Abstract
Matriptase is a type II transmembrane protease that is characterized by an N-terminal transmembrane and multiple extracellular domains, in addition to the conserved extracellular serine protease catalytic domain. The expression pattern of matriptase suggests that this protease may play broad roles in the biology of surface lining epithelial cells. In this study we report that α1-antitrypsin (AAT), an endogenous inhibitor of serine proteases, inhibits the catalytic domain of human recombinant matriptase in vitro. Co-incubation of AAT with matriptase (at a molar ratio 1:2) resulted in the formation of heat stable complexes, clearly seen in sodium dodecyl sulfate electrophoresis and Western blots. AAT was found to be a slow, tight-binding inhibitor of the catalytic domain of matriptase with a second order reaction rate constant of 0.31 × 103 M−1s−1. Notably, the oxidized form of AAT, which lacks serine protease inhibitor activity, failed to generate matriptase complexes and to inhibit matriptase activity. Since matriptase is involved in a number of physiologic processes, including activation of epithelial sodium channels, our findings offer considerable new insights into new regulatory function of AAT in vivo.
Keywords: serine proteases, α1-antitrypsin, matriptase, complex formation, kinetics
Matriptase (also known as MT-SP1, TADG15, suppressor of tumorigenicity 14, epithin; EC 3.4.21) was first isolated by Shi and coworkers (1), and was initially designated a gelatinase because of its gelatinolytic properties and gelatinase-like molecular weight. However, isolation and sequencing of its cDNA revealed a 683-residue multi-domain protein with a C-terminal serine proteinase domain. The enzyme was then named matriptase to emphasize its matrix degrading properties and trypsin-like specificity (2). The mouse, rat, and Xenopus laevis orthologs of matriptase have been cloned (3–5) and shown to share significant sequence homology with the human protein.
Matriptase is a complex trypsin-like type II transmembrane serine protease found in a wide range of human tissues (6). Indeed, matriptase has been specifically detected on cell surfaces at cell–cell junctions in normal epithelial cells, where its activation occurs (7). Studies in knockout mice revealed that matriptase is involved in the development of the epidermis, hair follicles, and cellular immune system (8). Hepatocyte growth factor (HGF), urokinase-type plasminogen activator (uPA), and protease-activated receptor 2 (PAR-2) have all been identified as in vitro substrates of matriptase (8, 9). Recently it has been shown that matriptase is co-localized with prostasin, a glycosylphosphatidylinositol-anchored membrane serine protease believed to be critical for the regulation of epithelial sodium channel activity, and it is likely that matripase is a physiologic activator of prostasin (10). Since matriptase is an auto-activating protease, it may also serve as the initiator of an epidermal proteolytic cascade system, similar to the closely related protease type II transmembrane serine protease, enteropeptidase (11). Matriptase is expressed in prostate, breast, and colorectal cancers in vitro and in vivo (12). Inhibition of matriptase appears to suppress both primary tumor growth and metastasis (9), leading to considerable interest in the development of potent matriptase inhibitors as anti-cancer drugs (13–17).
A number of endogenous matriptase inhibitors have been identified. Jiang and colleagues reported that the sunflower trypsin inhibitor (SFTI-1), isolated from sunflower seeds, inhibits matriptase with a Ki of 0.92 nM (18). In addition, matriptase has been shown to be inhibited by hepatocyte growth factor activator inhibitor-1 (HAI-1), a Kunitz-type protease inhibitor (19).
In this study we report for the first time that the catalytic domain of recombinant human matriptase is inhibited by the human serine protease inhibitor, α1-antitrypsin (AAT), in vitro. AAT is a broad-spectrum serine protease inhibitor used for augmentation therapy in patients with emphysema with inherited AAT deficiency. Recent clinical trials examining the responses of patients with cystic fibrosis to inhaled AAT suggest improvement in several markers of disease activity, including airway inflammation and reduced pseudomonas (20). Thus, it is possible that inhibition of matriptase may contribute to the beneficial effects of AAT in patients with cystic fibrosis and chronic obstructive pulmonary disease.
MATERIALS AND METHODS
AAT Preparations
Purified human plasma AAT was obtained from Calbiochem (Cat nr 178251; San Diego, CA). The purity of AAT preparations was ≥ 95% as determined by SDS-PAGE; the inhibitory activity was > 75%, determined by active-site titration using porcine trypsin as the protease and benzoyl-Arg-p-nitroanilide (Bz-Arg-pNA) as the substrate. Native AAT was diluted in phosphate-buffered saline (PBS), pH 7.4. Native AAT was oxidized with N-chlorosuccinimide (Sigma Chemical Co., St. Louis, MO). Briefly, reaction between AAT and N-chlorosuccinimide in a molar ratio 1:25 in a 0.1-M Tris-HCl buffer (pH 8.0) was allowed to proceed at room temperature for 30 minutes, and oxidized AAT was recovered after passing the reaction mixture through a centrifugal microconcentrator Centricon-30 (Amicon; Millipore Corp., Bedford, MA). The quality of oxidized AAT was analyzed by 7.5% SDS-PAGE. The oxidized AAT was also tested for capacity to form covalent complex with pancreatic elastase (EC 3.4.21.36) (Sigma-Aldrich, St. Louis, MO). Samples of oxidized or native AAT were incubated with pancreatic elastase at a 1.2:1 molar ratio for 30 minutes at room temperature. The reaction was stopped by adding SDS sample buffer, and mixtures were analyzed by 7.5% SDS-PAGE and stained with Coomassie Blue.
Human Matriptase/ST14
Recombinant human matriptase/ST14 (0.4 mg/ml) catalytic domain (Cat No 3946-SE) was purchased from R&D Systems, Inc. (San Diego, CA). Amino acid residues 596 to 855 and 615 to 855, corresponding to the active sites and catalytic domains of the peptide, respectively, were expressed with a N-terminal His tag in Eschrichia coli. The inclusion material was highly purified through a metal chelating column, refolded, and allowed to auto-activate (separating the activation peptide and catalytic domain). No exogenous enzyme was added during the activation process. The activated enzyme was further purified through a gel filtration column, which separated the active enzyme from unfolded fractions. The solution of the purified enzyme was sequenced, and we found that it contains the correct N-terminus amino acid (aa) 615, corresponding to the start of the catalytic domain. The buffer used in these assays contained 50 mM Tris.
Incubation of the catalytic domain of matriptase (13.5 nM) with the recombinant HAI-1 and HAI-2 (specific matriptase inhibitors; R&D Systems), at molar ratios 1:1 and 1:2 (HAI:matriptase) resulted in approximately 50% inhibition of matriptase activity (http://www.rndsystems.com/DAM_public/5886.pdf).
Protein Sequence and Structure Analysis
The protein sequences and structures were obtained from RSCB Protein Data Bank (PDB; http://www.rcsb.org/pdb/). Primary sequence alignment was performed using the ClustalW (21), a general purpose multiple sequence alignment program for proteins (available at the EBI web site http://www.ebi.ac.uk/Tools/clustalw/index.html). Default parameters were used for the analysis. Secondary structure alignment was done using PyMOL version 0.99rc6, a molecular visualization tool available online at http://pymol.sourceforge.net/.
Electrophoresis and Western Blotting
Matriptase (Mw = 26,000 kD, stock 0.5 mg/ml, 19.2 μM) and AAT (Mw = 52,000, stock 1 mg/ml, 19.2 μM) were used for electrophoresis studies. AAT/matriptase complexes were generated by incubating AAT with matriptase at 1:2 or 1:1 molar ratios in PBS (pH 7.4), from 5 minutes to 18 hours at room temperature. The reactions were stopped by the addition of an equal volume of SDS-PAGE sample buffer and boiling for 2 minutes. SDS-PAGE was performed on 7.5% polyacrylamide gels as described previously (22). In some cases, the resultant gels were stained with Coomassie Brilliant Blue R-250; alternatively, the proteins were transferred to a polyvinylidene fluoride (PVDF) membrane (Millipore) using a semi-dry blot electrophoretic transfer system and immunostained with an anti-human AAT monoclonal antibody (B9) IgG1 (1:200) (Santa Cruz Biotechnology, Santa Cruz, CA). The immunocomplexes were visualized with secondary horseradish peroxidase conjugated rabbit anti-mouse antibodies (1:800) (DAKO, A/S, Denmark). DAB (3,3-diaminobenzidine tetrahydrochloride; Sigma) was used as a peroxidase substrate.
Enzymatic Activity Assays
Enzyme activities were monitored by measuring the release of fluorescent AMC from peptide substrate Boc-Gln-Ala-Arg-AMC (Boc: t-Butyloxycarbonyl; AMC: 7-amino-4-methylcoumarin; cat no 1–1550; Bachem, Weil am Rhein, Germany) in a F16 black maxisorp plate using a fluorescent plate reader (FluoStar, Galaxy; Labtech GmbH, Offenburg, Germany). Matriptase was diluted to 5 nM in the assay buffer (50 mM NaCl, 0.01% Tween 20, pH 9.0) and transferred into wells of a black 96-well plate (50 μl/well). The reaction was started by adding 50 μl of substrate (100 μM/well). Fluorescence was continuously monitored for 20 minutes (excitation and emission wavelengths of 380 nm and 460 nm, respectively). The initial rate of the fluorescence increase (AMC generation) was calculated as the enzyme acitivity. These experiments were repeated using a spectrophotometer (Luminescence Spectrometer LS 50B; Perkin Elmer, Waltham, MA) under the same conditions of excitation and emission wavelengths (380 nm and 460 nm, respectively), and the reaction was monitored for 10 minutes with readings every 60 seconds.
Determination of Stoichiometry of Inhibition
Solutions of 19.2 μM matriptase were incubated with varying amounts of AAT (matriptase:AAT molar ratio: 1 to 0.25, 0.75, 0.5, 1, 2, 4, 8, 10, respectively) in the standard assay buffer at room temperature for 18 hours. After incubation, samples were diluted to final matriptase concentrations of 5 nM, and 2.6 μl of 100 μM substrate was added to the 100-μl reaction mixture. The enzyme activities were measured and calculated as described above. Activities were expressed as fractions of matriptase acitivity without AAT. The fraction of active matriptase in each sample was plotted against the ratio of [AAT]/[Matriptase]; results were fitted with linear regression; the X-intercept of this line was the stoichiometry of inhibition (SI).
Determination of the Reaction Rate Constant
The kinetics of inhibition of matriptase by AAT were measured by pre-incubating constant amount of matriptase (5 nM) with AAT (960 nM) for 5 minutes, 15 minutes, 30 minutes, 1 hour, 2 hours, 4 hours, and 18 hours. The matriptase activities were then measured and calculated as described above after the 100-μM substrate addition. Measurements were performed at least three times, with five repeats each.
When [AAT]0 > > [matriptase]0 (the initial concentration of AAT and matriptase, respectively) as used here, the reaction becomes pseudo–first-order for matriptase and kobs = k × [AAT]0. Based on first-order kinetics, the kobs is the slope of the plot of the exponential matriptase activity versus time of the matriptase incubation with AAT. Thus, k was solved using 960 nM of [AAT]0 here. Data within six half-lives were used for the plot of ln(activity) versus time.
Statistical Analysis
Data are shown as means ± 1 SD (or SEM) unless otherwise noted. The differences in the means in experimental results were analyzed for their statistical significance with independent-samples two-sided t test and/or one-way ANOVA combined with a multiple comparisons procedure (Scheffé multiple range test), with the overall significance level of α = 0.05. A Statistical Package for Social Sciences (SPSS for Windows, release 6.0; SPSS Inc., Chicago, IL) was used for the calculations.
RESULTS
AAT is the prototypic member of the serpin super family, an acute phase protein, and a major inhibitor of neutrophil elastase and protease 3. AAT, like other serpins, forms denatured-stable complexes with its target proteases and behaves kinetically as an irreversible (suicide) inhibitor (23). AAT is mainly synthesized in the liver, occurs in high concentrations in plasma (∼ 26 μM), and transudes from plasma into the lung epithelial lining fluid, where its concentration is approximately 4 μM (24). Under inflammatory conditions the concentration of AAT can rise by 3- to 5-fold above normal, and therefore the control of the excessive proteolytic activity by AAT has long been recognized to be crucial in protecting the lung parenchyma and other tissues in many inflammatory diseases (25). AAT has also been shown to regulate alveolar fluid clearance because of its ability to inhibit trypsin-like protease (20–28 kD), which is secreted into alveolar fluid by epithelial and inflammatory cells and which can activate ENaC (26). Furthermore, recent studies show that AAT can form complexes and inhibit activity of caspase-3 (a cysteine protease) in vitro and in vivo, and can protect various cells, including lung endothelial cells, against apoptosis (27).
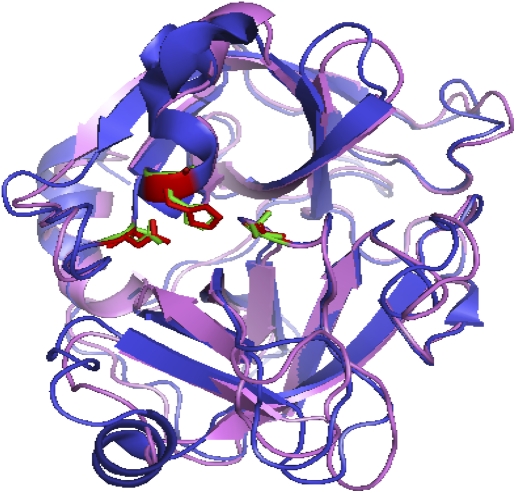
When the protein sequences of the three serine proteases, namely human neutrophil elastase (1B0F_A), porcine pancreatic elastase (1B0E_A), and catalytic domain of human matriptase (1EAX_A) were aligned with the ClustalW (21) program, a high sequence alignment score of 1,124 was obtained for all three proteins. Figure 1 shows that the sequences are well aligned with the catalytic triad (His 57, Asp 102, and Ser 195 highlighted in yellow in Figure 1) located in the active site of these enzymes well preserved. The catalytic triad is a coordinated structure consisting of these 3 amino acids located near the heart of the enzyme and each amino acid plays a key role in the cleaving ability of the proteases (28). As shown in Figure 2, crystal structure superimposition of the catalytic domains of human matriptase and neutrophil elastase structures confirms the alignment of the catalytic triad. The RMS score for this superimposition was 1.14 using the PyMOL molecular visualization tool suited for secondary structure alignments and production of high-quality three-dimensional images of proteins. The catalytic domain of human matriptase reveals the overall structural similarity to a (chymo)trypsin-like serine protease, specifically to elastase (Figures 1 and 2), but displays unique properties (such as the hydrophobic/acidic S2/S4 sub-sites) which considerably affect its substrate recognition and binding properties. Thus, the catalytic domain of human matriptase has a potential to bind to and form complexes with AAT. Therefore, in this study we examined for reaction between AAT and catalytic domain of matriptase.
Figure 1.
ClustalW alignment of the sequences of human neutrophil elastase (1B0F_A), porcine pancreatic elastase (1B0E_A), and human matriptase (1EAX_A). Sequence identities are marked with an asterisk underneath and shown in red. The catalytic triad consisting of amino acids His 57, Asp 102, and Ser 195 is shown in yellow. Residues with “:” or “.” underneath denote conserved amino acids.
Figure 2.
Superimposed crystal structures of human neutrophil elastase (violet) and human matriptase (blue) with the catalytic triad amino acids (His 57, Asp 102, and Ser 195) highlighted in red (for matriptase) and green (for elastase).
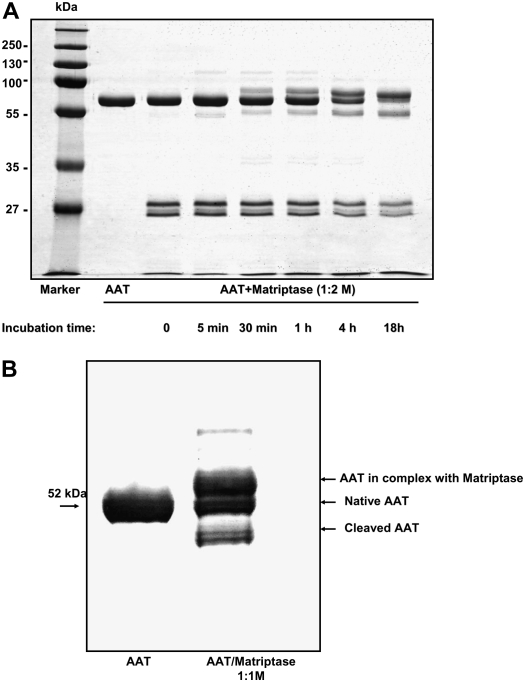
Native AAT was incubated with matriptase at a molar ratio 1:2 for variable time periods before SDS-polyacrylamide gel electrophoresis. There was a clear evidence for the time-dependent formation of AAT/matriptase complex. Figure 3A shows that the native AAT/matriptase mixtures exhibited three major protein bands corresponding to a complex between AAT and matriptase (∼ 78 kD), unreacted AAT (52 kD), and the cleaved AAT (45 kD). After 18 hours of AAT/matriptase incubation, all AAT appeared to be reacted with matriptase. Of note, the autoactivation process generated the mature form and a small pro-sequence of catalytic domain of matriptase (Figure 3A). In addition, AAT/matriptase mixtures (1:1), incubated for 18 hours at room temperature, were immunostained with a specific monoclonal antibody against AAT. As illustrated in Figure 3B, human AAT forms SDS-resistant complexes with catalytic domain of matriptase, suggestive of a covalent interaction. These findings indicate that AAT is a slow-binding inhibitor of matriptase catalytic domain that requires a certain time to build a inhibitor–protease complex. It is important to point out that the AAT/matriptase complex, similar to other AAT serine protease complexes, remained SDS-stable after incubation at room temperature for 24 and 48 hours, suggestive of a covalent interaction (data not shown).
Figure 3.
(A) 7.5% SDS-PAGE and (B) Western blot of native AAT and matriptase catalytic domain mixtures. (A) Native AAT/matriptase mixtures were prepared by incubating native AAT (1 mg/ml, 19.2 μM) with matriptase (0.5 mg/ml, 19.2 μM) at 1:2 molar ratio in PBS, pH 7.4, at room temperature for variable periods of time. AAT alone was incubated under the same experimental conditions. The reaction was stopped by the addition of an equal volume of SDS-PAGE sample buffer and boiling. Representative gel which was repeated seven times with identical results. (B) Native AAT/matriptase complex was prepared by incubating AAT with matriptase at 1:1 molar ratio in PBS, pH 7.4, at room temperature for 18 hours. Native AAT alone or in mixture with matriptase were separated on 7.5% SDS-PAGE and transferred to a polyvinylidene fluoride (PVDF) membrane using a semi-dry blot electrophoretic transfer system, and were immunostained with an anti-human AAT monoclonal antibody (B9) IgG1 (1:2,000 dilution). Three forms of AAT were detected: AAT in complex with matriptase, unreacted, and cleaved AAT. Representative Western blot, which was repeated three times, is shown.
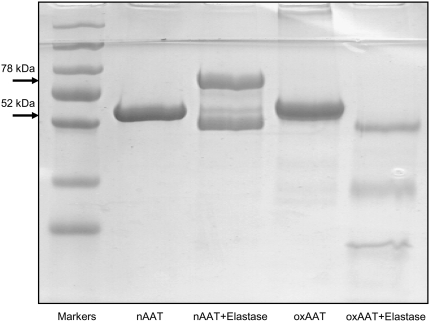
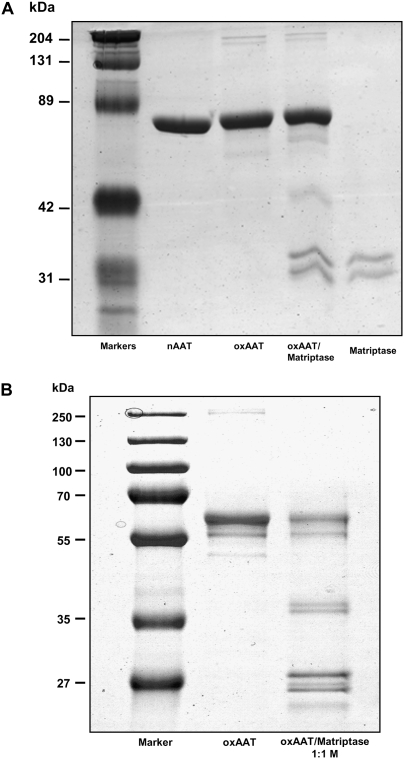
The structural properties that confer protease inhibitor activity of AAT make it also susceptible to post-translational modifications by oxidation, nitration, and polymerization (29). For instance, the oxidation of AAT by the neutrophil-derived myeloperoxidase/H2O2 system (30), activated neutrophils (31), or oxidation by components of cigarette smoke (32) result in oxidation of two of the nine methionines in AAT (358 and 351), leading to loss of inhibitory activity (33). We showed that oxidation with N-chlorosuccinimide (0.05 to 1 mg/ml) abolished the ability of AAT to form SDS-stable complexes with both neutrophil elastase (Figure 4) and with the catalytic domain of matripase (Figure 5A). In addition, the intensity of the 52-kD band of oxidized AAT decreased with time, and lower-molecular-size fragments were detected, indicating that the oxidized AAT is a substrate of matriptase (Figure 5B). Since fragments of AAT show a variety of biological functions (34), further studies are warranted to identify and characterize the fragments of oxidized AAT cleavage with matriptase.
Figure 4.
SDS-PAGE (7.5%) of oxidized and native AAT incubated with human elastase. Lane 1, molecular size markers; lane 2, native (n) AAT; lane 3, native AAT + elastase incubated for 30 minutes at a molar ratio 1.2:1; lane 4, oxidized (ox) AAT; lane 5, oxidized AAT + elastase. Figure is characteristic of an experiment that was repeated twice, with identical results.
Figure 5.
SDS-PAGE (7.5%) of oxidized AAT and matriptase catalytic domain mixtures. Oxidized AAT/matriptase complex was prepared by incubating AAT with matriptase at 1:1 molar ratio in PBS, pH 7.4, for (A) 30 minutes and (B) 18 hours at room temperature. Figure is characteristic of an experiment that was repeated twice, with similar results.
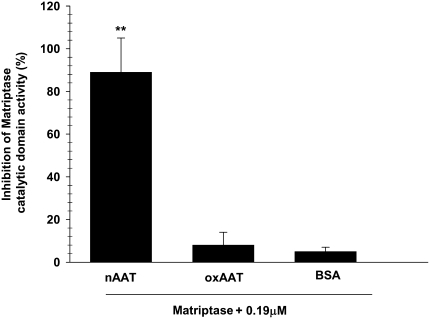
Parallel studies for functional effects of AAT on matriptase catalytic domain activity have shown that native, but not oxidized, AAT inhibits enzyme activity. Bovine serum albumin used as a negative control showed no significant effect on matriptase activity (Figure 6). Also, native but not oxidized AAT has been shown to inhibit amiloride-sensitive currents across Xenopus oocytes, heterologously transfected with α, β, and γ ENaC (35).
Figure 6.
Inhibition of matriptase activity by AAT. Effects of native (n) ad oxidized (ox) AAT, as well as bovine serum albumin (0.19 μM each), on the enzymatic activity of matriptase catalytic domain (2.5 nM). Results are shown as percent inhibition of maximum activity. Values are mean ± SD (n = 3); * P < 0.001 compared with nAAT (by t test).
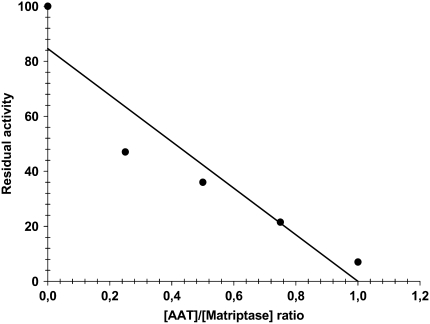
From the relatively slow formation of AAT–matriptase SDS-stable complexes, it became clear that a large-molar excess of AAT over the matriptase catalytic domain is required to observe full inhibition of enzyme in relatively shorter time period. A plot of the residual matriptase activity versus AAT/matriptase molar ratios is shown in Figure 7. Fitting the data with a straight line using linear regression (r2 = 0.882; P < 0.01) resulted in an x-intercept corresponding to the SI. Under our experimental conditions (18 h incubation of AAT and matriptase) the SI is equal to 1.
Figure 7.
Stoichiometry of inhibition. Matriptase (19.2 μM) was pre-incubated with various concentrations of AAT (from 1:0.25 to 1:10, [Matriptase]:[AAT] molar ratio) for 18 hours before assay of matriptase inhibition. Data were fitted with linear regression (r2 = 0.882; P < 0.01; X intercept = 1). Figure is characteristic of two experiments with similar results.
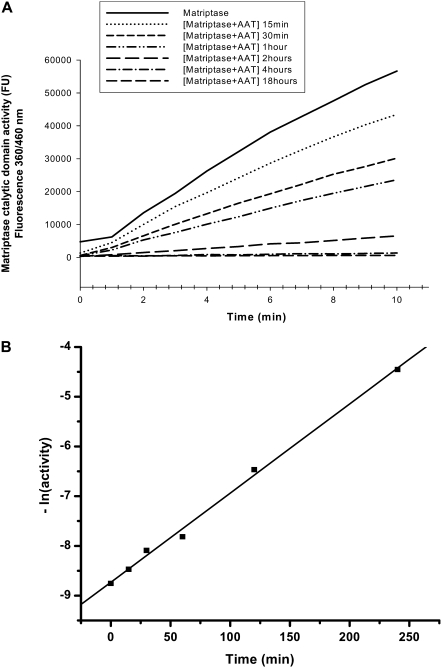
We also calculated the reaction rate constant (k) by plotting the exponential free matriptase activity versus time of incubation with AAT, as described in Materials and Methods (Figure 8B). A good linear fit was obtained (R = 0.99812), and from the slope of the fitted line the k was calculated as 0.31 × 103 M−1s−1 (kobs = k × [AAT]0 = 0.01794 min−1).
Figure 8.
Determination of reaction rate constant (k). (A) Kinetic curves of matriptase activity (cleavage of substrate) measured after pre-incubating matriptase catalytic domain (5 nM) with vehicle or with native AAT (960 nM) for 15 minutes, 30 minutes, 1 hour, 2 hours, 4 hours, and 18 hours. Three independent experiments were performed, with five repeats each. Figure shows one representative experiment of five repeats. (B) As described in Materials and Methods, the matriptase activity was calculated from A, and the exponential free enzyme activity was plotted versus time of incubation with AAT. A good linear fit was obtained (R = 0.99812). The k was calculated as 0.31 × 103 M−1s−1 from the slope of the plot (kobs = k × [AAT]0 = 0.01794 min−1). This experiment was repeated twice, with identical results.
This slow rate constant as well as our SDS-gel electrophoresis results confirm that AAT is a slow-binding matripase catalytic domain inhibitor. We speculate that this low inhibition rate, compared with other extracellular serine proteases, such as elastase (k = 6.5 × 107 M−1 s−1) (36), may reflect the requirement for specific microenvironments and effective (large) concentrations of the AAT at intracellular versus extracellular locations. A recent study, however, has shown matriptase complexes in human milk, which contain no HAI-1 (specific inhibitor of matriptase) and which are resistant to dissociation in boiling SDS in the absence of reducing agents. Proteomic and immunologic analysis identified the 45-kD fragment as the noncatalytic domains of matriptase and the 80-kD fragment as the matriptase serine protease domain covalently linked to serine protease inhibitors such as AAT, antithrombin III, and α2-antiplasmin. This finding for the first time provides evidence in vivo that the proteolytic activity of matriptase may be controlled by secreted serpins such as AAT (37). These data are in agreement with our findings and point out a potentially new function of AAT. Furthermore, our kinetics data indicate that even the small amount of AAT in the extracellular space is sufficient to inhibit matriptase.
We would like to offer a few examples to illustrate the putative physiologic importance of AAT–matriptase interaction. For example, the leading hypothesis regarding the cause of airway disease in cystic fibrosis is that excessive Na+ absorption leads to inadequate hydration, resulting in mucus stasis and recurrent infection. Prostasin (CAP1/PRSS8) is a glycosylphosphatidylinositol-anchored membrane serine protease believed to be critical for the regulation of epithelial sodium channel activity in cystic fibrosis. Prostasin is synthesized as an inactive zymogen that requires a site-specific endoproteolytic cleavage to be converted to an active protease. It has been recently reported that matriptase is necessary for this prostasin activation (11). Therefore, the inhibition of matriptase by AAT may provide a pharmacologic means of improving Na+ current in cystic fibrosis airway. As mentioned above, AAT decreases amiloride-sensitive transport across both Xenopus oocytes, heterologously injected with ENaC, H441 cells (a human Clara cell line expressing native ENaC) and across the alveolar epithelium of rats in vivo (25, 35).
On the other hand, the overexpression of matriptase is a consistent feature of human epithelial tumors. It has been shown that matriptase possesses a strong oncogenic potential when unopposed by its endogenous inhibitor, HAI-1 (38). Therefore, inhibition of proteolytic activity as a result of various kinds of proteinase–proteinase inhibitor interactions is an important aspect of tumor cell biology. Serpins such as maspin, AAT, α1-antichymotrypsin, and Kunitz-type inhibitors such as urinary trypsin inhibitor (39) have been previously implicated in suppression of cancer invasion. It has been reported that cultured human tumor cell lines produce serine protease inhibitors, including AAT, which may play a role in controlling tumor growth and invasion by regulating activity of specific enzymes.
In summary, our data provide new evidence for the formation of irreversible interactions among AAT, the most important serine-protease inhibitor, and matriptase, a serine-bound matriptase with diverse biological functions.
Acknowledgments
The authors thank Dr. Peter Andreasen, Department of Molecular Biology, University of Aarhus, Denmark, for expertise and assistance with enzymatic activity studies.
This work was supported by grants from the Swedish Research Council, the Grifol's Institute, Crafoords Foundation, and the Lundström Foundation to S.J., and by PHS grants HL-071189 to J.R.L. and HL-31197, HL-51173, and NIEHS-1U01ES015676 to S.M.
Originally Published in Press as DOI: 10.1165/rcmb.2008-0015RC on August 21, 2008
Conflict of Interest Statement: S.J. and S.M. received $55,100 from Talecris Biotherapeutics, Inc. for research. None of the other authors has a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Shi YE, Torri J, Yieh L, Wellstein A, Lippman ME, Dickson RB. Identification and characterization of a novel matrix-degrading protease from hormone-dependent human breast cancer cells. Cancer Res 1993;53:1409–1415. [PubMed] [Google Scholar]
- 2.Lin CY, Anders J, Johnson M, Sang QA, Dickson RB. Molecular cloning of cDNA for matriptase, a matrix-degrading serine protease with trypsin-like activity. J Biol Chem 1999;274:18231–18236. [DOI] [PubMed] [Google Scholar]
- 3.Kim MG, Chen C, Lyu MS, Cho EG, Park D, Kozak C, Schwartz RH. Cloning and chromosomal mapping of a gene isolated from thymic stromal cells encoding a new mouse type II membrane serine protease, epithin, containing four LDL receptor modules and two CUB domains. Immunogenetics 1999;49:420–428. [DOI] [PubMed] [Google Scholar]
- 4.Satomi S, Yamasaki Y, Tsuzuki S, Hitomi Y, Iwanaga T, Fushiki T. A role for membrane-type serine protease (MT-SP1) in intestinal epithelial turnover. Biochem Biophys Res Commun 2001;287:995–1002. [DOI] [PubMed] [Google Scholar]
- 5.Yamada K, Takabatake T, Takeshima K. Isolation and characterization of three novel serine protease genes from Xenopus laevis. Gene 2000;252:209–216. [DOI] [PubMed] [Google Scholar]
- 6.Takeuchi T, Shuman MA, Craik CS. Reverse biochemistry: use of macromolecular protease inhibitors to dissect complex biological processes and identify a membrane-type serine protease in epithelial cancer and normal tissue. Proc Natl Acad Sci USA 1999;96:11054–11061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hung RJ, Hsu Ia W, Dreiling JL, Lee MJ, Williams CA, Oberst MD, Dickson RB, Lin CY. Assembly of adherens junctions is required for sphingosine 1-phosphate-induced matriptase accumulation and activation at mammary epithelial cell-cell contacts. Am J Physiol Cell Physiol 2004;286:C1159–C1169. [DOI] [PubMed] [Google Scholar]
- 8.List K, Haudenschild CC, Szabo R, Chen W, Wahl SM, Swaim W, Engelholm LH, Behrendt N, Bugge TH. Matriptase/MT-SP1 is required for postnatal survival, epidermal barrier function, hair follicle development, and thymic homeostasis. Oncogene 2002;21:3765–3779. [DOI] [PubMed] [Google Scholar]
- 9.Takeuchi T, Harris JL, Huang W, Yan KW, Coughlin SR, Craik CS. Cellular localization of membrane-type serine protease 1 and identification of protease-activated receptor-2 and single-chain urokinase-type plasminogen activator as substrates. J Biol Chem 2000;275:26333–26342. [DOI] [PubMed] [Google Scholar]
- 10.List K, Hobson JP, Molinolo A, Bugge TH. Co-localization of the channel activating protease prostasin/(CAP1/PRSS8) with its candidate activator, matriptase. J Cell Physiol 2007;213:237–245. [DOI] [PubMed] [Google Scholar]
- 11.Netzel-Arnett S, Currie BM, Szabo R, Lin CY, Chen LM, Chai KX, Antalis TM, Bugge TH, List K. Evidence for a matriptase-prostasin proteolytic cascade regulating terminal epidermal differentiation. J Biol Chem 2006;281:32941–32945. [DOI] [PubMed] [Google Scholar]
- 12.Oberst M, Anders J, Xie B, Singh B, Ossandon M, Johnson M, Dickson RB, Lin CY. Matriptase and HAI-1 are expressed by normal and malignant epithelial cells in vitro and in vivo. Am J Pathol 2001;158:1301–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galkin AV, Mullen L, Fox WD, Brown J, Duncan D, Moreno O, Madison EL, Agus DB. CVS-3983, a selective matriptase inhibitor, suppresses the growth of androgen independent prostate tumor xenografts. Prostate 2004;61:228–235. [DOI] [PubMed] [Google Scholar]
- 14.Enyedy IJ, Lee SL, Kuo AH, Dickson RB, Lin CY, Wang S. Structure-based approach for the discovery of bis-benzamidines as novel inhibitors of matriptase. J Med Chem 2001;44:1349–1355. [DOI] [PubMed] [Google Scholar]
- 15.Stoop AA, Craik CS. Engineering of a macromolecular scaffold to develop specific protease inhibitors. Nat Biotechnol 2003;21:1063–1068. [DOI] [PubMed] [Google Scholar]
- 16.Sun J, Pons J, Craik CS. Potent and selective inhibition of membrane-type serine protease 1 by human single-chain antibodies. Biochemistry 2003;42:892–900. [DOI] [PubMed] [Google Scholar]
- 17.Desilets A, Longpre JM, Beaulieu ME, Leduc R. Inhibition of human matriptase by eglin c variants. FEBS Lett 2006;580:2227–2232. [DOI] [PubMed] [Google Scholar]
- 18.Jiang S, Li P, Lee SL, Lin CY, Long YQ, Johnson MD, Dickson RB, Roller PP. Design and synthesis of redox stable analogues of sunflower trypsin inhibitors (SFTI-1) on solid support, potent inhibitors of matriptase. Org Lett 2007;9:9–12. [DOI] [PubMed] [Google Scholar]
- 19.Szabo R, Molinolo A, List K, Bugge TH. Matriptase inhibition by hepatocyte growth factor activator inhibitor-1 is essential for placental development. Oncogene 2007;26:1546–1556. [DOI] [PubMed] [Google Scholar]
- 20.Griese M, Latzin P, Kappler M, Weckerle K, Heinzlmaier T, Bernhardt T, Hartl D. alpha1-Antitrypsin inhalation reduces airway inflammation in cystic fibrosis patients. Eur Respir J 2007;29:240–250. [DOI] [PubMed] [Google Scholar]
- 21.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 1994;22:4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moraga F, Janciauskiene S. Activation of primary human monocytes by the oxidized form of alpha1-antitrypsin. J Biol Chem 2000;275:7693–7700. [DOI] [PubMed] [Google Scholar]
- 23.Gettins PG. Serpin structure, mechanism, and function. Chem Rev 2002;102:4751–4804. [DOI] [PubMed] [Google Scholar]
- 24.Ogushi F, Hubbard RC, Fells GA, Casolaro MA, Curiel DT, Brantly ML, Crystal RG. Evaluation of the S-type of alpha-1-antitrypsin as an in vivo and in vitro inhibitor of neutrophil elastase. Am Rev Respir Dis 1988;137:364–370. [DOI] [PubMed] [Google Scholar]
- 25.Carrell RW, Lomas DA. Alpha1-antitrypsin deficiency–a model for conformational diseases. N Engl J Med 2002;346:45–53. [DOI] [PubMed] [Google Scholar]
- 26.Swystun V, Chen L, Factor P, Siroky B, Bell PD, Matalon S. Apical trypsin increases ion transport and resistance by a phospholipase C-dependent rise of Ca2+. Am J Physiol Lung Cell Mol Physiol 2005;288:L820–L830. [DOI] [PubMed] [Google Scholar]
- 27.Petrache I, Fijalkowska I, Medler TR, Skirball J, Cruz P, Zhen L, Petrache HI, Flotte TR, Tuder RM. alpha-1 antitrypsin inhibits caspase-3 activity, preventing lung endothelial cell apoptosis. Am J Pathol 2006;169:1155–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rawlings ND, Barrett AJ. Evolutionary families of peptidases. Biochem J 1993;290:205–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Janciauskiene S. Conformational properties of serine proteinase inhibitors (serpins) confer multiple pathophysiological roles. Biochim Biophys Acta 2001;1535:221–235. [DOI] [PubMed] [Google Scholar]
- 30.Matheson NR, Wong PS, Travis J. Enzymatic inactivation of human alpha-1-proteinase inhibitor by neutrophil myeloperoxidase. Biochem Biophys Res Commun 1979;88:402–409. [DOI] [PubMed] [Google Scholar]
- 31.Padrines M, Schneider-Pozzer M, Bieth JG. Inhibition of neutrophil elastase by alpha-1-proteinase inhibitor oxidized by activated neutrophils. Am Rev Respir Dis 1989;139:783–790. [DOI] [PubMed] [Google Scholar]
- 32.Carp H, Miller F, Hoidal JR, Janoff A. Potential mechanism of emphysema: alpha 1-proteinase inhibitor recovered from lungs of cigarette smokers contains oxidized methionine and has decreased elastase inhibitory capacity. Proc Natl Acad Sci USA 1982;79:2041–2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taggart C, Cervantes-Laurean D, Kim G, McElvaney NG, Wehr N, Moss J, Levine RL. Oxidation of either methionine 351 or methionine 358 in alpha 1-antitrypsin causes loss of anti-neutrophil elastase activity. J Biol Chem 2000;275:27258–27265. [DOI] [PubMed] [Google Scholar]
- 34.Congote LF. The C-terminal 26-residue peptide of serpin A1 is an inhibitor of HIV-1. Biochem Biophys Res Commun 2006;343:617–622. [DOI] [PubMed] [Google Scholar]
- 35.Janciauskiene S, Nita I, Lazrak A, Ji H-L, Matalon S. Antitrypsin decreases amiloride-sensitive currents across Xenopus oocytes injected with alpha beta gamma ENaC cRNAs. American Thoracic Society International Conference, Toronto, Canada, May 16–18, 2008. Am J Respir Crit Care Med 2008;177:A329. (Abstract). [Google Scholar]
- 36.Beatty K, Bieth J, Travis J. Kinetics of association of serine proteinases with native and oxidized alpha-1-proteinase inhibitor and alpha-1-antichymotrypsin. J Biol Chem 1980;255:3931–3934. [PubMed] [Google Scholar]
- 37.Tseng IC, Chou FP, Su SF, Oberst MD, Madayiputhiya N, Lee MS, Wang JK, Sloane DE, Johnson MD, Lin CY. Purification from human milk of matriptase complexes with secreted serpins: mechanism for inhibition of matriptase in the absence of HAI-1. Am J Physiol Cell Physiol 2008;295:C423–C431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.List K, Szabo R, Molinolo A, Sriuranpong V, Redeye V, Murdock T, Burke B, Nielsen BS, Gutkind JS, Bugge TH. Deregulated matriptase causes ras-independent multistage carcinogenesis and promotes ras-mediated malignant transformation. Genes Dev 2005;19:1934–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kataoka H, Itoh H, Koono M. Emerging multifunctional aspects of cellular serine proteinase inhibitors in tumor progression and tissue regeneration. Pathol Int 2002;52:89–102. [DOI] [PubMed] [Google Scholar]