scheijen, J.W. Voncken, K. Kieboom, A. Berns, and M. van Lohuizen. 1999. Bmi-1 collaborates with c-Myc in tumorigenesis by inhibiting c-Myc-induced apoptosis via INK4a/ARF. Genes Dev. 13:2678–2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maser, R.S., B. Choudhury, P.J. Campbell, B. Feng, K.K. Wong, A. Protopopov, J. O'Neil, A. Gutierrez, E. Ivanova, I. Perna, et al. 2007. Chromosomally unstable mouse tumours have genomic alterations similar to diverse human cancers. Nature. 447:966–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karlsson, A., S. Giuriato, F. Tang, J. Fung-Weier, G. Levan, and D.W. Felsher. 2003. Genomically complex lymphomas undergo sustained tumor regression upon MYC inactivation unless they acquire novel chromosomal translocations. Blood. 101:2797–2803. [DOI] [PubMed] [Google Scholar]
- 32.Ferrando, A.A., D.S. Neuberg, J. Staunton, M.L. Loh, C. Huard, S.C. Raimondi, F.G. Behm, C.H. Pui, J.R. Downing, D.G. Gilliland, et al. 2002. Gene expression signatures define novel oncogenic pathways in T cell acute lymphoblastic leukemia. Cancer Cell. 1:75–87. [DOI] [PubMed] [Google Scholar]
- 33.Yeoh, E.J., M.E. Ross, S.A. Shurtleff, W.K. Williams, D. Patel, R. Mahfouz, F.G. Behm, S.C. Raimondi, M.V. Relling, A. Patel, et al. 2002. Classification, subtype discovery, and prediction of outcome in pediatric acute lymphoblastic leukemia by gene expression profiling. Cancer Cell. 1:133–143. [DOI] [PubMed] [Google Scholar]
- 34.von Boehmer, H., and H.J. Fehling. 1997. Structure and function of the pre-T cell receptor. Annu. Rev. Immunol. 15:433–452. [DOI] [PubMed] [Google Scholar]
- 35.Bellavia, D., A.F. Campese, S. Checquolo, A. Balestri, A. Biondi, G. Cazzaniga, U. Lendahl, H.J. Fehling, A.C. Hayday, L. Frati, et al. 2002. Combined expression of pTalpha and Notch3 in T cell leukemia identifies the requirement of preTCR for leukemogenesis. Proc. Natl. Acad. Sci. USA. 99:3788–3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Campese, A.F., A.I. Garbe, A.F. Zhang, F. Grassi, I. Screpanti, and H. von Boehmer. 2006. Notch1-dependent lymphomagenesis is assisted by but does not essentially require pre-TCR signaling. Blood. 108:305–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ciofani, M., and J.C. Zuniga-Pflucker. 2005. Notch promotes survival of pre-T cells at the beta-selection checkpoint by regulating cellular metabolism. Nat. Immunol. 6:881–888. [DOI] [PubMed] [Google Scholar]
- 38.Zhu, C., K.D. Mills, D.O. Ferguson, C. Lee, J. Manis, J. Fleming, Y. Gao, C.C. Morton, and F.W. Alt. 2002. Unrepaired DNA breaks in p53-deficient cells lead to oncogenic gene amplification subsequent to translocations. Cell. 109:811–821. [DOI] [PubMed] [Google Scholar]
- 39.Maillard, I., A.P. Weng, A.C. Carpenter, C.G. Rodriguez, H. Sai, L. Xu, D. Allman, J.C. Aster, and W.S. Pear. 2004. Mastermind critically regulates Notch-mediated lymphoid cell fate decisions. Blood. 104:1696–1702. [DOI] [PubMed] [Google Scholar]
- 40.Aifantis, I., C. Borowski, F. Gounari, H.D. Lacorazza, J. Nikolich-Zugich, and H. von Boehmer. 2002. A critical role for the cytoplasmic tail of pTalpha in T lymphocyte development. Nat. Immunol. 3:483–488. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.