SUMMARY
A hallmark of morphogenesis is the orchestrated assembly of complex, supramolecular structures. One such structure is the proteineous coat that surrounds spores of the bacterium Bacillus subtilis. The coat is a multilayered shell that is composed of over fifty proteins. These proteins assemble around a basement layer composed of the morphogenetic protein SpoIVA. We show that SpoIVA harbors a Walker A box that is required for the proper deployment of the protein to the surface of the developing spore and proper assembly of the entire coat. We further show that purified SpoIVA both binds and hydrolyzes ATP and that the protein self-assembles into cable-like structures in a manner that depends on ATP hydrolysis. Self-assembly driven by ATP is an unusual mechanism for the construction of a large cellular structure.
Keywords: spore coat, subcellular localization, sporulation
INTRODUCTION
A culminating feature of development is the formation of complex supramolecular structures that define the endpoints of morphogenesis. Examples are bone, eggshells, flagella, teeth and virions (Hino et al., 2004; Waring, 2000; Macnab, 2003; Paine et al., 2001; Duda et al., 1995). A fundamental challenge in the field of development is to elucidate the mechanisms by which such structures are formed. An attractive model system for tackling this problem is the proteinaceous coat that surrounds spores of the Gram-positive bacterium Bacillus subtilis. The spores of B. subtlis and its relatives are perhaps the most durable cell types in nature, being capable of surviving extremes of time and environment (Setlow, 2006). The coat is a complex, multilayered assemblage of over fifty proteins that encases the spore and helps to protect it from noxious environmental agents (Driks, 2002; Henriques and Moran Jr., 2006). Here we address the question of how at the start of morphogenesis the innermost or basement layer of the coat is assembled as a shell around the developing spore.
Spores are produced in a sporangium that consists of a cell-within-a-cell. The inner cell is the forespore (or prespore) and it is destined to become the mature spore when development is complete. The outer cell is known as the mother cell, and it nurtures the forespore. When development is complete, the mother cell lyses, liberating the mature and now dormant spore (Losick et al., 1986; Stragier and Losick, 1996). Proteins that constitute the coat are produced in the mother cell and deposited around the outer membrane surface of the forespore.
The spore coat is built atop two morphogenetic proteins, SpoIVA (henceforth simply IVA) (Piggot and Coote, 1976; Roels et al., 1992; Driks et al., 1994), the subject of this investigation, and a small, amphipathic alpha helical peptide called SpoVM (hereafter VM) (Levin et al., 1993; Prajapati et al., 2000; Ramamurthi et al., 2006). VM is embedded in the outer membrane of the forespore and exposes its largely hydrophilic face to the mother cell cytosol, where it contacts the C-terminus of IVA and tethers it to the surface of the forespore (Ramamurthi et al., 2006). IVA, in turn, assembles into the basement layer of the coat and is directly or indirectly responsible for the subsequent recruitment of the myriad protein components of the coat (Kim et al., 2006).
Here we investigate how IVA assembles into a shell-like structure at the base of the coat. We report that IVA is an ATPase and that it self-assembles irreversibly into higher order structures in a process that is driven by ATP hydrolysis. We propose that energy from ATP hydrolysis is used to create a durable and stable platform upon which morphogenesis of the coat can take place.
RESULTS
The Walker A motif of IVA is required for efficient sporulation
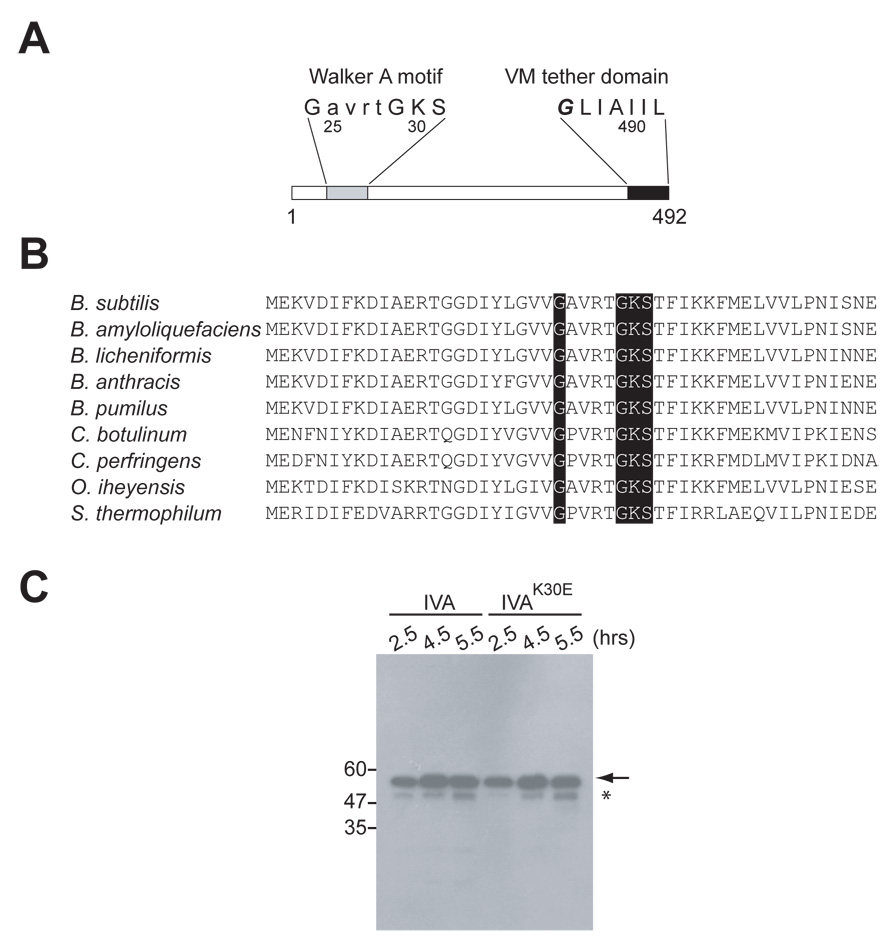
An alignment search (BLAST) revealed that IVA resembles nucleotide-binding proteins. Subsequent scanning of the amino acid sequence revealed a highly conserved sequence conforming to a Walker A nucleotide-binding motif located between residues 24 and 31 (Fig. 1A and 1B). A canonical Walker A box consists of two glycine residues separated by four amino acids, followed by a lysine and then either threonine or serine. The lysine is typically implicated in binding the γ- phosphoryl group of nucleoside triphosphates (typically ATP or GTP) (Walker et al., 1982).
Figure 1. IVA harbors a canonical Walker A box.
(A) Schematic representation of the primary structure of IVA. Residues 24 – 30 (gray) conform to a Walker A nucleotide binding motif. The seven C-terminal residues (black) target and anchor IVA to the surface of the forespore via a direct interaction with the peptide VM.
(B) Conservation of the Walker A box among IVA orthologs. N-terminal amino acid residues of IVA from Bacillus subtilis, B. amyloliquefaciens, B. licheniformis, B. anthracis, B. pumulis, Clostridium botulinum, C. perfringens, Oceanobacillus iheyensis, and Symbiobacterium thermophilum. Conserved residues of the GXXXXGKS Walker A box are shaded in gray.
(C) Immunoblot analysis of cell extracts taken at various times (hrs.) after the induction of sporulation using antibodies to IVA. Analyzed were extracts from wild type cells (KR394) and cells producing IVAK30E (KR438). The mobility of molecular weight markers (kD) is shown on the left. The arrow indicates full length IVA or IVAK30E. Accumulation of a truncated IVA species is indicated by the asterisk.
To investigate the significance of the apparent Walker A box, we converted codon 30, which specifies the lysine residue of the box, to an alanine or glutamic acid codon, and measured the sporulation efficiency of strains harboring the mutant alleles. Cells harboring a deletion in IVA were unable to sporulate (less than 1 in 108 cells; Table S1, strain A and B), but regain the capacity to sporulate efficiently when a wild type copy of IVA is inserted into the chromosome at the thr locus (thr::IVA). However, complementation of the IVA null mutation with thr::IVAK30A resulted in a 20-fold reduction in sporulation efficiency (Table S1, strains C–D) and an almost complete block in spore formation with thr::IVAK30E (sporulation efficiency of 1.1 × 10−8; Table S1, strain E). Complementation analysis with strains harboring both wild type IVA at the native locus and the mutant alleles at thr revealed that IVAK30A and IVAK30E were recessive to IVAwt (Table S1, strains G–H; compare to strains D–E).
Conceivably, the severe sporulation block caused by the replacement of Lys30 with Glu was simply due to loss of the mutant protein due to proteolysis. To investigate this possibility, we carried out immunoblot analysis using antibodies to IVA. In cells expressing wild type IVA, IVA levels gradually increased over time (Fig. 1C, arrow). At late times, a band of increased mobility appeared that reacted with the anti-IVA antiserum (Fig. 1C, asterisk). This band corresponded to a truncated IVA species that is the product of hydrolysis by the protease YabG (Takamatsu et al., 2000). In comparison, cells expressing IVAK30E accumulated both the full length product of the mutant gene as well as the truncated species. Moreover, no additional degradation product of IVA was evident. Taken together, it appears replacement of Lys30 abolishes the activity, but not the stability, of IVA, and dramatically reduces the ability of cells harboring the substitution to sporulate.
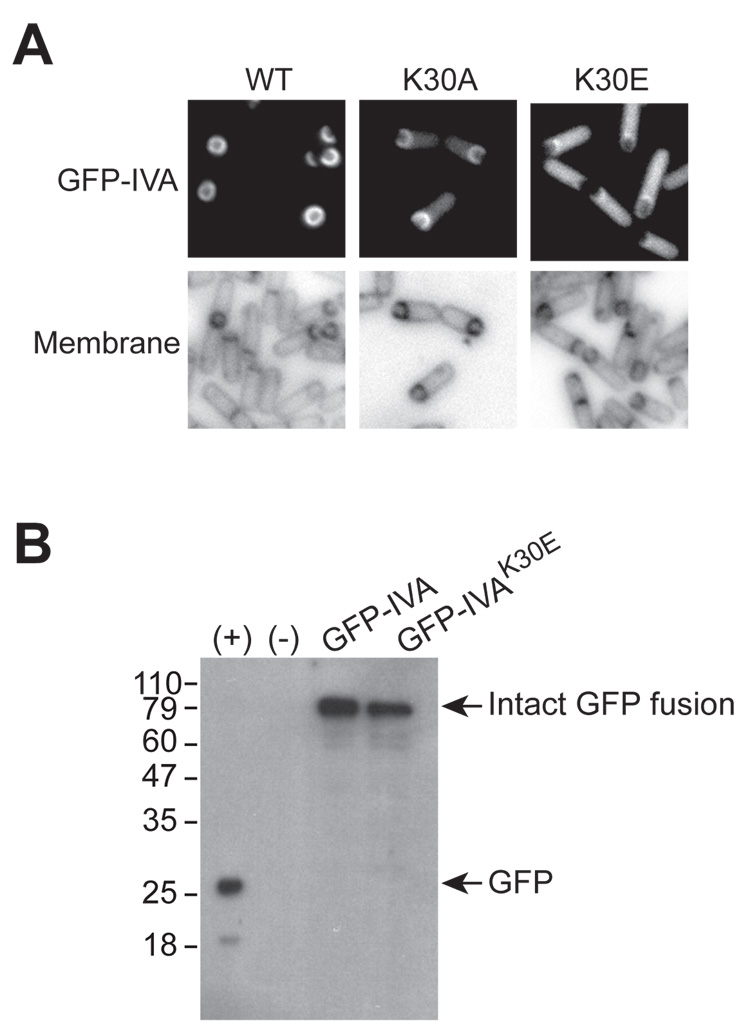
Proper subcellular localization of IVA depends on the Walker A motif
Formation of the spore coat is dependent on the proper deployment of IVA to the surface of the forespore. In light of the contribution of the IVA Walker A box to sporulation, we wondered if disruption of this motif would cause mislocalization of IVA. To test this, we employed a previously described fusion of the Green Fluorescent Protein to the N-terminus of IVA and measured the subcellular distribution of this fusion protein in cells that harbored this construct. As reported previously, GFP-IVA colocalized with the outer forespore membrane, consistent with its role in forming the basement layer of the spore coat (Fig. 2A, left; Price and Losick, 1999). Disruption of the Walker A motif by substituting Lys30 with Ala resulted in increased levels of fluorescence in the cytosol. Nevertheless, GFP-IVAK30A was largely enriched at the forespore periphery (Figure 2A, middle). Substitution of Lys30 with Glu, however, led to the dramatic mislocalization of the fluorescent signal, as GFP-IVAK30E was found principally in the mother cell cytosol. Thus, the severity of the effects of IVAK30A or IVAK30E on sporulation (the latter having the more severe effect) roughly correlated with the severity of the effects of each substitution on the localization the GFP-tagged proteins. To ensure that the aberrant localization of GFP-IVAK30E was not simply due to degradation of the protein and liberation of free GFP, we examined the stability of GFP-IVA and GFP-IVAK30E by immunoblotting. Extracts of cells expressing gfp-IVA or gfp-IVAK30E were separated and probed using anti-GFP antibodies (Fig. 2B). Both GFP-IVA and GFP-IVAK30E accumulated to similar levels, and migrated similarly. Furthermore, no additional degradation was evident (Fig. 2B, third and fourth lanes). Thus, the diffuse localization of GFP-IVAK30E was not due to instability of the protein.
Figure 2. Disruption of the Walker A box results in mislocalization of GFP-IVA.
(A) Top panels: localization of GFP-IVA (strain KR160, left), GFP-IVAK30A (KR391, middle), and GFP-IVAK30E (KR446, right) 2.5 hours after the induction of sporulation. Bottom panels: membranes of cells in the corresponding panels above visualized with the dye FM4-64.
(B) Immunoblot analysis of extracts from cells producing GFP (first lane, +, strain MF339), no GFP (second lane, -, PY79), GFP-IVA (third lane, KR160), or GFP-IVAK30E (fourth lane, KR446) using antibodies to GFP. The mobility of molecular weight markers (kD) is shown on the left, and mobility of free GFP and intact IVA-GFP fusions are indicated on the right (arrows).
In earlier work we showed that IVA is anchored to the outer surface of the forespore through an interaction near its C-terminus with the membrane-bound peptide VM (Ramamurthi et al., 2006). When this interaction is blocked, IVA misassembles into a large structure near the forespore, rather than encircling the forespore. In sharp contrast, the K30E substitution, as we have seen, caused IVA to become uniformly dispersed in the mother cell cytoplasm. These observations led us to hypothesize that the Walker box mediates polymerization of IVA into the basement layer of the coat whereas the IVA-VM interaction is responsible for targeting IVA to the outer surface of the forespore.
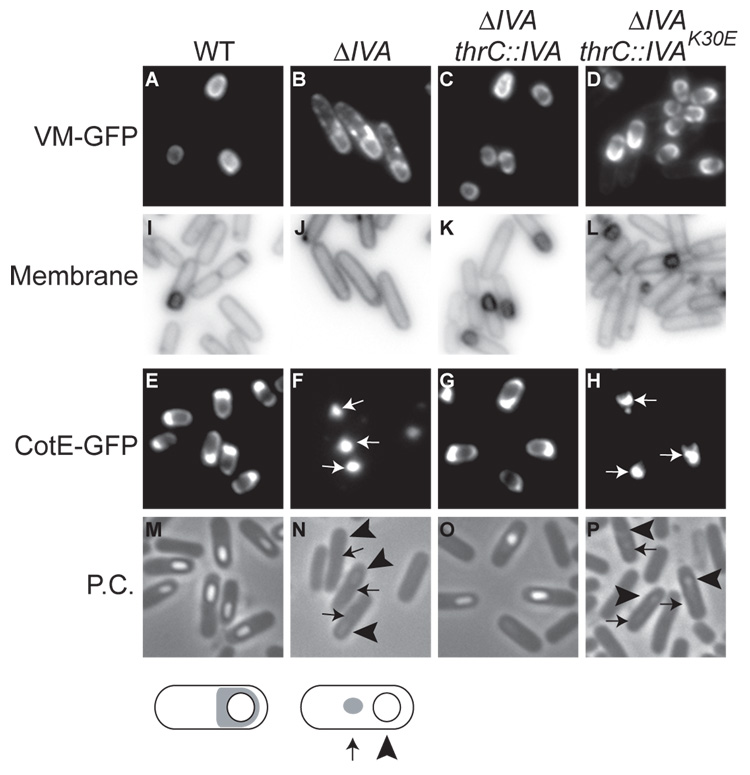
IVAK30E misassembles the spore coat
Given the potential involvement of the Walker A motif in the polymerization of IVA, we wished to test the effect that disrupting this motif has on the assembly of the spore coat. We know from earlier work that not only does the proper localization of IVA depend on VM (Price and Losick, 1999) but the localization of VM depends on IVA. Thus, in wild type cells, VM-GFP localized to the periphery of the forespore (van Ooij and Losick, 2003; Fig. 3A), whereas in the absence of IVA, VM-GFP, although enriched at the outer forespore membrane, mislocalized to patches of cytoplasmic membrane surrounding the mother cell (Ramamurthi et al., 2006; Fig. 3B). This mislocalization was corrected not only by a wild type copy of IVA that had been inserted at the thr locus, but also by a copy of IVAK30E (Fig 3 C–D). Thus, the IVA Walker A box is not needed for proper subcellular localization of VM-GFP.
Figure 3. IVAK30E mis-localizes CotE but not VM.
(A–D) VM-GFP localization 2.5 hours after the induction of sporulation (A) in the wild type strain (CVO1195), (B) in a IVA deletion mutant (KR119), (C) in a IVA deletion mutant harboring a complementing copy of IVA at the thr locus (KR219), and (D) in a IVA deletion mutant harboring IVAK30E at the thr locus (KR444). (I–L) Membranes of cells in (A) – (D) visualized with the dye FM4-64.
(E–H) CotE-GFP localization 4 hours after the induction of sporulation (E) in the wild type (CW271), (F) in a IVA deletion mutant (KR455), (G) in a IVA deletion mutant harboring a complementing copy of IVA at the thr locus (KR448), and (H) in a IVA deletion mutant harboring IVAK30E at the thr locus (KR449).
(M–P) Phase contrast images of cells in (E–H). Note the characteristic phase-bright forespores in (M) and (O) in properly sporulating cells.
Below, schematic representations of CotE-GFP distribution (gray) in wild type cells (left) show the fluorescent signal encircling the forespore, whereas in a IVA deletion mutant (right), CotE-GFP is seen as a spot in the mother cell cytosol.
Next, we examined the localization of CotE, which is recruited by IVA to the assembling spore coat and is in turn required for the recruitment of proteins that comprise the outer layers of the coat (Zheng et al., 1988). As reported previously, in wild type cells, CotE-GFP completely encircled the forespore, with a biased accumulation on the mother cell-proximal face of the forespore (Webb et al., 1995; Fig. 3E). In the absence of IVA, CotE-GFP mislocalized as a focus in the mother cell cytosol (Webb et al., 1995; Fig. 3F; Compare location of focus (arrows) to forespore (arrowheads)). While this mislocalization was corrected by wild type IVA, the IVAK30E mutant gene failed to do so (Fig. 3G–H). Taken together, we conclude that the polymerization defect of IVA resulting from the disruption of the Walker A box impairs the assembly of IVA-dependent outer layers of the spore coat, but not the innermost layer comprised of VM.
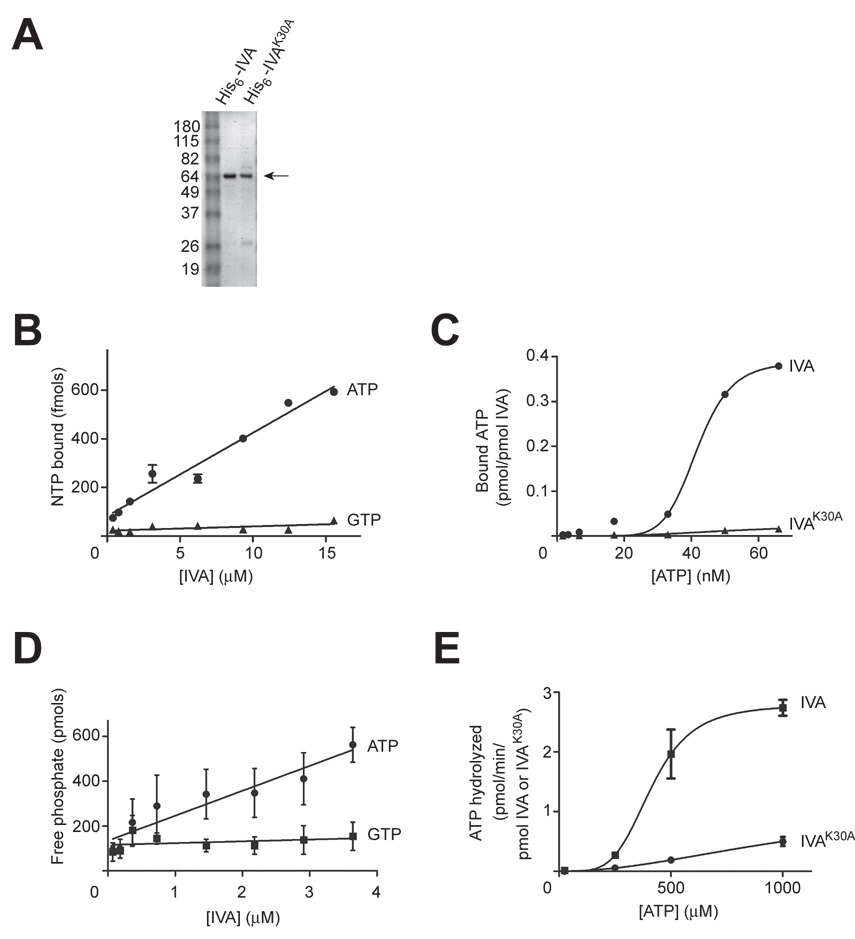
IVA binds and hydrolyzes ATP
We next investigated whether, in fact, IVA is a nucleotide-binding protein. First, we produced in E. coli and then purified either IVA or IVAK30A harboring a six-histidinyl affinity tag appended to the N-terminus (Fig. 4A; the K30E variant when overproduced in E. coli was largely insoluble. Therefore, the biochemical analysis was performed using IVAK30A). We then employed an assay for nucleotide binding that exploits the capacity of a nitrocellulose membrane to bind quantitatively protein, but not free nucleotides, when a mixture of protein and nucleotides is rapidly filtered through the membrane (Allen and Parsons, 1979; Carey et al., 1983). In this assay, radioactively-labeled nucleotides are retained on the membrane during filtration if they are bound to protein. Incubation of radioactively labeled ATP or GTP with increasing concentrations of purified IVA resulted in the concentration-dependent retention of ATP, but not GTP, on the filter (Fig. 4B). Thus, IVA appears to bind specifically ATP. We then measured the affinity of purified IVA or IVAK30A for ATP. With increasing concentrations of ATP, the fraction of nucleotide bound to IVA increased until it reached saturation at 70 nM (Fig. 4C). Analysis of the data by non-linear regression revealed that half-maximal binding was achieved at 42 nM, indicating that IVA has a relatively strong affinity for ATP. The Bmax for this curve was approximately 0.4 pmol ATP bound/pmol IVA, suggesting that each IVA molecule did not bind more than one molecule of ATP. In contrast, binding of ATP by IVAK30A at the highest concentration of ATP tested was reduced over 23-fold. Thus, efficient binding of ATP by IVA requires an intact Walker A box.
Figure 4. IVA binds and hydrolyzes ATP.
(A) Coomassie-stained polyacrylamide gel of purified His6-IVA and His6-IVAK30A used for ATP binding and hydrolysis studies. The arrow to the right indicates purified protein. The molecular weights (kD) corresponding to the size markers in lane 1 are indicated to the left.
(B) Concentration-dependent binding of IVA to radiolabeled ATP (circles), but not GTP (triangles), using a nitrocellulose filter binding assay.
(C) Saturation binding curve for IVA-ATP interaction. Varying concentrations of radiolabeled ATP were incubated with IVA (circles) or IVAK30A (triangles) and complexes were isolated using a nitrocellulose filter binding assay.
(D) Concentration-dependent hydrolysis of ATP by IVA (circles), but not GTP (squares) using the malachite green assay to measure liberated phosphate.
(E) Kinetic analysis of ATPase activity by IVA. Varying concentrations of ATP were incubated with IVA (squares) or IVAK30A (circles) and ADP production was measured.
Values are means ± SEM (n = 3).
Next, we wished to determine if IVA subsequently hydrolyzes the bound ATP. ATPase activity was determined by measuring liberated inorganic phosphate, utilizing a colorimetric assay which relies on the complex formation of the dye malachite green with phosphomolybdate. At 37°C, incubation of ATP with IVA resulted in the production of free phosphate in a concentration-dependent manner (Fig. 4D). In contrast, incubation of purified IVA with GTP did not result in the accumulation of phosphate, indicating that IVA specifically hydrolyzes ATP. We next investigated the kinetic properties of this ATPase activity by employing a more sensitive assay in which IVA was incubated with radiolabeled ATP (α-32P) and production of ADP was directly measured by separation of nucleotides by thin layer chromatography. Using a concentration of IVA in the linear region of the plot in Fig. 4D and varying the concentration of ATP produced the saturation curve shown in Fig. 4E. The hydrolysis of ATP reached saturation at approximately 1 mM. Analysis of the data by non-linear regression revealed a Km of 412 µM and a turnover rate of 2.7 pmols/min/pmol IVA at Vmax. Since the intracellular concentration of ATP is about 2–4 mM (Liberek et al., 1991), IVA can be expected to work efficiently in vivo despite its high Km value. However, its low turnover rate suggests that IVA is a slow enzyme. Incubation of ATP with IVAK30A, in contrast, led to the diminished accumulation of ADP, indicating that an intact Walker A box is required for the efficient hydrolysis of ATP by IVA. Taken together, we conclude that IVA harbors a functional Walker A box that is required for the specific binding and hydrolysis of ATP.
IVA multimerizes in vivo
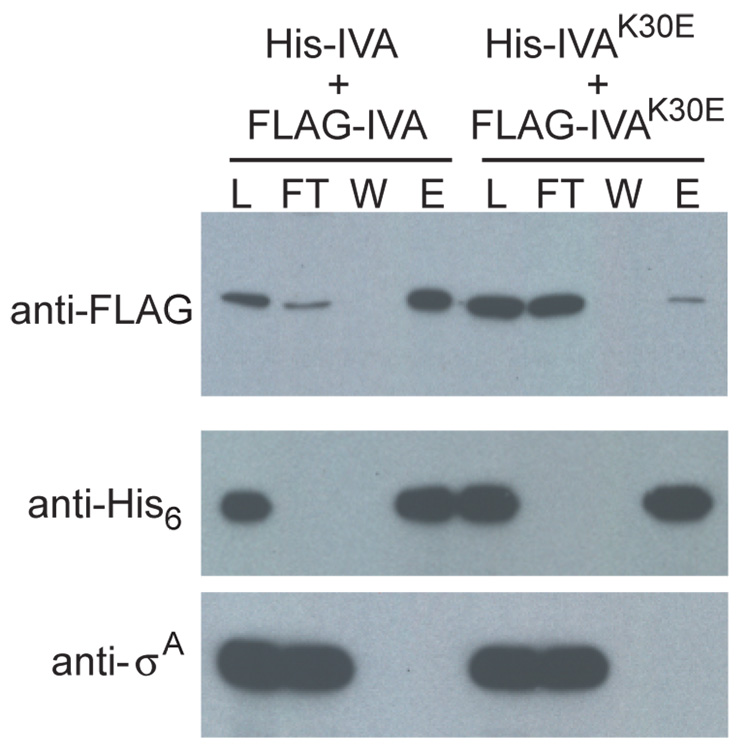
An appealing possibility raised by these results is that ATP-hydrolysis drives interaction between IVA molecules form the basement layer of the coat. As a first step in investigating this idea, we engineered cells that produced IVA with an N-terminal His6 affinity tag (His-IVA) and IVA with an N-terminal FLAG epitope (FLAG-IVA). Cells producing both proteins were induced to sporulate and cell extract was prepared 2.5 hours after the onset of sporulation. His-IVA was then purified by affinity chromatography and elution fractions were examined by immunoblotting for the co-purification of FLAG-IVA. The results of our analysis in Fig. 5 (left side) show that FLAG-IVA was retained on the column in the presence of His-IVA and was eluted with imidazole. As a control for specificity, an abundant non-sporulation protein (σA) was not found in the eluate. When the experiment was repeated with extract from cells producing His-IVAK30E and FLAG-IVAK30E, FLAG-IVAK30E was not efficiently retained on the column and was not appreciably detectable in the eluate (Fig. 5, right side). As a control, immunoblot analysis with antibodies against the His6 tag show that that His-IVA and His-IVAK30E were efficiently retained on the column. Thus, IVA molecules interact in vivo, and this interaction is dependent on an intact Walker A box.
Figure 5. IVA but not IVAK30E multimerizes in vivo.
Extracts from cells producing both His-IVA and FLAG-IVA (strain KR466), or His-IVAK30E and FLAG-IVAK30E (strain KR481), were prepared and purified by Ni2+-affinity chromatography. The presence of FLAG-tagged proteins (top panel), and as controls, His-tagged proteins (middle), and a non-sporulation protein σA (bottom) was monitored in the load (L), the flow-through (FT), the wash (W), and elution (E) fractions by immunoblotting using specific antibodies. Elution fractions were concentrated 4.5-fold relative to the load.
ATP-dependent self assembly of IVA
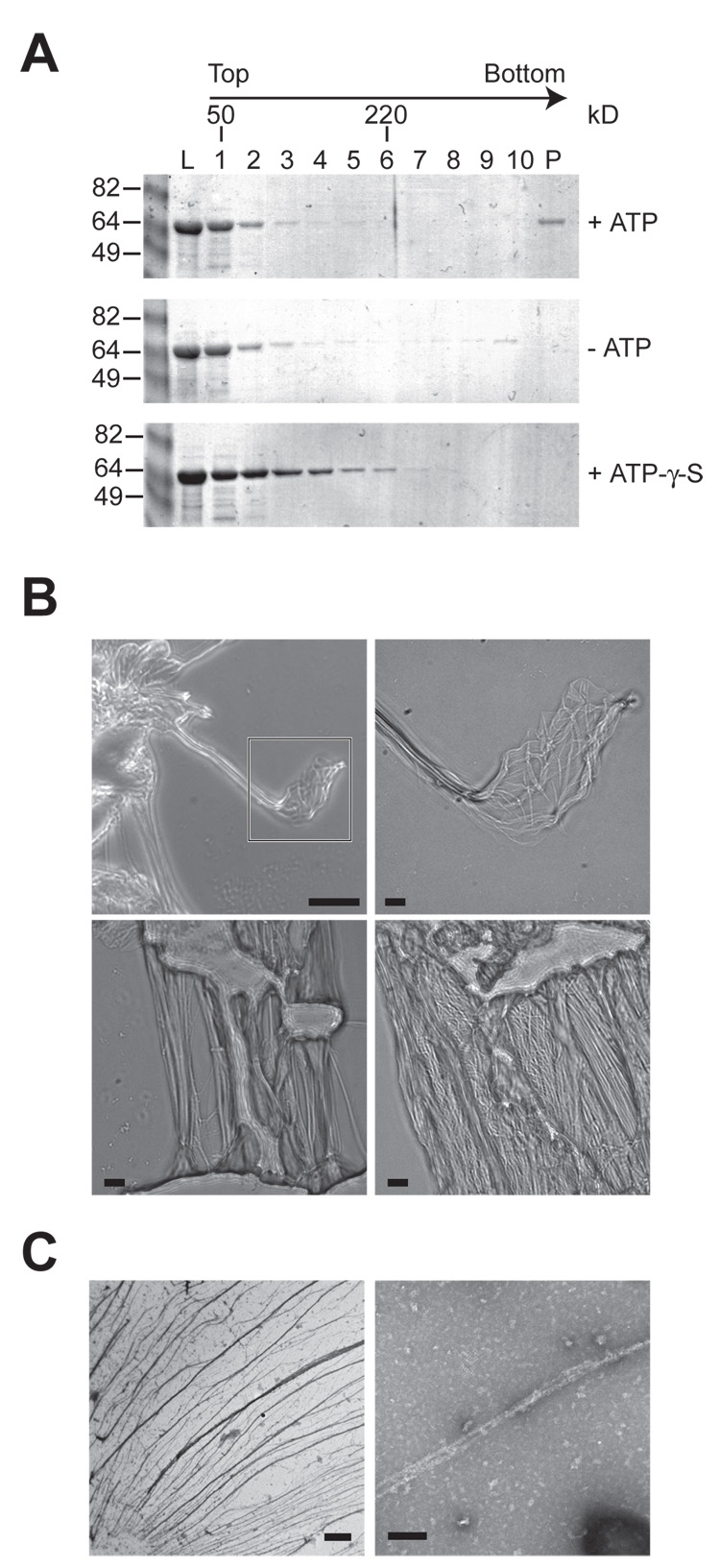
We next asked if the interaction between IVA molecules is driven by ATP hydrolysis. To test this, we incubated purified His-IVA in a buffered solution containing MgCl2, in the presence or absence of ATP, for 15 minutes at 37°C. Next, we separated this reaction by sucrose gradient centrifugation and analyzed fractions of the gradient by Coomassie staining. In the presence of ATP, purified IVA was detected near the top of gradient, but also in the pellet fraction, suggesting that some of the protein was present in a very large complex (Fig 6A, top). In the absence of ATP, however, IVA was found primarily near the top of the gradient (Fig 6A, middle). To test if ATP hydrolysis was required for multimerization, we incubated purified IVA with a non-hydrolyzable ATP analogue, ATP-γ-S (control experiments demonstrated that IVA binds ATP-γ-S with a similar affinity to that of ATP, data not shown). This resulted in the appearance of a population of IVA molecules that migrated near the middle of the gradient, suggesting that ATP binding was sufficient for some level of oligomerization (likely dimerization, judging from the molecular weight standards; Fig 6A, bottom). Nonetheless, incubation of IVA with ATP-γ-S did not result in the appearance of IVA in the pellet of the gradient, indicating that a higher order structure was not formed in the absence of ATP hydrolysis.
Figure 6. IVA polymerizes in vitro in an ATP-dependent manner.
(A) Purified His6-IVA was incubated in the presence or absence of ATP, or ATP-γ-S and separated by sucrose gradient centrifugation. Fractions were removed from the top of the gradient. The amount of protein in the load (L, 20% of total), gradient fractions (1–10), and resuspended pellet (P) were analyzed by Coomassie staining. Positions of molecular weight standards (50 kD and 220 kD) are indicated above.
(B) Gallery of light microscope images of polymerized IVA. Top left, polymerized IVA viewed against the background of a clean glass slide. Top right, magnification of the area indicated in the first panel of a IVA cable composed of individual filaments. Bottom left, individual IVA cables of varying diameters. Bottom right, meshwork of IVA filaments and cables associated through lateral interactions. Scale bar: 31 µm (top left) or 5 µm (all others).
(C) Electron micrographs of polymerized IVA. Left, field of negatively stained polymerized IVA filaments. Right, magnified view of two IVA filaments. Scale bar: 2 µm (left) or 100 nm (right).
As the incubation of purified IVA with ATP progressed, we noticed the formation of a visible stringy aggregate in the reaction vessel, which was not present in the absence of ATP or in the presence of ATP-γ-S. To test if this aggregate displayed any obvious structural features, we removed it from the reaction (without centrifugation) and examined it by light microscopy. The polymerized material at low magnification was readily apparent against the background of the glass slide and appeared to consist of striking filamentous structures that displayed a propensity to bundle into larger cables (Fig. 6B, upper left). At higher magnification the thinnest filaments we observed were less than 0.25 µm in diameter (Fig. 6B, upper right), and were found in bundles of varying diameters, even up to 5 µm (Fig. 6B, lower left). The majority of the material was present as a meshwork of layered cables of varying thicknesses (Fig. 6B, lower right). We also examined the polymerized IVA filaments by electron microscopy. Negative staining readily revealed webs of filaments which were often found in bundles as thick as 0.5 µm in diameter and tens of microns long (Fig. 6C, left). At higher magnification, smooth filaments as thin as 10 nm in diameter were apparent (Fig. 6C, right).
Taken together, the results indicate that IVA undergoes self assembly into supramolecular filamentous structures and does so in a manner that is largely driven by ATP hydrolysis.
DISCUSSION
This investigation has provided evidence that the formation of the basement layer of the spore coat occurs by a process of self assembly that is driven by hydrolysis of ATP. This conclusion is based on two findings. First, IVA harbors a canonical Walker A box that mediated ATP binding and hydrolysis. Disruption of the box impaired the assembly of IVA into a shell around the developing forespore (and, by extension, disrupted the assembly of the entire spore coat) and ultimately prevented the production of a mature spore. Second, IVA multimerization in vivo required an intact Walker A box, and ATP hydrolysis by IVA was needed for the self assembly of IVA into macroscopic filaments in vitro. This polymerization did not appear to require any additional factors, as purified IVA assembled spontaneously in an ATP-dependent manner. Of course, an unknown protein might augment polymerization in vivo.
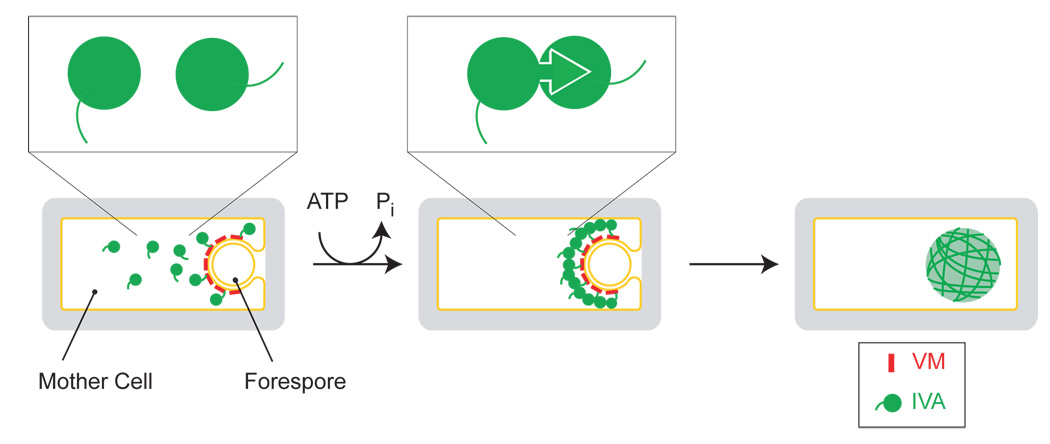
These and other findings lead us to the following model for the formation of the basement layer of the coat (Fig. 7). For simplicity, we have represented the stages of assembly sequentially, but we envision that one or more of these steps likely occur concurrently. First, the outer forespore membrane is decorated with molecules of VM, whose hydrophilic face recruits the C-terminus of IVA (the tails in the cartoons) and tethers it to the surface of the forespore. Second, multimerization of IVA is initiated by the binding of ATP, and subsequent ATP hydrolysis induces a conformational change that locks IVA molecules to each other. We posit that this step is irreversible and results in long polymers. Third, the polymers interact with each other laterally to create supramolecular filaments or cables. The filaments encapsulate the forespore and provide a platform for the subsequent addition of the other, proteinaceous layers of the coat.
Figure 7. Model for the ATP-dependent assembly of the spore coat basement layer.
Depicted is a sporangium in which the mother cell is engulfing the forespore. The amphipathic alpha helical peptide VM (red) is shown as marking the surface of the forespore and recruiting individual IVA molecules (green) via the C-terminal interaction domain (left). Upon hydrolyzing ATP, IVA molecules are shown to polymerize (middle). Above, the polymerization of IVA driven by ATP hydrolysis is depicted as a conformational change that irreversibly locks two molecules of IVA together. On the right, lateral associations between IVA filaments create supramolecular cables which envelope the forespore.
Two features of our model merit discussion. First, our model predicts that there are two kinds of interaction sites on the surface of IVA for other IVA molecules. One site is used for ATP-dependent polymerization and the second for lateral interactions between the polymers. If this prediction is correct, it should be possible to identify mutants of IVA that polymerize irreversibly but fail to form the thick cables observed by microscopy. Second, in our model not every molecule of IVA need be directly and individually tethered to the surface of the forespore by VM. Because IVA molecules assemble in higher order structures, VM need only contact some of the IVA molecules to hold the entire structure at the surface of the forespore.
If, as our results indicate, polymerization is driven by hydrolysis of ATP, it is noteworthy, as mentioned above, that the turnover of the reaction was relatively slow. We suggest two possible explanations for this observation. One explanation is that inefficient ATP hydrolysis might be a mechanism to allow formation of the supramolecular cables to occur slowly so as to prevent premature misassembly of IVA into aberrant structures off the outer surface of the developing spore. Alternatively, IVA is more active as an ATPase in vivo than we observe in vitro. Conceivably, another protein component of the basement layer of the coat plays a regulatory role in stimulating hydrolysis of ATP by IVA.
Elements of IVA assembly are reminiscent of other assembly processes in biological systems. The nucleotide-dependent multimerization of IVA is at first glance similar to the polymerization of actin and tubulin, whereby ATP and GTP binding, respectively, promotes the assembly of filaments (Schuler, 2001; Romberg and Levin, 2003). However, unlike these cytoskeletal proteins, for which hydrolysis of the bound nucleotide destabilizes the filaments, IVA appears to use ATP hydrolysis to drive the formation of higher order structures. Likewise, the nucleotide-dependent multimerization of various proteins that shuttle on and off the membrane (e.g., Sar1p, Arf family of proteins, MinD) (Lee et al., 2005; Pasqualato et al., 2002; Lutkenhaus, 2007) using an attached amphipathic alpha helical domain is analogous to the attachment of IVA (which multimerizes upon ATP binding) to the membrane via VM (an amphipathic alpha helix). Again, though ATP hydrolysis is typically used as a regulatory mechanism to disengage from the membrane, IVA hydrolyzes ATP to irreversibly associate with the forespore. Aspects of the assembly of retroviral capsids, which are composed of over 1000 copies of a single protein Pr55, are also functionally and mechanistically similar to coat assembly in that assembly of the capsid at the membrane requires the hydrolysis of ATP (Lingappa et al., 1997). However, Pr55 is unable to bind or hydrolyze ATP and requires at least two cellular factors to provide the energy for assembly. Unlike the assembly of retroviral capsids, then, the polymerization of IVA is mechanistically speaking, self-sufficient.
A central challenge in developmental biology is to elucidate the mechanisms by which proteins assemble into the complex structures. Here we have uncovered a previously unknown mechanism of morphogenesis: self assembly driven by ATP hydrolysis. Like many other examples of morphogenesis in biology assembly of the spore coat is an irreversible process that culminates in a structure that is stable for long periods of time. The use of energy from ATP hydrolysis to drive an irreversible assembly process to create a structure that is resilient against environmental stresses may not be limited to IVA. It will be interesting to see whether other morphogenetic proteins also self assemble by ATP-driven processes.
EXPERIMENTAL PROCEDURES
Strain construction and cell growth
Strains are otherwise isogenic derivatives of B. subtilis PY79 (Youngman et al., 1984). B. subtilis competent cells were prepared as described previously (Wilson and Bott, 1968). Sporulation efficiency was determined in 24 hour cultures induced to sporulate in DS medium by heat killing at 80°C for 20 minutes (Harwood and Cutting, 1990). Integration plasmids pKR13 and pKR5, designed to place spoIVA and gfp-spoIVA, respectively, at thrC have been described previously (Ramamurthi et al., 2006). FLAG-spoIVA and His6-spoIVA at thrC and amyE, respectively, were created by PCR amplifying the spoIVA open reading frame with primers spoIVA5’FlagHind (5’ CCCAAGCTTATGGACTACAAAGACGATGACGACAAGGAAAAGGTCGATATTTTCAAGG) or spoIVA5’HisHind (5’ CCCAAGCTTATGCATCACCATCACCATCACGAAAAGGTCGATATTTTCAAGG) and spoIVA3’Bam (5’ CGCGGATCCTTACAGGATGATGGCGATTAAG), digesting the PCR product with HindIII and BamHI, and cloning into the integration vector pKR5 or pKR129 (which harbor the spoIVA promoter upstream of the HindIII site), respectively.
Site-directed mutations in spoIVA or gfp-spoIVA were created using the Quikchange site directed mutagenesis kit (Stratagene) using complementary primers that harbored the desired mutation flanked by approximately 15 bp on either side and using either pKR13 or pKR5 as a template. Mutagenesis was confirmed by DNA sequencing.
Plasmids pKR145 and pKR151 harboring His6-spoIVA and His6-spoIVAK30A, respectively, under control of the T7 promoter were created by PCR amplifying either the spoIVA or spoIVAK30A open reading frame using primers spoIVA5’Nhe (5’ CTAGCTAGCATGGAAAAGGTCGATATTTTCAAG) and spoIVA3’Bam, digesting the PCR product with NheI and BamHI, and cloning into pET28a (Novagen) which appends a 23 amino acid-long extension at the N-terminus of SpoIVA that harbors a His6 tag and a thrombin cleavage site. pKR145 and pKR151 were transformed into Escherichia coli BL21(DE3).
Microscopy
Overnight cultures of cells grown at 22°C were diluted 1:20 into 20 ml CH medium (Sterlini and Mandelstam, 1969) and induced for sporulation by resuspension of exponential phase cells in SM medium. Harvested cells were resuspended in PBS containing 1µg/ml of the membrane dye FM4-64. Three microliters of the suspension was placed on a 1% agarose pad and cells were immobilized by using a freshly prepared coverslip treated briefly with poly-L-lysine. The equipment used and analysis of images were as previously described (Fujita and Losick, 2002).
Nucleotide binding and hydrolysis assays
His6-SpoIVA and His6-SpoIVAK30A were purified from E. coli BL21(DE3) pKR145 and E. coli BL21(DE3) pKR151, respectively, as follows. Overnight cultures grown at 37°C in LB supplemented with 50 µg/ml kanamycin for plasmid maintenance were diluted into 500 ml LB/Kan and grown for 2 hours at 37°C. IPTG was added to 1 mM final concentration to induce expression of the his-tagged construct and the culture was grown for 3 hours at 37°C. Harvested cells were resuspended in 15 ml ice cold buffer A (50 mM Tris at pH 7.5, 150 mM NaCl) and disrupted by sonication. All subsequent steps were performed at 4°C. Unbroken cells and cell debris were removed by centrifugation at 10,000 × g, and the cleared lysate was placed on 500 µl (bed volume) Ni2+-NTA agarose beads (Qiagen). The column was washed with 20 ml buffer B (buffer A containing 20 mM imidazole) and eluted in 4 ml buffer C (buffer A containing 200 mM imidazole). Purified protein was dialyzed for 1 hour against 4L buffer D (buffer A containing 1 mM DTT) and aliquots were stored promptly at −80°C. Protein aliquots were quick-thawed at 37°C immediately before use and kept on ice. Purified protein began to precipitate within days when stored at 4°C.
As verification that IVA itself was responsible for the ATPase activity, we showed that the capacity to hydrolyze ATP co-purified with the proteins during ion-exchange chromatography on MonoQ (Pharmacia) (data not shown).
Nucleotide binding assays were adapted from a previously described protocol (Carey et al., 1983). Briefly, purified His6-IVA was incubated in 50 µl reaction volumes of buffer E (buffer D containing 1 mM MgCl2). For saturation experiments, the final concentration of His6-IVA was 39 mM. Binding reactions were initiated with the addition of γ32P-ATP or γ32P-GTP (approximately 3.3 pmols per reaction for the nucleotide specificity experiments) and incubated at 30°C for 30 seconds. The binding reaction was stopped by the addition of 1 ml buffer E and the entire diluted reaction was immediately filtered through a HAWP nitrocellulose filter (Millipore) and washed with an additional 1 ml buffer E. We observed that further washing did not reduce the amount of unspecific radioactivity retained on the filter. Dilution and filtration of the binding reaction were typically complete in about 5 seconds. Radioactivity retained on the filters was quantified in 5 ml scintillation fluid.
Nucleotide hydrolysis assays were performed using either the Malachite Green Phosphate assay kit (BioAssay Systems), according to the manufacturer’s instructions, in 100 µl reaction volumes in buffer E containing 10 mM MgCl2 and 4 mM ATP or GTP at 37°C for 10 minutes, or by measuring ADP production as described previously (Field et al., 1996; Kogan et al., 2002). Briefly, purified His6-IVA (960 nM final concentration) was incubated in buffer E containing 10 mM MgCl2 with ATP (ATP:ATP (α-32P) ratio of 25000:1) in a 25 µl reaction volume for 30 minutes at 37°C. Reactions were stopped by the addition of 1% SDS (final concentration) and nucleotides were analyzed by thin layer chromatography on PEI cellulose plates (pre-spotted with a 5 mM ATP/ADP mix as a carrier) in 0.5M LiCl and 1 M formic acid. Migration of unlabeled ATP and ADP standards was visualized by UV adsorption. Labeled ATP and ADP on dried TLC plates were imaged on X-ray film, scanned, and quantitated using Image J software (NIH).
Non-linear regression analyses to fit data to curves were performed using Prism 5 software (GraphPad).
Co-purification of His-IVA and FLAG-IVA
Overnight cultures of KR466 and KR481 grown in CH medium were induced to sporulate in 30 ml resuspension medium as described above. 2.5 hours after induction of sporulation, cells were harvested and resuspended in 1 ml protoplast buffer (0.5 M sucrose, 20 mM MgCl2, 10 mM potassium phosphate at pH 6.8, 0.1 mg/ml lysozyme) and incubated at 37°C for 25 minutes. Protoplasts were collected by centrifugation and lysed by resuspending in 1 ml buffer D containing 4 mM ATP and incubating at 37°C for 10 minutes. Viscosity was reduced by passaging the lysate through a 23 gauge syringe needle, and cell debris was removed by centrifugation. 900 µl of the cleared lysate was added to 250 µl (bead volume) Ni2+-NTA agarose (equilibrated with buffer D) and incubated at room temperature for 10 minutes, gently mixing by inversion. Beads were extensively washed with buffer D containing 20 mM imidazole, and eluted by heating at 50°C for 10 minutes in 200 µl SDS sample buffer. Fractions were analyzed by immunoblotting using antisera raised against FLAG epitope (Sigma), His6 epitope (GeneTex), or σA.
In vitro polymerization of IVA
Purified His6-SpoIVA (approximately 250 µg) was incubated in a 300 µl reaction volume in buffer E in the presence or absence of 4 mM ATP or 4 mM ATP-γ-S at 37°C for 15 minutes. Control experiments indicated that ATP-γ-S was not hydrolyzed by IVA (data not shown). 250 µl of the reaction was layered on 5 ml 5%–20% continuous sucrose gradient made in buffer D and centrifuged at 236,500 × g for 1 hour at 4°C. Benchmark Protein Ladder (Invitrogen) was loaded onto a parallel sucrose gradient as a size standard. 525 µl fractions were removed from the top of the gradient, and the pellet was resuspended in 525 µl buffer D. All fractions were separated by 10% polyacrylamide gel electrophoresis and analyzed by Coomassie staining. Molecular weight markers were detected by silver staining. For light microscopy, purified His6-SpoIVA was incubated for 1 hour at 37°C. The visible polymerized material was removed with a pipet tip (without centrifugation) and placed on a clean glass microscope slide and immobilized with a clean glass coverslip. For electron microscopy, purified His6-SpoIVA was incubated for about 24 hours at 37°C. Specimens were prepared for EM using the conventional negative staining procedure: a 5 microliter drop of sample solution was adsorbed to a glow-discharged carbon-coated copper grid, washed with one drop of deionized water, and stained with freshly prepared 0.75% uranyl formate. In order to avoid unspecific aggregation, the polymerized material was not centrifuged. Samples were imaged using a Tecnai G2 Spirit BioTWIN electron microscope at an acceleration voltage of 80 kV. Images were taken with an AMT 2k CCD camera.
Supplementary Material
ACKNOWLEDGEMENTS
We thank members of the laboratory and Guocheng Yuan for helpful discussions, A. Driks, P. Eichenberger, and D. Needleman for advice on the manuscript, and Maria Ericsson of the Harvard Medical School EM Facility for assistance with electron microscopy. This work is supported by Ruth L. Kirschstein National Research Service Award GM072408 to K.S.R. and NIH grant GM18568 to R.L.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Allen JD, Parsons SM. Nitrocellulose filter binding: quantitation of the histidyl-tRNA-ATP phosphoribosyltransferase complex. Analytical biochemistry. 1979;92:22–30. doi: 10.1016/0003-2697(79)90620-1. [DOI] [PubMed] [Google Scholar]
- Carey J, Lowary PT, Uhlenbeck OC. Interaction of R17 coat protein with synthetic variants of its ribonucleic acid binding site. Biochemistry. 1983;22:4723–4730. doi: 10.1021/bi00289a017. [DOI] [PubMed] [Google Scholar]
- Driks A. Maximum shields: the assembly and function of the bacterial spore coat. Trends Microbiol. 2002;10:251–254. doi: 10.1016/s0966-842x(02)02373-9. [DOI] [PubMed] [Google Scholar]
- Driks A, Roels S, Beall B, Moran CP, Jr, Losick R. Subcellular localization of proteins involved in the assembly of the spore coat of Bacillus subtilis. Genes Dev. 1994;8:234–244. doi: 10.1101/gad.8.2.234. [DOI] [PubMed] [Google Scholar]
- Duda RL, Martincic K, Xie Z, Hendrix RW. Bacteriophage HK97 head assembly. FEMS microbiology reviews. 1995;17:41–46. doi: 10.1111/j.1574-6976.1995.tb00186.x. [DOI] [PubMed] [Google Scholar]
- Field CM, al-Awar O, Rosenblatt J, Wong ML, Alberts B, Mitchison TJ. A purified Drosophila septin complex forms filaments and exhibits GTPase activity. The Journal of cell biology. 1996;133:605–616. doi: 10.1083/jcb.133.3.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M, Losick R. An investigation into the compartmentalization of the sporulation transcription factor sigmaE in Bacillus subtilis. Mol Microbiol. 2002;43:27–38. doi: 10.1046/j.1365-2958.2002.02732.x. [DOI] [PubMed] [Google Scholar]
- Harwood CR, Cutting SM. Molecular Biological Methods for Bacillus. Modern Microbiological Methods. 1990 [Google Scholar]
- Henriques AO, Moran CP., Jr Structure, Assembly, and Function of the Spore Surface Layers. Annual review of microbiology. 2006 doi: 10.1146/annurev.micro.61.080706.093224. [DOI] [PubMed] [Google Scholar]
- Hino J, Kangawa K, Matsuo H, Nohno T, Nishimatsu S. Bone morphogenetic protein-3 family members and their biological functions. Front Biosci. 2004;9:1520–1529. doi: 10.2741/1355. [DOI] [PubMed] [Google Scholar]
- Kim H, Hahn M, Grabowski P, McPherson DC, Otte MM, Wang R, Ferguson CC, Eichenberger P, Driks A. The Bacillus subtilis spore coat protein interaction network. Mol Microbiol. 2006;59:487–502. doi: 10.1111/j.1365-2958.2005.04968.x. [DOI] [PubMed] [Google Scholar]
- Kogan I, Ramjeesingh M, Li C, Bear CE. Studies of the molecular basis for cystic fibrosis using purified reconstituted CFTR protein. Methods in molecular medicine. 2002;70:143–157. doi: 10.1385/1-59259-187-6:143. [DOI] [PubMed] [Google Scholar]
- Lee MC, Orci L, Hamamoto S, Futai E, Ravazzola M, Schekman R. Sar1p N-terminal helix initiates membrane curvature and completes the fission of a COPII vesicle. Cell. 2005;122:605–617. doi: 10.1016/j.cell.2005.07.025. [DOI] [PubMed] [Google Scholar]
- Levin PA, Fan N, Ricca E, Driks A, Losick R, Cutting S. An unusually small gene required for sporulation by Bacillus subtilis. Mol Microbiol. 1993;9:761–771. doi: 10.1111/j.1365-2958.1993.tb01736.x. [DOI] [PubMed] [Google Scholar]
- Liberek K, Marszalek J, Ang D, Georgopoulos C, Zylicz M. Escherichia coli DnaJ and GrpE heat shock proteins jointly stimulate ATPase activity of DnaK. Proc Natl Acad Sci U S A. 1991;88:2874–2878. doi: 10.1073/pnas.88.7.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingappa JR, Hill RL, Wong ML, Hegde RS. A multistep, ATP-dependent pathway for assembly of human immunodeficiency virus capsids in a cell-free system. The Journal of cell biology. 1997;136:567–581. doi: 10.1083/jcb.136.3.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losick R, Youngman P, Piggot PJ. Genetics of endospore formation in Bacillus subtilis. Annu Rev Genet. 1986;20:625–669. doi: 10.1146/annurev.ge.20.120186.003205. [DOI] [PubMed] [Google Scholar]
- Lutkenhaus J. Assembly dynamics of the bacterial MinCDE system and spatial regulation of the Z ring. Annual review of biochemistry. 2007;76:539–562. doi: 10.1146/annurev.biochem.75.103004.142652. [DOI] [PubMed] [Google Scholar]
- Macnab RM. How bacteria assemble flagella. Annual review of microbiology. 2003;57:77–100. doi: 10.1146/annurev.micro.57.030502.090832. [DOI] [PubMed] [Google Scholar]
- Paine ML, White SN, Luo W, Fong H, Sarikaya M, Snead ML. Regulated gene expression dictates enamel structure and tooth function. Matrix Biol. 2001;20:273–292. doi: 10.1016/s0945-053x(01)00153-6. [DOI] [PubMed] [Google Scholar]
- Pasqualato S, Renault L, Cherfils J. Arf, Arl, Arp and Sar proteins: a family of GTP-binding proteins with a structural device for 'front-back' communication. EMBO Rep. 2002;3:1035–1041. doi: 10.1093/embo-reports/kvf221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piggot PJ, Coote JG. Genetic aspects of bacterial endospore formation. Bacteriol Rev. 1976;40:908–962. doi: 10.1128/br.40.4.908-962.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prajapati RS, Ogura T, Cutting SM. Structural and functional studies on an FtsH inhibitor from Bacillus subtilis. Biochim Biophys Acta. 2000;1475:353–359. doi: 10.1016/s0304-4165(00)00089-1. [DOI] [PubMed] [Google Scholar]
- Price KD, Losick R. A four-dimensional view of assembly of a morphogenetic protein during sporulation in Bacillus subtilis. J Bacteriol. 1999;181:781–790. doi: 10.1128/jb.181.3.781-790.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramamurthi KS, Clapham KR, Losick R. Peptide anchoring spore coat assembly to the outer forespore membrane in Bacillus subtilis. Mol Microbiol. 2006;62:1547–1557. doi: 10.1111/j.1365-2958.2006.05468.x. [DOI] [PubMed] [Google Scholar]
- Roels S, Driks A, Losick R. Characterization of spoIVA, a sporulation gene involved in coat morphogenesis in Bacillus subtilis. J Bacteriol. 1992;174:575–585. doi: 10.1128/jb.174.2.575-585.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romberg L, Levin PA. Assembly dynamics of the bacterial cell division protein FTSZ: poised at the edge of stability. Annual review of microbiology. 2003;57:125–154. doi: 10.1146/annurev.micro.57.012903.074300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler H. ATPase activity and conformational changes in the regulation of actin. Biochim Biophys Acta. 2001;1549:137–147. doi: 10.1016/s0167-4838(01)00255-2. [DOI] [PubMed] [Google Scholar]
- Setlow P. Spores of Bacillus subtilis: their resistance to and killing by radiation, heat and chemicals. J Appl Microbiol. 2006;101:514–525. doi: 10.1111/j.1365-2672.2005.02736.x. [DOI] [PubMed] [Google Scholar]
- Sterlini JM, Mandelstam J. Commitment to sporulation in Bacillus subtilis and its relationship to development of actinomycin resistance. Biochem J. 1969;113:29–37. doi: 10.1042/bj1130029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stragier P, Losick R. Molecular genetics of sporulation in Bacillus subtilis. Annu Rev Genet. 1996;30:297–241. doi: 10.1146/annurev.genet.30.1.297. [DOI] [PubMed] [Google Scholar]
- Takamatsu H, Imamura A, Kodama T, Asai K, Ogasawara N, Watabe K. The yabG gene of Bacillus subtilis encodes a sporulation specific protease which is involved in the processing of several spore coat proteins. FEMS Microbiol Lett. 2000;192:33–38. doi: 10.1111/j.1574-6968.2000.tb09355.x. [DOI] [PubMed] [Google Scholar]
- van Ooij C, Losick R. Subcellular localization of a small sporulation protein in Bacillus subtilis. J Bacteriol. 2003;185:1391–1398. doi: 10.1128/JB.185.4.1391-1398.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker JE, Saraste M, Runswick MJ, Gay NJ. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. Embo J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waring GL. Morphogenesis of the eggshell in Drosophila. International review of cytology. 2000;198:67–108. doi: 10.1016/s0074-7696(00)98003-3. [DOI] [PubMed] [Google Scholar]
- Webb CD, Decatur A, Teleman A, Losick R. Use of green fluorescent protein for visualization of cell-specific gene expression and subcellular protein localization during sporulation in Bacillus subtilis. J Bacteriol. 1995;177:5906–5911. doi: 10.1128/jb.177.20.5906-5911.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson GA, Bott KF. Nutritional factors influencing the development of competence in the Bacillus subtilis transformation system. J Bacteriol. 1968;95:1439–1449. doi: 10.1128/jb.95.4.1439-1449.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngman P, Perkins JB, Losick R. Construction of a cloning site near one end of Tn917 into which foreign DNA may be inserted without affecting transposition in Bacillus subtilis or expression of the transposon-borne erm gene. Plasmid. 1984;12:1–9. doi: 10.1016/0147-619x(84)90061-1. [DOI] [PubMed] [Google Scholar]
- Zheng LB, Donovan WP, Fitz-James PC, Losick R. Gene encoding a morphogenic protein required in the assembly of the outer coat of the Bacillus subtilis endospore. Genes Dev. 1988;2:1047–1054. doi: 10.1101/gad.2.8.1047. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.