Abstract
In eukaryotes, proteins enter the secretory pathway through the translocon pore of the endoplasmic reticulum. This protein translocation channel is composed of three major subunits, called Sec61α, β and γ in mammals. Unlike the other subunits, the β subunit is dispensable for translocation and cell viability in all organisms studied. Intriguingly, the knockout of the Sec61β encoding genes results in different phenotypes in different species. Nevertheless, the β subunit shows a high level of sequence homology across species, suggesting the conservation of a biological function that remains ill-defined. To address its cellular roles, we characterized the homolog of Sec61β in the fission yeast Schizosaccharomyces pombe (Sbh1p). Here, we show that the knockout of sbh1 + results in severe cold sensitivity, increased sensitivity to cell-wall stress, and reduced protein secretion at 23°C. Sec61β homologs from Saccharomyces cerevisiae and human complement the knockout of sbh1 + in S. pombe. As in S. cerevisiae, the transmembrane domain (TMD) of S. pombe Sec61β is sufficient to complement the phenotypes resulting from the knockout of the entire encoding gene. Remarkably, the TMD of Sec61β from S. cerevisiae and human also complement the gene knockouts in both yeasts. Together, these observations indicate that the TMD of Sec61β exerts a cellular function that is conserved across species.
Introduction
Protein secretion is an essential process in all organisms. In eukaryotes, entry of proteins into the secretory pathway requires passage across the endoplasmic reticulum (ER) lipid bilayer. Translocation of proteins into the ER is accomplished through a protein conduction channel (PCC) [1], [2]. The PCC is composed of a highly conserved heterotrimeric core called the Sec61αβγ complex in eukaryotes, and the SecYEG complex in bacteria [3], [4], [5]. The PCC associates with several soluble proteins to form the translocon, the ER translocation machinery [6], [7]. The translocon functions in co-translational translocation of nascent proteins, as well as in the retro-translocation of misfolded polypeptides for degradation in the cytosol by the ER-associated degradation (ERAD) mechanism [3], [8], [9], [10], [11]. In addition, the association of the translocon with the tetrameric Sec62/63 complex and the BiP molecular chaperone forms the Sec complex, which is involved in post-translational translocation [12], [13], [14]. The genomes of all organisms sequenced to date encode at least one homolog of each Sec61αβγ/SecYEG subunits, suggesting that these proteins have a universal cellular role [15].
So far, the functions of the α and γ subunits of the PCC are the best understood. The α subunit consists of ten transmembrane segments assembled in an hourglass shape, forming the pore of the channel [16], [17] and acting as the main ribosome receptor [18], [19]. The γ subunit is a single-span membrane protein that binds and stabilizes the translocon pore by linking the two halves of the α subunit [16], [20]. By contrast, the precise function of the β subunit, a 10 kD C-tail-anchored transmembrane protein, remains elusive. Interestingly, Sec61β and its prokaryotic counterpart SecG are the only translocon subunits that are not essential for cell viability in yeast and bacteria [21], [22], [23], [24]. Moreover, mammalian Sec61β is not required for protein translocation in vitro, but has a kinetic effect on co-translational translocation [25]. Likewise, bacterial SecG is dispensable for translocation in reconstituted membrane vesicles, but has a stimulatory effect in this process [26]. In S. cerevisiae, the double knockout of the genes encoding the two paralogs of the translocon β subunits (SBH1 and SBH2) has only a moderate effect on translocation of reporter proteins [22], [24], [27], [28]. Regarding protein secretion, the relevance of the function of Sec61β in co- and post-translational translocation remains unclear as the knockout of the translocon β subunits reduces α-amylase secretion in S. cerevisiae, but has no effect in Yarrowia lipolytica [23], [27].
Based on the phenotypes resulting from its knockout, several cellular roles for the translocon β subunit have been proposed. In prokaryotes, SecG facilitates the membrane insertion/de-insertion cycle of the SecA ATPase, the bacterial translocation motor [29], [30]. In S. cerevisiae, Sbh1p was proposed to act as the guanine nucleotide exchange factor for the signal recognition particle receptor (SR) [31], whereas Sbh2p was suggested to mediate recognition of vacant translocons by functional interactions with the SR [28]. Sbh1p also rescues several exocyst mutants in yeasts and coimmunoprecipitates with Sec10p, Sec6p and Sec8p in mammalian cells, three components of the exocyst complex, suggesting a role in vesicular transport [24], [32], [33]. The translocon β subunit was also shown to be recruited to the ER-derived quality control compartment in ERAD [34], and to have an accessory function in the unfolded protein response in different organisms [23], [35], [36], [37]. The diversity of functions proposed for the translocon β subunit suggests that different homologs of this protein have species-specific roles.
Recently, Feng et al. [27] demonstrated that the transmembrane domain (TMD) of S. cerevisiae Sbh1p or Sbh2p are necessary and sufficient to suppress the phenotypes of sbh1Δsbh2Δ cells, including heat sensitivity at 38°C, and co-translational translocation defects. This was further confirmed in another study, where N-terminal truncation mutants of Sbh2p restored normal integration of DPAPB and translocation of CPY in the sbh1Δsbh2Δ strain [28]. Furthermore, the TMD of Sbh1p is sufficient for co-immunoprecipitation with the translocon α and γ subunits, indicating correct integration and interaction at the Sec61 translocon site [27]. The TMD of Sbh1p also interacts with Rtn1p, a protein involved in structuring the cortical ER, in absence of other translocon subunits, suggesting a role for the β subunit outside the translocation apparatus [27]. Taken together, these results suggest that the TMD is the critical region responsible for the undefined function of Sec61β.
Here, we analyzed the functional conservation of the translocon β subunit and its TMD. We show that the translocon β subunit is not essential in fission yeast at 30°C. However, it is required for growth at low temperature, and its deletion causes SDS sensitivity and reduced protein secretion at 23°C. Overexpression of sbh1 + also diminishes protein secretion of a model protein, increases sensitivity to cell-wall stress, and causes morphological abnormalities. The phenotypes associated with sbh1 + deletion are suppressed by the 26 amino acids of the TMD. Remarkably, the TMDs from both S. cerevisiae homologs of the translocon β subunit and that from human also complement the deletion phenotypes in fission and budding yeast. These results demonstrate that the TMD function of Sec61β is conserved and sufficient to exert most biological roles of the full-length protein. These observations pose Sec61β as a model to study alternative functions of TMDs apart from the anchoring to the ER membrane.
Results
S. pombe encodes a single Sec61β homolog
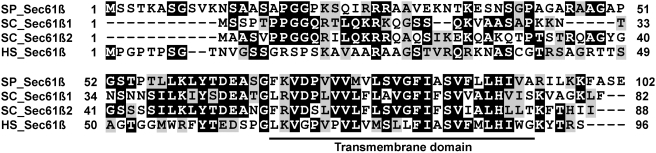
Based on Wu-Blast2 analyses of the fission yeast proteome [38], S. pombe encodes a single Sec61β homolog at ORF SPBC2G2.03c (sbh1 +), hereafter designed as SP_Sec61β for the sake of clarity. The deduced primary sequence of the protein is presented in Figure 1. As expected for a translocon subunit, the predicted protein displays a high level of sequence conservation with both paralogs of S. cerevisiae (SC) and that of human (HS). SP_Sec61β is 35% identical (66% similar) to SC_Sec61β1 (Sbh1p), 43% identical (65% similar) to SC_Sec61β2 (Sbh2p), and 29% identical (58% similar) to HS_Sec61β (human Sec61β). Multiple sequence alignment using Clustal W2 [39] revealed that SP_Sec61β is evolutionary closer to HS_Sec61β than its S. cerevisiae counterparts (not shown), thus validating our choice of S. pombe as a model organism to study the translocon β subunit of higher eukaryotes. Analyses of SP_Sec61β using various ExPASy proteomic tools [40] allowed the identification of a single domain, constituted of a continuous stretch of 26 amino acids near the C-terminus. This hydrophobic tail is predicted to form an alpha helix crossing the lipid bilayer once, with the N-terminus of the protein exposed to the cytoplasm. Thus, as in S. cerevisiae and human, SP_Sec61β is predicted to be a tail-anchored protein. Interestingly, the TMDs of Sec61β homologs are much more conserved than their N-terminal domains, 53–61% identical from yeast to human as compared to only 18–41% for the cytosolic part. In comparison, the tail anchor of the translocon γ subunit is only 40–50% identical from yeast to human, a level of sequence homology comparable to the entire protein, which is 44–48% identical. This conservation suggests a critical function for the TMD region of Sec61β homologs, not solely explained by its role as a membrane anchor, especially as almost any stretch of hydrophobic amino acids can perform this function [41].
Figure 1. The translocon β subunit is conserved from yeast to human.
Amino-acid sequence comparison between translocon beta subunits of S. pombe (SP_Sec61β), S. cerevisiae (SC_Sec61β1 and SC_Sec61β2) and human (HS_Sec61β). Identical amino acids are shaded in black, similar amino acids are shaded in grey. The predicted conserved transmembrane domain is underlined.
Overexpression of SP_Sec61β induces morphological changes and cell-wall sensitivity
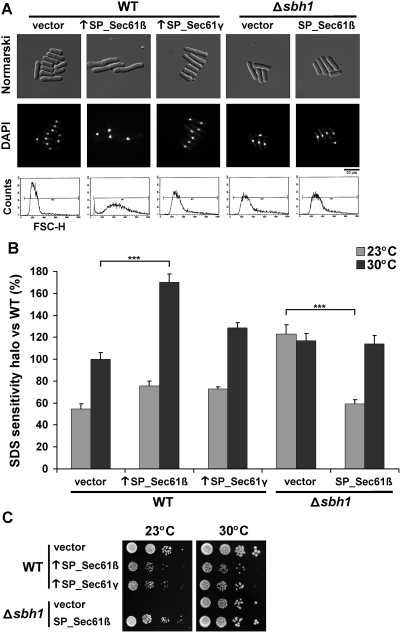
To explore its cellular roles in S. pombe, we overexpressed the translocon β subunit expecting to exacerbate its function and induce a revealing phenotype. The ORF of SP_Sec61β was cloned under the control of the strong thiamine-repressible nmt promoter in a pREP2 vector [42]. As shown in Figure 2A (upper panel), overexpression of SP_Sec61β resulted in a elongated phenotype with a single nucleus per cell (see DAPI staining, middle panel). This phenotype was specific to the translocon β subunit because the overexpression of S. pombe γ subunit (SP_Sec61γ) had no effect on cell morphology. Moreover, the elongated phenotype was reverted by addition of thiamine, which represses the promoter; and the phenotype was not observed when expression was driven by a weaker nmt* promoter (not shown). The elongated phenotype caused by overproduction of SP_Sec61β was further confirmed by global population analysis using flow cytometry (FACS). As shown in Figure 2A (lower panel), cells overexpressing SP_Sec61β had an average size (FSC-H) value 1.9 higher than WT. The wider size distribution can be explained by the presence of cells with various altered morphologies, which were either more granulated, had bigger vesicles or displayed an aberrant number of septa in ∼10% of cases (not shown).
Figure 2. Phenotypes associated with the knockout and overexpression of SP_Sec61β.
(A) Morphologic comparison between strains. WT cells (SP556), cells overexpressing S. pombe translocon β subunit (↑SP_Sec61β) or γ subunit (↑SP_Sec61γ) and cells deleted for the SP_Sec61β encoding gene (SP15039, Δsbh1) with or without a plasmid encoding SP_Sec61β were cultured to exponential phase, stained with DAPI (as nuclear marker) and analyzed by Normarski (upper panel) or fluorescent microscopy (middle panel). Cell size distribution was assessed by cell sorting of 10,000 cells (lower panel). (B) SDS sensitivity comparison between strains. Three exponential cultures of WT and Δsbh1 cells bearing the indicated plasmids were spread on top-agar MM+AL, following which a drop of 10 µL of SDS 10% was added on the center of each plate. After 3–5 days of incubation at the indicated temperatures, the area of the dead-cell halo was measured and reported to the WT strain at 30°C (100%). Data shown are mean±standard deviation of three or more independent experiments. *** indicates p<0.001 for Student's t-test. (C) Growth of WT and Δsbh1 strains devoid or overexpressing SP_Sec61β or SP_Sec61γ at different temperatures. Exponential cultures were serially-diluted (10−1–10−4) and spotted on EMM+AL plates. Growth was monitored for 7 days at the indicated temperatures. Results are representative of three or more independent experiments.
To assess if the elongated phenotype is associated with membrane and/or cell-wall defects, we performed an SDS resistance assay at various temperatures (Figure 2B). SDS is a strong detergent that disturbs the cytoplasmic membrane when the cell-wall is compromised, thus resulting in a dead-cell halo that can be measured [43]. In these conditions, cells overexpressing SP_Sec61β were up to 1.8 more sensitive to SDS as compared to the vector control at 30°C. In comparison, cells overexpressing SP_Sec61γ were only 1.3 times more sensitive at this temperature. At 23°C, the strains overproducing the β or γ subunits were both 1.4 times more sensitive than the vector control. As addition of 1.2 M sorbitol, an osmotic stabilizer [44], did not rescue the SDS sensitivity nor the elongated cell size (not shown), it is unlikely that these phenotypes are only due to an incorrect cell-wall formation. These results demonstrate that the overexpression of SP_Sec61β alters cellular morphology, accompanied by a higher sensitivity to membrane stress, suggesting a potential role of this protein in the coordination between cell-membrane growth and the cell-cycle in fission yeast.
Cells depleted of SP_Sec61β are cold-sensitive
To investigate whether SP_Sec61β is essential for viability, a sbh1 +/Δsbh1::kan R diploid strain was sporulated to analyze its meiotic progeny. All spores were viable on YE medium, with a 2:2 segregation of the kan R marker. Deletion of sbh1 + was confirmed by Northern blotting, Southern blotting and PCR, demonstrating that SP_Sec61β is not essential for viability (not shown). In contrast to the overexpression of SP_Sec61β, the deletion of sbh1 + did not induce morphological changes (Figure 2A). As it was shown in S. cerevisiae that deletion of the genes encoding the two translocon β subunits (SBH1 and SBH2) results in heat sensitivity at 38°C [22], [24], we measured the growth of S. pombe Δsbh1 cells at different temperatures. In contrast to S. cerevisiae, Δsbh1 cells from S. pombe were not heat-sensitive (not shown), but the deletion resulted in severe cold sensitivity at 23°C (Figure 2C). Normal growth was not restored by replacing the cells at 30°C after incubation at 23°C for 7 days, indicating that growth is permanently arrested or that most Δsbh1 cells are dead (not shown). In addition, Δsbh1 cells were 2.2 times more sensitive to SDS than WT cells at 23°C, while sensitivity was similar at 30°C (Figure 2B). Transformation of sbh1 + on a plasmid under the medium strength nmt* promoter was sufficient to rescue to WT levels the cold- and SDS-sensitivity (Figure 2B–C). Globally, these results demonstrate that while SP_Sec61β is not essential for cell viability at 30°C, it is critical at low temperature.
The levels of SP_Sec61β modulate secretion
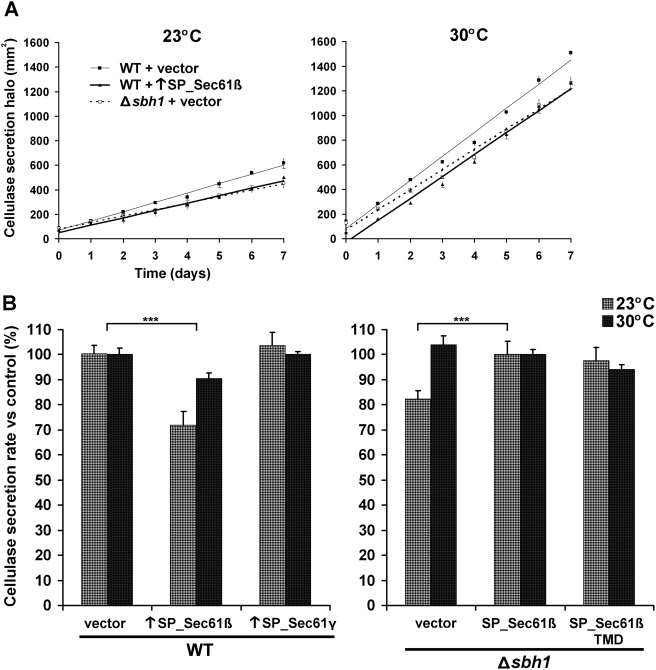
Because the knockout of Sec61β was reported to lower protein secretion in S. cerevisiae, but not in Y. lipolytica [23], [27], we wanted to assess the importance of SP_Sec61β in protein secretion. To this end, we used a previously characterized genomic cassette expressing the reporter protein cellulase from Aspergillus aculeatus [45]. The activity of secreted cellulase was measured during 7 days at different temperatures to evaluate the rate of secretion of each strain. As shown in Figure 3B, the secretion rate was not reduced in Δsbh1 cells at 30°C, but was reduced to 82% of the control strain at 23°C. Thus, the kinetic effect of the translocon β subunit on secretion is only observed in conditions where secretion is already considerably diminished, such as at low temperature (secretion rate 2.6-fold lower than at 30°C, Figure 3A).
Figure 3. Effects of SP_Sec61β levels on protein secretion.
(A) Cellulase secretion efficiency at different temperatures. WT (SP15073) and Δsbh1 (SP15074) cells expressing A. aculeatus cellulase from a genomic cassette under the control of the adh1p promoter, and bearing the empty vector or a plasmid overexpressing (↑) SP_Sec61β were analyzed for secretion. Cells were grown to exponential phase and spotted on EMM+ALH plates supplemented with 0.1% AZCL-HE-cellulose as substrate. The area of the blue halo created after cleavage of the chromogenic substrate by secreted cellulase was monitored for 7 days at 30°C or at 23°C. Time zero represents the moment when area of the halo exceeds the area under the colony. Each point is the mean±standard deviation of three independent cultures. (B) Rate of cellulase secretion at different temperatures. Secretion rates for strains presented in (A) were calculated as the mean slope of three independent cultures, using the WT value (left panel) or the complemented knockout (right panel) at the corresponding temperature as 100% (± standard deviation). WT cells overexpressing SP_Sec61γ and Δsbh1 cells bearing a plasmid encoding SP_Sec61β or its transmembrane domain only (TMD) are shown as controls. *** indicates p<0.001 for Student's t-test.
In S. cerevisiae, overexpression of the endogenous translocon β subunit or the Kluyveromyces lactis homolog increases the secretion efficiency of α-amylase [46]. By contrast, in fission yeast, we show that overproduction of the translocon β subunit reduces the secretion rate of cellulase (Figure 3B). Again, this effect was more pronounced at 23°C (72% of the WT value) than at 30°C (91% of the WT value). This reduction is not an unspecific effect caused by saturation of the ER, as overexpression of the translocon γ subunit did not alter the secretion rate. These results support the idea that even though SP_Sec61β is not essential for growth at normal temperature, a correct SP_Sec61β level is important for efficient protein secretion, especially at low temperature.
Sec61β is functionally conserved between species
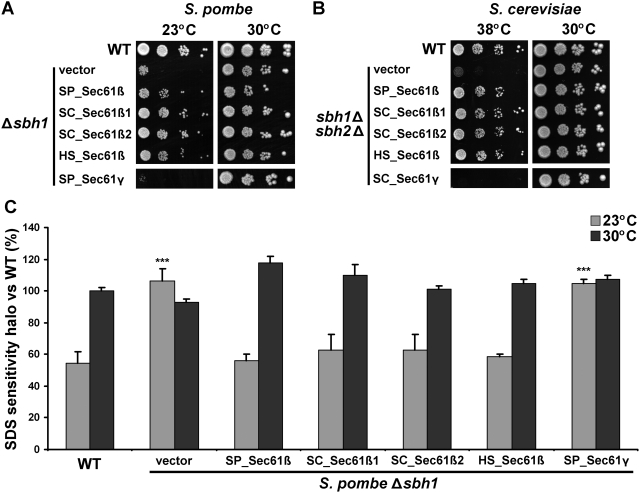
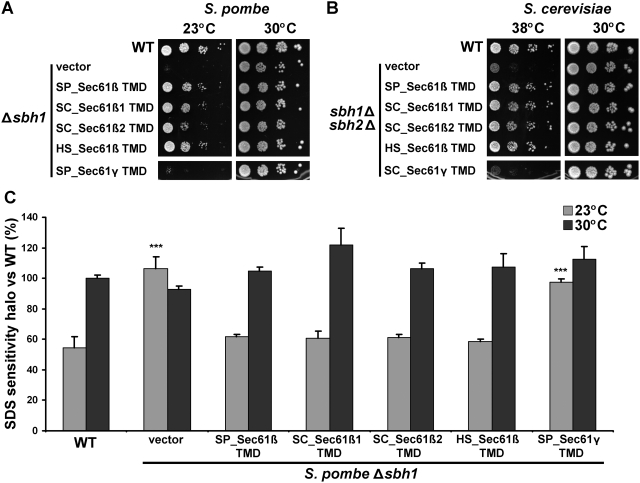
As described above, the knockout of sbh1 + results in cold-sensitive phenotypes in S. pombe. Interestingly, in budding yeast, the double deletion of both paralogs of Sec61β, hereafter designated as SC_Sec61β1 and SC_Sec61β2, results in heat sensitivity instead of cold sensitivity [22], [24]. Nevertheless, we reasoned that if the function of Sec61β is conserved there should be cross-species complementation in spite of the opposite temperature phenotypes found in S. pombe versus S. cerevisiae. As shown in Figure 4A, both Sec61β paralogs of S. cerevisiae complemented the cold sensitivity of the S. pombe Δsbh1 strain as efficiently as the endogenous translocon β subunit. Moreover, SC_Sec61β1 and SC_Sec61β2 also fully reversed the SDS sensitivity of the S. pombe Δsbh1 strain (Figure 4C). Similarly, SP_Sec61β complemented the heat sensitivity of the sbh1Δsbh2Δ S. cerevisiae strain (Figure 4B). This effect was specific to the translocon β subunit, as the γ subunit homologs of S. pombe (SP_Sec61γ) and S. cerevisiae (SC_Sec61γ) were unable to complement the temperature-dependent phenotypes in either yeast.
Figure 4. Different homologs complement the phenotypes of the Sec61β knockout in fission and budding yeast.
(A) Cold sensitivity of S. pombe Δsbh1 cells is rescued by different Sec61β homologs. S. pombe Δsbh1 cells (SP15039) expressing different Sec61β homologs from S. pombe (SP), S. cerevisiae (SC) or human (HS) were serially-diluted (10−1–10−4) and spotted on MM+AL plates. Growth was monitored during 7 days at the indicated temperatures. SP_Sec61γ was used as a negative control. Results are representative of three or more independent experiments. (B) Heat sensitivity of S. cerevisiae sbh1Δsbh2Δ cells is rescued by different Sec61β homologs. S. cerevisiae sbh1Δsbh2Δ cells (SC3232) expressing Sec61β homologs from S. pombe (SP), S. cerevisiae (SC) or human (HS) were serially-diluted (10−1–10−4) and spotted on SD-L plates. Growth was monitored during 5 days at the indicated temperatures. SC_Sec61γ was used as a negative control. Results are representative of three or more independent experiments. (C) Suppression of S. pombe Δsbh1 SDS sensitivity by different Sec61β homologs. SDS-sensitivity halo was measured after 3–5 days of incubation at the indicated temperatures for cultures of the strains presented in (A). Data shown are mean±standard deviation of three or more independent experiments. *** indicates p<0.001 for Student's t-test versus WT.
To investigate the possibility of a functional conservation with higher eukaryotes, we transformed the human homolog of Sec61β (HS_Sec61β) in the Sec61β knockout strain of both S. pombe and S. cerevisiae. Strikingly, HS_Sec61β also complemented the cold sensitivity and SDS sensitivity of the S. pombe Δsbh1 strain, as well as the heat sensitivity of the S. cerevisiae sbh1Δsbh2Δ strain (Figure 4, line 5). Hence, despite distinct phenotypes resulting from the deletion of translocon β subunit in two different yeast species, the function of Sec61β appears to be conserved from yeast to human.
The transmembrane domain of Sec61β is sufficient for heterologous complementation
Next, we wanted to define the domain of Sec61β responsible for the inter-species complementation. Bioinformatic analyses revealed that the most conserved region of Sec61β in all species studied is mapped to its TMD [47]. Moreover, the TMD of SC_Sec61β1 is sufficient to rescue the heat sensitivity of sbh1Δsbh2Δ cells, and to associate with the translocon α and γ subunits [27]. Therefore, we tested if the TMD of different Sec61β homologs could rescue the temperature-sensitive phenotypes in S. pombe and S. cerevisiae knockout strains. We found that the 26 amino acids of the tail anchor of SP_Sec61β are sufficient to complement the cold sensitivity (Figure 5A), SDS sensitivity (Figure 5C) and secretion defects (Figure 3B) of the S. pombe Δsbh1 strain to levels equivalent to the full-length protein. The TMDs of the other Sec61β homologs tested were also able to fully complement the defects of the Δsbh1 strain, indicating that the fundamental function of the TMD is conserved between species (Figure 5A–C). Likewise, all Sec61β TMDs complemented the heat sensitivity of the sbh1Δsbh2Δ S. cerevisiae strain (Figure 5B). Again, complementation was specific to the TMD of Sec61β, as the TMD from Sec61γ (Figure 5, line 5) and calnexin (not shown) failed to suppress the phenotypes of the sbh1Δsbh2Δ S. cerevisiae strain. Collectively, these results indicate that the tail anchor of Sec61β is sufficient to complement the knockout of the entire protein, and that this can be indistinctively accomplished by homologous transmembrane domains ranging from yeast to human. This further confirms that the diverse phenotypes associated with the deletion of Sec61β in distinct species are not due to divergence in the amino acid sequences, but rather they reflect different downstream effects of a conserved Sec61β function supported by the TMD.
Figure 5. The transmembrane domain (TMD) of different Sec61β homologs is sufficient to complement the knockout of the whole gene in fission and budding yeast.
(A) Cold sensitivity of S. pombe Δsbh1 cells (SP15039) and (B) heat sensitivity of S. cerevisiae sbh1Δsbh2Δ cells (SC3232) are rescued by the TMD of different Sec61β homologs. Δsbh1 and sbh1Δsbh2Δ cells expressing the 26 amino acids of the TMD of Sec61β homologs from S. pombe (SP), S. cerevisiae (SC) and human (HS) were serial-diluted (10−1–10−4) and spotted on MM+AL plates or SD-L plates. Growth was monitored during 5 days for S. cerevisiae and 7 days for S. pombe at the indicated temperatures. Sec61γ from S. pombe (SP) or S. cerevisiae (SC) were used as negative controls. Results are representative of three or more independent experiments. (C) Suppression of S. pombe Δsbh1 SDS sensitivity by the TMD of different Sec61β homologs. SDS-sensitivity halo was measured after 3–5 days of incubation at the indicated temperatures for cultures of the strains presented in (A). Data shown are mean±standard deviation of three or more independent experiments. *** indicates p<0.001 for Student's t-test versus WT.
Discussion
We have characterized for the first time the Sec61β homolog from fission yeast. We describe S. pombe-specific phenotypes resulting from the overproduction or the lack of this protein, revealing the involvement of SP_Sec61β in cellular morphology, stress resistance and protein secretion. These S. pombe phenotypes are distinct from those observed in S. cerevisiae [22], [23], [24], [27]. Nevertheless, the Sec61β homologs from S. pombe, S. cerevisiae and human complement the phenotypes of knockout strains of both fission yeast and budding yeast, to a level comparable to that of the endogenous proteins. Remarkably, the transmembrane domains of Sec61β homologs are sufficient for heterologous complementation, demonstrating that the TMD region supports a fundamental biological function that is conserved across species.
Sec61β is not essential for survival in microorganisms, but is required for development in Drosophila [22], [23], [24], [48]. In S. pombe, we show that Sec61β is dispensable for growth at optimal temperature, but becomes critical at low temperature (Figure 2–3). Cold sensitivity was also observed for the Escherichia coli SecG deletion [21], although it was dependent on a second mutation in glpR, involved in glycerol phosphate regulation [49]. In bacteria, the translocation process itself is cold-sensitive, presumably because membrane insertion of the SecA ATPase motor, which is mediated by SecG, is reduced at low temperature [30], [50]. In eukaryotes, a role for Sec61β in cold resistance was proposed in the Gymnadenia conopsea psychrophile plant [51]. Thus, the heat-sensitive phenotype observed in sbh1Δsbh2Δ cells of S. cerevisiae [22], [24] does not reflect a universal phenotype. A possibility is that the differences between the two organisms reflect the prevalence of co- vs post-translational translocation processes in each yeast.
Interestingly, the SDS sensitivity of Δsbh1 cells in S. pombe contrasts with the SDS resistance of Δsbh1 cells observed in Y. lipolytica [23]. In addition, in Y. lipolytica, cells genetically depleted of Sec61β form colonies displaying morphological changes, while in S. pombe, only the overexpression of Sec61β affected cellular morphology and not colony formation. As morphology and SDS resistance are linked to cell-wall biosynthesis, these opposite effects could be due to differences in the composition of the cell-wall of these two yeasts [52]. Since we observed that the translocon β subunit levels also affect cell size, it remains possible that SP_Sec61β is involved in the coordination of cell-membrane growth in fission yeast.
Regarding the effect of Sec61β on secretion, it appears correlated with the efficiency of this process in each yeast. As S. cerevisiae is somewhat inefficient for secretion [53], the impact of Sec61β deletion is more apparent in this yeast (47% diminution) [27], while in Y. lipolytica, which is more competent for secretion, no difference are observed [23]. In fission yeast, the importance of Sec61β is revealed at 23°C, when the secretion rate of the WT strain is already reduced by 62% as compared to 30°C (Figure 3). The different phenotypes observed for the Sec61β-knockout in S. pombe as compared to S. cerevisiae and Y. lipolytica suggested that different homologs could have distinct functions. However, the lack of the translocon β subunit could affect more severely the secretion of certain proteins, thus providing an alternative explanation for the diverse, and sometimes contrasting, phenotypes associated with deletion or overexpression of Sec61β among species.
Importantly, we show that despite these phenotypic differences, SP_Sec61β is able to complement sbh1Δsbh2Δ in budding yeast. Similarly, both SC_Sec61β paralogs complement the cold and SDS sensitivity in S. pombe. This is in accordance with previous reports that Sec61β homologs from Yarrowia lipolytica, Kluyveromyces lactis, Antonospora locustae and Encephalitozoon cuniculi rescue the heat sensitivity of S. cerevisiae, indicating that they are functional homologs [23], [46], [54]. Here we demonstrate that this functional complementation is possible even with higher eukaryotes, as the human homolog of Sec61β also complements the knockout in S. pombe and S. cerevisiae. These results further suggest that the distinct phenotypes associated with the Sec61β deletion are caused by downstream effects resulting from loss of a common function of the translocon β subunit that can be performed by any homolog.
Phylogenetic studies on Sec61β/Sbh1p/SecG revealed that the consensus region for this protein is located at the hydrophobic peak corresponding to its TMD, while the N-terminal domain is more divergent and disordered [15], [16]. In many archaea, the N-terminal portion of Sec61β homologs is almost twice smaller than their mammalian counterparts. In some eukaryotes, such as parasitic microsporidia, the N-terminal region of Sec61β is practically as short as in archaea, and the protein still complements heat sensitivity of S. cerevisiae sbh1Δsbh2Δ cells [54]. Domain mapping of SC_ Sec61β showed that the N-terminal part is dispensable for normal rates of co-translational translocation, heat resistance, association with other translocon subunits and interaction with the SR [27], [28]. This suggests that the TMD of Sec61β is the critical region for protein function, while the larger N-terminal cytosolic part plays secondary roles. In this paper, we show that the fundamental function of the TMD of Sec61β is conserved among distant species. Indeed, the S. pombe, S. cerevisiae and human TMD homologs complemented various defects caused by Sec61β deletion in fission and budding yeasts, to levels comparable to the entire protein. This property is inherent to the TMD of Sec61β, since other transmembrane sequences did not suppress the phenotypes associated with the Sec61β-knockout in fission and budding yeast.
To date, the only precise function ascribed to the TMD of Sec61β is its role as a tail anchor and ER localization signal. Disruption of this domain results in accumulation of Sec61β in large cytoplasmic puncta [55], [56], while N-terminal fragments of Sec61β devoid of the TMD are unable to complement any known function of the protein in S. cerevisiae [27]. Insertion of Sec61β into the ER is not spontaneous, but requires recognition of the TMD by a post-translational targeting machinery, TRC40/Asna1 in mammals or the GET complex in yeast [55], [56]. Thus, it is conceivable that specific residues of the TMD are essential to recruit this complex to the ER membrane. Yet, other tail-anchored proteins, including the essential translocon γ subunit, display lower conservation of the TMD than the β subunit, suggesting a key function for this domain in Sec61β. Indeed, apart from its adaptor role in the translocon, the TMD of SC_Sec61β1 was also found in a distinct complex including Rtn1p, an exocyst-associated protein involved in ER reticulation [57], suggesting additional localizations of Sec61β in the ER [27]. Nevertheless, the TMD of SC_Sec61β2, which is part of the second translocon complex in budding yeast (the Ssh1 complex) [22], [58], [59], also complements cold- and SDS-sensitivity in S. pombe (Figure 5). This suggests a redundant and conserved role for this domain. Considering the small size of these hydrophobic sequences, it is unlikely that it exerts a catalytic function, but interactions with conserved transmembrane proteins could mediate the central TMD function. As Sec61β was also shown in crosslinking experiments to form dimers [25], a role of the TMD in translocon oligomerization cannot be excluded [60]. The demonstration of a scaffolding or a recruiting function for the TMD of Sec61β awaits further experimentation.
In sum, our results demonstrate a conserved biological role for the TMD of Sec61β. Our observations support the notion that single-span TMDs have other functions than anchoring in the lipid bilayer. Indeed, many TMDs have been implicated in protein complex assembly, oligomerization, helix packing, transmembrane interactions and signal transduction [61]. Sec61β from S. pombe appears as an interesting model for elucidation of the cellular roles of a highly conserved TMD present in every living kingdom.
Materials and Methods
Strains and media
S. pombe strains were cultured in Edinburgh Minimal Medium (EMM) supplemented with required nutrients [62]. S. cerevisiae strains were grown in synthetic defined media (SD) lacking appropriate amino acids [63]. All strains were grown aerobically at 30°C unless stated otherwise. G418 was used at a concentration of 150 µg/mL. Yeast strains used in this study are listed in Table 1.
Table 1. Yeast strains used in this study.
| Strain | Genotype | Source |
| S. pombe | ||
| SP556 | h− ade6-M216 leu1-32 ura4-D18 | Paul Nurse Lab |
| SP15039 | h+ ade6-M210 leu1-32 ura4-D18 Δsbh1::kan R | Bioneer |
| SP15073 | h+ ade6-M210 his3-D1 leu1-32::adh1p_cel1_3HA_adh1t_kan R ura4-D18 | This study |
| SP15074 | h− ade6-M216 his3-D1 leu1-32::adh1p_cel1_3HA_adh1t_kan R ura4-D18 Δsbh1::kan R | This study |
| S. cerevisiae | ||
| NY179 | MATα leu2-3,112 ura3-52 | Peter Novick Lab |
| SC3232 | MATα sbh1Δ::kan R sbh2Δ::hph R leu2-3,112 ura3-52 | Jussi Jäntti Lab |
Plasmid constructions and transformation
Plasmids used in this study are described in Table 2. All oligonucleotide sequences are available upon request. The coding sequences of sbh1 + and sss1 + were amplified by PCR from a S. pombe cDNA library. Forward primers contained a NdeI restriction site followed by a sequence encoding the V5 epitope tag. Reverse primers contained a BamHI restriction site. The entire ORFs were cloned into the pREP42 and pREP2 vectors [42], at the NdeI and BamHI sites. pREP42 contains a medium strength thiamine-repressible nmt* promoter, while pREP2 contains a strong nmt promoter for overexpression [64]. As previously described in S. cerevisiae [27], addition of a N-terminal tag on Sec61β had no effect on overexpression phenotypes nor complementation phenotypes (not shown). Amplification of the SBH1 and SBH2 genes was similarly achieved except that S. cerevisiae genomic DNA was used and no V5 tag was added. Amplification of SEC61B was carried out using a human cDNA bank as template. No V5 tag was added, and a BglII site was used instead of BamHI. Cloning of SBH1, SBH2 and SEC61B in pREP42 was performed as for sbh1 +. For TMD constructs, the sequences encoding the 26 amino acids of the conserved transmembrane domains of each protein, as mapped by HMMTOP [65], were cloned (see Figure 1 for amino-acid sequences). All TMD sequences were amplified to encode a V5 tag on the N-terminal side of the protein, and inserted into pREP42 using NdeI and BamHI restriction sites. For cloning in p415, amplification of sbh1 +, SBH1, SBH2, SEC61B and TMD constructs was carried out as before but with primers containing BamHI and XhoI restriction sites instead of NdeI and BamHI, respectively. SSS1 and SSS1_TMD were amplified from S. cerevisiae genomic DNA, adding a c-MYC tag instead of a V5 tag. The amplified sequences were inserted into the BamHI/XhoI restriction sites of p415. All constructs were verified using standard DNA sequencing methods. Transformations were done by the PEG (Polyethylene glycol)/lithium acetate procedure [66]. Plasmids derived from pREP42 were transformed into SP15039 and SP15074 (see below), while plasmids derived from pREP2 were transformed into SP556 and SP15073. Plasmids derived from p415 were transformed into SC3232. Transformations were confirmed by prototrophy and Western blotting.
Table 2. Plasmids used in this study.
| Plasmid | Features | Source |
| S. pombe | ||
| pREP42 | Multicopy S. pombe expression vector with ura4 + selectable marker and mild nmt* promoter | [42], [70] |
| pREP42-SP_sbh1 | pREP42 containing S. pombe sbh1 + tagged with V5 (N-terminal) | This study |
| pREP42-SC_SBH1 | pREP42 containing S. cerevisiae SBH1 | This study |
| pREP42-SC_SBH2 | pREP42 containing S. cerevisiae SBH2 | This study |
| pREP42-HS_SEC61B | pREP42 containing human SEC61B | This study |
| pREP42-SP_sss1 | pREP42 containing S. pombe sss1 + tagged with V5 (N-terminal) | This study |
| pREP42-SP_sbh1TMD | pREP42 containing S. pombe V5-sbh1_TMD (amino acids 68 to 93) | This study |
| pREP42-SC_ SBH1TMD | pREP42 containing S. cerevisiae V5-SBH1_TMD (amino acids 50 to 75) | This study |
| pREP42-SC_ SBH2TMD | pREP42 containing S. cerevisiae V5-SBH2_TMD (amino acids 57 to 82) | This study |
| pREP42-HS_ SEC61BTMD | pREP42 containing human V5-SEC61B_TMD (amino acids 66 to 91) | This study |
| pREP42-SP_sss1TMD | pREP42 containing S. pombe V5- sss1_TMD (amino acids 30 to 55) | This study |
| pREP2 | Multicopy S. pombe expression vector with ura4 + selectable marker and strong nmt promoter | [42], [70] |
| pREP2-SP_sbh1 | pREP2 containing S. pombe sbh1 + tagged with V5 (N-terminal) | This study |
| pREP2-SP_sss1 | pREP2 containing S. pombe sss1 + tagged with V5 (N-terminal) | This study |
| S. cerevisiae | ||
| p415 | pRS415 containing Venus fluorescent protein under ADH1 promoter and LEU2 marker. (Venus was removed in the other constructions) | Pascal Chartrand Lab |
| p415-SP_sbh1 | p415 containing S. pombe sbh1 + tagged with V5 (N-terminal) | This study |
| p415-SC_SBH1 | p415 containing S. cerevisiae SBH1 | This study |
| p415-SC_SBH2 | p415 containing S. cerevisiae SBH2 | This study |
| p415-HS_SEC61B | p415 containing human SEC61B | This study |
| p415-SC_SSS1 | p415 containing S. cerevisiae SSS1 tagged with c-MYC (N-terminal) | This study |
| p415-SP_sbh1TMD | p415 containing S. pombe V5-sbh1_TMD (amino acids 68 to 93) | This study |
| p415-SC_ SBH1TMD | p415 containing S. cerevisiae V5-SBH1_TMD (amino acids 50 to 75) | This study |
| p415-SC_ SBH2TMD | p415 containing S. cerevisiae V5-SBH2_TMD (amino acids 57 to 82) | This study |
| p415-HS_ SEC61BTMD | p415 containing human V5-SEC61B_TMD (amino acids 66 to 91) | This study |
| p415-SC_SSS1TMD | p415 containing S. cerevisiae c-MYC-SSS1_TMD (amino acids 49 to 71) | This study |
Sporulation and mating
Strain SP15039 (Δsbh1 +) was obtained by sporulation of the heterozygous diploid BG_3365 (h+/h+ ade6-M210/ade6-M216 leu1-32/leu1-32 ura4-D18/ura4-D18 sbh1 + /Δsbh1::kan R) purchased from Bioneer (Daejeon, Korea). After identification of sporulating diploids by iodine staining, tetrads were dissected on EMM plates supplemented with adenine, uracil, leucine and histidine (AULH), using a Nikon Eclipse E400 micromanipulator. Δsbh1 cells were identified as G418 resistant, and the knockout was confirmed by PCR, Northern blotting and Southern blotting. Strains SP15073 and SP15074 were obtained by mating SP15039 with SP11033 (h− his3-D1 leu1-32::adh1p_cel1_3HA_adh1t_kan R ura4-D18 Δcnx1::his3 + + pREP41cnx1 +) [45], which expresses Aspergillus aculeatus cellulase I under the control of the adh1p promoter. After mating on EMM+AULH plates, sporulation was induced in liquid EMM+G418 medium with low amounts of nitrogen. Asci were then subjected to random spore analysis [67]. Clones bearing the his3-D1 deletion were identified by histidine auxotrophy. Clones expressing the essential calnexin gene at the cnx1 + locus instead of the pREP41cnx1 + plasmid (leucine marker) were identified by leucine auxotrophy. Clones expressing cellulase were identified by the blue halo created on plates containing 0.1% (w/v) AZCL-HE-Cellulose chromogenic substrate (MegaZyme, North Rocks, N.S.W., Australia). Δsbh1 clones (SP15074) were identified by their cold sensitivity at 23°C. The genomic background of each strain was further confirmed by multiple PCR analyses.
Microscopy imaging
Differential interference contrast (DIC) Nomarski imaging and DAPI-staining microscopy were carried out as previously described [68]. Microscopy analyses were performed with a fluorescence inverted microscope Nikon TE2000U. Images were acquired using a motion-picture camera CCD coolSnapFX M® 12 bit, and treated with UIC Metamorph® software. Flow cytometry analyses (FACS) were done using a FACS Calibur device (Becton Dickinson Biosciences) on 10,000 cells after dilution of exponential cultures to OD595 = 0.5 in PBS buffer (136 mM NaCl, 25 mM KCl, 12 mM NaHPO4, 18 mM KH2PO4 pH 7.4).
Phenotypic assays
The sodium dodecyl sulfate (SDS) sensitivity measurements were carried out as described previously [69]. Briefly, exponentially growing cells were spread on EMM+AL top agar (0.7%), following which 10 µL of 10% SDS was added on a small 3 M paper at the center of each plate. After 3–5 days of incubation at 30°C or 23°C, area of the sensitivity halo was measured and reported on the WT value at 30°C. For temperature sensitivity assays, exponential cultures of each strain were diluted to OD595 = 0.5, serially diluted (10−1–10−4), spotted on solid media (EMM+AL for S. pombe or SD-L for S. cerevisiae), and incubated at the indicated temperatures (7 days for S. pombe or 5 days for S. cerevisiae). All results are representative of at least 3 independent experiments.
Cellulase secretion assays
Cellulase secretion [45] was quantified by activity halo measurements at 30°C or 23°C. An equivalent of 0.2 OD595 of exponentially growing cells derived from strains SP15073 (WT) or SP15074 (Δsbh1) were spotted on EMM+ALH supplemented with 0.1% (w/v) AZCL-HE-Cellulose. The area of the blue halo created by secreted cellulase activity was measured each day during 7 days after time zero, which was adjusted for every strain as the moment when the halo exceeds area of the colony (all colonies were of equal size). Each measure is the mean of 3 plates. Secretion rate was calculated for each strain by averaging the slope of independent experiments and divided them by the control strain value (100%) at the corresponding temperature.
Acknowledgments
We thank Bernhard Dobberstein and Randy Schekman for kind gifts of antibodies. We thank Tom A. Rapoport, Jaana Toikkanen and Jussi Jäntti for S. cerevisiae strains, antibodies and useful ideas. Pascal Chartrand is acknowledged for fruitful discussions as well as for plasmids and antibodies. Finally, we thank all the members of the Luis A. Rokeach lab for critical reading of the manuscript and technical assistance.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by a a NSERC grant (#171325) to Luis A. Rokeach and by a NSERC scholarship to Alexandre Leroux. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Simon SM, Blobel G. A protein-conducting channel in the endoplasmic reticulum. Cell. 1991;65:371–380. doi: 10.1016/0092-8674(91)90455-8. [DOI] [PubMed] [Google Scholar]
- 2.Rapoport TA. Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature. 2007;450:663–669. doi: 10.1038/nature06384. [DOI] [PubMed] [Google Scholar]
- 3.Gorlich D, Rapoport TA. Protein translocation into proteoliposomes reconstituted from purified components of the endoplasmic reticulum membrane. Cell. 1993;75:615–630. doi: 10.1016/0092-8674(93)90483-7. [DOI] [PubMed] [Google Scholar]
- 4.Brundage L, Fimmel CJ, Mizushima S, Wickner W. SecY, SecE, and band 1 form the membrane-embedded domain of Escherichia coli preprotein translocase. J Biol Chem. 1992;267:4166–4170. [PubMed] [Google Scholar]
- 5.Manting EH, Driessen AJ. Escherichia coli translocase: the unravelling of a molecular machine. Mol Microbiol. 2000;37:226–238. doi: 10.1046/j.1365-2958.2000.01980.x. [DOI] [PubMed] [Google Scholar]
- 6.Walter P, Lingappa VR. Mechanism of protein translocation across the endoplasmic reticulum membrane. Annu Rev Cell Biol. 1986;2:499–516. doi: 10.1146/annurev.cb.02.110186.002435. [DOI] [PubMed] [Google Scholar]
- 7.Wickner W, Driessen AJ, Hartl FU. The enzymology of protein translocation across the Escherichia coli plasma membrane. Annu Rev Biochem. 1991;60:101–124. doi: 10.1146/annurev.bi.60.070191.000533. [DOI] [PubMed] [Google Scholar]
- 8.Hartmann E, Sommer T, Prehn S, Gorlich D, Jentsch S, et al. Evolutionary conservation of components of the protein translocation complex. Nature. 1994;367:654–657. doi: 10.1038/367654a0. [DOI] [PubMed] [Google Scholar]
- 9.Wiertz EJ, Tortorella D, Bogyo M, Yu J, Mothes W, et al. Sec61-mediated transfer of a membrane protein from the endoplasmic reticulum to the proteasome for destruction. Nature. 1996;384:432–438. doi: 10.1038/384432a0. [DOI] [PubMed] [Google Scholar]
- 10.Pilon M, Schekman R, Romisch K. Sec61p mediates export of a misfolded secretory protein from the endoplasmic reticulum to the cytosol for degradation. EMBO J. 1997;16:4540–4548. doi: 10.1093/emboj/16.15.4540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakatsukasa K, Brodsky JL. The recognition and retrotranslocation of misfolded proteins from the endoplasmic reticulum. Traffic. 2008;9:861–870. doi: 10.1111/j.1600-0854.2008.00729.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deshaies RJ, Sanders SL, Feldheim DA, Schekman R. Assembly of yeast Sec proteins involved in translocation into the endoplasmic reticulum into a membrane-bound multisubunit complex. Nature. 1991;349:806–808. doi: 10.1038/349806a0. [DOI] [PubMed] [Google Scholar]
- 13.Panzner S, Dreier L, Hartmann E, Kostka S, Rapoport TA. Posttranslational protein transport in yeast reconstituted with a purified complex of Sec proteins and Kar2p. Cell. 1995;81:561–570. doi: 10.1016/0092-8674(95)90077-2. [DOI] [PubMed] [Google Scholar]
- 14.Roitsch T, Lehle L. Post-translational translocation of polypeptides across the mammalian endoplasmic reticulum membrane is size and ribosome dependent. Eur J Biochem. 1988;174:699–705. doi: 10.1111/j.1432-1033.1988.tb14154.x. [DOI] [PubMed] [Google Scholar]
- 15.Cao TB, Saier MH., Jr The general protein secretory pathway: phylogenetic analyses leading to evolutionary conclusions. Biochim Biophys Acta. 2003;1609:115–125. doi: 10.1016/s0005-2736(02)00662-4. [DOI] [PubMed] [Google Scholar]
- 16.Van den Berg B, Clemons WM, Jr, Collinson I, Modis Y, Hartmann E, et al. X-ray structure of a protein-conducting channel. Nature. 2004;427:36–44. doi: 10.1038/nature02218. [DOI] [PubMed] [Google Scholar]
- 17.Mothes W, Prehn S, Rapoport TA. Systematic probing of the environment of a translocating secretory protein during translocation through the ER membrane. EMBO J. 1994;13:3973–3982. doi: 10.1002/j.1460-2075.1994.tb06713.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kalies KU, Gorlich D, Rapoport TA. Binding of ribosomes to the rough endoplasmic reticulum mediated by the Sec61p-complex. J Cell Biol. 1994;126:925–934. doi: 10.1083/jcb.126.4.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prinz A, Hartmann E, Kalies KU. Sec61p is the main ribosome receptor in the endoplasmic reticulum of Saccharomyces cerevisiae. Biol Chem. 2000;381:1025–1029. doi: 10.1515/BC.2000.126. [DOI] [PubMed] [Google Scholar]
- 20.Esnault Y, Feldheim D, Blondel MO, Schekman R, Kepes F. SSS1 encodes a stabilizing component of the Sec61 subcomplex of the yeast protein translocation apparatus. J Biol Chem. 1994;269:27478–27485. [PubMed] [Google Scholar]
- 21.Nishiyama K, Hanada M, Tokuda H. Disruption of the gene encoding p12 (SecG) reveals the direct involvement and important function of SecG in the protein translocation of Escherichia coli at low temperature. EMBO J. 1994;13:3272–3277. doi: 10.1002/j.1460-2075.1994.tb06628.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Finke K, Plath K, Panzner S, Prehn S, Rapoport TA, et al. A second trimeric complex containing homologs of the Sec61p complex functions in protein transport across the ER membrane of S. cerevisiae. EMBO J. 1996;15:1482–1494. [PMC free article] [PubMed] [Google Scholar]
- 23.Boisrame A, Chasles M, Babour A, Beckerich JM, Gaillardin C. Sbh1p, a subunit of the Sec61 translocon, interacts with the chaperone calnexin in the yeast Yarrowia lipolytica. J Cell Sci. 2002;115:4947–4956. doi: 10.1242/jcs.00187. [DOI] [PubMed] [Google Scholar]
- 24.Toikkanen J, Gatti E, Takei K, Saloheimo M, Olkkonen VM, et al. Yeast protein translocation complex: isolation of two genes SEB1 and SEB2 encoding proteins homologous to the Sec61 beta subunit. Yeast. 1996;12:425–438. doi: 10.1002/(SICI)1097-0061(199604)12:5%3C425::AID-YEA924%3E3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 25.Kalies KU, Rapoport TA, Hartmann E. The beta subunit of the Sec61 complex facilitates cotranslational protein transport and interacts with the signal peptidase during translocation. J Cell Biol. 1998;141:887–894. doi: 10.1083/jcb.141.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nishiyama K, Mizushima S, Tokuda H. A novel membrane protein involved in protein translocation across the cytoplasmic membrane of Escherichia coli. EMBO J. 1993;12:3409–3415. doi: 10.1002/j.1460-2075.1993.tb06015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feng D, Zhao X, Soromani C, Toikkanen J, Romisch K, et al. The transmembrane domain is sufficient for Sbh1p function, its association with the Sec61 complex, and interaction with Rtn1p. J Biol Chem. 2007;282:30618–30628. doi: 10.1074/jbc.M701840200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang Y, Cheng Z, Mandon EC, Gilmore R. An interaction between the SRP receptor and the translocon is critical during cotranslational protein translocation. J Cell Biol. 2008;180:1149–1161. doi: 10.1083/jcb.200707196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Duong F, Wickner W. Distinct catalytic roles of the SecYE, SecG and SecDFyajC subunits of preprotein translocase holoenzyme. EMBO J. 1997;16:2756–2768. doi: 10.1093/emboj/16.10.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsumoto G, Mori H, Ito K. Roles of SecG in ATP- and SecA-dependent protein translocation. Proc Natl Acad Sci U S A. 1998;95:13567–13572. doi: 10.1073/pnas.95.23.13567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Helmers J, Schmidt D, Glavy JS, Blobel G, Schwartz T. The {beta}-Subunit of the Protein-conducting Channel of the Endoplasmic Reticulum Functions as the Guanine Nucleotide Exchange Factor for the {beta}-Subunit of the Signal Recognition Particle Receptor. J Biol Chem. 2003;278:23686–23690. doi: 10.1074/jbc.C300180200. [DOI] [PubMed] [Google Scholar]
- 32.Lipschutz JH, Lingappa VR, Mostov KE. The exocyst affects protein synthesis by acting on the translocation machinery of the endoplasmic reticulum. J Biol Chem. 2003;278:20954–20960. doi: 10.1074/jbc.M213210200. [DOI] [PubMed] [Google Scholar]
- 33.Toikkanen JH, Miller KJ, Soderlund H, Jantti J, Keranen S. The {beta} Subunit of the Sec61p Endoplasmic Reticulum Translocon Interacts with the Exocyst Complex in Saccharomyces cerevisiae. J Biol Chem. 2003;278:20946–20953. doi: 10.1074/jbc.M213111200. [DOI] [PubMed] [Google Scholar]
- 34.Kondratyev M, Avezov E, Shenkman M, Groisman B, Lederkremer GZ. PERK-dependent compartmentalization of ERAD and unfolded protein response machineries during ER stress. Exp Cell Res. 2007;313:3395–3407. doi: 10.1016/j.yexcr.2007.07.006. [DOI] [PubMed] [Google Scholar]
- 35.Wang B, Heath-Engel H, Zhang D, Nguyen N, Thomas DY, et al. BAP31 interacts with Sec61 translocons and promotes retrotranslocation of CFTRDeltaF508 via the derlin-1 complex. Cell. 2008;133:1080–1092. doi: 10.1016/j.cell.2008.04.042. [DOI] [PubMed] [Google Scholar]
- 36.Yamaguchi A, Hori O, Stern DM, Hartmann E, Ogawa S, et al. Stress-associated endoplasmic reticulum protein 1 (SERP1)/Ribosome-associated membrane protein 4 (RAMP4) stabilizes membrane proteins during stress and facilitates subsequent glycosylation. J Cell Biol. 1999;147:1195–1204. doi: 10.1083/jcb.147.6.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hori O, Miyazaki M, Tamatani T, Ozawa K, Takano K, et al. Deletion of SERP1/RAMP4, a component of the endoplasmic reticulum (ER) translocation sites, leads to ER stress. Mol Cell Biol. 2006;26:4257–4267. doi: 10.1128/MCB.02055-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lopez R, Silventoinen V, Robinson S, Kibria A, Gish W. WU-Blast2 server at the European Bioinformatics Institute. Nucleic Acids Res. 2003;31:3795–3798. doi: 10.1093/nar/gkg573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, et al. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, et al. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31:3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vergeres G, Waskell L. Expression of cytochrome b5 in yeast and characterization of mutants of the membrane-anchoring domain. J Biol Chem. 1992;267:12583–12591. [PubMed] [Google Scholar]
- 42.Maundrell K. Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene. 1993;123:127–130. doi: 10.1016/0378-1119(93)90551-d. [DOI] [PubMed] [Google Scholar]
- 43.Sirisattha S, Momose Y, Kitagawa E, Iwahashi H. Toxicity of anionic detergents determined by Saccharomyces cerevisiae microarray analysis. Water Res. 2004;38:61–70. doi: 10.1016/j.watres.2003.08.027. [DOI] [PubMed] [Google Scholar]
- 44.Osumi M, Yamada N, Kobori H, Taki A, Naito N, et al. Cell wall formation in regenerating protoplasts of Schizosaccharomyces pombe: study by high resolution, low voltage scanning electron microscopy. J Electron Microsc (Tokyo) 1989;38:457–468. [PubMed] [Google Scholar]
- 45.Hajjar F, Beauregard PB, Rokeach LA. The 160 N-terminal residues of calnexin define a novel region supporting viability in Schizosaccharomyces pombe. Yeast. 2007;24:89–103. doi: 10.1002/yea.1440. [DOI] [PubMed] [Google Scholar]
- 46.Toikkanen JH, Sundqvist L, Keranen S. Kluyveromyces lactis SSO1 and SEB1 genes are functional in Saccharomyces cerevisiae and enhance production of secreted proteins when overexpressed. Yeast. 2004;21:1045–1055. doi: 10.1002/yea.1151. [DOI] [PubMed] [Google Scholar]
- 47.Kinch LN, Saier MH, Jr, Grishin NV. Sec61beta–a component of the archaeal protein secretory system. Trends Biochem Sci. 2002;27:170–171. doi: 10.1016/s0968-0004(01)02055-2. [DOI] [PubMed] [Google Scholar]
- 48.Valcarcel R, Weber U, Jackson DB, Benes V, Ansorge W, et al. Sec61beta, a subunit of the protein translocation channel, is required during Drosophila development. J Cell Sci. 1999;112(Pt 23):4389–4396. doi: 10.1242/jcs.112.23.4389. [DOI] [PubMed] [Google Scholar]
- 49.Flower AM. SecG function and phospholipid metabolism in Escherichia coli. J Bacteriol. 2001;183:2006–2012. doi: 10.1128/JB.183.6.2006-2012.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pogliano KJ, Beckwith J. The Cs sec mutants of Escherichia coli reflect the cold sensitivity of protein export itself. Genetics. 1993;133:763–773. doi: 10.1093/genetics/133.4.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang ZJ, Zhou JP, Li GR, Zhang Y, Ren ZL. [Cloning and expression of Gymnadenia conopsea GcSec61beta gene encoding endosplasmic reticulum membrane translocation channel protein]. Zhi Wu Sheng Li Yu Fen Zi Sheng Wu Xue Xue Bao. 2007;33:354–360. [PubMed] [Google Scholar]
- 52.Vega R, Dominguez A. Cell wall composition of the yeast and mycelial forms of Yarrowia lipolytica. Arch Microbiol. 1986;144:124–130. [Google Scholar]
- 53.Muller S, Sandal T, Kamp-Hansen P, Dalboge H. Comparison of expression systems in the yeasts Saccharomyces cerevisiae, Hansenula polymorpha, Klyveromyces lactis, Schizosaccharomyces pombe and Yarrowia lipolytica. Cloning of two novel promoters from Yarrowia lipolytica. Yeast. 1998;14:1267–1283. doi: 10.1002/(SICI)1097-0061(1998100)14:14<1267::AID-YEA327>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 54.Slamovits CH, Burri L, Keeling PJ. Characterization of a divergent Sec61beta gene in microsporidia. J Mol Biol. 2006;359:1196–1202. doi: 10.1016/j.jmb.2006.04.028. [DOI] [PubMed] [Google Scholar]
- 55.Stefanovic S, Hegde RS. Identification of a targeting factor for posttranslational membrane protein insertion into the ER. Cell. 2007;128:1147–1159. doi: 10.1016/j.cell.2007.01.036. [DOI] [PubMed] [Google Scholar]
- 56.Schuldiner M, Metz J, Schmid V, Denic V, Rakwalska M, et al. The GET complex mediates insertion of tail-anchored proteins into the ER membrane. Cell. 2008;134:634–645. doi: 10.1016/j.cell.2008.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.De Craene JO, Coleman J, Estrada de Martin P, Pypaert M, Anderson S, et al. Rtn1p is involved in structuring the cortical endoplasmic reticulum. Mol Biol Cell. 2006;17:3009–3020. doi: 10.1091/mbc.E06-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wilkinson BM, Tyson JR, Stirling CJ. Ssh1p determines the translocation and dislocation capacities of the yeast endoplasmic reticulum. Dev Cell. 2001;1:401–409. doi: 10.1016/s1534-5807(01)00043-0. [DOI] [PubMed] [Google Scholar]
- 59.Wittke S, Dunnwald M, Albertsen M, Johnsson N. Recognition of a subset of signal sequences by Ssh1p, a Sec61p-related protein in the membrane of endoplasmic reticulum of yeast Saccharomyces cerevisiae. Mol Biol Cell. 2002;13:2223–2232. doi: 10.1091/mbc.01-10-0518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hanein D, Matlack KE, Jungnickel B, Plath K, Kalies KU, et al. Oligomeric rings of the Sec61p complex induced by ligands required for protein translocation. Cell. 1996;87:721–732. doi: 10.1016/s0092-8674(00)81391-4. [DOI] [PubMed] [Google Scholar]
- 61.Moore DT, Berger BW, DeGrado WF. Protein-protein interactions in the membrane: sequence, structural, and biological motifs. Structure. 2008;16:991–1001. doi: 10.1016/j.str.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Alfa C, Fantes P, Hyams J, McLeod M, Warbrick E. Experiments with Fission Yeasts: Laboratory Course Manual; Cold Spring Harbor Laboratory Press; 1993. CSH, NY, editor. [Google Scholar]
- 63.Sherman F, Fink GR, Hicks JB. Methods in Yeast Genetics. Cold Spring Harbor Laboratory; 1981. CSH, NY, editor. [Google Scholar]
- 64.Forsburg SL, Sherman DA. General purpose tagging vectors for fission yeast. Gene. 1997;191:191–195. doi: 10.1016/s0378-1119(97)00058-9. [DOI] [PubMed] [Google Scholar]
- 65.Tusnady GE, Simon I. Principles governing amino acid composition of integral membrane proteins: application to topology prediction. J Mol Biol. 1998;283:489–506. doi: 10.1006/jmbi.1998.2107. [DOI] [PubMed] [Google Scholar]
- 66.Elbe R. A simple and efficient procedure for transformation of yeasts. Biotechniques. 1992;13:18–19. [PubMed] [Google Scholar]
- 67.Jannatipour M, Rokeach LA. The Schizosaccharomyces pombe homologue of the chaperone calnexin is essential for viability. J Biol Chem. 1995;270:4845–4853. doi: 10.1074/jbc.270.9.4845. [DOI] [PubMed] [Google Scholar]
- 68.Guerin R, Arseneault G, Dumont S, Rokeach LA. Calnexin Is Involved in Apoptosis Induced by ER Stress in the Fission Yeast. Mol Biol Cell. 2008 doi: 10.1091/mbc.E08-02-0188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Turcotte C, Roux A, Beauregard PB, Guerin R, Senechal P, et al. The calnexin-independent state does not compensate for all calnexin functions in Schizosaccharomyces pombe. FEMS Yeast Res. 2007;7:196–208. doi: 10.1111/j.1567-1364.2006.00145.x. [DOI] [PubMed] [Google Scholar]
- 70.Basi G, Schmid E, Maundrell K. TATA box mutations in the Schizosaccharomyces pombe nmt1 promoter affect transcription efficiency but not the transcription start point or thiamine repressibility. Gene. 1993;123:131–136. doi: 10.1016/0378-1119(93)90552-e. [DOI] [PubMed] [Google Scholar]