Abstract
In Vibrio cholerae, expression of the quorum sensing regulator HapR is induced by the accumulation of a major autoinducer synthesized by the activity of CqsA. Here we show that the cAMP-cAMP receptor protein complex regulates cqsA expression at the post-transcriptional level. This conclusion is supported by the analysis of cqsA-lacZ fusions, the ectopic expression of cqsA in Δcrp mutants and by Northern blot analysis showing that cqsA mRNA is unstable in Δcrp and Δcya (adenylate cyclase) mutants. Addition of cAMP to the culture of a Δcya mutant restored cqsA mRNA stability and CAI-1 production. Lowering intracellular cAMP levels by addition of D-glucose increased the cell density required to activate HapR. These results indicate that cAMP acts as a quorum modulator.
Keywords: Vibrio cholerae, signal transduction, quorum sensing, cAMP, cAMP receptor protein, carbon catabolite repression
1. Introduction
Vibrio cholerae is a Gram-negative highly motile bacterium that colonizes the human small intestine and produce cholera toxin (CT) which causes the profuse watery diarrhea typical of cholera [1, 2]. The expression of CT is regulated by quorum sensing. Quorum sensing is a process by which bacterial cells communicate with one another by secreting extracellular signaling molecules termed autoinducers. Two autoinducer systems function in V. cholerae. The cholera autoinducer 1 (CAI-1) synthase CqsA is responsible for the biosynthesis of CAI-1 [(S)-3-hydroxytridecan-4-one] which at high cell density binds to its receptor CqsS [3]. Autoinducer 2 (AI-2) is a furanosyl borate diester [(2S, 4S)-2-methyl-2,3,3,4-tetrahydroxytetrahydrofuran borate] synthesized by the activity of LuxS [4] and recognized by receptor LuxPQ. Accumulation of CAI-1 and AI-2 activates the expression of the master quorum sensing regulator HapR [5]. HapR plays a pivotal role in regulating virulence gene expression, protease production and biofilm formation [6-8] by modulating intracellular cyclic diguanylate (c-di-GMP) levels, as well as by acting at specific promoters that control virulence gene expression and exopolysaccharide biosynthesis [9, 10].
The cAMP receptor protein (CRP) is a global regulator well known for its role in carbon catabolite repression: the inhibition of gene expression by the presence of a rapidly metabolizable carbon source (i.e. glucose) in the growth medium [11-13]. In carbon catabolite repression, the presence of glucose indirectly inhibits adenylate cyclase through a component of the posphoenolpyruvate-dependent phosphotransferase system (PTS) leading to low intracellular cAMP levels [14]. Conversely, when glucose is limited (or replaced by a poor carbon source), the activity of adenylate cyclase increases leading to high intracellular levels of cAMP and formation of the cAMP-CRP complex [12]. The cAMP-CRP complex binds as a dimer to the consensus sequence TGTGA-(N6)-TCACA which can be found within, adjacent or upstream of responsive promoters [11-13].
In a previous study we demonstrated that CRP is required for expression of the CAI-1 synthase CqsA [15]. Consequently, deletion of crp was shown to impact the expression of HapR, multiple HapR-regulated genes and phenotypes such as production of hemagglutinin (HA)/protease, CT, exopolysaccharide biosynthesis and biofilm formation [15, 16]. In this study we show that cAMP levels post-transcriptionally activate the expression of CqsA by enhancing the stability of the cqsA mRNA. Thus, cAMP acts upstream of quorum sensing to regulate CAI-1 biosynthesis and modulate the cell density required to activate HapR. Finally, this is the first report of the cAMP-CRP complex modulating gene expression at the level of mRNA stability to integrate carbon catabolite repression and quorum sensing.
2. Materials and methods
2.1. Strains and media
V. cholerae mutants used in this study were constructed from the El Tor biotype strain C7258 (Perú isolate, 1991). Construction of strains AJB61 (C7258ΔcqsA), WL7258 (C7258Δcrp), C7258ΔlacZ and WL7258ΔlacZ have been described previously [15, 17]. E. coli TOP10 (Invitrogen) and S17−1λpir [18] were used for cloning purposes. V. cholerae strains were grown in Bacto tryptic soy broth (TSB) (Becton, Dickinson & Co.) or LB at 37°C with agitation (250 rpm). When necessary, culture media were supplemented with ampicillin (Amp, 100-μg/ml), tetracycline (Tet, 5-μg/ml), kanamycin (Km, 25-μg/ml), X-gal (20-μg/ml) or polymyxin B (100-units/ml). Plasmid DNA was introduced in V. cholerae by electroporation [19].
2.2 Construction of mutants
Strain WL7259 (C7258Δcya) containing a deletion of cya encoding adenylate cyclase was constructed by transferring the suicide vector pCVDΔcya-km [16] to C7258 followed by sucrose selection as described previously [16]. To construct the ΔcrpΔluxO double mutant SZS013, chromosomal DNA fragments flanking the luxO open reading frame (ORF) were amplified from C7258 genomic DNA with the Advantage 2 PCR kit (Clontech) using the primer sets 5′-GCGCGAGCTCGTGATTTACGATTAGGCG/5′-GCGCGGATCCAAACATCACACATCTAGAC and 5′-GCGCGGATCCGTGAACTCAATGATTAC/5′-GCGCGTCGACAGATAACCTTTCGGTGC and the amplicons were sequentially cloned in pCVD442 [20]. The resulting suicide vector containing the chromosomal luxO deletion was transferred to strain WL7258ΔlacZ by conjugation and the ΔluxO mutant was obtained by sucrose selection as described previously [15, 16].
2.3. Construction of plasmids and chromosomally integrated lacZ fusions
A 1.6-kb DNA fragment encompassing the cqsA ORF flanked by 5′ and 3′ non-coding (intergenic) sequences was amplified using primers 5′-GGGGATCCTGACCGTGATGTATTGCTA and 5′-GAACTGCAGCGCTCAGTAAACTCCTAA and cloned as a BamHI-SphI fragment in pBR322 to yield pBRCqsA2383. To construct a cqsA-lacZ transcriptional fusion we sequentially cloned the rrnBT1T2 transcription terminator [21] and a 214-bp fragment containing the cqsA promoter region amplified with primers 5′-GGCCAAGCTTTCGCAATATATCCTAGTT and 5′-GAACTGCAGCGCTCAGTAAACTCCTAA in pUC19 to generate pTTcqsA. A 1-kb internal fragment of the C7258 lacZ gene (lacZVC) was amplified using primers 5′-CGAAGGTACCAATCCCCGATTCA and 5′-GCCTCTAGATCGCCACCGTTTTACACTG and cloned as a KpnI-XbaI fragment in pTTcqsA upstream of the rrnBT1T2 transcription terminator to generate pZTC. Next, a 1.6-kb KpnI-HindIII fragment containing the lacZVC fragment, the rrnBT1T2 terminator and the cqsA promoter was sub-cloned upstream of the promoterless E. coli lacZ gene (lacZEC) in plasmid pKRZ1 [22] to yield pWLZTC. Finally, a 5.6-kb PstI-KpnI DNA fragment containing the lacZVC-rrnBT1T2 - cqsA - lacZEC unit was cloned as a PstI-KpnI fragment in the suicide vector pCVD442 [20] to yield pCVDZTC. The suicide vector pCVDZTC containing a cqsA-lacZEC transcriptional fusion was mobilized from E. coli S17−1λpir to C7258 and WL7258 by conjugation to generate strains C72ZTC and WL72ZTC. Integration of pCVDZTC by homologous recombination results in disruption of V. cholerae chromosomal lacZ gene. Correct integration was confirmed by PCR and DNA sequencing. To construct cqsA-lacZ translational fusions, primer pairs 5′-CGGAATTCCGAGTCTACGACAATGAT/5′-CGGAATTCTATCGCTATCTATTTCGTC and 5′-CCGAATTCCCAGATTGAGATAATAGACA/5′-CGGAATTCTATCGCTATCTATTTCGTC were used to amplify DNA fragments starting within the upstream locus VCA0524 and containing different length of cqsA coding sequence, respectively. The PCR products, of size 1183- and 984-bp, were confirmed by DNA sequencing and cloned as XbaI-EcoRI fragments in the suicide vector pVIK111 [23] to generate the translational cqsA-lacZ (in-frame) fusions contained in plasmids pVIK1648 and pVIK1847, respectively. Plasmid pVIK1648 contains DNA encoding the first 174 amino acid of CqsA while pVIK1847 contains DNA encoding the first 107 amino acids. Plasmids pVIK1648 and pVIK1847 were transferred by conjugation from S17−1λpir to C7258ΔlacZ and WL7258ΔlacZ. Stable integration within the cqsA locus was confirmed by PCR and DNA sequencing. To express cqsA from a heterologous Tac promoter, primers 5′-CGGGATCCGATGAACAAGCCTCAACT and 5′- GCTCTAGATGACCGTGATGTATTGCT were used to amplify the cqsA ORF and 3’-UTR. The PCR product was confirmed by DNA sequencing and cloned as a BamHI-PstI fragment in pALTER-Ex2 (Promega) to create pTac-CqsA. Construction of plasmid pHapRLac2 containing a hapR-lacZ fusion has been described previously [24].
2.4. RNA techniques
For total RNA preparation, cells were treated with the RNA stabilizing reagent RNA Protect (QIAGEN Inc.) and total RNA was isolated using the RNeasy kit and RNase-free DNAse set (QIAGEN Inc.). Total RNA was fractionated in a 1% agarose - 20 mM MOPS - 5 mM sodium acetate - 2 mM EDTA - 2% formaldehyde gel and transferred to positively charged nylon membranes (Roche Applied Sciences) by capillary transfer. The membranes were hybridized to a cqsA probe in DIG Easy Hyb (Roche) at 50°C overnight, washed and developed with an alkaline phosphatase-conjugated anti-digoxigenin antibody and CSDP (Roche). To determine the stability of cqsA in different genetic backgrounds, cells were grown in TSB to a predetermined cell density, treated with rifampicin (200-μg/ml) to block transcription and samples were taken subsequently at different time points for Northern blot analysis. The primer pairs 5′-CGGGATCCCTAGGATATATTGCGATG/5′-GCTCTAGATGACCGTGATGTATTGCT and 5′-GTGCTGTGGATGTCATCGTTGTTG/5′-CGCTTTACCTTGGCCGATTT were used to generate DIG-labeled cqsA and recA DNA probes, respectively. To this end, the resulting PCR products were gel-purified and labeled by random priming using the DIG-random prime kit (Roche).
2.5 Measurement of β-galactosidase and protease activities
β-Galactosidase activity was measured as described by Miller [25] using the substrate o-nitrophenyl-β-D-galctotopyranoside (ONPG). Specific activities are given in Miller units [1000 (OD420 / t .v .OD600)] where t is reaction time and v is the volume of enzyme extract per reaction. Expression of HA/protease [26] was measured using an azocasein assay as described previously [27]. One azocasein unit is the amount of enzyme producing an increase of 0.01 OD442 units per h.
2.6. Assay of CAI-1 activity
To measure the production of CAI-1 activity, cultures were centrifuged at 12,000 rpm for 10-min and the supernatants were filtered through a 0.22-μm syringe filter. Cell-free culture supernatants were tested for the presence of CAI-1 activity by inducing light production in the V. cholerae reporter strain MM920 containing the cosmid pBB1, which carries the V. harveyi lux operon [5]. The reporter strain was grown overnight with shaking at 30°C, diluted 1:10 in fresh medium, and 70-μL aliquots transferred to an opaque-wall 96-well microtiter plate. Cell-free culture fluids were added to a final concentration of 30 % (v/v). The plates were incubated at 30°C with agitation and light production was measured at 30-min intervals in a Genios Plus Tecan luminometer. Results are expressed as light fold induction relative to a sterile medium control.
3. Results
In a previous study we showed that production of CAI-1 requires an active crp allele [15]. According to common knowledge, the activity of E. coli CRP is determined by the intracellular concentration of cAMP [14]. However, genetic variants of CRP have been identified exhibiting significant cAMP-independent activity [28]. Contrary to E. coli, the V. cholerae CRP protein has not been extensively characterized. To determine if the strong cqsA dependency on CRP can be fully accounted by cAMP binding to CRP, we constructed an adenylate cyclase deletion mutant. In Fig. 1 we show that no CAI-1 could be detected in strain WL7259 lacking adenylate cyclase. The CAI-1 defect of strain WL7259 could be complemented by introducing the wild type (WT) cya gene on plasmid pTTCya [16] (Fig. 1, shadowed bar). Furthermore, production of CAI-1 in strain WL7259 could be restored by supplementing the medium with 2.5- or 5.0-mM cAMP. As expected, addition of cAMP to the medium did not restore CAI-1 expression in strain WL7258 lacking CRP (Fig. 1). These results indicate that expression of CqsA is under positive regulation by the cAMP-CRP complex (Fig. 1). Since CRP is a global regulator which affects the expression of many genes [15], we found it important to confirm that CRP regulation of CAI-1 is the major mechanism by which CRP modulates HapR. CAI-1 acts to inactivate phospho-LuxO, a repressor of HapR [5]. We have shown that Δcrp mutants do not make HapR-dependent HA/protease [15, 27]. As expected, deletion of luxO from strain WL7258ΔlacZ (Δcrp) to generate SZS013 (ΔcrpΔluxO) fully restored HA/protease production (azocasein units/OD600: C7258ΔlacZ, 18.2 ± 0.4; WL758ΔlacZ, 0.9 ± 0.4; SZS013, 24.3 ± 1.6) suggesting that CRP regulation of CAI-1 is the major regulatory event by which CRP controls the quorum sensing master regulator HapR. We used the software virtual footprint ((http://www.prodoric.de/vfp/) to scan the 5′ un-translated DNA preceding cqsA for putative cAMP-CRP binding sites. This analysis did not return high-scoring putative binding sites suggesting that CRP regulates cqsA indirectly and/or post-transcriptionally.
Fig. 1. Exogenous cAMP restores the capacity of an adenylate cyclase mutant to produce CAI-1.

Overnight cultures of WL7259 (Δcya) were diluted 100-fold in fresh TSB and grown to OD600 0.5. Each culture was divided in three portions and cAMP (Sigma Chemical Co.) was added to a final concentrations of zero (control), 2.5- and 5-mM respectively. The cultures were incubated at 37°C for 1-h with shaking and the amount of CAI-1 in the cell-free supernatant was determined as described in methods. Strain WL7258 (Δcrp) and strain WL7259 (Δcya) containing the complementing plasmid pTTCya (shadowed bar) were used as controls. Each value represents the average of three independent cultures. Error bars indicate the standard deviation.
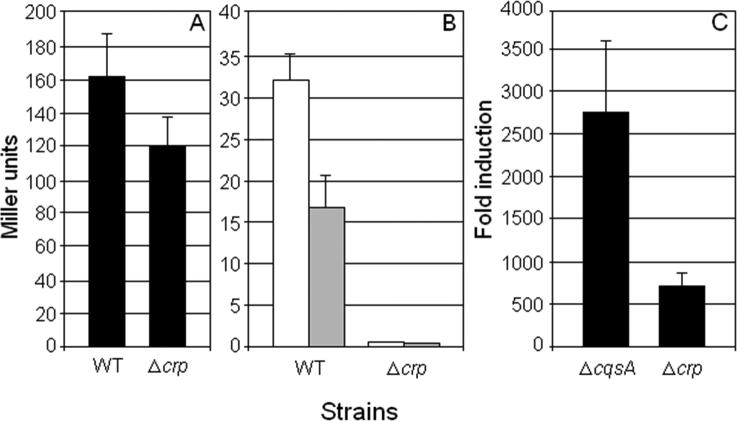
To determine if cAMP-CRP regulation of cqsA is at the level of transcription or post-transcriptional, we constructed strains harboring chromosomally integrated transcriptional and translational cqsA-lacZ fusions. In the transcriptional fusions, cqsA 5′ un-translated DNA was ligated to a promoterless E. coli lacZ gene in plasmid pCVDZTC and recombined into the chromosome of strains C7258 (WT) and WL7258 (Δcrp). No significant differences in β-galactosidase expression were detected between the WT and the Δcrp mutant suggesting that the regulation of cqsA by cAMP-CRP does not involve the 5′ un-translated DNA preceding the cqsA ORF (Fig. 2A). Next, we constructed two chromosomally integrated translational (in-frame) fusions containing DNA encoding the first 107 (pVIK1847) and 174 (pVIK1648) amino acids of CqsA ligated to a lacZ gene devoid of translation initiation signals. As shown in Fig. 2B, the expression of both translational fusions was strongly CRP-dependent indicating that the cqsA region responsive to CRP resides within the DNA encoding the first 107 N-terminal amino acids of CqsA. To further confirm that regulation of cqsA by CRP is post-transcriptional, we tested whether ectopic expression of cqsA using a heterologous promoter was still CRP-dependent. To this end, we constructed pTac-CqsA expressing cqsA from the strong Tac promoter. The Tac promoter lacks the CRP responsive region of the WT lac promoter and is not sensitive to cAMP [29]. In addition, V. cholerae does not produce a lac repressor resulting in constitutive expression of genes placed under the control of this promoter. In Fig. 2C we show that pTac-CqsA effectively complemented a ΔcqsA (crp-positive) mutant. However, expression of cqsA from pTac-CqsA was still significantly diminished in the isogenic Δcrp mutant (Fig. 2C). Taken together, the above results strongly suggest that cAMP-CRP post-transcriptionally regulates the expression of CqsA.
Fig. 2. Gene fusion analysis of cAMP-CRP regulation of cqsA expression.
(A). Transcriptional fusion. β-Galactosidase expression in strains C72ZTC (WT background) and WL72ZTC (Δcrp background) containing a cqsA-lacZ transcriptional fusion. Strains were grown at 37°C with agitation in LB overnight. (B). Translational fusions. β-Galactosidase expression in strains C7258ΔlacZ (WT background) and WL7258ΔlacZ (Δcrp background) containing the cqsA-lacZ translational fusions pVIK1847 (open bar) or pVIK1648 (shadowed bar). Strains were grown at 37°C with agitation in LB overnight. (C). Ectopic expression of cqsA. Strains AJB61 (C7258ΔcqsA) and WL7258 (C7258Δcrp) containing pTac-CqsA were grown at 37°C with agitation in TSB overnight and production of CAI-1 activity was determined as described in methods. In all cases, each value represents the average of three independent experiments. Error bars indicate standard deviations.
The level of cqsA mRNA produced in a Δcrp mutant is below the level of detection of the chemiluminescence detection system described in methods. To determine the size of the cqsA transcript produced in the Δcrp mutant, we decided to boost the amount of cqsA mRNA by placing the cqsA gene on plasmid pBRCqsA2383. The plasmid was introduced by electroporation in ΔcqsA and Δcrp deletion mutants and the resulting transformants were analyzed for production of CAI-1. Although pBRCqsA2383 fully complemented the ΔcqsA mutant for CAI-1 production (fold Induction > 1000), very little CAI-1 (fold induction < 50) was detected in the Δcrp mutant suggesting that placing the cqsA gene on a multicopy plasmid can not titrate off its requirement for the cAMP-CRP complex. Production of cqsA mRNA by these transformants was investigated by Northern blot analysis. In the WT strain (C7258), a single cqsA mRNA band which migrated below a 1517-bp marker could be detected (Fig. 3, lane 1) suggesting that the 1170-bp cqsA ORF is not co-transcribed with other genes. A ribonuclease protection assay indicated that the cqsA mRNA produced in C7258 starts approximately 39-bp upstream the cqsA translational start (data not shown). Introduction of pBRCqsA2383 in strain AJB61 (ΔcqsA) resulted in over expression of the cqsA full length transcript (Fig. 3, lane 3). However, very little amount of full length transcript could be detected in the Δcrp mutant (Fig. 3, lane 2). Instead, a diffuse material of smaller molecular weight hybridizing to the cqsA probe was detected (Fig. 3, lane 2, arrow). This result suggests that a low level of a modified (shorter) cqsA transcript might be produced in the Δcrp mutant either by de novo synthesis or degradation of the full length transcript.
Fig. 3. Analysis of cqsA transcription in ΔcqsA and Δcrp mutants containing plasmid pBRCqsA2383.

Samples containing 20-μg of RNA were subjected to Northern blot analysis using a DIG-labeled cqsA probe. The recA mRNA was used as loading control. Lane 1, C7258; lane 2, WL7258/pBRCqsA2383; lane3, AJB61/pBRCqsA2383.
We decided to analyze the longevity of the cqsA mRNA in WT, Δcrp and Δcya genetic backgrounds using non-steady state Northern blot analysis. To visualize sufficient full length cqsA transcript for this analysis, we introduced pBRCqsA2383 in strain AJB61 (ΔcqsA) and WL7258 (Δcrp). The transformants were grown in TSB to OD600 1.0 and rifampicin was added to block transcription. Next, samples were taken at different time points to determine cqsA mRNA decay. As shown in Fig. 4A, no cqsA mRNA could be detected 5-min after rifampicin addition in the Δcrp mutant. In contrast, an identical experiment using the complemented strain AJB61 (ΔcqsA) showed that the cqsA mRNA could be detected 20-min after the addition of rifampicin (Fig. 4A). Since addition of cAMP to TSB medium restored the ability of an adenylate cyclase mutant to produce CAI-1 (Fig. 1), we examined if exogenous cAMP acted by stabilizing the cqsA mRNA. To this end, strain WL7259 (Δcya) was grown in TSB to OD600 0.5, the culture was divided in halves and 5-mM cAMP was added to one half (the other used as a control). One h later, rifampicin was added to block transcription and samples were taken from each culture to assess the longevity of cqsA mRNA. As shown in Fig. 4B, no cqsA mRNA could be detected in the Δcya mutant either at time zero or 5-min after the addition of rifampicin. Contrastingly, supplementation of the WL7259 culture with 5-mM cAMP resulted in production of a more stable cqsA mRNA that could be detected 30-min after the addition of rifampicin (Fig. 4B). The time-dependent decay of recA mRNA was used as an independent positive control of the rifampicin-induced transcription block. In summary, the above results strongly suggest that formation of the cAMP-CRP complex positively affects cqsA expression at the level of mRNA stability. Thus, inactivation of either crp or cya leads to the production of an unstable cqsA mRNA that does not sustain CqsA expression and production of CAI-1. Based on the finding that cAMP controls the rate of CAI-1 production, we hypothesized that lowering intracellular cAMP levels by supplementation of the culture medium with D-glucose [14, 30] should increase the cell density required to activate expression of HapR. To test this hypothesis, we introduced plasmid pHapRLac2 containing a hapR-lacZ fusion that displays the typical cell density-dependent (U-shaped) pattern of quorum sensing regulated phenotypes in strain C7258ΔlacZ [24]. As shown in Fig. 5, the glucose-supplemented culture required a higher cell density to activate HapR expression.
Fig. 4. Longevity of cqsA mRNA in WT, Δcrp and Δcya genetic backgrounds.
(A). Overnight cultures of WL7258 ( crp) and AJB61 (ΔcqsA) containing pBRcqsA2383 were diluted 100-fold in fresh TSB and grown at 37°C with agitation to OD600 1.0. Rifampicin was added and samples were taken at the indicated time points for Northern blots using a DIG-labeled cqsA probe. Each lane contains 25-μg of total RNA. (B). Overnight cultures of WL7259 (C7258Δcya) were diluted 100-fold in fresh TSB and grown at 37°C with agitation to OD600 0.5. Each culture was divided in halves, one half was used as a control and the other half was transferred to a new flask containing cAMP (5-mM). The cultures were incubated 1-h as described above and rifampicin was added to block transcription. Samples were taken at the indicated time points for Northern blot analysis. Each lane was loaded with 25-μg of total RNA. After stripping the cqsA probe, the membrane was re-probed with a DIG-labeled recA DNA fragment.
Fig.5. Carbon catabolite repression regulates the cell density or quorum required to activate HapR.
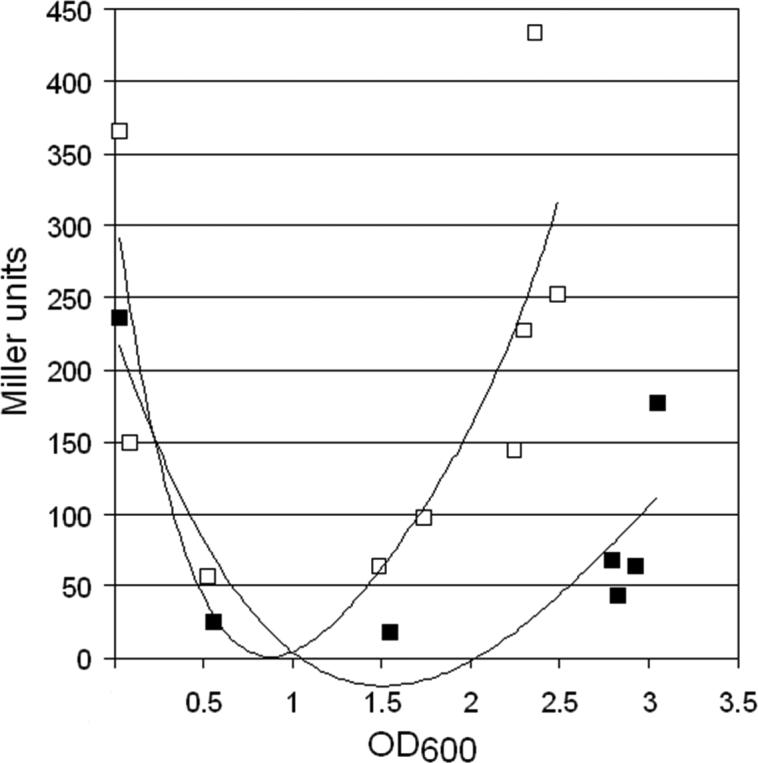
Strain C7258ΔlacZ containing plasmid pHapRLac2 was grown overnight in LB at 37°C. On the next morning the culture was diluted in fresh LB (□) or LB containing 0.2% glucose (■) and samples taken at different times to determine the expression of β-galactosidase activity. Each point is the average of three independent experiments.
4. Discussion
Results reported in this study are relevant to how bacteria integrate and respond to multiple sensory information. Here we focus on the transduction of two major external signals: carbon source and population density. Global gene expression profiling of a V. cholerae Δcrp mutant showed that CRP is required for V. cholerae to make CAI-1 [15]. Results shown in Fig. 1 further demonstrate that CRP acts through the conventional cAMP activation mechanism. The significance of this result is that it establishes a novel small molecule signaling pathway by which the primary messenger cAMP activates the production of (S)-3-hydroxytridecan-4-one which functions as an autoinducer to turn on quorum sensing signal and activate HapR. In turn, HapR has been shown to regulate the intracellular concentration of the second messenger c-di-GMP [9]. Therefore, our data suggests the existence of a cAMP – (S)-3-hydroxytridecan-4-one – c-di-GMP signaling pathway modulating a broad range of cellular activities. Our results provide a new paradigm for the integration of carbon catabolite repression and quorum sensing. V. cholerae cells resolve integrating carbon source and population density sensory information by placing the biosynthesis of a major autoinducer under cAMP control. Consequently, the intracellular concentration of cAMP acts upstream of quorum sensing to control the major input signal required for cell-to-cell communication in response to carbon source. This interpretation is consistent with previous observations showing that glucose strongly represses the production of CAI-1, hapR mRNA and the HapR-dependent hemagglutinin/protease [15, 24,27]. In fact, we here demonstrate that a higher cell density is required to activate HapR under conditions that lower intracellular cAMP such as supplementation of the culture medium with D-glucose (Fig. 5). We therefore define cAMP as acting as a quorum modulator. It has been reported that production of AI-2 in E. coli [31] and more recently in Edwardsiella tarda [32] is modulated by cAMP. Thus, cAMP regulation of the biosynthesis of an autoinducer molecule could be a general strategy by which bacteria integrate population density with other features of the extracellular milieu.
The result of our cqsA-lacZ transcriptional and translational fusion analysis is consistent with cAMP-CRP acting at the post-transcriptional level. This conclusion is further sustained by the finding that ectopic expression of cqsA from a heterologous cAMP-independent constitutive promoter is still diminished in a Δcrp mutant. Moreover, non-steady state Northern blot analysis demonstrated that the cqsA mRNA is rapidly degraded in Δcrp and Δcya mutants but can be rescued (in the Δcya mutant) by supplementing the medium with pure cAMP. To our knowledge this is the first report of cAMP-CRP controlling gene expression at the level of mRNA stability. It should be recognized that the instability of cqsA mRNA in crp and cya mutants could result from a translation defect. Since CRP is not known to have RNA binding activity, it is likely that it acts by modulating the expression of an unknown protein or small RNA (sRNA) that could interact with cqsA mRNA to exert the expected regulation. We have recently observed that in vitro transcribed cqsA does not cleave itself in the absence of added protein or RNA factors (unpublished results). This observation favors a mechanism in which CRP is required to express a protein or sRNA factor that protects cqsA mRNA from catalytic rather than autocatalytic degradation.
We have used the program TargetRNA (http://snowwhite.wellesley.edu/targetRNA/index_2.html) to find potential sRNAs in the V. cholerae genome capable of base pairing with the cqsA mRNA encoding the CqsA N-terminal region required for CRP regulation. However, none of the hits obtained with this software identified known sRNAs or exhibited additional predictive features of sRNAs. The RNA analyzer software RegRNA (http://regrna.mbc.nctu.edu) revealed two sm-sites (RAU3−6GR, R = purine nucleotide) within the cqsA N-terminal coding sequence. These sites occur in small nuclear ribonucleoproteins (snRNP) that participate in eukaryotic pre-mRNA splicing. One small bacterial sm-like protein that regulates quorum sensing is Hfq [33]. However, deletion of hfq in a Δcrp background did not restore cqsA expression (unpublished results). Therefore, it is likely that CRP regulation of cqsA mRNA stability represents a novel pathway independent of Hfq and known sRNAs. We have used the program RNAfold (http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi) to examine the secondary structure of the cqsA mRNA region which, according to our deletion and cqsA-lacZ fusion data, should contain the cAMP-CRP responsive element. This analysis revealed that the mRNA encoding the N-terminal sequence responsible for cAMP-CRP regulation can adopt a stable secondary structure with several simple and complex multipartite stem loops including the translational start. Analysis of this sequence using riboswitch finder (http://riboswitch.bioapps.biozentrum.uni-wuerzburg.de/server.html) or RNA analyzer (http://rnaanalyzer.bioapps.biozentrum.uni-wuerzburg.de/server.html) did not reveal any known motif. Furthermore, the region of cqsA capable of forming a stable secondary structure was analyzed for suboptimal conformations using the vRNAsubopt software of the Vienna RNA package (http://emboss.bioinformatics.nl/cgi-bin/emboss/vrnasubopt). Suboptimal structures calculated with this program differed only slightly and did not affect the cqsA translational start. Since cAMP-CRP regulation of cqsA mRNA stability and CAI-1 production appears to involve unknown protein and/or sRNA regulators not predictable by current computational approaches, a genetic screening is underway in our laboratory to identify regulatory factors linking cAMP-CRP to cqsA mRNA expression.
5. Acknowledgements
This work was supported by PHS Grant RO1AI063187 and 2S06GM008248-20 from the National Institutes of Health to J.A.B. and A.J.S. respectively. We are grateful to the National BioResource Project (NIG, Japan) for providing plasmid pVIK111.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
6. References
- 1.Finkelstein RA. Cholera enterotoxin (choleragen): a historical perspective. In: Barua DM, Greenough WB, editors. Cholera. Plenum Medical Book Company; New York: 1992. pp. 155–187. [Google Scholar]
- 2.Kaper JB, Morris G, Jr., Levine MM. Cholera. Clin. Microbiol. Rev. 1995;8:48–86. doi: 10.1128/cmr.8.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Higgins DA, Pomianek ME, Kraml CM, Taylor RK, Semmelhack MF, Bassler BL. The major Vibrio cholerae autoinducer and its role in virulence factor production. Nature. 2007;450:883–886. doi: 10.1038/nature06284. [DOI] [PubMed] [Google Scholar]
- 4.Chen X, Schauder S, Potier N, Van Dorsselaer A, Pelczer I, Bassler BL, Hughson FM. Structural identification of a bacterial quorum-sensing signal containing boron. Nature. 2002;415:545–549. doi: 10.1038/415545a. [DOI] [PubMed] [Google Scholar]
- 5.Miller MB, Skorupski K, Lenz DH, Taylor RK, Bassler BL. Parallel quorum sensing systems converge to regulate virulence in Vibrio cholerae. Cell. 2002;110:303–314. doi: 10.1016/s0092-8674(02)00829-2. [DOI] [PubMed] [Google Scholar]
- 6.Jobling MG, Holmes RK. Characterization of hapR, a positive regulator of the Vibrio cholerae HA/protease gene hap, and its identification as a functional homologue of the Vibrio harveyi luxR gene. Mol. Microbiol. 1997;26:1023–1034. doi: 10.1046/j.1365-2958.1997.6402011.x. [DOI] [PubMed] [Google Scholar]
- 7.Hammer BK, Bassler BL. Quorum sensing controls biofilm formation in Vibrio cholerae. Mol. Microbiol. 2003;50:101–114. doi: 10.1046/j.1365-2958.2003.03688.x. [DOI] [PubMed] [Google Scholar]
- 8.Zhu J, Miller MB, Vance RE, Dziejman M, Bassler BL, Mekalanos JJ. Quorum-sensing regulators control virulence gene expression in Vibrio cholerae. Proc. Natl. Acad. Sci. U.S.A. 2002;99:3129–3134. doi: 10.1073/pnas.052694299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Waters CM, Lu W, Rabinowitz JD, Bassler BL. Quorum sensing controls biofilm formation in Vibrio cholerae through modulation of cyclic di-GMP levels and repression of vpsT. J. Bacteriol. 2008;190:2527–2536. doi: 10.1128/JB.01756-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kovacikova G, Skorupski K. Regulation of virulence gene expression in Vibrio cholerae by quorum sensing: HapR functions at the aphA promoter. Mol. Microbiol. 2002;46:1135–1147. doi: 10.1046/j.1365-2958.2002.03229.x. [DOI] [PubMed] [Google Scholar]
- 11.Brückner R, Titgemeyer F. Carbon catabolite repression in bacteria: choice of the carbon source and autoregulatory limitation of sugar utilization. FEMS Microbiol. Letters. 2002;209:141–148. doi: 10.1111/j.1574-6968.2002.tb11123.x. [DOI] [PubMed] [Google Scholar]
- 12.Kolb A, Busby S, Buc H, Garges S, Adhya S. Transcriptional regulation by cAMP and its receptor protein. Ann. Rev. Biochem. 1993;62:749–795. doi: 10.1146/annurev.bi.62.070193.003533. [DOI] [PubMed] [Google Scholar]
- 13.Stülke J, Hillen W. Carbon catabolite repression in bacteria. Curr. Opinion Microbiol. 1999;2:195–201. doi: 10.1016/S1369-5274(99)80034-4. [DOI] [PubMed] [Google Scholar]
- 14.Deutscher J, Francke C, Postma PW. How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolisms in bacteria. Microbiol. Molec. Biol. Rev. 2006;70:939–1031. doi: 10.1128/MMBR.00024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liang W, Pascual-Montano A, Silva AJ, Benitez JA. The cyclic AMP receptor protein modulates quorum sensing, motility and multiple genes that affect intestinal colonization in Vibrio cholerae. Microbiology. 2007;153:2964–2975. doi: 10.1099/mic.0.2007/006668-0. [DOI] [PubMed] [Google Scholar]
- 16.Liang W, Silva AJ, Benitez JA. The cyclic AMP receptor protein modulates colonial morphology in Vibrio cholerae. Appl. Environ. Microbiol. 2007;73:7482–7487. doi: 10.1128/AEM.01564-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Silva AJ, Zafar Sultan S, Liang W, Benitez JA. Role of the histone-like nucleoid structuring protein (H-NS) in the regulation of RpoS and RpoS-dependent genes in Vibrio cholerae. J. Bacteriol. 2008 Sep 12;190 doi: 10.1128/JB.00360-08. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Lorenzo V, Eltis L, Kessler B, Timmis KN. Analysis of the Pseudomonas gene products using lacIq/Ptrp-lac plasmids and transposons that confer conditional phenotypes. Gene. 1993;123:17–24. doi: 10.1016/0378-1119(93)90533-9. [DOI] [PubMed] [Google Scholar]
- 19.Marcus H, Ketley JM, Kaper JB, Holmes RK. Effect of DNAse production, plasmid size and restriction barrier on transformation of Vibrio cholerae by electroporation and osmotic shock. FEMS Microbiol. Letters. 1990;68:149–154. doi: 10.1111/j.1574-6968.1990.tb04139.x. [DOI] [PubMed] [Google Scholar]
- 20.Donnenberg MS, Kaper JB. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect Immun. 1991;59:4310–4317. doi: 10.1128/iai.59.12.4310-4317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Silva AJ, Pham K, Benitez JA. Hemaglutinnin/protease expression and mucin gel penetration in El Tor biotype Vibrio cholerae. Microbiology. 2003;149:1883–1891. doi: 10.1099/mic.0.26086-0. [DOI] [PubMed] [Google Scholar]
- 22.Rothmel RD, Shinabarger D, Parsek M, Aldrich T, Chakrabarty AM. Functional analysis of the Pseudomonas putida regulatory protein CatR: transcriptional studies and determination of the CatR DNA binding site by hydroxyl-radical footprinting. J. Bacteriol. 1991;173:4717–4724. doi: 10.1128/jb.173.15.4717-4724.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kalogeraki VS, Winans SC. Suicide plasmids containing promoterless reporter genes can simultaneously disrupt and create fusions to target genes of diverse bacteria. Gene. 1997;188:69–75. doi: 10.1016/s0378-1119(96)00778-0. [DOI] [PubMed] [Google Scholar]
- 24.Silva AJ, Benitez JA. Transcriptional regulation of Vibrio cholerae hemagglutinin/protease by the cyclic AMP receptor protein and RpoS. J. Bacteriol. 2004;186:6374–6382. doi: 10.1128/JB.186.19.6374-6382.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 1971. [Google Scholar]
- 26.Häse CC, Finkelstein RA. Cloning and nucleotide sequence of the Vibrio cholerae hemagglutinin/protease (HA/protease) gene and construction of an HA/protease-negative strain. J. Bacteriol. 1991;173:3311–3317. doi: 10.1128/jb.173.11.3311-3317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Benitez JA, Silva AJ, Finkelstein RA. Environmental signals controlling production of hemagglutinin/protease in Vibrio cholerae. Infect. Immun. 2001;69:6549–6553. doi: 10.1128/IAI.69.10.6549-6553.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Youn H, Kerby RL, Conrad M, Roberts GP. Study of Highly Constitutively Active Mutants Suggests How cAMP Activates cAMP Receptor Protein. J. Biol. Chem. 2006;281:1119–1127. doi: 10.1074/jbc.M509421200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.De Boer HA, Comstock LJ, Vasser M. The tac promoter: a functional hybrid derived from the trp and lac promoters. Proc. Natl. Acad. Sci. USA. 1983;80:21–25. doi: 10.1073/pnas.80.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ganguly U, Greennough WB., III Adenosine 3'5'- cyclic monophosphate in Vibrio cholerae. Infect. Immun. 1975;11:343–349. doi: 10.1128/iai.11.2.343-349.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang l., Hashimoto Y, Tsao CY, Valdes JJ, Bentley WE. Cyclic AMP (cAMP) and cAMP receptor protein influence both synthesis and uptake of extracellular autoinducer 2 in Escherichia coli. J. Bacteriol. 2005;187:2066–2076. doi: 10.1128/JB.187.6.2066-2076.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang M, Sun K, Sun L. Regulation of autoinducer 2 production and luxS expression in a pathogenic Edwardsiella tarda strain. Microbiology. 2008;154:2060–2069. doi: 10.1099/mic.0.2008/017343-0. [DOI] [PubMed] [Google Scholar]
- 33.Lenz DH, Mok KC, Lilley BN, Kulkarni RV, Wingreen NS, Bassler BL. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell. 2004;118:69–82. doi: 10.1016/j.cell.2004.06.009. [DOI] [PubMed] [Google Scholar]