SUMMARY
MicroRNAs (miRNAs) are crucial for normal embryonic stem (ES) cell self-renewal and cellular differentiation, but how miRNA gene expression is controlled by the key transcriptional regulators of ES cells has not been established. We describe here a new map of the transcriptional regulatory circuitry of ES cells that incorporates both protein-coding and miRNA genes, and which is based on high-resolution ChIP-seq data, systematic identification of miRNA promoters, and quantitative sequencing of short transcripts in multiple cell types. We find that the key ES cell transcription factors are associated with promoters for most miRNAs that are preferentially expressed in ES cells and with promoters for a set of silent miRNA genes. This silent set of miRNA genes is co-occupied by Polycomb Group proteins in ES cells and expressed in a tissue-specific fashion in differentiated cells. These data reveal how key ES cell transcription factors promote the miRNA expression program that contributes to normal self-renewal and cellular differentiation, and integrate miRNAs and their targets into an expanded model of the regulatory circuitry controlling ES cell identity.
IN THIS ISSUE STATEMENT
MicroRNAs (miRNAs) are crucial for normal embryonic stem (ES) cell self-renewal and cellular differentiation. Marson et al. describe how key ES cell transcription factors promote the miRNA expression program necessary for ES cell state using high-resolution ChIP-seq data, systematic identification of miRNA promoters, and quantitative sequencing of short transcripts in multiple cell types. They use this data to generate a model of ES cell regulatory circuitry that includes both protein-coding and miRNA genes. This information should prove useful as investigators continue to probe the role of miRNAs in pluripotency, cell state, disease and regenerative medicine.
INTRODUCTION
Embryonic stem (ES) cells hold significant potential for clinical therapies because of their distinctive capacity to both self-renew and differentiate into a wide range of specialized cell types. Understanding the transcriptional regulatory circuitry of ES cells and early cellular differentiation is fundamental to understanding human development and realizing the therapeutic potential of these cells. Transcription factors that control ES cell pluripotency and self-renewal have been identified (Chambers and Smith, 2004; Niwa, 2007) and a draft of the core regulatory circuitry by which these factors exert their regulatory effects on protein-coding genes has been described (Boyer et al., 2005; Loh et al., 2006; Lee et al., 2006; Boyer et al. 2006; Jiang et al., 2008; Cole et al., 2008; Kim et al., 2008; Tam et al., 2008). MicroRNAs (miRNAs) are also likely to play key roles in ES cell gene regulation (Kanellopoulou et al., 2005; Murchison et al., 2005; Wang et al., 2007), but little is known about how miRNAs participate in the core regulatory circuitry controlling self-renewal and pluripotency in ES cells.
Several lines of evidence indicate that miRNAs contribute to the control of early development. miRNAs appear to regulate the expression of a significant percentage of all genes in a wide array of mammalian cell types (Lewis et al., 2005; Lim et al., 2005; Krek et al., 2005; Farh et al., 2005). A subset of miRNAs is preferentially expressed in ES cells or embryonic tissue (Houbaviy et al., 2003; Suh et al., 2004; Houbaviy et al., 2005; Mineno et al., 2006). Dicer-deficient mice fail to develop (Bernstein et al., 2003) and ES cells deficient in miRNA processing enzymes show defects in differentiation and proliferation (Kanellopoulou et al., 2005; Murchison et al., 2005; Wang et al., 2007). Specific miRNAs have been shown to participate in mammalian cellular differentiation and embryonic development (Stefani and Slack, 2008). However, how transcription factors and miRNAs function together in the regulatory circuitry that controls early development has not yet been examined.
The major limitation in connecting miRNA genes to the core transcriptional circuitry of ES cells has been sparse annotation of miRNA gene transcriptional start sites and promoter regions. Mature miRNAs, which specify post-transcriptional gene repression, arise from larger transcripts that are then processed (Bartel, 2004). Over 400 mature miRNAs have been confidently identified in the human genome (Landgraf et al., 2007), but only a minority of the primary transcripts have been identified and annotated. Prior attempts to connect ES cell transcriptional regulators to miRNA genes have searched for transcription factor binding sites only close to the annotated mature miRNA sequences (Boyer et al., 2005; Loh et al., 2006; Lee et al., 2006). Additionally, studies of the core transcriptional circuitry of ES cells have compared transcription factor occupancy to mRNA expression data, but have not examined systematically miRNA expression in ES cells and differentiated cell types, limiting our knowledge of transcriptional regulation of miRNA genes in these cells (Boyer et al., 2005; Loh et al., 2006; Lee et al., 2006; Boyer et al. 2006; Jiang et al., 2008; Cole et al., 2008; Kim et al., 2008; Tam et al., 2008).
To incorporate miRNA gene regulation into the model of transcriptional regulatory circuitry of ES cells, we began by generating new, high-resolution, genome-wide maps of binding sites for key ES cell transcription factors using massive parallel sequencing of chromatin immunoprecipitation (ChIP-seq). These data reveal highly overlapping occupancy of Oct4, Sox2, Nanog and Tcf3 at the transcriptional start sites of miRNA transcripts, which we systematically mapped based on a method that uses chromatin landmarks and transcript data. We then carried out quantitative sequencing of short transcripts in ES cells, neural precursor cells (NPCs) and mouse embryonic fibroblasts (MEFs), which revealed that Oct4, Sox2, Nanog and Tcf3 occupy the promoters of most miRNAs that are preferentially or uniquely expressed in ES cells. Our data also revealed that a subset of the Oct4/Sox2/Nanog/Tcf3 occupied miRNA genes are silenced in ES cells by Polycomb Group proteins, but are expressed later in development in specific lineages. High-resolution transcription factor location analysis, systematic mapping of the primary miRNA transcriptional start sites in mouse and human, and quantitative sequencing of miRNAs in three different cell types provide a valuable data resource for studies of the gene expression program in ES and other cells and the regulatory mechanisms that control cell fate. The data also produce an expanded model of ES cell core transcriptional regulatory circuitry that now incorporates transcriptional regulation of miRNAs, and post-transcriptional regulation mediated by miRNAs, into the molecular understanding of pluripotency and early cellular differentiation.
RESULTS
High-resolution genome-wide location analysis in ES cells with ChIP-seq
To connect miRNA genes to the core transcriptional circuitry of ES cells, we first generated high-resolution genome-wide maps of Oct4, Sox2, Nanog, and Tcf3 occupancy (Figure 1; similar data was recently described for Oct4, Sox2, and Nanog (Chen et al,)). ChIP-seq allowed us to map transcription factor binding sites and histone modifications across the entire genome at high resolution (Barski et al., 2007; Johnson et al., 2007; Mikkelsen et al., 2007; Robertson et al., 2007), and we optimized the protocol to allow for robust analysis of transcription factor binding in murine ES cells (Supplementary Text). Oct4, Sox2, Nanog and Tcf3 were found to co-occupy 14,230 sites in the genome (Figures 1A, S1, S2 and Table S1–Table S3). Approximately one quarter of these occurred within 8kb of the transcription start site of 3,289 annotated genes, another one quarter occurred within genes but more than 8kb from the start site, and almost half occurred in intergenic regions distal from annotated start sites (Supplementary Text). Binding of the four factors at sites surrounding the Sox2 gene (Figure 1B) exemplified two key features of the data: all four transcription factors co-occupied the identified binding sites and the resolution was sufficient to determine the DNA sequence associated with these binding events to a resolution of <25bp. Composite analysis of all bound regions provided higher resolution and suggested how these factors occupy their common DNA-sequence motif (Figure S4, Table S4). Knowledge of these binding sites provided data necessary to map these key transcription factors to the promoters of miRNA genes.
Figure 1. High-resolution genome-wide mapping of core ES cell transcription factors with ChIP-seq.
A. Summary of binding data for Oct4, Sox2, Nanog and Tcf3. 14,230 sites are co-bound genome wide and mapped to either promoter proximal (TSS +/− 8kb, dark green, 27% of binding sites), genic (>8kb from TSS, middle green, 30% of binding sites), or intergenic (light green, 43% of binding sites). The promoter proximal binding sites are associated with 3,289 genes. B. (upper) Binding of Oct4 (blue), Sox2 (purple), Nanog (orange) and Tcf3 (red) across 37.5kb of mouse chromosome 3 surrounding the Sox2 gene (black box below the graph, arrow indicates transcription start site). Short sequences uniquely and perfectly mapping to the genome were extended to 200bp (maximum fragment length) and scored in 25bp bins. The score of the bins were then normalized to the total number of reads mapped. Oct4/Sox2 DNA binding motifs (Loh et al., 2006) were mapped across the genome and are shown as grey boxes below the graph. Height of the box reflects the quality of the motif. (lower) Detailed analysis of three enriched regions (Chromosome 3: 4,837,600-34,838,300, 34,845,300-34,846,000, and 34,859,900–34,860,500) at the Sox2 gene indicated with dotted boxes above. The 5’ most base from ChIP-seq were separated by strand and binned into 25bp regions. Sense (darker tone) and anti-sense (light tone) of each of the four factors tested are directed towards the binding site, which in each case occurs at a high-confidence Oct/Sox2 DNA binding motif indicated below.
Identification of miRNA promoters
Imperfect knowledge of the start sites of primary miRNA transcripts has limited our ability to identify the transcription factor binding events that control miRNA gene expression in vertebrates. Previous strategies to identify the 5’ ends of primary miRNAs have been hampered because they relied on isolation of transient primary miRNA transcript, required knowledge of the specific cell type in which each given miRNA is transcribed, or focused only on potential start sites proximal to mature miRNAs (Fukao et al., 2007; Mikkelsen et al., 2007; Zhou et al., 2007; Barrera et al., 2008). To identify systematically transcriptional start sites for miRNA genes in the mouse and human genomes, we took advantage of the recent observation that histone H3 is tri-methylated at its lysine 4 residue (H3K4me3) at the transcriptional start sites of most genes in the genome, even when genes are not productively transcribed, and knowledge that this covalent modification is restricted to sites of transcription initiation (Barski et al., 2007; Guenther et al., 2007). We used the genomic coordinates of the H3K4me3 enriched loci derived from multiple cell types (Table S5, Barski et al., 2007; Guenther et al., 2007; Mikkelsen et al., 2007) to create a library of candidate transcription start sites in both human and mouse (Figures 2 and S5).
Figure 2. Identification of miRNA promoters.
A. Description of algorithm for miRNA promoter identification. A library of candidate transcriptional start sites was generated with histone H3 lysine 4 tri-methyl (H3K4me3) location analysis data from multiple tissues (Barski et al., 2007; Guenther et al., 2007; Mikkelsen et al., 2007). Candidates were scored to assess likelihood that they represent true miRNA promoters. Based on scores, a list of mouse and human miRNA promoters was assembled. Additional details can be found in Supplemental Text. B. Examples of identified miRNA promoter regions. A map of H3K4me3 enrichment is displayed in regions neighbouring selected human and mouse miRNAs for multiple cell types: human ES cells (hES), REH human pro-B cell line (B cell), primary human hepatocytes (Liver), primary human T cells (T cell), mouse ES cells (mES), neural precursor cells (NPCs) and mouse embryonic fibroblasts (MEFs). miRNA promoter coordinates were confirmed by distance to mature miRNA genomic sequence, conservation and EST data (shown as solid line where available). Predicted transcriptional start site and direction of transcription are noted by an arrow, with mature miRNA sequences indicated (red). CpG islands, commonly found at promoters, are indicated (green). Dotted lines denote presumed transcripts. C. Confirmation of predicted transcription start sites for active miRNAs using chromatin modifications. Normalized ChIP-seq counts for H3K4me3 (red), H3K79me2 (blue) and H3K36me3 (green) are shown for two miRNA genes where EST data was unavailable. Predicted start site (arrow), CpG islands (green bar), presumed transcript (dotted lines) and miRNA positions (red bar) are shown. D. Most human and mouse miRNA promoters show evidence of H3K4me3 enrichment in multiple tissues.
High-confidence promoters were identified for over 80% of miRNAs in both mouse and human (Figures 2, S5 and Table S6 and Table S7). These promoters were associated with 185 murine primary microRNA transcripts (pri-miRNAs) (specifying 336 mature miRNAs), and 294 human pri-miRNAs (specifying 441 mature miRNAs) (Table S6 and Table S7). To identify promoters for miRNA genes, the association of candidate transcriptional start sites with regions encoding mature miRNAs was scored based on proximity to annotated mature miRNA sequences (Landgraf et al., 2007), available EST data, and conservation between species (Figures 2A, S5 and S6 and Supplementary Text).
Five lines of evidence indicated that this approach identified genuine transcriptional start sites for miRNA genes. Existing EST data provided evidence that the predicted transcripts do in fact originate at the identified start sites and continue through the annotated loci of mature miRNAs (Figures 2B, S5 and S6). In addition to the chromatin signature of promoters, a high fraction of these regions contained CpG islands, a DNA sequence element often associated with promoters (Figure 2B and Table S6 and Table S7). Third, in some instances where evidence of primary miRNA transcripts, which may be present only transiently before processing, was not available in published databases at the identified transcriptional start sites, chromatin marks associated with transcriptional elongation including nucleosomes methylated at H3 lysine 36 (H3K36me3) and H3 lysine 79 (H3K79me2), provided evidence that such transcripts are actively produced (Figure 2C and Mikkelsen et al., 2007). Fourth, most miRNA promoters showed evidence of H3K4me3 enrichment in multiple tissues, as observed at the promoters of most protein-coding genes (Barski et al., 2007; Guenther et al., 2007; Heintzman et al., 2007) (Figure 2D). Finally, there was a high degree of correlation (8/10) between the identified miRNA transcriptional start sites and those that have mapped previously by other methods (Supplementary Text).
Occupancy of miRNA promoters by core ES cell transcription factors
The binding sites of the ES cell transcription factors Oct4, Sox2, Nanog and Tcf3 were next mapped to these high-confidence miRNA promoters (Figure 3). In murine ES cells, Oct4, Sox2, Nanog, and Tcf3 co-occupied the promoters for 55 distinct miRNA transcription units, which included three clusters of miRNAs that are expressed as large polycistrons, thus suggesting that these regulators have the potential to directly control the transcription of 81 distinct mature miRNAs (Figure 3A and Table S6). This set of miRNAs occupied by Oct4/Sox2/Nanog/Tcf3 represented roughly 20 percent of annotated mammalian miRNAs, similar to the ~20 percent of protein-coding genes that were bound at their promoters by these key transcription factors (Table S2).
Figure 3. Oct4, Sox2, Nanog and Tcf3 occupancy and regulation of miRNA promoters.
A. Oct4 (blue), Sox2 (purple), Nanog (orange) and Tcf3 (red) binding is shown at four murine miRNA genes as in Figure 1A. H3K4me3 enrichment in ES cells is indicated by shading across genomic region. Presumed transcripts are shown as dotted lines. Coordinates for the mmu-mir-290-295 cluster are derived from NCBI build 37. B. Oct4 ChIP enrichment ratios (ChIP-enriched versus total genomic DNA) are shown across human miRNA promoter region for the hsa-mir-302 cluster. H3K4me3 enrichment in ES cells is indicated by shading across genomic region. C. Schematic of miRNAs with conserved binding by the core transcription factors in ES cells. Transcription factors are represented by dark blue circles and miRNAs are represented by purple hexagons. D. Quantitative RT-PCR analysis of RNA extracted from ZHBTc4 cells in the presence or absence of doxycycline treatment. Fold change was calculated for each pri-miRNA for samples from 12 hours and 24 hours of doxycyline treatment relative to those from untreated cells. Transcript levels were normalized to Gapdh levels. Error bars indicate standard deviation derived from triplicate PCR reactions.
To determine if transcription factor occupancy of miRNA promoters is conserved across species, we performed genome-wide location analysis for Oct4 in human ES cells using microarray-based analysis. We found extensive conservation of the set of miRNA genes that were occupied at their promoters by Oct4, as exemplified by the mir-302 cluster (Figures 3A, 3B and Table S7 and Table S8). Transcription factor occupancy does not mean necessarily that the adjacent gene is regulated by that factor; conserved transcription factor occupancy of a promoter, however, suggests gene regulation by that factor. Thus, our data identified a set of miRNA genes that are bound at their promoters by key ES cell transcription factors in mouse and human cells (Figure 3C), suggesting that core ES cell transcription factor occupancy of these particular miRNA genes has functional significance.
The dependence of Oct4/Sox2/Nanog/Tcf3-bound miRNA genes on the core ES cell transcription factors was assessed by examining changes in miRNA expression following perturbation of individual transcription factors. First, we studied the effects of Oct4 depletion in ZHBTc4 ES cells, which allow for conditional repression of Oct4 with doxycycline treatment (Niwa et al., Nature Genetics 2000). miRNA expression was examined at 12 and 24 hours after doxycycline treatment, when Oct4 was silenced but the cells remained ES-like morphologically and still expressed Sox2 (Figure S7). Effects on miRNA regulation were measured globally using quantitative sequencing of short RNAs (18–30 nucleotides) in these cells (Morin et al., 2008). Although mature miRNAs have exceptionally long half-lives, small reductions in the levels of the majority of Oct4/Sox2/Nanog/Tcf3-bound miRNAs were observed (Figure S7E). To measure more directly the transcriptional activity of Oct4/Sox2/Nanog/Tcf3-occupied miRNA genes, the levels of primary miRNA transcripts were assessed by quantitative PCR (Figure 3D and S7). All five of the primary miRNAs examined were reduced significantly upon Oct4 depletion (Figure 3D).
Tcf3 has been shown to repress the expression of key pluripotency protein-coding genes in ES cells under standard culture conditions (Cole et al., 2008; Tam et al., 2008; Yi et al., 2008). Following shRNA-mediated knockdown of Tcf3 in ES cells, qPCR revealed that levels of primary transcripts for ES cell miRNA genes mir-290-295 and the mir-302 cluster were elevated, though this effect was modest (Figure S8). In summary, the observation that these key ES cell miRNAs are generally down-regulated in response to Oct4 depletion and up-regulated upon Tcf3 depletion, as would be predicted based on published effects of these factors on protein-coding genes, demonstrates that the core transcriptional circuitry of ES cells can play a functional role in the regulation of miRNA genes.
Regulation of Oct4/Sox2/Nanog/Tcf3-bound miRNA genes during differentiation
Oct4 and Nanog are silenced as ES cells begin to differentiate (Chambers and Smith, 2004; Niwa, 2007). If the Oct4/Sox2/Nanog/Tcf3 complex is required for activation or repression of its target miRNAs, the targets should be differentially expressed when ES cells are compared to a differentiated cell-type. To test this hypothesis, Solexa sequencing of 18–30 nucleotide transcripts in ES cells, mouse embryonic fibroblasts (MEFs), and neural precursors (NPCs), was performed to obtain quantitative information on the abundance of miRNAs in pluripotent cells relative to two differentiated cell types (Figure 4A and Table S1). In each cell type examined, a small subset of miRNAs predominated, with pronounced changes in miRNA abundance observed among the cell types (Supplementary Text).
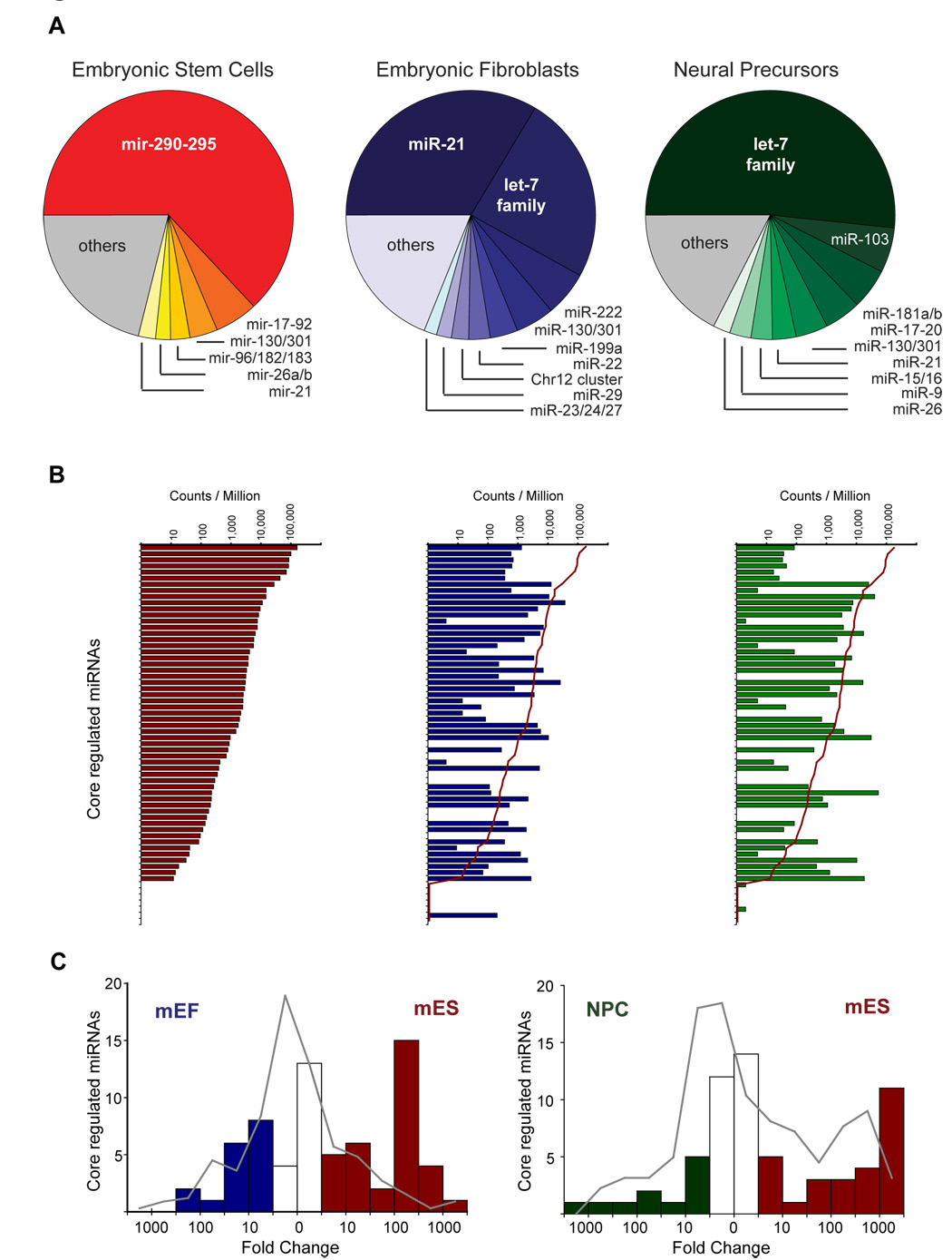
Figure 4. Regulation of Oct4/Sox2/Nanog/TCF3-bound miRNAs during differentiation.
A. Pie charts showing relative contributions of miRNAs to the complete population of miRNAs in mES cells (red), MEFs (blue) and neural precursors (NPCs, green) based on quantification of miRNAs from by small RNA sequencing. A full list of the miRNAs identified can be found in Table S9. B. Normalized frequency of detection of individual mature miRNAs whose promoters are occupied by Oct4/Sox2/Nanog/ Tcf3 in mouse. Red line in center and right panel show the level of detection in ES cells. C. Histogram of changes in frequency of detection. Changes for miRNAs whose promoters are occupied by Oct4, Sox2, Nanog and Tcf3 in mouse are shown as bars (red for ES enriched, blue for MEF enriched and green for NPC enriched). The background frequency for non-occupied miRNAs is shown as a grey line.
When the abundance of all miRNAs bound by Oct4/Sox2/Nanog/Tcf3 in ES cells was examined in the three cell types, approximately half of the miRNAs dropped more than an order of magnitude in abundance in MEFs and NPCs relative to ES cells, as predicted (Figure 4B). A small number of the Oct4/Sox2/Nanog/Tcf3-occupied miRNAs, which will be further discussed below, were scarce in ES cells and showed increased abundance in MEFs and NPCs.
Oct4/Sox2/Nanog/Tcf3-occupied miRNAs were, in general, preferentially expressed in embryonic stem cells, as demonstrated by the analysis shown in Figure 4C. Whereas most miRNAs are unchanged in expression in ES cells relative to MEFs or NPCs, a significant portion of Oct4/Sox2/Nanog/Tcf3 occupied miRNAs are 100 fold more abundant in ES cells than in MEFs (p<5 × 10−15), and 1,000 fold more abundant in ES cells than in NPCs (p<5 × 10−9). This group of Oct4/Sox2/Nanog/Tcf3 bound miRNAs that was significantly more abundant in ES cells than in NPCs and MEFs, was also found to be actively expressed in induced pluripotent stem (iPS) cells (generated as described in Wernig et al., 2007), at levels comparable to those in ES cells (Figure S9). This is consistent with our evidence (Figures 3, S7 and S8) that core ES cell transcription factors regulate the expression of these miRNAs in pluripotent cells.
Polycomb Group Proteins co-occupy tissue-specific miRNAs that are silenced in ES cells
Previous studies have revealed that core ES cell transcription factors occupy a set of transcriptionally active genes, but also occupy, with Polycomb Group proteins, a set of transcriptionally repressed genes that are poised for expression upon cellular differentiation (Lee et al., 2006; Bernstein et al., 2006; Boyer et al., 2006). As noted above, Oct4/Sox2/Nanog/Tcf3 similarly occupied a group of miRNA genes that were transcriptionally inactive in ES cells, but were activated selectively in particular differentiated cell types (Figure 4B). We reasoned that Polycomb complexes might also co-occupy Oct4/Sox2/Nanog/Tcf3-occupied miRNA genes that are inactive in ES cells. Indeed, new ChIP-seq data for the Polycomb Group protein Suz12 in murine ES cells supported this hypothesis (Figure 5A and Table S6, Table S7, Table S10). As expected, these promoters were also enriched for nucleosomes with histone H3K27me3, a chromatin modification catalyzed by Polycomb Group proteins (Figure 5A and Table S6 and Mikkelsen et al., 2007). In keeping with the repressive function of the Polycomb Group proteins reported at protein coding genes, miRNAs occupied at their promoters by Suz12 in ES cells were significantly less abundant in ES cells compared to all other miRNAs (Figure 5B). Approximately one quarter of the Oct4/Sox2/Nanog/Tcf3-occupied miRNAs belonged to the repressed set of miRNA genes bound by Suz12 in murine ES cells (Table S6 and Table S7).
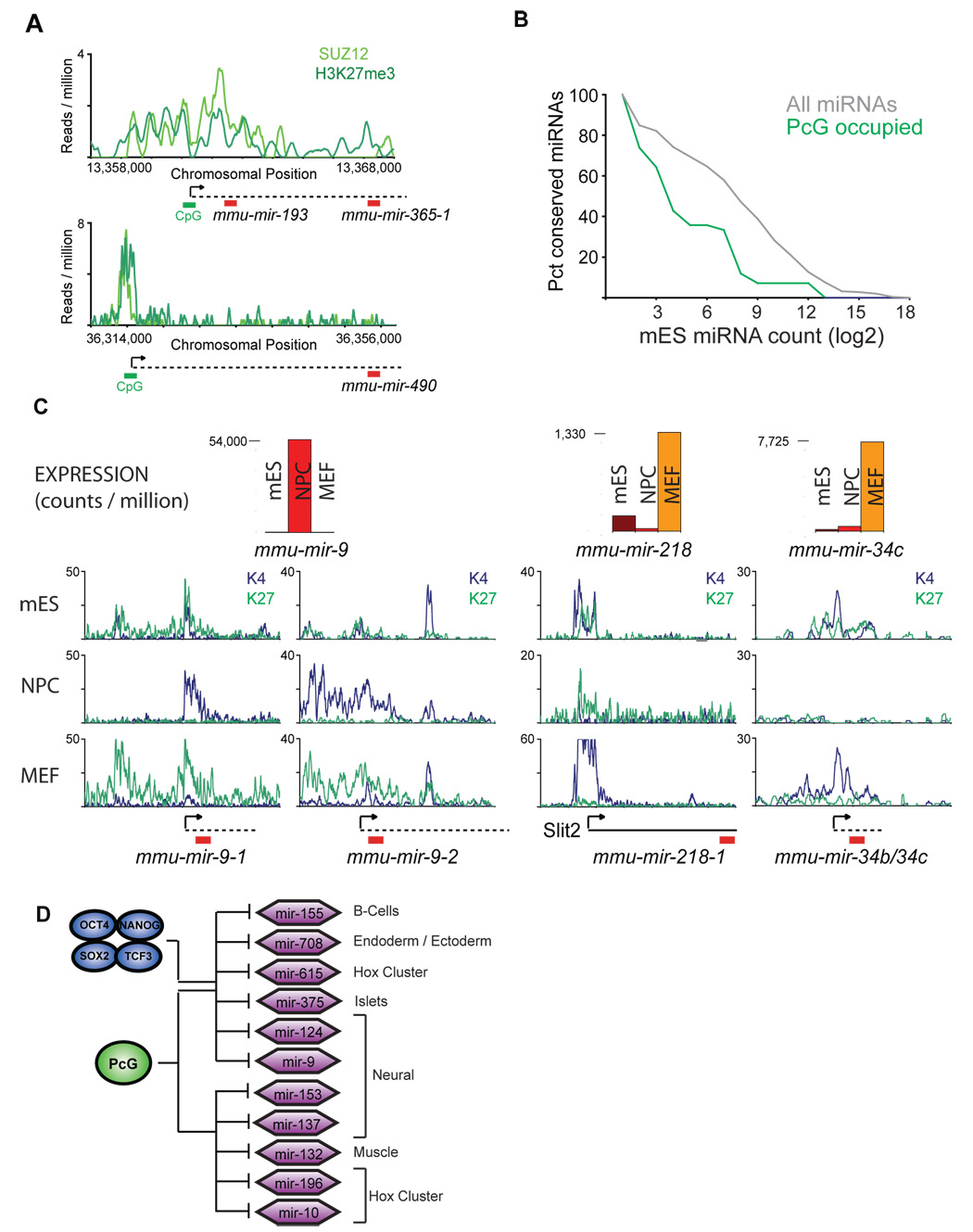
Figure 5. Polycomb represses lineage-specific miRNAs in ES cells.
A. Suz12 (light green) and H3K27me3 (dark green, Mikkelsen et al., 2007) binding are shown for two miRNA genes in murine ES cells. Predicted start sites (arrow), CpG islands (green bar), presumed miRNA primary transcript (dotted line) and mature miRNA (red bar) are shown. B. Expression analysis of miRNAs from mES cells based on quantitative small RNA sequencing. Cumulative distributions for Polycomb-bound miRNAs (green line) and all miRNAs (grey line) are shown. C. Expression analysis of miRNAs occupied by Suz12 in mES cells. Relative counts are shown for mES (red), NPCs (orange) and MEFs (yellow). H3K27me3 (green line) and H3K4me3 (blue line) mapped reads are shown for mES cells, MEFs and NPCs (Mikkelsen et al., 2007). D. Schematic of a subset of miRNA genes occupied by Suz12 in both mES and hES cells as in Figure 3C. miRNA genes where Oct4/Sox2/Nanog/Tcf3 are also present are indicated. Cells known to selectively express these miRNAs based on computation predictions (Farh et al., 2005) or experimental confirmation (Landgraf et al., 2007) are indicated. The Polycomb group (PcG) protein Suz12 is represented by a green circle.
To further examine the behavior of this set of miRNAs during embryonic cell-fate commitment, we returned to our quantitative sequencing data of short transcripts in ES cells, MEFs and NPCs (Figure 5C). Notably, miRNAs that were bound by Polycomb Group proteins in ES cells were among the transcripts that were specifically induced in each of the differentiated cell types. For example, transcript levels of miR-9, a miRNA previously identified in neural cells and which promotes neural differentiation (Lagos-Quintana et al., 2002; Krichevsky et al., 2006), were significantly elevated in NPCs relative to ES cells, but this miRNA remained scarce in MEFs. Similarly, miR-218 and miR-34b/34c expression was induced in MEFs, but remained at low levels in NPCs (Figure 5C). Consistent with Polycomb-mediated repression of these lineage-specific miRNAs, the repressive chromatin mark deposited by Polycomb Group proteins, H3K27me3, was selectively lost at the promoters of the miRNAs in the cells in which they were induced (Figure 5C and Mikkelsen et al., 2007).
The tissue-specific expression pattern of miRNAs repressed by Polycomb in ES cells is consistent with these miRNAs serving as determinants of cell-fate decisions in a manner analogous to the developmental regulators whose genes are repressed by Polycomb in ES cells (Lee et al., 2006; Bernstein et al., 2006; Boyer et al., 2006). Such a function in cell-fate determination would require that these miRNAs remain silenced in pluripotent ES cells. Indeed, the miRNAs that were repressed in ES cells by Polycomb Group proteins appear to be induced, later in development, in a highly restricted subset of differentiated tissues specific to each miRNA (Figure S10), unlike the majority of miRNAs identified in mouse (Landgraf et al., 2007). The miRNAs with promoters bound by Polycomb Group proteins in ES cells were significantly enriched (p<0.005) among the set of the most tissue-specific mammalian miRNAs (Figure S10 and Landgraf et al., 2007). This suggests a model whereby Polycomb Group proteins repress a set of tissue-specific miRNA genes in ES cells, a subset of which are co-occupied by Oct4, Sox2, Nanog and Tcf3 (Figure 5D).
DISCUSSION
Here we provide new high-resolution, genome-wide maps of core ES cell transcription factors, identify promoter regions for most miRNA genes, and deduce the association of the ES cell transcription factors with these miRNA genes. We also provide quantitative sequence data of short RNAs in ES cells, NPCs and MEFs to examine changes in miRNA transcription. The key transcriptional regulators in ES cells collectively occupied the promoters of many of the miRNAs that were most abundant in ES cells, including those that were down-regulated as ES cells differentiate. In addition, these factors also occupied the promoters of a second, smaller set of miRNAs that were repressed in ES cells and were selectively expressed in specific differentiated cell types. In ES cells, this second group of miRNAs were co-occupied by Polycomb group proteins, which are also known to silence key lineage-specific, protein-coding developmental regulators. Together these data reveal two key groups of miRNAs that are direct targets of Oct4/Sox2/Nanog/Tcf3: one group of miRNAs that is preferentially expressed in pluripotent cells, and a second, Polycomb-occupied group that is silenced in ES cells and is poised to contribute to cell fate-decisions during mammalian development.
miRNA contribution to ES cell identity
Several miRNA polycistrons, which encode the most abundant miRNAs in ES cells and which are silenced during early cellular differentiation (Houbaviy et al., 2003; Suh et al., 2004; Houbaviy et al., 2005), were occupied at their promoters by Oct4, Sox2, Nanog and Tcf3. The most abundant in murine ES cells was the mir-290-295 cluster, which contains multiple mature miRNAs with seed sequences similar or identical to those of the miRNAs in the mir-302 cluster and the mir-17-92 cluster. miRNAs with the same seed sequence also predominate in human embryonic stem cells (Laurent et al., 2008). miRNAs in this family have been implicated in cell proliferation (O’Donnell et al., 2005; He et al., 2005; Voorhoeve et al., 2006), consistent with the impaired self-renewal phenotype observed in miRNA-deficient ES cells (Kanellopoulou et al., 2005; Murchison et al., 2005; Wang et al., 2007). Additionally, the zebrafish homologue of this miRNA family, mir-430, contributes to the rapid degradation of maternal transcripts in early zygotic development (Giraldez et al., 2006), and mRNA expression data suggests that this miRNA family also promotes the clearance of transcripts in early mammalian development (Farh et al., 2005).
In addition to promoting the rapid clearance of transcripts as cells transition from one state to another during development, miRNAs also likely contribute to the control of cell identity by fine-tuning the expression of genes. mir-430, the zebrafish homologue of the mammalian mir-290-295 family, serves to precisely tune the levels of Nodal antagonists Lefty1 and Lefty 2 relative to Nodal, a subtle modulation of protein levels that has pronounced effects on embryonic development (Choi et al., 2007). Recently, a list of ~250 murine ES cell mRNAs that appear to be under the control of miRNAs in the mir-290-295 cluster was reported (Sinkkonen et al., 2008). This study reports that Lefty1 and Lefty2 are evolutionarily conserved targets of the mir-290-295 miRNA family. These miRNAs also maintain the expression of de novo DNA methyltransferases 3a and 3b (Dnmt3a and Dnmt3b), perhaps by dampening the expression of the transcriptional repressor Rbl2, helping to poise ES cells for efficient methylation of Oct4 and other pluripotency genes during differentiation.
Knowledge of how the core transcriptional circuitry of ES cells connects to both miRNAs and protein-coding genes reveals recognizable network motifs downstream of Oct4/Sox2/Nanog/Tcf3, involving both transcriptional and post-transcriptional regulation, that provide new insights into how this circuitry controls ES cell identity (Figure 6). Lefty1 and Lefty2, both actively expressed in ES cells, are directly occupied at their promoters by Oct4/Sox2/Nanog/Tcf3. mir-290-295, which is also directly occupied by Oct4/Sox2/Nanog/Tcf3, depends on Oct4 for proper expression (Figure 3D). Therefore, core ES cell transcription factors appear to promote the active expression of Lefty1 and Lefty2, but also fine-tune the expression of these important signaling proteins by activating a family of miRNAs that target the Lefty1 and Lefty2 3’UTRs. This network motif whereby a regulator exerts both positive and negative effects on its target, termed “incoherent feed-forward” regulation (Alon, 2007), provides a mechanism to fine-tune the steady-state level or kinetics of a target’s activation (Figure 6A). Over a quarter of the proposed targets of the mir-290-295 miRNAs (Sinkkonen et al., 2008) are likely under the direct transcriptional control of Oct4/Sox2/Nanog/Tcf3 based on our binding maps, suggesting that these miRNAs could participate broadly in tuning the effects of ES cell transcription factors (Figure 6A and Supplementary Text).
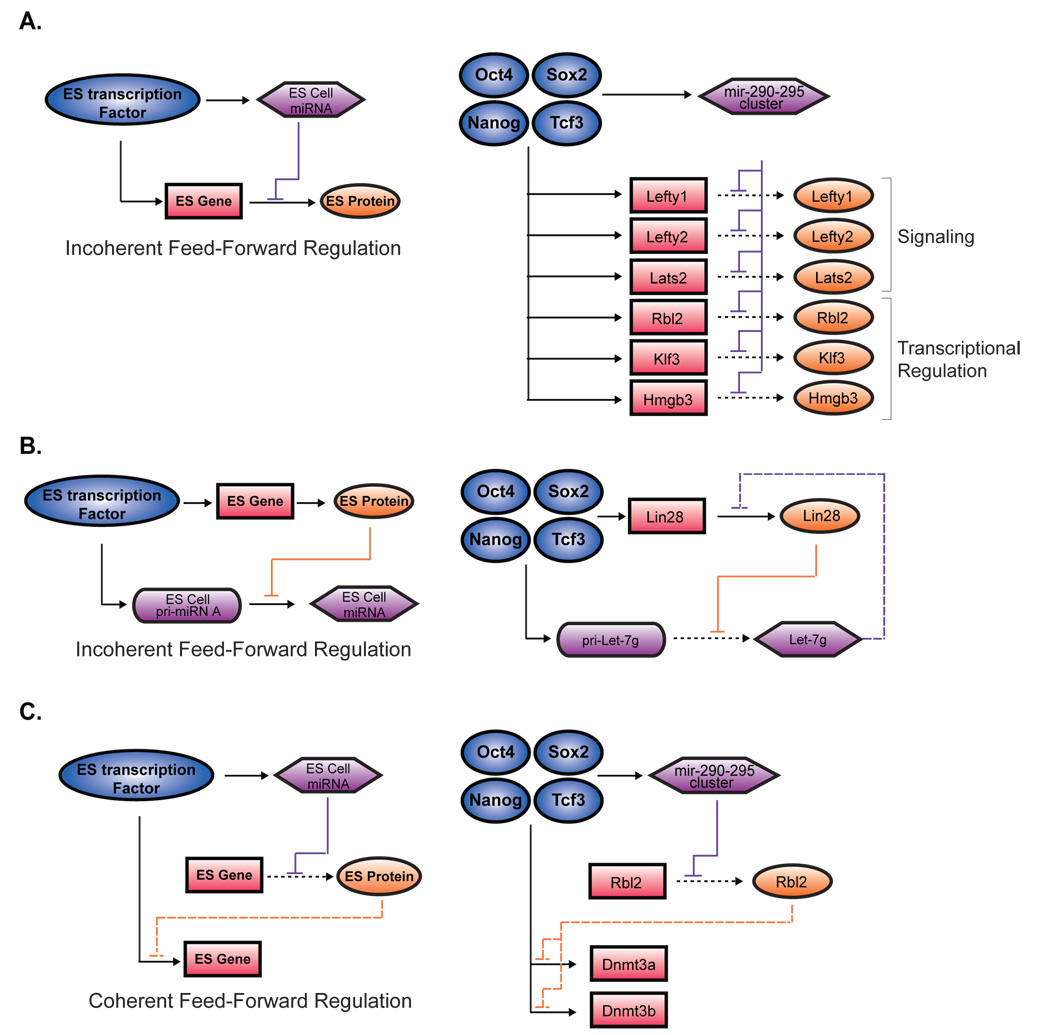
Figure 6. miRNA modulation of the gene regulatory network in ES cells.
A. An incoherent feed-forward motif (Alon, 2007) involving a miRNA repression of a transcription factor target gene is illustrated (left). Transcription factors are represented by dark blue circles, miRNAs in purple hexagons, protein-coding gene in pink rectangles and proteins in orange ovals. Selected instances of this network motif identified in ES cells based on data from Sinkkonen et al., 2008 or data in Figure S11 are shown (right). B. Second model of incoherent feed-forward motif (Alon 2007) involving protein repression of a miRNA is illustrated (left). In ES cells, Lin28 blocks the maturation of primary Let-7g (Visiwanthan et al., 2008). Lin28 and the Let-7g gene are occupied by Oct4/Sox2/Nanog/Tcf3. Targetscan prediction (Grimson et al., 2007), of Lin28 by mature Let-7g is noted (purple dashed line, right). C. A coherent feed-forward motif (Alon 2007) involving miRNA repression of a transcriptional repressor that regulates a transcription factor target gene is illustrated (left). This motif is found in ES cells, where mir-290-295 miRNAs repress Rbl2 indirectly maintaining the expression of Dnmt3a and Dnmt3a, which are also occupied at their promoters by Oct4/Sox2/Nanog/Tcf3 (right).
The miRNA expression program directly downstream of Oct4/Sox2/Nanog/Tcf3 could help poise ES cells for rapid and efficient differentiation, consistent with the phenotype of miRNA-deficient cells (Kanellopoulou et al., 2005; Murchison et al., 2005; Wang et al., 2007). Oct4/Sox2/Nanog/Tcf3 likely contributes to this poising by their occupancy of the Let-7g promoter. Mature Let-7 transcripts are scarce in ES cells, but were among the most abundant miRNAs in both MEFs and NPCs (Figure 3). Primary Let-7g transcript is abundant in ES cells, but its maturation is blocked by Lin28 (Viswanathan et al., 2008). We now report that the promoters of both Let-7g and Lin28 are occupied by Oct4/Sox2/Nanog/Tcf3, suggesting that the core ES cell transcription factors promote the transcription of both primary Let-7g and Lin28, which blocks the maturation of Let-7g. Indeed, proper expression of pri-Let-7g is dependent on Oct4 (Figure 3D). In this way Let-7 and Lin-28 participate in an incoherent feed-forward circuit downstream of Oct4/Sox2/Nanog/Tcf3 to contribute to rapid cellular differentiation (Figure 6B). Notably, ectopic expression of Lin28 in human fibroblasts promotes the induction of pluripotency (Yu et al., 2007), suggesting blocked maturation of pri-Let-7 transcripts plays an important role in the pluripotent state. Additionally, Dnmt3a and Dnmt3b, which are indirectly up-regulated by the mir-290-295 miRNAs (Sinkkonen et al., 2008), are also occupied at their promoters by Oct4/Sox2/Nanog/Tcf3, providing examples of “coherent” regulation of important target genes by ES cell transcription factors and the ES cell miRNAs maintained by those transcription factors (Figure 6C).
Multi-layer Regulatory Circuitry of ES cell identity
The regulatory circuitry we present for miRNAs in ES cells can now be integrated into the model of core regulatory circuitry of pluripotency we have proposed previously (Boyer et al., 2005; Lee et al., 2006; Cole et al., 2008), as illustrated in Figure 7. Our data reveal that Oct4, Sox2, Nanog and Tcf3 occupy the promoters of two key sets of miRNAs, similar to the two sets of protein-coding genes regulated by these factors: one set that is actively expressed in pluripotent ES cells and another that is silenced in these cells by Polycomb Group proteins and whose later expression might serve to facilitate establishment or maintenance of differentiated cell states.
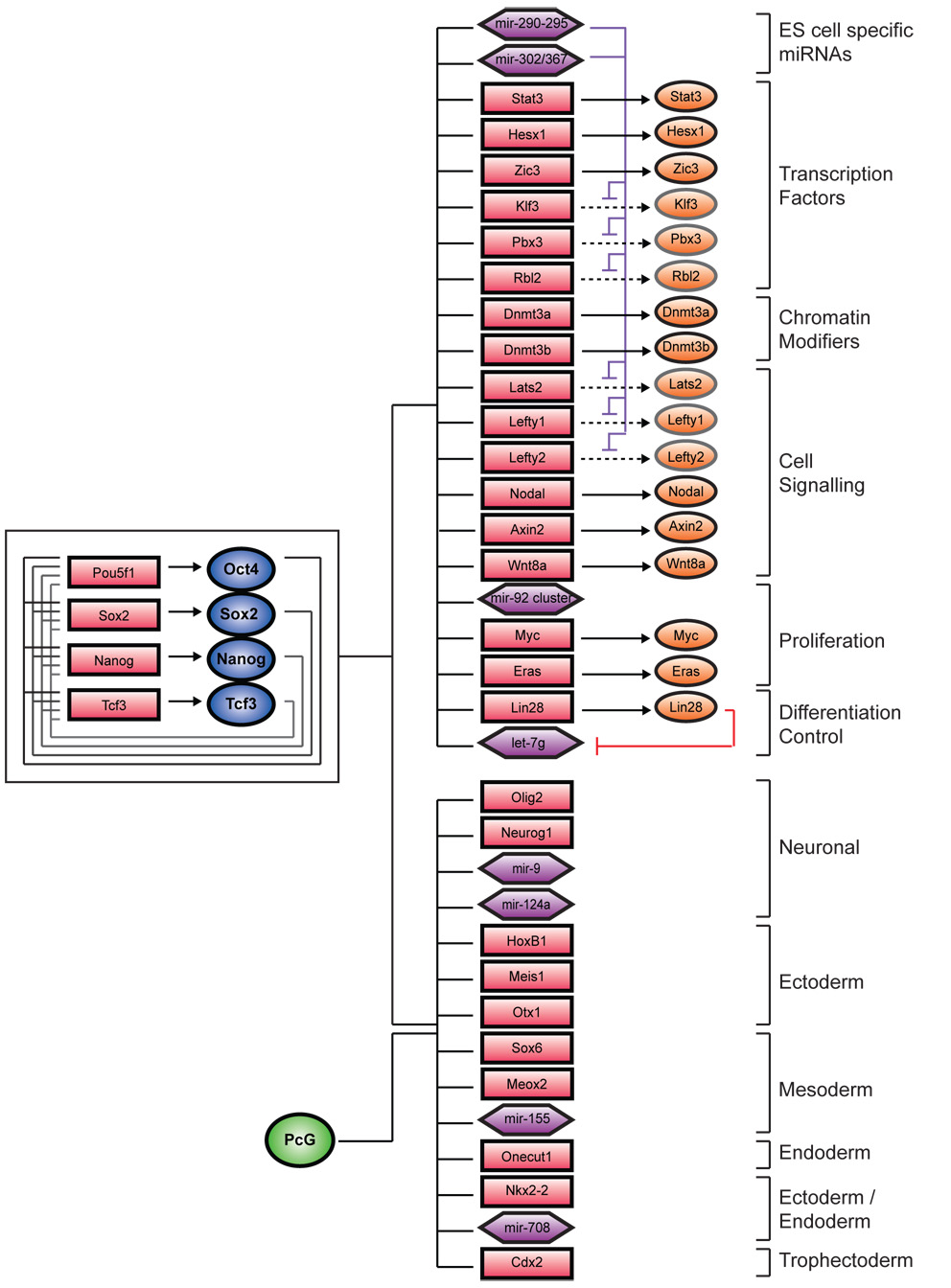
Figure 7. Multi-level regulatory network controlling ES cell identity.
Updated map of ES cell regulatory circuitry is shown. Interconnected auto-regulatory loop is shown to the left. Active genes are shown at the top right, and inactive genes are shown at the bottom right. Transcription factors are represented by dark blue circles, and Suz12 by a green circle. Gene promoters are represented by red rectangles, gene products by orange circles, and miRNA promoters are represented by purple hexagons.
The expanded circuit diagram presented here integrates transcription factor occupancy of miRNA genes and existing data on miRNA targets into our model of the molecular control of the pluripotent state. These data suggest that miRNAs that are activated in ES cells by Oct4/Sox2/Nanog/Tcf3, serve to modulate the direct effects of these transcription factors, participating in incoherent feed-forward regulation to tune levels of key genes, and modifying the gene expression program to help poise ES cells for efficient differentiation. Thus, the core ES cell transcription factors and the miRNAs under their control coordinately contribute transcriptional and post-transcriptional gene regulation to the network that maintains ES cell identity.
Concluding Remark
Knowledge of how protein-coding genes are controlled by key ES cell transcription factors and chromatin regulators has provided important insights into the molecular control of ES cell identity and cellular reprogramming (Jaenisch and Young, 2008). This knowledge also has begun to shed light on human disease, as elements of the ES cell gene expression program are recapitulated in cancer cells (Wong et al., 2008; Ben-Porath et al., 2008). We now connect miRNA genes to the core circuitry of ES cells with high-resolution genome-wide ChIP-seq data, and quantitative sequencing of short transcripts in multiple cell types. This information should prove useful as investigators continue to probe the role of miRNAs in pluripotency, cell state, disease and regenerative medicine.
EXPERIMENTAL PROCEDURES
A detailed description of all materials and methods used can be found in Supplementary Text.
Cell Culture
V6.5 (C57BL/6-129) murine ES cells were grown under typical ES conditions (see Supplementary Text) on irradiated mouse embryonic fibroblasts (MEFs). For location analysis, cells were grown for one passage off of MEFs, on gelatinized tissue-culture plates. Neural precursor cells (NPCs) derived from V6.5 ES cells and mouse embryonic fibroblasts prepared and cultured from DR-4 strain mice were grown using standard protocols as previously described (See Supplementary Text). ZHBTc4 cells harboring a doxycycline-repressible Oct4 allele (Niwa et al., 2000), a gift from A. Smith, were cultured under standard ES cell conditions on gelatin. Cultures were treated with 2µg/ml doxycycline (SIGMA, D-9891) for 12hrs or 24hrs.
ChIP-seq
Detailed descriptions of antibodies, antibody specificity and ChIP methods used in this study have been published previously and are provided in Supplementary Text. Purified immunoprecipitated DNA was prepared for sequencing according to a modified version of the Solexa Genomic DNA protocol. Fragmented DNA was end repaired and subjected to 18 cycles of linker-mediated (LM)-PCR using oligos purchased from Illumina. Amplified fragments between 150 and 300bp were isolated by agarose gel electrophoresis and purified. High quality samples were confirmed by the appearance of a smooth smear of fragments from 100–1000bp with a peak distribution between 150 and 300bp. 3ng of linker-ligated DNA was applied to the flow-cell using the Solexa Cluster Station fluidics device. Samples were then subjected to 26 bases of sequencing according to Illumina’s standard protocols.
Images acquired from the Solexa sequencer were processed through the bundled Solexa image extraction pipeline and aligned to both mouse NCBI build 36 and 37 using ELAND. Analysis is described in Supplementary Text. All sequence data is available at the NCBI GEO database under the accession designation GSE11724.
Whole genome array design and data extraction
The design of the oligonucleotide-based whole genome array set and data extraction methods are described in Lee et al., 2006. The microarrays used for location analysis in this study were manufactured by Agilent Technologies (http://www.agilent.com). All array data is available at Array Express under the accession designation E-MEXP-1687.
Quanititative short RNA sequencing
A method of cloning the 18–30nt transcripts previously described (Lau et al., 2001) was modified to allow for Solexa (Illumina) sequencing (manuscript submitted). RNA extraction was performed using Trizol, followed by RNeasy purification (Qiagen) for mES cells, MEFs and NPCs. Single-stranded cDNA libraries of short transcripts were generated using size selected RNA from mES cells NPCs, MEFs and ZHBTc4 cells (See Supplementary Text). Single-stranded DNA samples were resuspended in 10mM Tris (EB buffer)/0.1% Tween and used as indicated in the standard Solexa sequencing protocol (Illumina). All sequence data is available at to the NCBI GEO database under the accession designation GSE11724.
Quantitative PCR of primary miRNAs
Real time PCR primers were designed using the standard specifications of PrimerExpress (Applied Biosystems) to amplify regions within the ~200nt immediately upstream of the tested miRNA hairpins or in the middle of mir-290-295 polycistron, but outside of any miRNA hairpin regions (Supplementary Text and Figure S8B). Primers were used in SYBR Green quantitative PCR assays on the Appied Biosystems 7500 Real Time PCR system. Expression levels were calculated relative to Gapdh mRNA levels, which were quantified in parallel by Taqman analysis. Detailed methods and primer sequences can be found in Supplementary Information.
Supplementary Material
Acknowledgements
We thank members of the Young, Jaenisch and Bartel laboratories, especially T. Lee, for discussions and critical review of the manuscript. We also thank A. Ravi and A. Seila for their help and A. Smith for providing the ZHBT-c4 cells. We are grateful to L.A. Boyer, B. Chevalier, R. Kumar, and T. Lee who were instrumental in performing location analysis in hES cells. We also thank Biology and Research Computing (BaRC), especially Tom DiCesare, for computational and technical support and graphic assistance. This work was supported in part by NIH grants 5-RO1-HDO45022, 5-R37-CA084198, and 5-RO1- CA087869 to R.J. and by NIH grant HG002668 and a grant from the Whitehead Institute to R.A.Y, and partially by a Cancer Center Support (Core) grant P30-CA14051 from the NCI.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alon U. Network motifs: theory and experimental approaches. Nat Rev Genet. 2007;8:450–461. doi: 10.1038/nrg2102. [DOI] [PubMed] [Google Scholar]
- Barrera LO, Li Z, Smith AD, Arden KC, Cavenee WK, Zhang MQ, Green RD, Ren B. Genome-wide mapping and analysis of active promoters in mouse embryonic stem cells and adult organs. Genome Res. 2008;18:46–59. doi: 10.1101/gr.6654808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, Weinberg RA. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV, Hannon GJ. Dicer is essential for mouse development. Nat Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Chambers I, Smith A. Self-renewal of teratocarcinoma and embryonic stem cells. Oncogene. 2004;23:7150–7160. doi: 10.1038/sj.onc.1207930. [DOI] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;113:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- Choi WY, Giraldez AJ, Schier AF. Target protectors reveal dampening and balancing of Nodal agonist and antagonist by miR-430. Science. 2007;318:271–274. doi: 10.1126/science.1147535. [DOI] [PubMed] [Google Scholar]
- Farh KK, Grimson A, Jan C, Lewis BP, Johnston WK, Lim LP, Burge CB, Bartel DP. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- Fukao T, Fukuda Y, Kiga K, Sharif J, Hino K, Enomoto Y, Kawamura A, Nakamura K, Takeuchi T, Tanabe M. An evolutionarily conserved mechanism for microRNA-223 expression revealed by microRNA gene profiling. Cell. 2007;129:617–631. doi: 10.1016/j.cell.2007.02.048. [DOI] [PubMed] [Google Scholar]
- Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, Enright AJ, Schier AF. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, Hammond SM. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- Houbaviy HB, Dennis L, Jaenisch R, Sharp PA. Characterization of a highly variable eutherian microRNA gene. Rna. 2005;11:1245–1257. doi: 10.1261/rna.2890305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbaviy HB, Murray MF, Sharp PA. Embryonic stem cell-specific MicroRNAs. Dev Cell. 2003;5:351–358. doi: 10.1016/s1534-5807(03)00227-2. [DOI] [PubMed] [Google Scholar]
- Jaenisch R, Young R. Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell. 2008;132:567–582. doi: 10.1016/j.cell.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Chan YS, Loh YH, Cai J, Tong GQ, Lim CA, Robson P, Zhong S, Ng HH. A core Klf circuitry regulates self-renewal of embryonic stem cells. Nat Cell Biol. 2008;10:353–360. doi: 10.1038/ncb1698. [DOI] [PubMed] [Google Scholar]
- Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, Livingston DM, Rajewsky K. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, Rajewsky N. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- Krichevsky AM, Sonntag KC, Isacson O, Kosik KS. Specific microRNAs modulate embryonic stem cell-derived neurogenesis. Stem Cells. 2006;24:857–864. doi: 10.1634/stemcells.2005-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent LC, Chen J, Ulitsky I, Mueller FJ, Lu C, Shamir R, Fan JB, Loring JF. Comprehensive MicroRNA Profiling Reveals a Unique Human Embryonic Stem Cell Signature Dominated by a Single Seed Sequence. Stem Cells. 2008 doi: 10.1634/stemcells.2007-1081. [DOI] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mineno J, Okamoto S, Ando T, Sato M, Chono H, Izu H, Takayama M, Asada K, Mirochnitchenko O, Inouye M, Kato I. The expression profile of microRNAs in mouse embryos. Nucleic Acids Res. 2006;34:1765–1771. doi: 10.1093/nar/gkl096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin RD, O'Connor MD, Griffith M, Kuchenbauer F, Delaney A, Prabhu AL, Zhao Y, McDonald H, Zeng T, Hirst M, et al. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 2008;18:610–621. doi: 10.1101/gr.7179508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchison EP, Partridge JF, Tam OH, Cheloufi S, Hannon GJ. Characterization of Dicer-deficient murine embryonic stem cells. Proc Natl Acad Sci U S A. 2005;102:12135–12140. doi: 10.1073/pnas.0505479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H. How is pluripotency determined and maintained? Development. 2007;134:635–646. doi: 10.1242/dev.02787. [DOI] [PubMed] [Google Scholar]
- Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- Robertson G, Hirst M, Bainbridge M, Bilenky M, Zhao Y, Zeng T, Euskirchen G, Bernier B, Varhol R, Delaney A, et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat Methods. 2007;4:651–657. doi: 10.1038/nmeth1068. [DOI] [PubMed] [Google Scholar]
- Silva J, Smith A. Capturing pluripotency. Cell. 2008;132:532–536. doi: 10.1016/j.cell.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinkkonen L, Hugenschmidt T, Berninger P, Gaidatzis D, Mohn F, Artus-Revel CG, Zavolan M, Svoboda P, Filipowicz W. MicroRNAs control de novo DNA methylation through regulation of transcriptional repressors in mouse embryonic stem cells. Nat Struct Mol Biol. 2008;15:259–267. doi: 10.1038/nsmb.1391. [DOI] [PubMed] [Google Scholar]
- Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol. 2008;9:219–230. doi: 10.1038/nrm2347. [DOI] [PubMed] [Google Scholar]
- Suh MR, Lee Y, Kim JY, Kim SK, Moon SH, Lee JY, Cha KY, Chung HM, Yoon HS, Moon SY, et al. Human embryonic stem cells express a unique set of microRNAs. Dev Biol. 2004;270:488–498. doi: 10.1016/j.ydbio.2004.02.019. [DOI] [PubMed] [Google Scholar]
- Tam WL, Lim CY, Han J, Zhang J, Ang YS, Ng HH, Yang H, Lim B. Tcf3 Regulates Embryonic Stem Cell Pluripotency and Self-Renewal by the Transcriptional Control of Multiple Lineage Pathways. Stem Cells. 2008 doi: 10.1634/stemcells.2007-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan SR, Daley GQ, Gregory RI. Selective Blockade of MicroRNA Processing by Lin-28. Science. 2008 doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorhoeve PM, le Sage C, Schrier M, Gillis AJ, Stoop H, Nagel R, Liu YP, van Duijse J, Drost J, Griekspoor A, et al. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell. 2006;124:1169–1181. doi: 10.1016/j.cell.2006.02.037. [DOI] [PubMed] [Google Scholar]
- Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet. 2007;39:380–385. doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K, Bernstein BE, Jaenisch R. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- Wong DJ, Liu H, Ridky TW, Cassarino D, Segal E, Chang HY. Module map of stem cell genes guides creation of epithelial cancer stem cells. Cell Stem Cell. 2008;2:333–344. doi: 10.1016/j.stem.2008.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi F, Pereira L, Merrill BJ. Tcf3 Functions as a Steady State Limiter of Transcriptional Programs of Mouse Embryonic Stem Cell Self Renewal. Stem Cells. 2008 doi: 10.1634/stemcells.2008-0229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Zhou X, Ruan J, Wang G, Zhang W. Characterization and identification of microRNA core promoters in four model species. PLoS Comput Biol. 2007;3:e37. doi: 10.1371/journal.pcbi.0030037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.