Abstract
Mononuclear phagocytes play an important role in the removal of apoptotic cells by expressing cell surface receptors that recognize and remove apoptotic cells. Based on the knowledge that cigarette smoking is associated with increased lung cell turnover, we hypothesized that alveolar macrophages (AMs) of normal cigarette smokers may exhibit enhanced expression of apoptotic cell removal receptor genes. AMs obtained by bronchoalveolar lavage of normal nonsmokers (n = 11) and phenotypic normal smokers (n = 13; 36 ± 6 pack-years) were screened for mRNA expression of all known apoptotic cell removal receptors using Affymetrix HG-U133 Plus 2.0 microarray chips with TaqMan RT-PCR confirmation. Of the 14 known apoptotic receptors expressed, only MER tyrosine kinase (MERTK), a transmembrane tyrosine kinase receptor, was significantly up-regulated in smokers. MERTK expression was then assessed in AMs of smokers versus nonsmokers by TaqMan RT-PCR, immunocytochemistry, Western analysis, and flow analysis. Smoker AMs had up-regulation of MERTK mRNA levels (smoker vs. nonsmoker: 3.6-fold by microarray, P < 0.003; 9.5-fold by TaqMan RT-PCR, P < 0.02). Immunocytochemistry demonstrated a qualitative increase in MERTK protein expression on AMs of smokers. Increased protein expression of MERTK on AMs of smokers was confirmed by Western and flow analyses (P < 0.007 and P < 0.0002, respectively). MERTK, a cell surface receptor that recognizes apoptotic cells, is expressed on human AMs, and its expression is up-regulated in AMs of cigarette smokers. This up-regulation of MERTK may reflect an increased demand for removal of apoptotic cells in smokers, an observation with implications for the development of chronic obstructive pulmonary disease, a disorder associated with dysregulated apoptosis of lung parenchymal cells.
Keywords: apoptosis, MER tyrosine kinase, alveolar macrophages
CLINICAL RELEVANCE
The observation that MER receptor tyrosine kinase (MERTK), an apoptotic cell removal receptor on alveolar macrophages, is up-regulated may reflect an increased demand for removal of dying cells and this, in turn, has implications for the development of chronic obstructive pulmonary disease.
MER receptor tyrosine kinase (MERTK), a 110-kD transmembrane protein member of the receptor tyrosine kinase family of cell surface receptors, plays a role in the clearance of apoptotic cells (1–4). The MERTK gene is normally expressed on mononuclear phagocytes, dendritic cells, retinal pigment epithelial (RPE) cells, and reproductive tissue (5–8). In mice, deletion of MERTK results in delayed clearance of apoptotic cells and development of autoimmune disease (1, 9, 10). In humans, mutations in MERTK result in retinitis pigmentosa, a disease characterized by a defect in the RPE phagocytosis pathway, where the RPE cells are unable to ingest the shed apoptotic tips of photoreceptor cells (11–14).
In the lung, the clearance of apoptotic cells is the normal function of alveolar macrophages (AMs), the monocyte-derived pulmonary representative of the mononuclear phagocyte system (15–18). Because phagocytosis of apoptotic cells by mononuclear phagocytes invokes an array of receptors on the phagocyte that interacts with bridging molecules and cell-specific ligands on the apoptotic cells, the control of expression of apoptotic receptors on the phagocyte plays an important role in the clearance of apoptotic cells in the lung (19–23). Based on the knowledge that AMs from normal cigarette smokers are defective in the phagocytosis of apoptotic bronchial epithelial cells, together with a concomitant reduction in expression of some of the known apoptotic cell removal receptors (24), and that blood monocytes express MERTK (5), we asked the questions: (1) is MERTK expressed on human AMs, and (2) is the expression of MERTK modified by cigarette smoking? The data demonstrate that MERTK is expressed in AMs. Interestingly, the expression of MERTK is significantly up-regulated in AMs of smokers compared with nonsmokers, as assessed at the mRNA level by microarray and TaqMan RT-PCR, and at the protein level, as assessed by immunocytochemical staining, Western blot, and flow analysis. Similarly, AMs from smokers with chronic obstructive pulmonary disease (COPD) have increased expression of MERTK, as assessed by TaqMan RT-PCR and immunocytochemical analysis. Interestingly, this has functional consequences, as blocking the AM MERTK receptor in normal nonsmokers and normal smokers results in decreased clearance of apoptotic cells.
In the context that there is more cell turnover in smokers and individuals with COPD (25–34), the observation that MERTK is up-regulated and functional in the AMs of smokers may reflect several important findings. It may reflect not only an increased demand for removal of apoptotic cells but also an effort to compensate for the lack of up-regulation or dysfunction of other apoptotic cell removal receptors which would ultimately decrease the inflammatory burden in the lungs of smokers.
MATERIALS AND METHODS
Study Population
Normal nonsmokers, healthy smokers, and smokers with established COPD were evaluated at the Weill Cornell National Institutes of Health General Clinical Research Center and Department of Genetic Medicine Clinical Research Facility under protocols approved by the Weill Cornell Medical College Institutional Review Board. Written informed consent was obtained from each individual before enrollment in the study. Normal nonsmokers and normal smokers were determined to be phenotypically normal on the basis of clinical history, physical examination, routine blood screening tests, urinalysis, chest X-ray, electrocardiogram, and pulmonary function testing. Current smoking status was confirmed by history, venous carboxyhemoglobin levels, and urinalysis for levels of nicotine and its derivative, cotinine. Smokers with established COPD were defined according to Global Initiative for Chronic Obstructive Lung Disease (GOLD) criteria (35). Individuals who met the inclusion criteria underwent flexible fiberoptic bronchoscopy and bronchoalveolar lavage (BAL) to obtain AMs.
Collection of Alveolar Macrophages and Bronchoalveolar Fluid
Fiberoptic bronchoscopy was performed to obtain human AMs by BAL using methods described previously (34, 36). The total volume used per site was typically 100 ml, and a maximum of three sites were lavaged per individual. Recovery of the infused volume ranged from 45 to 65%. BAL fluid was filtered through gauze to remove debris and mucus and then centrifuged at 1,200 rpm for 5 minutes at 4°C. Cells were washed twice in RPMI 1,640 containing 10% FBS, 50 U/ml streptomycin, and 2 mM glutamine (Invitrogen, Carlsbad, CA) and plated in six-well tissue culture plates (2 × 106 in 2 ml/well). After overnight adherence at 37°C in a 5% CO2, humidified incubator, nonadherent cells were gently removed. Cell viability was estimated by Trypan blue exclusion and expressed as a percentage of the total cells recovered. Total cell number was determined by counting on a hemocytometer. Differential cell count was assessed on sedimented cells prepared by cytocentrifugation (Cytospin 3; Shandon Instruments, Pittsburgh, PA) stained with DiffQuik and performed by counting 500 cells on each slide. Light microscopy was used to assess the morphological features of the cells, and all samples tested contained greater than 88% AMs before the overnight adherence and greater than 98% AMs after adherence. Cells not processed for RNA extraction were used for immunocytochemical, Western blot, and flow cytometry studies. BAL supernatant was also collected, concentrated 10-fold with Centricon Plus-20 centrifugal filter units (Millipore, Billerica, MA), and stored in −80°C until use.
RNA Extraction and Microarray Processing
Total RNA was extracted using TRIzol reagent (Invitrogen) followed by RNeasy (Qiagen, Valencia, CA) to remove residual DNA, yielding 2–4 μg RNA/106 cells. An aliquot of each RNA sample was run on an Agilent Bioanalyzer (Agilent Technologies, Palo Alto, CA) to visualize and quantify the degree of RNA integrity. A NanoDrop ND-100 spectrophotometer (NanoDrop Technologies, Wilmington, DE) was used to determine the concentration of RNA. Quality control criteria used to accept an RNA sample for further processing included: (1) A260/A280 ratio between 1.7 and 2.3; (2) concentration within the range of 0.2 to 6 μg/ml; and (3) Agilent electropherogram displaying two distinct peaks corresponding to the 28S and 18S ribosomal RNA bands at a ratio of 28S:18S of greater than 0.5 with minimal or no degradation. Double-stranded cDNA was synthesized from 3 μg of total RNA using the GeneChip One-Cycle cDNA Synthesis Kit, followed by cleanup with GeneChip Sample Cleanup Module, in vitro transcription reaction using the GeneChip IVT Labeling Kit, and cleanup and quantification of the biotin-labeled cRNA yield by spectrophotometric analysis (all kits from Affymetrix, Santa Clara, CA). Hybridizations to test chips and to the HG-U133 Plus 2.0 array (54,000 probe sets) were done according to Affymetrix protocols, processed by the Affymetrix GeneChip Fluidics Station 450, and scanned with an Affymetrix GeneChip Scanner 3,000 7G. To maintain quality, only samples hybridized to test chips with a 3′:5′ ratio of less than 3 were deemed satisfactory.
Microarray Data Analysis
Analysis was performed on Affymetrix HG-U133 Plus 2.0 microarrays. Captured images were analyzed using Microarray Suite version 5.0 (MAS 5.0) algorithm (Affymetrix). These data were normalized using GeneSpring version 7.3 software (Agilent Technologies) as follows: (1) per array, by dividing the raw data by the 50th percentile of all measurements; and (2) per gene, by dividing the raw data by the median expression level for all the genes across all arrays in a data set.
TaqMan RT-PCR Confirmation of Microarray Expression Levels
cDNA was synthesized from 2 μg RNA in a 100 μl reaction volume using the TaqMan Reverse Transciptase Reaction Kit (Applied Biosystems, Foster City, CA) with random hexamers as primers. Two dilutions of 1:50 and 1:100 were made from each sample, and triplicate wells were run with each dilution. TaqMan PCR reactions were performed using premade kits from Applied Biosystems and 2 μl of cDNA was used in each 25-μl reaction volume. The endogenous control was 18S ribosomal RNA, and relative expression levels were determined using the ΔΔCt method (Applied Biosystems), with the average value for the nonsmokers as the calibrator. The rRNA probe was labeled with VIC dye and the probe for MERTK labeled with FAM (6-carboxy fluorescein). The PCR reactions were run in an Applied Biosystems Sequence Detection System 7500, and the relative quantity was determined using the algorithm provided by Applied Biosystems.
Cell Lines
Rat RPE cells were kindly provided by Ching-Hwa Sung from the Department of Cell and Developmental Biology, Weill Medical College of Cornell University. RPE cells were maintained in Dulbecco's modified Eagle medium with 4% heat-inactivated FBS (Cellect Gold FBS), and incubated at 32°C in 5% CO2.
The human Jurkat T cell line was obtained from the American Type Culture Collection (Manassas, VA) and cultured in RPMI 1,640 medium with 10% FBS, supplemented with glutamine, penicillin, and streptomycin, and incubated at 37°C in 5% CO2.
Immunocytochemistry
The immunocytochemical procedure was performed on samples obtained by BAL cytocentrifugation followed by adherence of AMs to culture well dishes (37). Alveolar macrophages were purified by allowing them to adhere to culture well dishes (3.5 × 104) for 2 hours, at 37°C in a 5% CO2, humidified incubator. Any nonadherent cells were removed by washing with PBS. The slides were then fixed with 4% paraformaldehyde for 10 minutes, washed with PBS five times, and then blocked with 5% donkey serum for 20 minutes at 23°C to inhibit nonspecific immunoreactivity. The slides were incubated overnight at 4°C with the primary antibodies, goat polyclonal anti-human MERTK (anti-human mer; Santa Cruz Biotechnology, Santa Cruz, CA) and goat IgG isotype control (Jackson ImmunoResearch Lab, West Grove, PA) at a 1:50 dilution. Slides were rinsed three times in PBS and incubated with donkey anti-goat alkaline phosphatase at a 1:250 dilution (Jackson ImmunoResearch Lab). The DAKO New Fuchsin Substrate System (DAKO Corporation, Carpinteria, CA) was used to visualize antibody binding, and levamisole was used to inhibit any endogenous alkaline phosphatase activity. The slides were counterstained with hematoxylin (Sigma Aldrich, St. Louis, MO) and mounted using GVA mounting medium (Zymed, San Francisco, CA). Brightfield microscopy was performed using a Nikon Microphot microscope equipped with a Plan 40× numerical aperture 0.70 objective lens. Images were captured with an Olympus DP70 CCD camera (Olympus America, Center Valley, PA).
Western Analysis
Western analysis was used to quantitatively assess MERTK protein expression in AMs from normal nonsmokers and normal smokers. AMs were isolated on six-well culture plates as described above. The adherent cells were scraped in PBS, pH 7.4 and centrifuged at 3,000 rpm for 5 minutes at 4°C, and the supernatant was discarded. Whole-cell lysates were prepared by homogenizing the cells with a cell lytic buffer and protease inhibitors (Sigma-Aldrich) on ice. The homogenate was then centrifuged at 10,000 rpm for 10 minutes at 4°C and the supernatant containing the protein of interest was collected. The protein concentration of all supernatants was assessed using a Nanodrop ND-100 spectrophotometer (NanoDrop Technologies, Wilmington, DE). In order to ensure equal protein loading of all gels, each sample was quantified and 20 μg of protein loaded in each lane of a 4–12% Nupage Bis-Tris gel (Invitrogen) after being mixed with NuPage LDS Sample Loading Buffer and reducing agent (Invitrogen). Protein electrophoresis was performed at 200 V for 1 hour at room temperature. Transfer of proteins to a polyvinylidene fluoride membrane (Invitrogen) was performed by electrophoresis at 30 V for 1 hour at 4°C in a Bio-Rad Western blot apparatus (Bio-Rad, Hercules, CA). Membranes were then blocked for 1.5 hours in 0.1% PBS-Tween 20 (PBS-T) with 5% nonfat dried milk on a shaker at 23°C. The anti-MERTK polyclonal antibody was used as the primary antibody at a 1:500 dilution, and the membranes were incubated overnight at 4°C. The membranes were washed in PBS-T and then incubated with horseradish peroxidase–conjugated donkey anti-goat IgG antibody at a 1:5,000 dilution (Santa Cruz Biotechnology) for 1 hour at 23°C, followed by further washing in PBS-T, and then visualization by enhanced chemiluminescence (ECL kit; GE Healthcare, Pittsburgh, PA) using Hyperfilm ECL (GE Healthcare). Immunoreactive proteins were measured quantitatively by inverting the digitally imaged film. The pixel intensity of each band was determined and the background pixel intensity for a negative area of the film of identical size subtracted using MetaMorph image analysis software (Universal Imaging, Downingtown, PA). The protein samples were assayed for GAPDH expression (Santa Cruz Biotechnology) as a control for equal protein concentration. Rat RPE cells served as the positive control.
Immunoprecipitation of GAS6 Ligand and Western Analysis of Soluble MER
Unconcentrated and concentrated BAL supernatant was incubated with anti-human GAS6 antibody (Santa Cruz Biotechnology) for 1 hour at 23°C. Protein G-Sepharose beads (GE Healthcare) were added and complexes rocked for an additional 1 hour at 23°C. Beads were collected by centrifugation and washed three times with cold 1× PBS and 0.02% Tween. Bound proteins were then subjected to protein electrophoresis similar to the method described above. Anti-human monoclonal MER antibody at a 1:1,000 dilution (Caveo Therapeutics, Aurora, CO) served as the primary antibody, whereas human serum served as the positive control (38). The protein samples were assayed for albumin expression (Santa Cruz Biotechnology) as a control for equal protein concentration.
Flow Cytometry
Cells freshly isolated by BAL were used to analyze the surface expression of MERTK. CD68 served as the AM marker. Cells (106) recovered by BAL were washed in PBS containing 2% BSA and resuspended in PBS with 2% BSA and 10% inactivated human serum for 15 minutes at 4°C to block nonspecific antibody binding. After incubation, the cells were stained with phycoerythrin-conjugated mouse monoclonal anti-human Mer or the control isotype mouse immunoglobulin (Ig) G (R&D Systems, Minneapolis, MN) for 30 minutes at 4°C. Subsequently, cells were fixed and permeabilized with Cytofix/Cytoperm reagent (BD Biosciences, San Diego, CA) for 20 min, 4°C, and washed twice with Perm/Wash solution. The cells were then incubated for intracellular staining with fluorescein-conjugated anti-human CD68 antibody and its isotype antibody control for 30 minutes at 23°C (R&D Systems). All samples were prepared and analyzed with and without quenching with 0.2% crystal violet (1 min, 4°C; Polyscientific, Bayshore, NY) to reduce autofluorescence (39–42). Cells were then washed, fixed with 2% paraformaldehyde, and acquired by a FACScalibur cytometer (BD Biosciences, Pharmingen, San Jose, CA) using Cell Quest software. A total of 10,000 events were collected for each sample. Flow cytometry data were analyzed with Flow Jo software (TreeStar, Ashland, OR). For analysis, a region was drawn to define AMs based on forward- and side-scatter characteristics and staining with CD68. All quadrants were set up according to matched isotype control antibodies. All results are reported as percentages of expression.
Induction of Apoptosis
Jurkat T cells were induced to undergo apoptosis by exposure to UV irradiation at 254 nm for 10 minutes (UV Stratalinker 1800; Stratagene, La Jolla, CA) followed by culturing the cells for 3.5 hours at 37°C in 5% CO2 before experiments. Percent apoptosis was determined by flow cytometry utilizing annexin V–fluorescein isothiocyanate and propidium iodide (BD Biosciences Pharmingen) and by evaluation of nuclear morphology under light microscopy. Flow cytometry revealed greater than 70% apoptotic Jurkat T cells, and these were used for the functional experiments. Jurkat T cells (live and apoptotic) were labeled with 5 μM green fluorescent marker 5-chloroethyl fluorescein diacetate (Cell Tracker Green CMFDA; Molecular Probes, Eugene OR).
Phagocytosis Assay
Human AMs were cultured in 12-well culture dishes at a concentration of 5 × 105 overnight at 37°C in 5% CO2. The following day, labeled apoptotic Jurkat T cells were added to AMs for 30 minutes at a ratio of 10:1 (apoptotic cell: AM) at 37°C in 5% CO2. For inhibition studies AMs were preincubated with monoclonal anti-human MERTK antibody (Caveo Therapeutics) for 30 minutes at a final concentration of 10 mg/ml before adding labeled apoptotic Jurkat T cells.
AMs were then washed three times with ice-cold PBS and briefly treated with 0.02% ethylenediaminetetraacetic acid (Invitrogen) to remove undigested Jurkat T cells before AMs were scraped from the plate. After removal, cells were treated with crystal violet as described in Flow Cytometry, and then analyzed by flow cytometry. Serum-free media and viable Jurkat cells were used as controls. Forward and side-scatter gates were set to include cells but exclude debris and unbound particles.
Flow cytometry was used to quantify the uptake of Celltracker Green labeled apoptotic cells into AMs. The data was normalized by using a phagocytic index given that there is individual to individual variability in the phagocytosis of apoptotic cells in nonsmokers and smokers. The phagocytic index was defined as the percentage of AMs containing apoptotic cells (% phagocytosis) multiplied by the mean fluorescence intensity of the cells.
Statistical Analysis
HG-U133 Plus 2.0 microarrays were analyzed using GeneSpring software. Statistical comparisons for microarray data were calculated using GeneSpring software and associated two-tailed t test assuming unequal variance. Average expression values for apoptotic cell removal receptor genes from AMs were calculated from normalized expression levels for nonsmokers and normal smokers. Fold change greater than 2.0 and P less than 0.05 was considered significant. TaqMan data for MERTK were normalized by dividing individual values by the median expression level of all values and, subsequently, the mean and SE were calculated for normalized values of expression. Statistical comparisons for categorical data was achieved using Chi-square test. All other statistical comparisons were calculated using a two-tailed t test, assuming unequal variance (Welch's t test).
Web Deposition of Data
All data has been deposited in the Gene Expression Omnibus (GEO) site, (http://www.ncbi.nlm.nih.gov/geo) curated by the National Center for Bioinformatics. The accession number is GSE8823.
RESULTS
Study Population
The study population of normal nonsmokers (n = 11) and phenotypic normal smokers (n = 13) had no significant prior medical history and a normal physical examination (Table 1). Normal nonsmokers and normal smokers were similar with regard to age (P > 0.2), sex (P > 0.4), and self-reported ancestry (P > 0.2). All individuals were HIV negative, with blood and urine parameters within normal ranges. Normal smokers had an average smoking history of 36 (±6) pack-years, with urine nicotine and cotinine and venous blood carboxyhemoglobin levels confirming current smoking status. Pulmonary function testing revealed normal lung function in both groups.
TABLE 1.
STUDY POPULATION AND ALVEOLAR MACROPHAGE SAMPLES
| Parameter | Normal Nonsmokers | Normal Smokers |
|---|---|---|
| n | 11 | 13 |
| Sex, male/female | 9/2 | 11/2 |
| Age, yr | 44 ± 2 | 35 ± 6 |
| Race, black/white/Hispanic | 4/5/2 | 6/7/0 |
| Smoking history, pack-years | 0 | 36 ± 6 |
| Urine nicotine, ng/ml | Negative | 994 ± 394 |
| Urine cotinine, ng/ml | Negative | 1198 ± 258 |
| Venous CO-Hb* | 0.7 ± 3 | 2.9 ± 0.8 |
| Pulmonary function | ||
| FVC % predicted | 104 ± 2 | 107 ± 3 |
| FEV1% predictedtrun -1 | 104 ± 2 | 104 ± 4 |
| FEV1/FVC % observed | 82 ± 1 | 81 ± 1 |
| TLC % predicted | 96 ± 2 | 98 ± 4 |
| DlCO % predicted | 93 ± 2 | 94 ± 3 |
| BAL | ||
| Total no. recovered × 106 | 1.8 ± 3 | 4.4 ± 5 |
| % Viability | 99 ± 1 | 99 ± 1 |
| % AMs | 89 ± 2 | 94 ± 1 |
| % Lymphocytes | 9 ± 2 | 3 ± 1 |
| % Polymorphonuclear cells | 1 ± 1 | 1 ± 1 |
| % Epithelial cells | 2 ± 1 | 2 ± 1 |
Definition of abbreviations: AM, alveolar macrophage; CO-Hb, carboxy hemoglobin; DlCO, diffusing capacity of carbon monoxide; TLC, total lung capacity.
Demographic characteristics of 24 subjects for whom microarray data was obtained; bronchoalveolar cell differentials at the bottom of the table represent the samples used to obtain the data for the 24 microarrays; all data are mean ± SE.
Venous carboxy hemoglobin, a secondary marker of current smoking; nonsmokers, <1.5%.
The smokers with COPD (n = 7) had an average smoking history of 44 (±2) pack-years and were classified according to the GOLD guidelines (Table 2) with pulmonary function tests demonstrating nonreversible post-bronchodilator reduced FEV1/FVC ratio (35).
TABLE 2.
SMOKERS WITH CHRONIC OBSTRUCTIVE PULMONARY DISEASE AND ALVEOLAR MACROPHAGE SAMPLES
| Parameter | Smokers with COPD |
|---|---|
| n | 7 |
| Sex, male/female | 6/1 |
| Age, yr | 44 ± 2 |
| Race, black/white/Hispanic | 3/3/1 |
| Smoking history, pack-years | 42 ± 6 |
| Current smoking status (n) | 7 |
| Urine nicotine, ng/ml | 448 ± 169 |
| Urine cotinine, ng/ml | 900 ± 25 |
| Venous CO-Hb | 4.1 ± 07 |
| GOLD stage | |
| I | 4 |
| II | 3 |
| Pulmonary function | |
| Prebronchodilator | |
| FVC % predicted | 97 ± 10 |
| FEV1% predicted | 86 ± 9 |
| FEV1/FVC % observed | 68 ± 1 |
| TLC % predicted | 101 ± 8 |
| DlCO % predicted | 72 ± 6 |
| Postbronchodilator | |
| FVC % predicted | 103 ± 10 |
| FEV1% predicted | 90 ± 8 |
| FEV1/FVC % observed | 66 ± 1 |
| TLC % predicted | ND |
| DlCO % predicted | ND |
| BAL | |
| Total no. recovered × 106 | 9.0 ± 2.3 |
| % Viability | 89 ± 1 |
| % AMs | 97 ± 0 0.5 |
| % Lymphocytes | 2.0 ± 0.6 |
| % Polymorphonuclear cells | 0.3 ± 0.6 |
| % Epithelial cells | 0.2 ± 0.1 |
Definition of abbreviations: AM, alveolar macrophage; CO-Hb, carboxy hemoglobin; DlCO, diffusing capacity of carbon monoxide; GOLD, Global Initiative for Chronic Obstructive Lung Disease; ND, not done; TLC, total lung capacity.
Demographic characteristics of seven individuals with COPD for whom TaqMan RT-PCR was performed; all data are mean ± SE.
Sampling of Alveolar Macrophages
The total AMs obtained ranged from 1.0 to 3.2 × 106 for nonsmokers and 1.3 to 8.5 × 106 for normal smokers. On average, more AMs were recovered from normal smokers compared with normal nonsmokers (P < 0.01). Cell viability was, on average, 99% in BAL samples from both groups and cell differential counts were as expected (>90% AM), with no differences between normal nonsmokers and normal smokers (P > 0.05, all comparisons [Table 1]). The total AMs obtained in smokers with COPD ranged from 5.3 to 18.2 × 106, with an average cell viability of 89% and a cell differential count with a predominant AM population (Table 2).
Expression of Apoptotic Cell Removal Receptor Genes
To assess the expression profile of apoptotic cell removal receptor genes in AMs, a list of 14 known apoptotic cell removal receptor–related genes was compiled from the literature, including complement receptor (CR3), CR4, CD14, CD31, the thrombospondin receptor (CD36), CD44, CD93, integrin, α V vitronectin receptor (ITGAV), low-density lipoprotein–receptor related protein (LRP), oxidized low-density lipoprotein receptor 1, MERTK, macrophage scavenger receptor 1, phosphatidylserine receptor, and β2-glycoprotein I. Using an expression criteria of having Affymetrix Detection Call of “Present” in 50% or more of either nonsmoker or smoker samples, all of these apoptotic cell removal receptor genes were expressed in AMs in nonsmokers and smokers except β2-glycoprotein 1 (Table 3).
TABLE 3.
EXPRESSION OF APOPTOTIC CELL REMOVAL RECEPTOR GENES IN ALVEOLAR MACROPHAGES OF NORMAL SMOKERS COMPARED TO NORMAL NONSMOKERS
| Probe Set ID | Gene Symbol | Gene Title | Nonsmoker, % Present | Smoker, % Present | Smoker/Nonsmoker | P Value |
|---|---|---|---|---|---|---|
| 205786_s_at | CR3 | Complement component receptor 3 | 100 | 100 | 1.31 | 0.231 |
| 210184_at | CR4 | Complement component receptor 4 | 100 | 100 | 1.36 | 0.112 |
| 201743_at | CD14 | CD14 | 100 | 100 | −1.86 | 0.003 |
| 208981_at | CD31 | CD31 | 100 | 100 | −1.01 | 0.946 |
| 206488_s_at | CD36 | CD36 (thrombospondin receptor) | 100 | 100 | 1.24 | 0.363 |
| 1565868_at | CD44 | CD44 | 100 | 100 | 1.73 | 0.142 |
| 202878_s_at | CD93 | CD93 | 82 | 100 | −1.01 | 0.932 |
| 232797_at | ITGAV | Integrin, α V (vitronectin receptor) | 100 | 100 | −1.08 | 0.724 |
| 200784_s_at | LRP | Low-density lipoprotein–related protein (α-2-macroglobulin receptor) | 100 | 100 | 1.18 | 0.494 |
| 242397_at | OLR1 | Oxidized low density lipoprotein receptor | 100 | 100 | −1.46 | 0.081 |
| 206028_s_at | MERTK | c-Mer protooncogene tyrosine kinase | 64 | 100 | 3.60 | 0.003 |
| 214770_at | MSR1 | Macrophage scavenger receptor 1 | 100 | 100 | −1.04 | 0.768 |
| 212723_at | PTDSR | Phosphatidylserine receptor | 100 | 100 | 1.05 | 0.594 |
| 205216_s_at | APOH | β2-glycoprotein I | 0 | 0 | — | — |
Expression was assessed by Affymetrix HG U133 Plus 2.0 microarray; listed are all apoptotic cell removal receptor genes considered “expressed” based on the criteria of % present in ≥ 50% samples of either normal nonsmokers or normal smokers. MERTK (in boldface) was the only apoptotic receptor most significantly up-regulated in smokers compared with nonsmokers.
Smoking-Related Changes in mRNA Levels
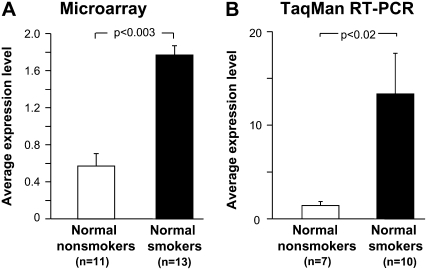
Of the 13 apoptotic cell removal receptor genes expressed, there were no significant differences in the relative gene expression levels among normal smokers compared with normal nonsmokers for most genes (Table 3). The expression of one receptor, CD14, was mildly down-regulated in normal smokers (−1.86-fold; P < 0.003). In contrast, the microarray survey showed that the expression of MERTK was significantly up-regulated in normal smokers compared with normal nonsmokers (3.6-fold increase, P < 0.003; Figure 1A).
Figure 1.
Gene expression levels of the MER tyrosine kinase (MERTK) apoptotic cell removal receptor in alveolar macrophages (AMs). (A) Affymetrix HG-U133 Plus 2.0 microarray assessment of MERTK expression in normal nonsmokers (n = 11) and in normal smokers (n = 13). The ordinate shows the average gene expression levels for each group. (B) Confirmation of microarray results with TaqMan RT-PCR in normal nonsmokers (n = 7) and normal smokers (n = 10). The ordinate shows average gene expression levels. For both panels, each bar represents the mean expression level ± SE for each group, with P values represented in brackets above the bars.
In order to validate the results obtained from the microarray analysis, TaqMan RT-PCR was used to assess RNA samples from AMs of 7 normal nonsmokers and 10 normal smokers. The individuals used to perform TaqMan RT-PCR were the same individuals assessed by microarray. The TaqMan analysis demonstrated that there were no significant differences in expression levels of CD14 between normal smokers and nonsmokers (P > 0.05). In marked contrast, the TaqMan data confirmed that MERTK was significantly up-regulated in AMs of normal smokers compared with normal nonsmokers (9.44-fold increase, P < 0.02; Figure 1B). On this basis, the study focused on MERTK expression.
Smoking-Related Changes at the Protein Level
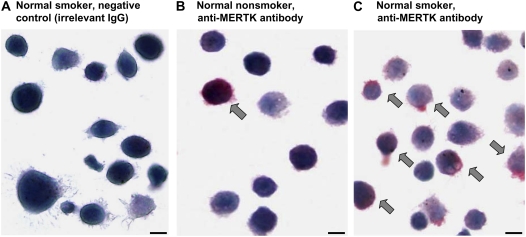
Immunocytochemistry was used to assess expression of MERTK on AMs cytopreparations obtained from nonsmokers and smokers (Figure 2). MERTK was observed on the surface of AMs from nonsmokers and normal smokers, with qualitatively greater expression of MERTK at the protein level on AMs from normal smokers.
Figure 2.
Immunocytochemistry assessment of expression of MERTK in AMs of normal nonsmokers and normal smokers (n = 6 in each group). (A) Normal smoker, IgG isotype control; (B) normal nonsmoker, anti-human MERTK antibody; (C) normal smoker, anti-human MERTK antibody. Arrows, positive staining is indicated by the red fuchsin color. Scale bar = 10 μm.
Western analysis was performed to quantitatively assess MERTK expression on AM whole-cell lysate samples from normal nonsmokers (n = 5) and normal smokers (n = 4). This analysis demonstrated that MERTK expression is significantly increased in smokers compared with nonsmokers (P < 0.007; Figure 3).
Figure 3.
Western analysis of MERTK protein expression in AMs of normal nonsmokers (n = 5) and normal smokers (n = 4). (A) Upper panel, MERTK protein expression in nonsmokers (lanes 1–5) and smokers (lanes 6–9). Lower panel, protein samples assayed for GAPDH expression, control for protein loading. (B) Quantification of MERTK protein expression by densitometry. Data expressed as mean ± SEM.
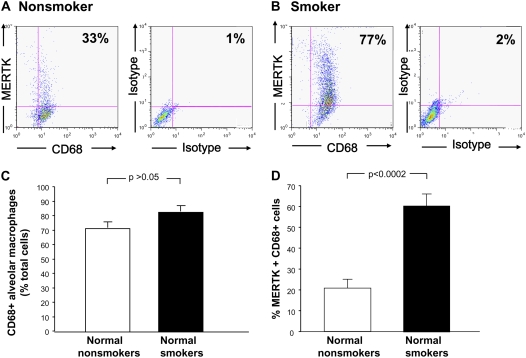
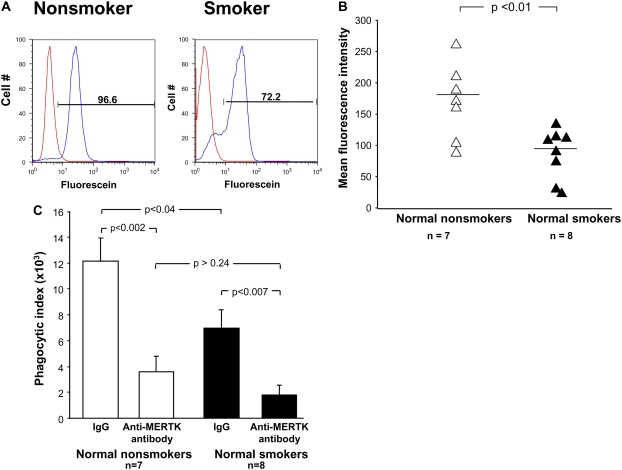
Due to autofluorescence of AMs, quenching with crystal violet was performed before assessment of protein expression by flow cytometry. In order to ensure that crystal violet did not interfere with MERTK detection, samples were analyzed from the same normal nonsmokers (n = 7), with and without quenching. These data revealed no interference with MERTK expression (P > 0.05, data not shown). Flow cytometry analysis of MERTK expression on the surface of AMs demonstrated a higher level of expression in normal smokers (n = 7) compared with normal nonsmokers (n = 6). Although there was no difference in expression of the macrophage marker CD68 between normal nonsmokers and normal smokers (P > 0.05), there was an increased percentage of MERTK on CD68+ AMs from normal smokers compared with normal nonsmokers (60% vs. 21%, P < 0.0002; Table 4 and Figure 4).
TABLE 4.
FLOW CYTOMETRY ASSESSMENT OF CELL SURFACE EXPRESSION OF MERTK AND CD68 ON HUMAN ALVEOLAR MACROPHAGES OF NORMAL SMOKERS COMPARED WITH NORMAL NONSMOKERS*
| Phenotype
|
|||
|---|---|---|---|
| Normal Nonsmokers | Normal Smokers | P Value | |
| Cells, % | |||
| CD68+ | 72 ± 4 | 83 ± 4 | P > 0.05 |
| MERTK+ | 21 ± 3 | 52 ± 4 | P < 0.0001 |
| CD68+ MERTK+ | 21 ± 4 | 60 ± 6 | P < 0.0002 |
Definition of abbreviation: MERTK, c-mer protooncogene tyrosine kinase.
See Figure 4 for examples.
Figure 4.
Flow cytometry analysis of MERTK surface cell expression on AMs. Cells recovered by lavage were gated on CD68+ cells and according to size and granularity on forward scatter and side scatter. Quadrants were set according to respective isotype controls (n = 6 normal nonsmoker; n = 7 normal smokers). (A) Normal nonsmoker; left panel, ordinate–MERTK+ phycoerythrin–conjugated, abscissa–CD68+, fluorescein-conjugated; right panel, same individual with respective negative isotype controls. (A) Normal smoker, ordinate and abscissa same as panels (A) and (C). Percent CD68+ cells among normal nonsmokers compared with normal smokers. (D) Percentage of MERTK+, CD68+, cells among normal nonsmokers compared with normal smokers. Data expressed as mean ± SEM.
MERTK Up-Regulation in Individuals with COPD
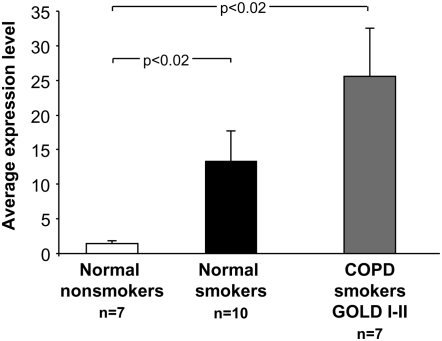
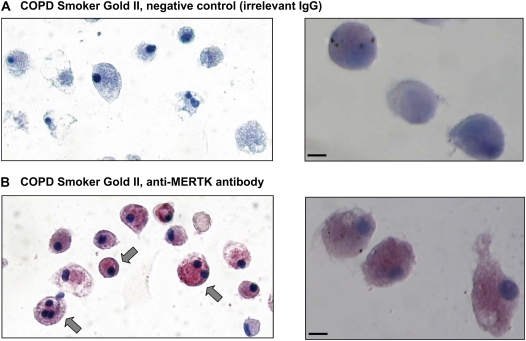
Assessment of smokers with COPD (n = 7; Table 2) by TaqMan RT-PCR analysis demonstrated a significant up-regulation of MERTK gene expression compared with normal nonsmokers (18-fold increase, P < 0.02; Figure 5). Immunocytochemical staining also revealed a greater expression of MERTK at the protein level (Figure 6).
Figure 5.
Gene expression levels of MERTK in AMs of individuals with chronic obstructive pulmonary disease (COPD). TaqMan RT PCR, The ordinate shows the average gene expression levels for each group in normal nonsmokers (n = 7), normal smokers (n = 10), and COPD smokers (n = 7). Data expressed as mean ± SEM.
Figure 6.
Immunocytochemistry assessment of MERTK expression in AMs of smokers with COPD. (A) COPD individual GOLD II, IgG control. Right panel, higher magnification 60x under oil immersion. (B) COPD individual GOLD II, anti-human MERTK antibody. Right panel, higher magnification 60x under oil immersion. Arrows, positive staining is indicated by the red fuchsin color. Scale bar = 10 μm.
MERTK Plays a Role in the Uptake of Apoptotic Cells
Investigation of the phagocytic capacity of AMs demonstrated a significant decrease in the percent phagocytosis of apoptotic Jurkat T cells in normal smokers compared with normal nonsmokers, as shown by Hodge and coworkers (41). The average percent phagocytosis of apoptotic cells in normal smokers was reduced compared with nonsmokers (70 vs. 83%; Figure 7A, representative of one individual from each group). Furthermore, the mean fluorescence intensity revealed a similar downward trend in smokers compared with nonsmokers (P < 0.01; Figure 7B). Prior studies have established that the defect in the engulfment of apoptotic cells was due to a decreased expression of certain receptors involved in this pathway, and raised the possibility that targeted therapy to increase expression of AM recognition molecules may counteract this defect (24).
Figure 7.
Flow cytometric evaluation of apoptotic cell phagocytosis by MERTK in AMs. (A) Representative histograms showing the fluorescence of AMs (red) at baseline and then with internalization of apoptotic Jurkat T cells (blue) from a normal nonsmoker and a normal smoker. On average the % phagocytosis was higher in nonsmokers (n = 7 normal nonsmokers, n = 8 normal smokers). (B) Phagocytosis of apoptotic cells represented as mean fluorescence intensity (ordinate) for normal nonsmokers (n = 7) and normal smokers (n = 8). Each triangle depicts the result from one individual. Scale bar = median for each group. (C) Blocking of MERTK with anti-MERTK antibody inhibits apoptotic cell phagocytosis. Phagocytic index expression on the ordinate in normal nonsmokers (n = 7) and normal smokers (n = 8) in the presence of IgG control or anti-MERTK antibody.
The unexpected finding that MERTK was the only receptor up-regulated in normal smokers compared with normal nonsmokers was further investigated by competitive inhibition with anti-MERTK antibody. Incubation of AMs from normal nonsmokers and normal smokers with anti-MERTK antibody significantly reduced phagocytosis of apoptotic Jurkat T cells (Table 5 and Figure 7). The reduction in the phagocytic index in normal nonsmokers between AMs treated with IgG control antibody versus anti-MERTK antibody was 70% (P < 0.002; Figure 7C). Similarly, AMs of smokers treated with IgG control antibody versus anti-MERTK antibody exhibited a similar decrease of 82% in the phagocytic index (P < 0.007, Figure 7C). Because UV does not render all Jurkat T cells apoptotic, viable labeled Jurkat T cells served as a control. When viable Jurkat T cells were added to AMs and analyzed by flow cytometry, there was no difference observed in phagocytosis as compared with AMs in serum-free media without incubation of Jurkat cells (data not shown). Interestingly, there was no significant difference in percent phagocytosis and the phagocytic index between nonsmokers and smokers in the presence of anti-MERTK (P > 0.5 and P > 0.2 respectively; Table 5 and Figure 7C).
TABLE 5.
PHAGOCYTIC INDEX OF HUMAN ALVEOLAR MACROPHAGES WITH APOPTOTIC CELLS IN NORMAL SMOKERS COMPARED WITH NORMAL NONSMOKERS*
| Phenotype
|
|||
|---|---|---|---|
| Normal Nonsmokers (n = 7) | Normal Smokers (n = 8) | P Value | |
| Phagocytic index† | 12,150 ± 1,766 | 6,978 ± 1,376 | P < 0.04 |
| Phagocytic index+ anti-MERTK Ab | 3,580 ± 1,184 | 1,775 ± 769 | P = 0.24 |
| P Value | P < 0.002 | P < 0.008 | — |
Definition of abbreviations: Ab, antibody; MERTK, c-mer protooncogene tyrosine kinase.
See Figure 7 for examples.
Phagocytic index defined as % phagocytosis of apoptotic cells x mean fluorescence intensity. All data are mean ± SE.
Soluble MERTK in Bronchoalveolar Fluid
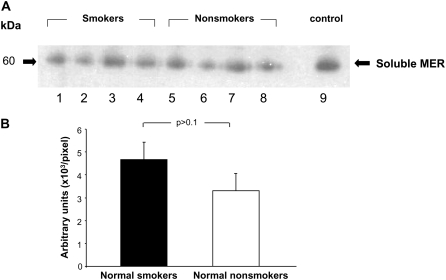
The recent novel finding of a cleaved form of the MERTK receptor (soluble MER) in human and cell lines which binds to Gas6 ligand and prevents apoptotic cell engulfment (38) may serve as a potential mechanism as to why there is a decrease in phagocytosis of dying cells in the AMs of smokers. With this observation, it would be expected that smokers and individuals with COPD would have an abundance of the cleaved receptor in the local lung milieu and thus we evaluated human bronchoalveolar fluid for soluble MER. The data demonstrate that soluble MER/Gas6 complex is present in human bronchoalveolar fluid, but there is no significant difference between smokers (n = 4) and nonsmokers (n = 4) (P > 0.1; Figure 8).
Figure 8.
Western analysis of the soluble form the MERTK receptor in bronchoalveolar fluid of normal smokers and normal nonsmokers. (A) Soluble MER protein expression in normal smokers (lanes 1–4) and normal nonsmokers (lanes 4–8). (B) Quantification of soluble MER protein expression by densitometry. Data are expressed as mean ± SEM.
DISCUSSION
Mononuclear phagocytes play an important role in clearing apoptotic cells, with the mononuclear phagocyte recognizing apoptotic cells via an array of specific receptors on the phagocyte that interact with bridging molecules and apoptotic-specific ligands on the cells undergoing apoptosis (16, 18–24, 43). The present study demonstrates that MERTK, an apoptotic cell surface removal receptor (3, 5, 44, 45), is expressed on normal human AMs, and the expression of MERTK is markedly up-regulated on AMs of normal cigarette smokers, as demonstrated by mRNA analysis by microarray and TaqMan RT-PCR, and protein analysis with immunocytochemistry, Western analysis, and flow cytometry. In the context that smoking is associated with increased pulmonary cell turnover, the finding that expression of MERTK is up-regulated in normal smokers may reflect an attempt to enhance clearance of apoptotic cells in the lung burdened with the stress of smoking (17, 18, 24–33, 46–48).
Expression of MERTK on AMs
Of the 14 apoptotic removal receptors assessed by microarray analysis, 13 were observed to be expressed on AMs of both normal nonsmokers and normal smokers, including CR3 and 4, CD14, CD31, CD36, CD44, CD93, LRP, macrophage scavenger receptors 1, vitronectin receptor, phosphatidylserine receptor, oxidized low-density lipoprotein receptor 1, and MERTK (16, 18–24, 43). Of these, CR3, CD14, CD31, CD36, CD44, LRP, vitronectin receptor, and the phosphatidylserine receptor have been previously observed to be expressed on human AMs (24, 49–52), and MERTK has been noted to be expressed on cultured blood monocytes, the cells from which AMs are derived (5). In the present study, expression by MERTK by AMs was observed by microarray, TaqMan RT-PCR, immunocytochemistry, Western analysis, and flow cytometry.
MERTK, a member of the receptor tyrosine kinase family of cell surface receptors, is a 110-kD protein that consists of an extracellular domain with two N-terminal Ig-like and two membrane proximal fibronectin III regions, a transmembrane region, and an intracellular tyrosine kinase domain (3, 5, 44, 45). In addition to its role in the clearance of apoptotic cells, as demonstrated in MERTK knockout mice and in individuals with retinitis pigmentosa who display mutations in the MERTK gene, MERTK is overexpressed in several human cancers, including mantle cell lymphoma, gastric cancer, and T-cell acute lymphoblastic leukemia (53–56).
Despite the evidence from MERTK knockout mice and patients with retinitis pigmentosa that MERTK functions as an apoptotic cell removal receptor, in human cancers where MERTK is overexpressed, there appears to be suppressed apoptosis (55, 57). MERTK may also have other functions, in that up-regulation of MERTK appears to lower the activation state of murine 32D myelomonocytic cells (58), and, when MERTK knockout mice are challenged with LPS, peritoneal macrophages up-regulate NF-κB and are susceptible to endotoxic shock through up-regulation of tumor necrosis factor-α (59).
Relevance to the Response of the Lung to Cigarette Smoking
There is evidence that AMs from individuals with COPD and normal smokers ingest less apoptotic cells than normal nonsmokers (24). Although there are a significant number of possibilities to explain this defect in the engulfment of apoptotic cells, an obvious mechanism may lie in the apoptotic cell–ligand–receptor interaction. The AM receptors are just one step in the pathway involved in apoptotic cell clearance and, although their redundancy allows this process to be efficient, it is a potential site where a defect can lead to a dysregulated inflammatory response and ultimately result in COPD. This intriguing finding of an up-regulation in a receptor known to be involved in the clearance of apoptotic cells in smokers suggests that it plays a critical role in this pathway. In order to further investigate the function of MERTK, we performed blocking experiments and found that phagocytosis of apoptotic cells is indeed further reduced, indicating that, in fact, the receptor is working. Interestingly, when AMs from normal nonsmokers and normal smokers were blocked with anti-MERTK antibody, their ability to phagocytose apoptotic cells did not differ significantly. This observation may signify that MERTK plays a more significant role than the other receptors, or that it may be one of the last apoptotic cell removal receptors to be affected by smoking before disease occurs. Therefore, up-regulation of MERTK in AMs of normal cigarette smokers may represent a response of the lung to the stress of smoking with an attendant increased parenchymal cell turnover and an increase in the number of apoptotic cells (18, 20, 23, 27–29, 31–33, 60). However, as demonstrated by Hodge and colleagues (16, 24), the clearance of apoptotic cells appears to be defective in normal smokers and given that MERTK expression is significantly up-regulated, it must be that the system is overwhelmed by other processes and this results in the integrated consequence of overall smoking-induced dysfunction of the apoptotic clearance pathway of AMs.
Implications of MERTK in COPD
In all likelihood, the pathway involved in the phagocytosis of apoptotic cells is complex, and MERTK is just one of the many molecules that play a critical role. It would be intriguing to see if knockout mice have any lung pathology at baseline, and if it is made worse with exposure to tobacco smoke. In our study we observed that individuals with COPD (GOLD I/II, smokers) had an up-regulation of MERTK gene expression at the mRNA and protein level. Future studies will need to investigate the modulation of the inflammatory response with regard to MERTK and specifically look at how this receptor functions in smokers with COPD GOLD III/IV and emphysema.
Acknowledgments
The authors thank Igor Dolgalev, Barbara Ferris, Tina Raman, Bishnu De, and Ralf Huebner for expert technical assistance; and Nahla Mohamed for help in preparing this manuscript.
This work was supported, in part, by National Institutes of Health grants R01 HL074326, P50 HL084936 (R.G.C.), and UL1RR024996 (principal investigator: Julianne Imperato-McGinley), and by the Will Rogers Memorial Fund, Los Angeles, California.
Originally Published in Press as DOI: 10.1165/rcmb.2007-0306OC on June 27, 2008
Conflict of Interest Statement: None of the authors has a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Scott RS, McMahon EJ, Pop SM, Reap EA, Caricchio R, Cohen PL, Earp HS, Matsushima GK. Phagocytosis and clearance of apoptotic cells is mediated by MER. Nature 2001;411:207–211. [DOI] [PubMed] [Google Scholar]
- 2.Hu B, Jennings JH, Sonstein J, Floros J, Todt JC, Polak T, Curtis JL. Resident murine alveolar and peritoneal macrophages differ in adhesion of apoptotic thymocytes. Am J Respir Cell Mol Biol 2004;30:687–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hafizi S, Dahlback B. Signalling and functional diversity within the Axl subfamily of receptor tyrosine kinases. Cytokine Growth Factor Rev 2006;17:295–304. [DOI] [PubMed] [Google Scholar]
- 4.Seitz HM, Camenisch TD, Lemke G, Earp HS, Matsushima GK. Macrophages and dendritic cells use different Axl/Mertk/Tyro3 receptors in clearance of apoptotic cells. J Immunol 2007;178:5635–5642. [DOI] [PubMed] [Google Scholar]
- 5.Graham DK, Dawson TL, Mullaney DL, Snodgrass HR, Earp HS. Cloning and mRNA expression analysis of a novel human protooncogene, c-mer. Cell Growth Differ 1994;5:647–657. [PubMed] [Google Scholar]
- 6.Graham DK, Bowman GW, Dawson TL, Stanford WL, Earp HS, Snodgrass HR. Cloning and developmental expression analysis of the murine c-mer tyrosine kinase. Oncogene 1995;10:2349–2359. [PubMed] [Google Scholar]
- 7.Behrens EM, Gadue P, Gong SY, Garrett S, Stein PL, Cohen PL. The mer receptor tyrosine kinase: expression and function suggest a role in innate immunity. Eur J Immunol 2003;33:2160–2167. [DOI] [PubMed] [Google Scholar]
- 8.Wang H, Chen Y, Ge Y, Ma P, Ma Q, Ma J, Wang H, Xue S, Han D. Immunoexpression of Tyro 3 family receptors–Tyro 3, Axl, and Mer–and their ligand Gas6 in postnatal developing mouse testis. J Histochem Cytochem 2005;53:1355–1364. [DOI] [PubMed] [Google Scholar]
- 9.Lu Q, Lemke G. Homeostatic regulation of the immune system by receptor tyrosine kinases of the Tyro 3 family. Science 2001;293:306–311. [DOI] [PubMed] [Google Scholar]
- 10.Cohen PL, Caricchio R, Abraham V, Camenisch TD, Jennette JC, Roubey RA, Earp HS, Matsushima G, Reap EA. Delayed apoptotic cell clearance and lupus-like autoimmunity in mice lacking the c-mer membrane tyrosine kinase. J Exp Med 2002;196:135–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.D'Cruz PM, Yasumura D, Weir J, Matthes MT, Abderrahim H, LaVail MM, Vollrath D. Mutation of the receptor tyrosine kinase gene Mertk in the retinal dystrophic RCS rat. Hum Mol Genet 2000;9:645–651. [DOI] [PubMed] [Google Scholar]
- 12.Gal A, Li Y, Thompson DA, Weir J, Orth U, Jacobson SG, Apfelstedt-Sylla E, Vollrath D. Mutations in MERTK, the human orthologue of the RCS rat retinal dystrophy gene, cause retinitis pigmentosa. Nat Genet 2000;26:270–271. [DOI] [PubMed] [Google Scholar]
- 13.Vollrath D, Feng W, Duncan JL, Yasumura D, D'Cruz PM, Chappelow A, Matthes MT, Kay MA, LaVail MM. Correction of the retinal dystrophy phenotype of the RCS rat by viral gene transfer of Mertk. Proc Natl Acad Sci USA 2001;98:12584–12589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McHenry CL, Liu Y, Feng W, Nair AR, Feathers KL, Ding X, Gal A, Vollrath D, Sieving PA, Thompson DA. MERTK arginine-844-cysteine in a patient with severe rod-cone dystrophy: loss of mutant protein function in transfected cells. Invest Ophthalmol Vis Sci 2004;45:1456–1463. [DOI] [PubMed] [Google Scholar]
- 15.Bezdicek P, Crystal RG. Pulmonary macrophages. In: Crystal RG, West JB, Weibel ER, Barnes PJ, editors. The lung: scientific foundations, Philadelphia: Lippincott-Raven Publishers; 1997. pp. 859–975.
- 16.Hodge S, Hodge G, Scicchitano R, Reynolds PN, Holmes M. Alveolar macrophages from subjects with chronic obstructive pulmonary disease are deficient in their ability to phagocytose apoptotic airway epithelial cells. Immunol Cell Biol 2003;81:289–296. [DOI] [PubMed] [Google Scholar]
- 17.Barnes PJ. Alveolar macrophages in chronic obstructive pulmonary disease (COPD). Cell Mol Biol (Noisy.-le-grand) 2004;50: OL627–OL637. Available from: http://www.cellmolbiol.com [accessed on 16 April 2008]. [PubMed]
- 18.Vandivier RW, Henson PM, Douglas IS. Burying the dead: the impact of failed apoptotic cell removal (efferocytosis) on chronic inflammatory lung disease. Chest 2006;129:1673–1682. [DOI] [PubMed] [Google Scholar]
- 19.Savill J, Fadok V. Corpse clearance defines the meaning of cell death. Nature 2000;407:784–788. [DOI] [PubMed] [Google Scholar]
- 20.Fadok VA, Bratton DL, Henson PM. Phagocyte receptors for apoptotic cells: recognition, uptake, and consequences. J Clin Invest 2001;108:957–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Henson PM, Bratton DL, Fadok VA. Apoptotic cell removal. Curr Biol 2001;11:R795–R805. [DOI] [PubMed] [Google Scholar]
- 22.Savill J, Dransfield I, Gregory C, Haslett C. A blast from the past: clearance of apoptotic cells regulates immune responses. Nat Rev Immunol 2002;2:965–975. [DOI] [PubMed] [Google Scholar]
- 23.Taylor PR, Martinez-Pomares L, Stacey M, Lin HH, Brown GD, Gordon S. Macrophage receptors and immune recognition. Annu Rev Immunol 2005;23:901–944. [DOI] [PubMed] [Google Scholar]
- 24.Hodge S, Hodge G, Ahern J, Jersmann H, Holmes M, Reynolds PN. Smoking alters alveolar macrophage recognition and phagocytic ability: implications in COPD. Am J Respir Cell Mol Biol 2007;37:748–755. [DOI] [PubMed] [Google Scholar]
- 25.Bowden DH. Cell turnover in the lung. Am Rev Respir Dis 1983;128:S46–S48. [DOI] [PubMed] [Google Scholar]
- 26.Rennard SI. Inflammation and repair processes in chronic obstructive pulmonary disease. Am J Respir Crit Care Med 1999;160:S12–S16. [DOI] [PubMed] [Google Scholar]
- 27.Segura-Valdez L, Pardo A, Gaxiola M, Uhal BD, Becerril C, Selman M. Upregulation of gelatinases A and B, collagenases 1 and 2, and increased parenchymal cell death in COPD. Chest 2000;117:684–694. [DOI] [PubMed] [Google Scholar]
- 28.Kasahara Y, Tuder RM, Cool CD, Lynch DA, Flores SC, Voelkel NF. Endothelial cell death and decreased expression of vascular endothelial growth factor and vascular endothelial growth factor receptor 2 in emphysema. Am J Respir Crit Care Med 2001;163:737–744. [DOI] [PubMed] [Google Scholar]
- 29.Aoshiba K, Yokohori N, Nagai A. Alveolar wall apoptosis causes lung destruction and emphysematous changes. Am J Respir Cell Mol Biol 2003;28:555–562. [DOI] [PubMed] [Google Scholar]
- 30.Tuder RM, Petrache I, Elias JA, Voelkel NF, Henson PM. Apoptosis and emphysema: the missing link. Am J Respir Cell Mol Biol 2003;28:551–554. [DOI] [PubMed] [Google Scholar]
- 31.Yokohori N, Aoshiba K, Nagai A. Increased levels of cell death and proliferation in alveolar wall cells in patients with pulmonary emphysema. Chest 2004;125:626–632. [DOI] [PubMed] [Google Scholar]
- 32.Imai K, Mercer BA, Schulman LL, Sonett JR, D'Armiento JM. Correlation of lung surface area to apoptosis and proliferation in human emphysema. Eur Respir J 2005;25:250–258. [DOI] [PubMed] [Google Scholar]
- 33.Demedts IK, Demoor T, Bracke KR, Joos GF, Brusselle GG. Role of apoptosis in the pathogenesis of COPD and pulmonary emphysema. Respir Res 2006;7:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heguy A, O'Connor TP, Luettich K, Worgall S, Cieciuch A, Harvey BG, Hackett NR, Crystal RG. Gene expression profiling of human alveolar macrophages of phenotypically normal smokers and nonsmokers reveals a previously unrecognized subset of genes modulated by cigarette smoking. J Mol Med 2006;84:318–328. [DOI] [PubMed] [Google Scholar]
- 35.Global Strategy for the Diagnosis, Management and Prevention of COPD, Global Initiative for Chronic Obstructive Lung Disease (GOLD) 2007. [accessed April 11, 2008]. Available from: http://www.goldcopd.org.
- 36.Russi TJ, Crystal RG. Use of bronchoalveolar lavage and airway brushing to investigate the human lung. In: Crystal RG, West JB, Weibel ER, Barnes PJ., editors. The lung: scientific foundations, 2nd ed. Philadelphia: Lippincott-Raven Publishers; 1997. pp. 371–382.
- 37.Leopold PL, Ferris B, Grinberg I, Worgall S, Hackett NR, Crystal RG. Fluorescent virions: dynamic tracking of the pathway of adenoviral gene transfer vectors in living cells. Hum Gene Ther 1998;9:367–378. [DOI] [PubMed] [Google Scholar]
- 38.Sather S, Kenyon KD, Lefkowitz JB, Liang X, Varnum BC, Henson PM, Graham DK. A soluble form of the Mer receptor tyrosine kinase inhibits macrophage clearance of apoptotic cells and platelet aggregation. Blood 2007;109:1026–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hallden G, Skold CM, Eklund A, Forslid J, Hed J. Quenching of intracellular autofluorescence in alveolar macrophages permits analysis of fluorochrome labelled surface antigens by flow cytofluorometry. J Immunol Methods 1991;142:207–214. [DOI] [PubMed] [Google Scholar]
- 40.Umino T, Skold CM, Pirruccello SJ, Spurzem JR, Rennard SI. Two-colour flow-cytometric analysis of pulmonary alveolar macrophages from smokers. Eur Respir J 1999;13:894–899. [DOI] [PubMed] [Google Scholar]
- 41.Hodge SJ, Hodge GL, Holmes M, Reynolds PN. Flow cytometric characterization of cell populations in bronchoalveolar lavage and bronchial brushings from patients with chronic obstructive pulmonary disease. Cytometry B Clin Cytom 2004;61:27–34. [DOI] [PubMed] [Google Scholar]
- 42.Garn H. Specific aspects of flow cytometric analysis of cells from the lung. Exp Toxicol Pathol 2006;57:21–24. [DOI] [PubMed] [Google Scholar]
- 43.Lauber K, Blumenthal SG, Waibel M, Wesselborg S. Clearance of apoptotic cells: getting rid of the corpses. Mol Cell 2004;14:277–287. [DOI] [PubMed] [Google Scholar]
- 44.Lemke G, Lu Q. Macrophage regulation by Tyro 3 family receptors. Curr Opin Immunol 2003;15:31–36. [DOI] [PubMed] [Google Scholar]
- 45.Hafizi S, Dahlback B. Gas6 and protein S: vitamin K–dependent ligands for the Axl receptor tyrosine kinase subfamily. FEBS J 2006;273:5231–5244. [DOI] [PubMed] [Google Scholar]
- 46.Aoshiba K, Tamaoki J, Nagai A. Acute cigarette smoke exposure induces apoptosis of alveolar macrophages. Am J Physiol Lung Cell Mol Physiol 2001;281:L1392–L1401. [DOI] [PubMed] [Google Scholar]
- 47.Jiao ZX, Ao QL, Xiong M. Cigarette smoke extract inhibits the proliferation of alveolar epithelial cells and induces apoptosis. Sheng Li Xue Bao 2006;58:244–254. [PubMed] [Google Scholar]
- 48.Heguy A, Harvey BG, Leopold PL, Dolgalev I, Raman T, Crystal RG. Responses of the human airway epithelium transcriptome to in vivo injury. Physiol Genomics 2007;29:139–148. [DOI] [PubMed] [Google Scholar]
- 49.Skold CM, Lundahl J, Hallden G, Hallgren M, Eklund A. Chronic smoke exposure alters the phenotype pattern and the metabolic response in human alveolar macrophages. Clin Exp Immunol 1996;106:108–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pons AR, Noguera A, Blanquer D, Sauleda J, Pons J, Agusti AG. Phenotypic characterisation of alveolar macrophages and peripheral blood monocytes in COPD. Eur Respir J 2005;25:647–652. [DOI] [PubMed] [Google Scholar]
- 51.Lofdahl JM, Wahlstrom J, Skold CM. Different inflammatory cell pattern and macrophage phenotype in chronic obstructive pulmonary disease patients, smokers and non-smokers. Clin Exp Immunol 2006;145:428–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hodge S, Hodge G, Brozyna S, Jersmann H, Holmes M, Reynolds PN. Azithromycin increases phagocytosis of apoptotic bronchial epithelial cells by alveolar macrophages. Eur Respir J 2006;28:486–495. [DOI] [PubMed] [Google Scholar]
- 53.Ek S, Hogerkorp CM, Dictor M, Ehinger M, Borrebaeck CA. Mantle cell lymphomas express a distinct genetic signature affecting lymphocyte trafficking and growth regulation as compared with subpopulations of normal human B cells. Cancer Res 2002;62:4398–4405. [PubMed] [Google Scholar]
- 54.Graham DK, Salzberg DB, Kurtzberg J, Sather S, Matsushima GK, Keating AK, Liang X, Lovell MA, Williams SA, Dawson TL, et al. Ectopic expression of the proto-oncogene Mer in pediatric T-cell acute lymphoblastic leukemia. Clin Cancer Res 2006;12:2662–2669. [DOI] [PubMed] [Google Scholar]
- 55.Keating AK, Salzberg DB, Sather S, Liang X, Nickoloff S, Anwar A, Deryckere D, Hill K, Joung D, Sawczyn KK, et al. Lymphoblastic leukemia/lymphoma in mice overexpressing the Mer (MerTK) receptor tyrosine kinase. Oncogene 2006;25:6092–6100. [DOI] [PubMed] [Google Scholar]
- 56.Wu CW, Li AF, Chi CW, Lai CH, Huang CL, Lo SS, Lui WY, Lin WC. Clinical significance of AXL kinase family in gastric cancer. Anticancer Res 2002;22:1071–1078. [PubMed] [Google Scholar]
- 57.Georgescu MM, Kirsch KH, Shishido T, Zong C, Hanafusa H. Biological effects of c-Mer receptor tyrosine kinase in hematopoietic cells depend on the Grb2 binding site in the receptor and activation of NF-κB. Mol Cell Biol 1999;19:1171–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guttridge KL, Luft JC, Dawson TL, Kozlowska E, Mahajan NP, Varnum B, Earp HS. Mer receptor tyrosine kinase signaling: prevention of apoptosis and alteration of cytoskeletal architecture without stimulation or proliferation. J Biol Chem 2002;277:24057–24066. [DOI] [PubMed] [Google Scholar]
- 59.Camenisch TD, Koller BH, Earp HS, Matsushima GK. A novel receptor tyrosine kinase, Mer, inhibits TNF-α production and lipopolysaccharide-induced endotoxic shock. J Immunol 1999;162:3498–3503. [PubMed] [Google Scholar]
- 60.Henson PM, Vandivier RW, Douglas IS. Cell death, remodeling, and repair in chronic obstructive pulmonary disease? Proc Am Thorac Soc 2006;3:713–717. [DOI] [PMC free article] [PubMed] [Google Scholar]