Abstract
A derivative of rhodamine 110 has been designed and assessed as a probe for cytochrome P450 activity. This probe is the first to utilize a “trimethyl lock” that is triggered by cleavage of an ether bond. In vitro, fluorescence was manifested by the CYP1A1 isozyme with kcat/KM = 8.8 × 103 M−1s−1 and KM = 0.09 µM. In cellulo, the probe revealed the induction of cytochrome P450 activity by the carcinogen 2,3,7,8-tetrachlorodibenzo-p-dioxin, and its repression by the chemoprotectant resveratrol.
Keywords: Carcinogen, CYP1A1 Isozyme, Cytochrome P450, Dioxin, Fluorogenic Substrate, Prodrug, Resveratrol, Rhodamine 110, Trimethyl Lock
The cytochrome P450 (P450) family of enzymes is responsible for the oxidative metabolism of a wide variety of compounds, including chemotherapeutic agents and environmental toxins.1,2 The catalytic activity of P450 enzymes controls the rate of xenobiotic metabolism, and can produce undesirable byproducts.3 Originally, this activity had been assessed by using HPLC or other methods to separate and quantify metabolites. In the 1970’s, 7-ethoxycoumarin and 7-ethoxyresorufin were introduced as the first fluorogenic substrates for assays of P450 activity.4 Although these and other fluorogenic substrates have been used to assay P450 activity in vitro and enable assays in cellulo,5,6 they suffer from background fluorescence.7 For example, alkoxycoumarins exhibit moderate fluorescence and are used frequently as fluorophores in peptidase substrates based on Forster resonance energy transfer (FRET).8 In addition, both 7-ethoxyresorufin and resorufin fluoresce brightly.6 This problem arises because O-alkylation of the hydroxyl group of fluorophores such as coumarin and resorufin does little to deter the oxygen electrons from participating in the resonance that gives rise to fluorescence.
Here, we report on a superior small-molecule probe for assessing P450 activity. Our probe employs the trimethyl lock.9–11 The trimethyl lock is an o-hydroxycinnamic acid derivative in which severe crowding of three methyl groups induces rapid lactonization to form a hydrocoumarin.12 In this strategy, the phenolic oxygen of the o-hydroxycinnamic acid is modified to create a functional group that is a substrate for a designated enzyme, and the carboxyl group is condensed with the amino group of a dye. Unmasking of the phenolic oxygen leads to rapid lactonization with concomitant release of the dye. An important attribute of this strategy is that the fluorescence/absorbance of the dye is masked completely by amidic resonance and the resulting lactonization within the rhodamine moiety.9,10
The human genome contains 27 genes encoding P450 isozymes, along with many pseudogenes.13 Of these isozymes, cytochrome P450 1A1 (CYP1A1) is known to be especially important in the metabolism of xenobiotics.14 Unlike most P450 isoforms, which are found primarily in the liver, CYP1A1 is present mainly in the lungs, where it plays an important role in the metabolic activation of chemical carcinogens.1,15 The lung is a primary site of exposure for inhaled toxins along with carcinogens that can ultimately yield lung carcinomas.16 CYP1A1 is strongly induced by cigarette smoking.15 Many compounds, including some found in cigarette smoke, are not hazardous until metabolized by CYP1A1.16,17 Accordingly, CYP1A1 levels could be correlated with human disease.
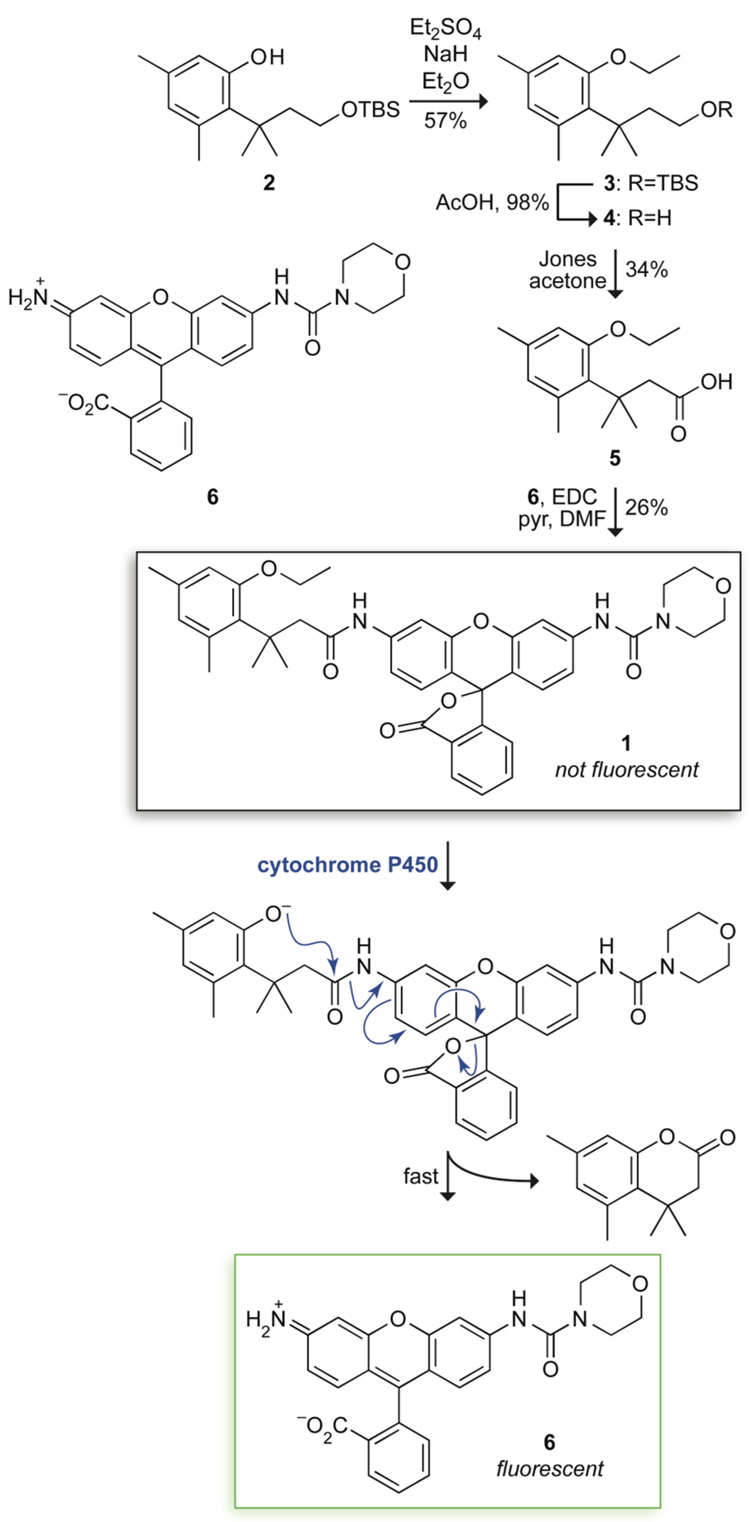
We suspected that the trimethyl lock could provide the basis for a useful probe for CYP1A1 activity. As a dye, we chose a morpholino urea derivative of rhodamine 110 (Rh110) that is bright (ε × Φ = 2.38 × 104 M−1cm−1) but has no measurable fluorescence after N-acylation.10 We installed an ethyl group on the phenolic oxygen of the trimethyl lock because ethyl ethers are especially effective substrates for CYP1A1.15 The synthetic route to fluorogenic probe 1 is shown in Figure 1. Briefly, known intermediate 2 18 was alkylated with diethyl sulfate to give ethyl ether 3. Removal of the silyl group followed by Jones oxidation afforded carboxylic acid 5. Condensation with urea–rhodamine 6 gave fluorogenic probe 1 in 5% overall yield.
Figure 1.
Scheme for the synthesis and use of fluorogenic probe 1.
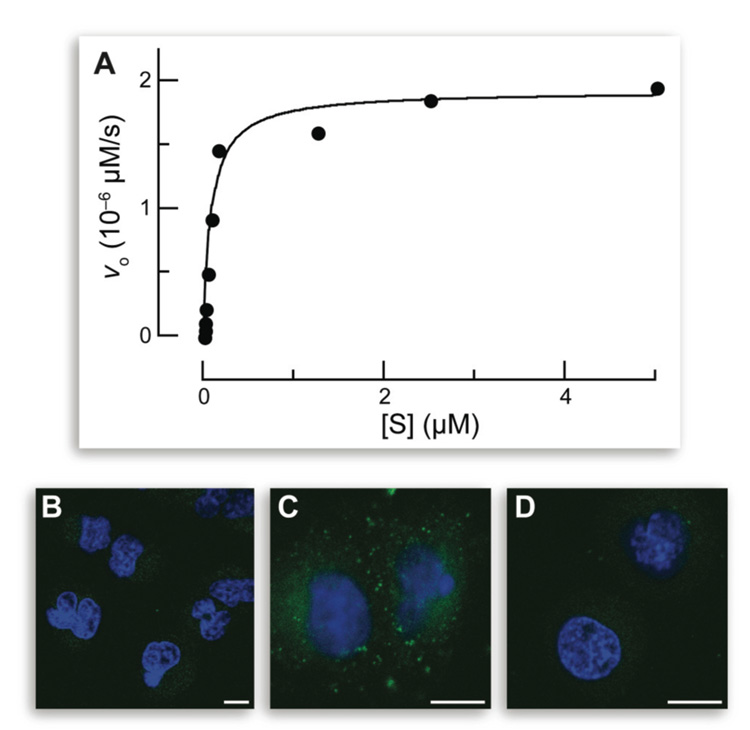
Fluorogenic probe 1 was first assayed as a substrate for human CYP1A1 in vitro. Fluorogenesis was rapid, with kcat/KM = 8.8 × 103 M−1s−1 and KM = 0.09 µM (Figure 2A). These values are comparable to the highest values obtained with other fluorogenic substrates.1,2 These data are the first to demonstrate that the trimethyl lock can be activated by the cleavage of an ether bond.
Figure 2.
Fluorogenesis from fluorogenic probe 1 in vitro and in cellulo. (A) Data for the in vitro cleavage of fluorogenic probe 1 by human CYP1A1 (5.0 pM) in PBS containing NADPH (8 mM) and MgCl2 (8 mM). (B–D) Images of the in cellulo cleavage of fluorogenic probe 1. A549 cells were incubated with fluorogenic probe 1 (10 µM) and an additive for 1 h and counterstained with Hoechst 33342. (B) No additive. (C) TCDD (10 nM). (D) TCDD (10 nM) and resveratrol (50 µM). Scale bars: 10 µm.
Next, fluorogenic probe 1 was assayed as a substrate for CYP1A1 in live human cells. These experiments employed human lung adenocarcinoma cell line A549, which is especially well suited for studying the expression of the pulmonary CYP system.16,19 A low but observable level of CYP1A1 was apparent after a 1-h incubation with fluorogenic probe 1 (Figure 2B).
Then, fluorogenic probe 1 was evaluated as a means to detect an increase in CYP1A1 levels. To do so, A549 cells were incubated with 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD; 10 nM), which is the notorious contaminant in the herbicide Agent Orange and the most potent known inducer of CYP1A1.20 The effect of TCDD on fluorogenesis within A549 cells was dramatic (Figure 2C).
Finally, fluorogenic probe 1 was used to reveal a more complex modulation of P450 activity. Levels of P450 are highly variable in individuals, and there are many known P450 polymorphisms.21 Inhibitors of P450 activity have potential as chemotherapeutic agents.22 For example, resveratrol (3,5,4′-trihydroxystilbene), which is a natural phytoalexin present in grapes and other foods, has been proposed to have a chemoprotective effect against lung cancer by virtue of its ability to decrease CYP1A1 activity.23 To test this hypothesis with fluorogenic probe 1, live A549 cells were treated with both TCDD and resveratrol, along the probe. After a 1-h incubation, cells exhibited a dramatic decrease in fluorescence compared with cells treated with TCDD (Figure 2D). The levels appeared to be even lower than those in untreated cells. These and other data23 provide direct and conclusive evidence that resveratrol decreases CYP1A1 activity in cellulo.
In conclusion, fluorogenic probe 1 is the first to utilize a “trimethyl lock” that is triggered by cleavage of an ether bond. This probe has numerous desirable attributes. Its chemical and photophysical properties allow for real-time imaging of P450 levels in cellulo. The modularity of this probe enables its extension to enzymes throughout the P450 family, and its success indicates that the trimethyl lock strategy can be applied to P450-activated prodrugs. Finally, appending the urea group with a trichloromethyl ketone or other weak electrophile would allow the probe to react with an intracellular thiol and enable its retention within a cell, providing additional utility.24
Supplementary Material
Detailed procedures for the synthesis, analysis, and use of fluorogenic probe 1 can be found, in the online version, at doi:10.1016/j.bmcl.2008
Acknowledgements
This paper is dedicated to Professor Benjamin F. Cravatt on the occasion of his winning the 2008 Tetrahedron Young Investigator Award. This work was supported by grant CA073808 (NIH). NMRFAM was supported by grant P41 RR02301 (NIH). M.M.Y. was a Pfizer Undergraduate Summer Fellow in Chemistry and a Hilldale Undergraduate/Faculty Research Fellow. L.D.L was supported by Biotechnology Training Grant 08349 (NIH) and an ACS Division of Organic Chemistry Graduate Fellowship sponsored by the Genentech Foundation. T.-Y.C. was supported by the Dr. James Chieh-Hsia Mao Wisconsin Distinguished Graduate Fellowship.
References and notes
- 1.Ortiz de Montellano PR, editor. Cytochrome P450: Structure, Mechanism, and Biochemistry. New York: Kluwer Academic/Plenum Publishers; 2005. [Google Scholar]
- 2.(a) Gungerich FP. Chem. Res. Toxicol. 2008;21:70. doi: 10.1021/tx700079z. [DOI] [PubMed] [Google Scholar]; (b) Johnson WW. Drug Metab. Rev. 2008;40:101. doi: 10.1080/03602530701836704. [DOI] [PubMed] [Google Scholar]
- 3.Nebert DW, Dalton TP. Nat. Rev. Cancer. 2006;6:947. doi: 10.1038/nrc2015. [DOI] [PubMed] [Google Scholar]
- 4.(a) Ullrich V, Weber P. Hoppe-Seyler’s X. Physiol. Chem. 1972;353:1171. doi: 10.1515/bchm2.1972.353.2.1171. [DOI] [PubMed] [Google Scholar]; (b) Burke MD, Mayer RT. Drug Metab. Dispos. 1974;2:583. [PubMed] [Google Scholar]; (c) Lavis LD, Raines RT. ACS Chem. Biol. 2008;3:142. doi: 10.1021/cb700248m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.(a) Burke MD, Murray GI, Lees GM. Biochem. J. 1983;212:15. doi: 10.1042/bj2120015. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) White IN. Anal. Biochem. 1988;172:304. doi: 10.1016/0003-2697(88)90449-6. [DOI] [PubMed] [Google Scholar]; (c) Mayer RT, Netter KJ, Heubel F, Hanhemann B, Buchheister A, Mayer GK, Burke MD. Biochem. Pharmacol. 1990;40:1645. doi: 10.1016/0006-2952(90)90467-y. [DOI] [PubMed] [Google Scholar]; (d) Buters JT, Schiller CD, Chou RC. Biochem. Pharmacol. 1993;46:1577. doi: 10.1016/0006-2952(93)90326-r. [DOI] [PubMed] [Google Scholar]; (e) Mayer RT, Dolence EK, Mayer GE. Drug Metab. Dispos. 2007;35:103. doi: 10.1124/dmd.106.011601. [DOI] [PubMed] [Google Scholar]
- 6.Ghosal A, Hapangama N, Yuan Y, Lu X, Horne D, Patrick JE, Zbaida S. Biopharm. Drug Dispos. 2003;24:375. doi: 10.1002/bdd.374. [DOI] [PubMed] [Google Scholar]
- 7.Wright AT, Cravatt BF. Chem. Biol. 2007;14:1043. doi: 10.1016/j.chembiol.2007.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lesner A, Wysocka M, Guzow K, Wiczk W, Legowska A, Rolka K. Anal. Biochem. 2008;15:306. doi: 10.1016/j.ab.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 9.(a) Chandran SS, Dickson KA, Raines RT. J. Am. Chem. Soc. 2005;127:1652. doi: 10.1021/ja043736v. [DOI] [PubMed] [Google Scholar]; (b) Huang S-T, Lin Y-L. Org. Lett. 2006;8:265. doi: 10.1021/ol052635e. [DOI] [PubMed] [Google Scholar]; (c) Lavis LD, Chao TY, Raines RT. ChemBioChem. 2006;7:1151. doi: 10.1002/cbic.200500559. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Levine MN, Lavis LD, Raines RT. Molecules. 2008;13:204. doi: 10.3390/molecules13020204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lavis LD, Chao T-Y, Raines RT. ACS Chem. Biol. 2006;1:252. doi: 10.1021/cb600132m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.(a) Johnson RJ, Chao TY, Lavis LD, Raines RT. Biochemistry. 2007;46:10308. doi: 10.1021/bi700857u. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Mangold SL, Carpenter RT, Kiessling LL. Org. Lett. 2008;10:xxxx. doi: 10.1021/ol800932w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.(a) Milstein S, Cohen LA. J. Am. Chem. Soc. 1972;94:9158. doi: 10.1021/ja00781a029. [DOI] [PubMed] [Google Scholar]; (b) Borchardt RT, Cohen LA. J. Am. Chem. Soc. 1972;94:9166. doi: 10.1021/ja00781a030. [DOI] [PubMed] [Google Scholar]
- 13.Nelson DR, Zeldin DC, Hoffman SMG, Maltais LJ, Wain HM, Nebert DW. Pharmacogenetics. 2004;14:1. doi: 10.1097/00008571-200401000-00001. [DOI] [PubMed] [Google Scholar]
- 14.Rooseboom M, Commandeur JNM, Vermeulen NPE. Pharmacol. Rev. 2004;56:53. doi: 10.1124/pr.56.1.3. [DOI] [PubMed] [Google Scholar]
- 15.Liu J, Ericksen SS, Besspiata D, Fisher CW, Szklarz GD. Drug Metab. Dispos. 2003;31:412. doi: 10.1124/dmd.31.4.412. [DOI] [PubMed] [Google Scholar]
- 16.Hukkanen J, Lassila A, Paivarinta K, Valanne S, Sarpo S, Hakkola J, Pelkonen O, Raunio H. Am. J. Respir. Cell. Mol. Biol. 2000;22:360. doi: 10.1165/ajrcmb.22.3.3845. [DOI] [PubMed] [Google Scholar]
- 17.(a) Ueng TH, Hu SH, Chen RM, Wang HW, Kuo ML. J. Toxicol. Environ. Health A. 2000;60:101. doi: 10.1080/009841000156529. [DOI] [PubMed] [Google Scholar]; (b) Lemm F, Wilhelm M, Roos PH. Int. J. Hyg. Environ. Health. 2004;207:325. doi: 10.1078/1438-4639-00298. [DOI] [PubMed] [Google Scholar]
- 18.Nicolaou MG, Yuan C-S, Borchardt RT. J. Org. Chem. 1996;61:8636. doi: 10.1021/jo961069l. [DOI] [PubMed] [Google Scholar]
- 19.Urani C, Doldi M, Crippa S, Camatini M. Chemosphere. 1998;37:2785. doi: 10.1016/s0045-6535(98)00321-x. [DOI] [PubMed] [Google Scholar]
- 20.Whitlock JP., Jr Annu. Rev. Pharmacol. Toxicol. 1999;39:103. doi: 10.1146/annurev.pharmtox.39.1.103. [DOI] [PubMed] [Google Scholar]
- 21.(a) Murray M, Petrovic N. Curr. Opin. Mol. Ther. 2006;8:480. [PubMed] [Google Scholar]; (b) Ingelman-Sundberg M, Sim SC, Gomez A, Rodriguez-Antona C. Pharmacol. Ther. 2007;116:496. doi: 10.1016/j.pharmthera.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 22.Schuster I, Bernhardt R. Drug Metab. Rev. 2007;39:481. doi: 10.1080/03602530701498455. [DOI] [PubMed] [Google Scholar]
- 23.(a) Ciolino HP, Daschner PJ, Yeh GC. Cancer Res. 1998;58:5707. [PubMed] [Google Scholar]; (b) Mollerup S, Ovrebo S, Haugen A. Int. J. Cancer. 2001;92:18. [PubMed] [Google Scholar]; (c) Chen Z-H, Hurh Y-J, Na H-K, Kim J-H, Chun Y-J, Kim D-H, Kang K-S, Cho M-H, Surh Y-J. Carcinogenesis. 2004;25:2005. doi: 10.1093/carcin/bgh183. [DOI] [PubMed] [Google Scholar]
- 24.Haugland RP, Spence MTZ, Johnson ID, Basey A. The Handbook: A Guide to Fluorescent Probes and Labeling Technologies. 10th ed. Eugene, OR: Molecular Probes; 2005. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Detailed procedures for the synthesis, analysis, and use of fluorogenic probe 1 can be found, in the online version, at doi:10.1016/j.bmcl.2008