Summary
The capsids of tailed DNA bacteriophages first assemble as procapsids, which mature by converting into a new form that is strong enough to contain a densely packed viral chromosome. We demonstrate that the intersubunit crosslinking occurring during maturation of HK97 capsids actually promotes the structural transformation. Small-angle X-ray scattering and crosslinking assays reveal that a shift in crosslink pattern accompanies conversion of a semi-mature particle, Expansion Intermediate-I/II, to a more mature state, Balloon. This transition occurs in a switch-like fashion. We find that crosslink formation shifts the global conformational balance to favor the Balloon state. A pseudoatomic model of EI-I/II derived from cryo-EM provides insight into the relationship between crosslink formation and conformational switching.
Introduction
A simplistic view of viral capsids is that they are merely containers that surround and protect their chromosomes until they can be delivered to new hosts. However a growing body of evidence shows that capsids are much more than that— they are also complex macromolecular machines that often carry out large scale transformations of their structures between their initial assembly and infection. In many cases global changes in capsid size or shape occur that are the result of the coordinated movements of hundreds of protein subunits. Such changes are observed during assembly, chromosome packaging, receptor binding, and cell entry in a broad range of virus families (e.g. (Belnap et al., 2000; Hewat and Blaas, 2004; Jardine and Coombs, 1998; Trus et al., 1996). Some of the most dramatic structural remodeling in viruses is observed during post-assembly capsid maturation processes (Steven et al., 2005). In most cases, such as in lentiviruses, papillomaviruses, insect tetraviruses, and dsDNA viruses such as herpesviruses and tailed bacteriophages, maturation converts a relatively fragile procapsid into a more stable form (e.g. Buck et al., 2005; Canady et al., 2000; Dokland and Murialdo, 1993; Heymann et al., 2003; Turner and Summers, 1999).
During bacteriophage maturation, the suite of interactions between subunits may change drastically as nearly every inter-subunit interface is remodeled (Wikoff et al., 2006). Our goal is to understand the structural and mechanical principles that underlie maturation of the bacteriophage capsids. Bacteriophage HK97 has emerged as a favorable system for investigating structural maturation. This was the first dsDNA phage for which a high-resolution capsid structure was determined (Figure 1A) (Wikoff et al., 2000). Subsequent studies of a number of phages including T4, T5, T7, P22, epsilon-15, and ϕ-29 have revealed the ubiquity of the mature HK97 capsid protein fold as a structural building block for capsids (Agirrezabala et al., 2007; Effantin et al., 2006; Fokine et al., 2005; Jiang et al., 2006; Jiang et al., 2003; Morais et al., 2005). Genome sequence analysis also suggests that proteins with the HK97 fold may be found in many other phages (Hendrix, 2005). In addition, it has recently been shown that the floor domain of the herpesvirus major capsid protein is consistent with the HK97 fold (Baker et al., 2005). An understanding of dynamic reorganization in HK97 capsids is likely to apply to these related systems that also undergo similar maturational events.
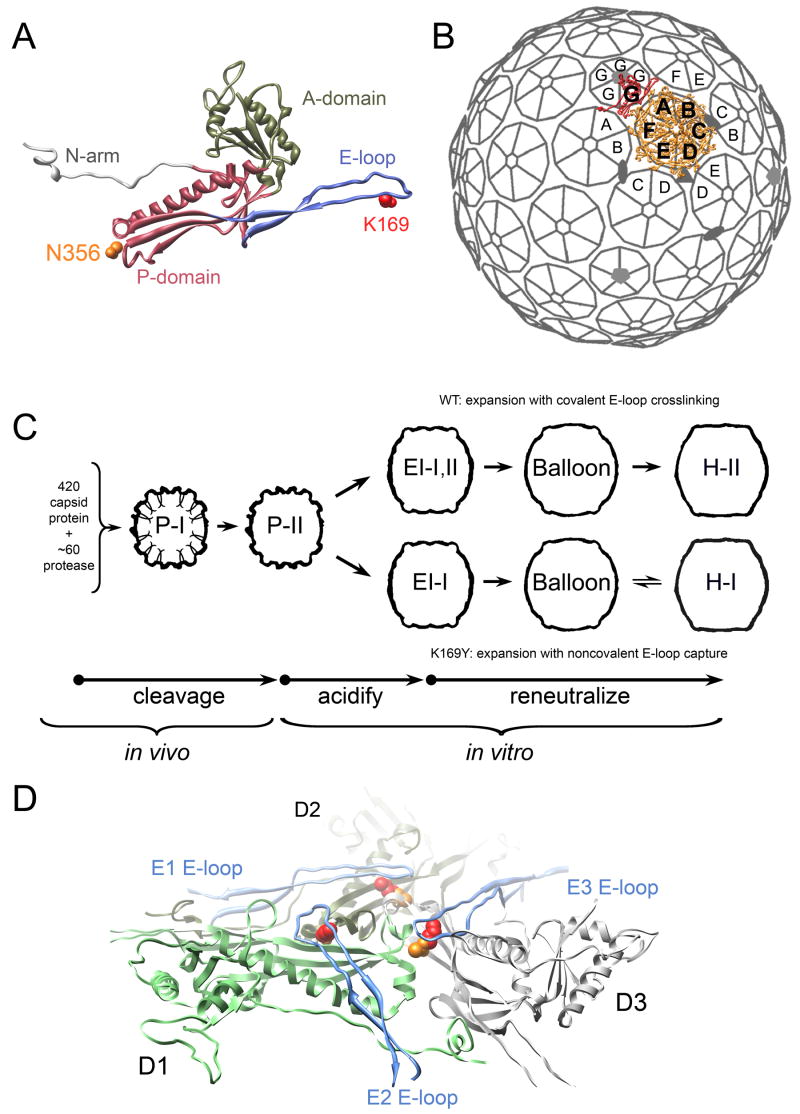
Figure 1. Bacteriophage HK97 capsid organization and maturation pathway.
(A) HK97 capsid protein structure (H-II A subunit shown) consists of four domains, with a core formed by A (pink ribbon) and P-domains (tan ribbon) and extended N-arm (light grey) and E-loop (blue ribbon) domains. The intersubunit crosslink formed by each subunit joins K169 (red) on the E-loop of one subunit and N356 (orange) on the P-domain of a neighboring subunit. (B) 420 copies of the capsid protein are organized into a T=7laveo lattice in HK97 particles produced in our expression system; subunits A-F (orange) form 60 hexons and G subunits (red) form 12 pentons; one asymmetric unit is rendered. (C) Prohead-I (P-I) particles are assembled from 420 copies of capsid protein and ~60 copies of phage protease in the expression system. Prohead-II (P-II), resulting from proteolytic processing of P-I, converts in vitro into Expansion Intermediate-I particles (EI-I) when perturbed by acidic pH. In WT, crosslinks begin to form once particles have converted to EI-I; particles that have the same quaternary structure as EI-I but bearing crosslinks are termed EI-II. A second maturation stage is triggered by reneutralization, which allows EI-I/II particles to convert to fully mature Head-II (H-II). Crosslink-defective K169Y particles follow a similar maturation pathway. The K169Y end-state is called Head-I (H-I) to distinguish it from crosslinked H-II. (D) The H-II structure reveald that three crosslink sites cluster around the intersection of three P-domains (D subunits shown at true 3-fold position); E-loops (blue) from adjacent subunits (E subunits shown) converge on the P-domain nexus, and their K169 residues (red) crosslink with N356 (orange) on the P-domains. One D subunit is colored green to show the extent of a single subunit.
HK97 capsids have T=7laevo quasiequivalent symmetry (Figure 1B). The 7 chemically identical subunits per icosahedral asymmetric unit (designated A–G) each reside in distinct local environments in the lattice. Subunits A–F are organized into hexons, of which there are 60 per capsid, and G subunits form pentons around the 5-fold vertices. In authentic HK97 proheads, one of the 12 pentagonal vertices is occupied by a portal complex, which mediates DNA packaging and tail attachment. The particles used in most in vitro studies of HK97 maturation are generated by an expression system that produces prohead particles that are indistinguishable from authentic proheads, but with the portal complex replaced by a twelfth penton (Duda et al., 1995a). If the viral protease is also expressed, ~60 copies co-assemble within the prohead form named Prohead-I (Figure 1C); they then digest 102 residues from the N-terminus of each capsid protein subunit. The proteases also auto-digest, leaving behind the processed prohead form named Prohead-II.
Prohead-II (P-II) is metastable; its maturation can be induced in vitro by exposing the particles to acidic pH, denaturants, or organic solvents (Duda et al., 1995a). The HK97 capsid exhibits discrete intermediate particle forms during in vitro maturation, which is separable into several segments (Figure 1C) (Lata et al., 2000). The first is triggered by acidification and converts P-II into particles that are ~15% larger, termed EI-I. We demonstrated that this stage of the capsid reorganization occurs as a cooperative, all-or-none transition (Lee et al., 2005). If pH is restored to neutral, maturation progresses all the way to the Head-II state. Particles that have attained the EI-I morphology become competent to form intersubunit crosslinks, which are essential for phage viability (Ross et al., 2005). Crosslink formation is autocatalytic and results in an isopeptide bond linking a lysine, K169, on one subunit and an asparagine, N356, on an adjacent subunit (Duda et al., 1995a; Wikoff et al., 2000). As shown in Figure 1D, a crosslink-competent organization of the residues coalesces at the intersection of three hexons (true 3-fold and quasi-3-fold vertices) and two hexons and one penton (quasi-3-fold vertex) only after P-II particles have reorganized into the EI-I structure. Particles that are essentially identical to EI-I by cryo-EM but that have begun to crosslink are termed EI-II; we refer to the shared, crosslink-independent quaternary structure of these particles as EI-I/II.
In fully mature Head-II particles, the complete network of crosslinks form interlocked rings of 5 and 6 crosslinked subunits (5 and 6-circles) that link the capsid protein subunits into a structure with the topology of molecular chainmail (Wikoff et al., 2000). Previous studies that followed the time-dependent accumulation of crosslinks along the acid-induced maturation pathway found that crosslinks accumulate up to a stable state in which ~60% of the total possible have formed and that by SDS-PAGE appears as a ladder extending from monomers to crosslinked, linear 2, 3, 4, 5, and 6-mers, and closed 5-circles (Duda et al., 1995a; Gan et al., 2004). Using selective solution conditions, these particles could be converted to a penultimate maturation state that has a larger (by ~9% compared with EI-I/II), rounder, thin-walled morphology, designated Balloon, in which all quasiequivalent classes of crosslinks but one had formed (crosslink state termed EI-IV) (Gan et al., 2004). In Balloon and Head-II particles, the hexon organization is nearly identical; only the position of the pentons differ. It was demonstrated that the formation of the final class of crosslinks locks the pentons, which have significant mobility in Balloon particles, into the conformation seen in the Head-II structure (Gan et al., 2006). Crosslinks thus were shown to have an effect on the quaternary structure during this late maturation stage.
In this report, we focus on the coupling between covalent crosslink formation and large-scale reorganizations of the capsid lattice during the middle (EI-I/II to Balloon) stage of maturation. It has been inferred that crosslink formation promotes this transition (Ross et al., 2005); however, the mechanistic role played by the crosslinks in this context has remained elusive. We applied time-resolved, small-angle x-ray scattering (SAXS) and biochemical assays to monitor the conformational change of particles in parallel with crosslink formation. SAXS provides structural information for particles in solution. We find that as crosslinks accumulate, leading up to a threshold of ~60% of total crosslinks, the conformational balance shifts increasingly to favor the more mature particle forms. We also found that the EI-I/II to Balloon transition depended on the integrity of the E-loop. This was tested using K169Y mutant capsids, trypsin-treated WT (trpWT) capsids, and capsids that have a genetically truncated E-loop tip (ΔEloop). K169Y capsids cannot crosslink because the critical K169 is replaced by a non-crosslinkable tyrosine (K169Y); nevertheless, they follow a maturation pathway similar to wild type (Figure 1C), ending in a state called Head-I that resembles Head-II in morphology, but lacking molecular chainmail (Conway et al., 1995). Both trpWT and ΔEloop particles can mature to EI-I, but are conformationally trapped in that state. Taken together, our studies reveal that in the EI-I/II to Balloon transition, crosslinking acts as a statistical bias on the capsid’s global morphological switch. This is a considerably different mechanism than the localized conformational effect of crosslinking on penton position that was observed previously in the late maturation stage (Balloon to Head-II) (Gan et al., 2006).
Results
EI-I/II particles can accommodate a diversity of crosslink states
We sought to first determine whether crosslink formation leads directly to changes in quaternary structure, such as by mechanically ratcheting the lattice into a more mature configuration. To address this, we used SAXS to compare the structures of minimally crosslinked EI-I particles to particles that had been acidified for 72 h, allowing ~60% of the total possible complement of crosslinks to form.
In order to obtain as homogeneous of a population of particles as possible, pH 3.9 was used in the acidification reaction. It had been shown previously by cryo-EM that the population distribution between EI-I/II and Balloon particles is highly sensitive to solution pH, with a greater accumulation of Balloon particles observed when WT or K169Y particles are incubated at pH 4.4 vs. pH<4.0 (Ross et al., 2005). At pH 3.9, essentially the entire population of particles (>98%) was observed by EM to be in the EI-I/II form. SAXS measurements for WT and K169Y particles acidified to pH 3.9 likewise confirmed that observation (data not shown). We estimate that no more than 5% of particles are in the larger Balloon state under these conditions.
We observed that WT particles, acidified at pH 3.9 for 72 h accumulated a stable ladder crosslink pattern (monomers and linear 2-, 3-, 4-, 5-, 6-mers, and closed 5-circles), but were indistinguishable by SAXS from the EI-I state, which lacks crosslinks (Figure 2). This demonstrated that the EI-I/II quaternary structure can accommodate a diversity of crosslink configurations, from no crosslinks up to an aggregate of nearly 60% of the total possible crosslinks, without changing quaternary structure.
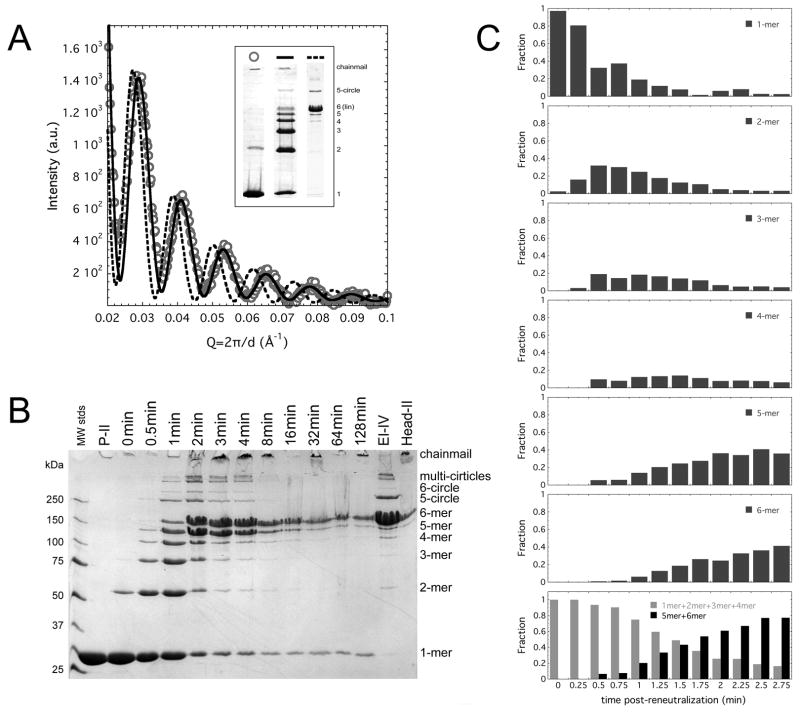
Figure 2. EI-I/II particles accommodate a broad diversity of crosslink states.
(A) SAXS patterns for un-crosslinked EI-I particles (circles) are superimposable with that measured for EI-II particles that bear ~60% of the total possible crosslinks (solid line); dashed line is the SAXS measured for the larger Balloon particles, which possess the nearly complete EI-IV crosslink pattern. Inset depicts typical crosslink patterns associated with the particle states whose SAXS patterns are shown in panel A. (B) Time-course of crosslink polymerization starting with un-crosslinked EI-I particles. When a population of EI-I particles is reneutralized (time post-reneutralization indicated above the gel), crosslinks accumulate progressively until they form the ladder pattern within 1 minute post-reneutralization. ~60% of all possible crosslinks have formed by this point. An abrupt transition from the ladder crosslink pattern to a pattern heavily weighted with linear 5 and 6-mers occurs between 1 and 3 min post-reneutralization. The crosslink patterns then gradually convert to the EI-IV pattern and to complete chainmail, which does not enter the gel matrix. The monomeric subunits are from residual unexpanded P-II. (C) Quantitation of a time-course of crosslinked oligomers observed by SDS-PAGE (15 s sampling) shows a bimodal shift from monomers and lower molecular weight linear oligomers (2-mer, 3-mer, and 4-mers) to higher molecular weight linear oligomers (5-mers and 6-mers) taking place between ~1 and 3 minutes post-reneutralization. 5-circles and 6-circles were not included due to difficulties in accurately estimating their abundance due to their incorporation into chainmail.
Crosslink patterns shift upon reneutralization
The time dependent formation of crosslinks under neutral pH conditions was examined by starting with a population of un-crosslinked EI-I particles, then rapidly reneutralizing the solution of particles. This allows the particles to mature completely, leading all the way to 100% crosslinked, chainmail-bearing H-II. As shown in Figure 2B, crosslinks accumulate initially in a progressive fashion. The conversion of monomers into linear oligomers of 2, 3, 4, 5, and 6 subunits mirrors the trend observed when particles are incubated for prolonged periods (hours) at acidic pH (Gan et al., 2004); at neutral pH the ladder pattern is reached within the first minute. Subsequently, a major oligomeric shift is observed between 1 and 3 min (Figure 2B, C). This shift converts the ladder pattern to one that is dominated by linear 5- and 6-mers with a small complement of circular 5-mers. Over the next several minutes the linear 5-mers convert to linear 6-mers or to closed 5-circles. The resulting pattern is similar to the EI-IV pattern that is associated with particles bearing a Balloon morphology (Gan et al., 2006), and eventually the pattern converts to full chainmail that is trapped in the gel loading wells.
A finer sampling of crosslink formation as a function of time reveals the bimodal nature of the crosslink pattern shift (Figure 2C). As the combination of monomers and smaller linear oligomers (2, 3, and 4-mers, approximating the ladder pattern) become depleted between 1 and 3 minutes, the linear 5-mer and 6-mer populations increase correspondingly. These trends suggest that a shift between two dominant populations (particles bearing crosslinking up to the ladder pattern and those bearing the linear 5-mer and 6-mer dominated pattern) best describes the progression of the crosslinking reaction.
Expansion is accelerated by crosslinking, but it remains a two-state transition
Time-resolved SAXS was used to monitor quaternary structural changes under neutral pH conditions that parallel those used in the time-dependent crosslinking assays for WT HK97 capsids (Figure 3). The transition from EI-I/II to Balloon (and at later time points, Head-II, which cannot be distinguished from Balloon using SAXS due to their similar structures) as monitored by SAXS was observed to occur primarily between 1 and 4 minutes following re-neutralization of WT particles. This coincides with the time frame in which we observed the shift in crosslink patterns (Figure 2). The clear presence of isoscattering points in the WT HK97 time-resolved SAXS indicates that the population of particles shifts from EI-I/II to Balloon in a two-state fashion. To confirm that the transition is two-state, Singular Value Decomposition (SVD) was used to decompose the scattering pattern time series into a set of orthogonal spectral components. SVD also yields a set of singular values that describe the relative significance of each spectral component. The product of the singular values, a time-dependent amplitude matrix, and the matrix of spectral components can be used to reconstruct the experimental SAXS pattern at any time point. The first five singular values are 38478, 1233, 279, 230, and 213. After the first two values, a baseline is reached, indicating that third and higher order components do not contribute to the time-resolved data. Likewise, the time-dependent amplitudes and scattering patterns for the third and higher order components fluctuated randomly around zero and hence are attributable to noise. We conclude that despite the presence and on-going formation of crosslinks in the wild type particles, only the two distinguishable quaternary structures, EI-I/II and Balloon/Head-II, exist as stable states during the transition. Intermediate states are populated transiently and comprise a very small fraction (<1% based on SVD analysis) of the total population at any given time.
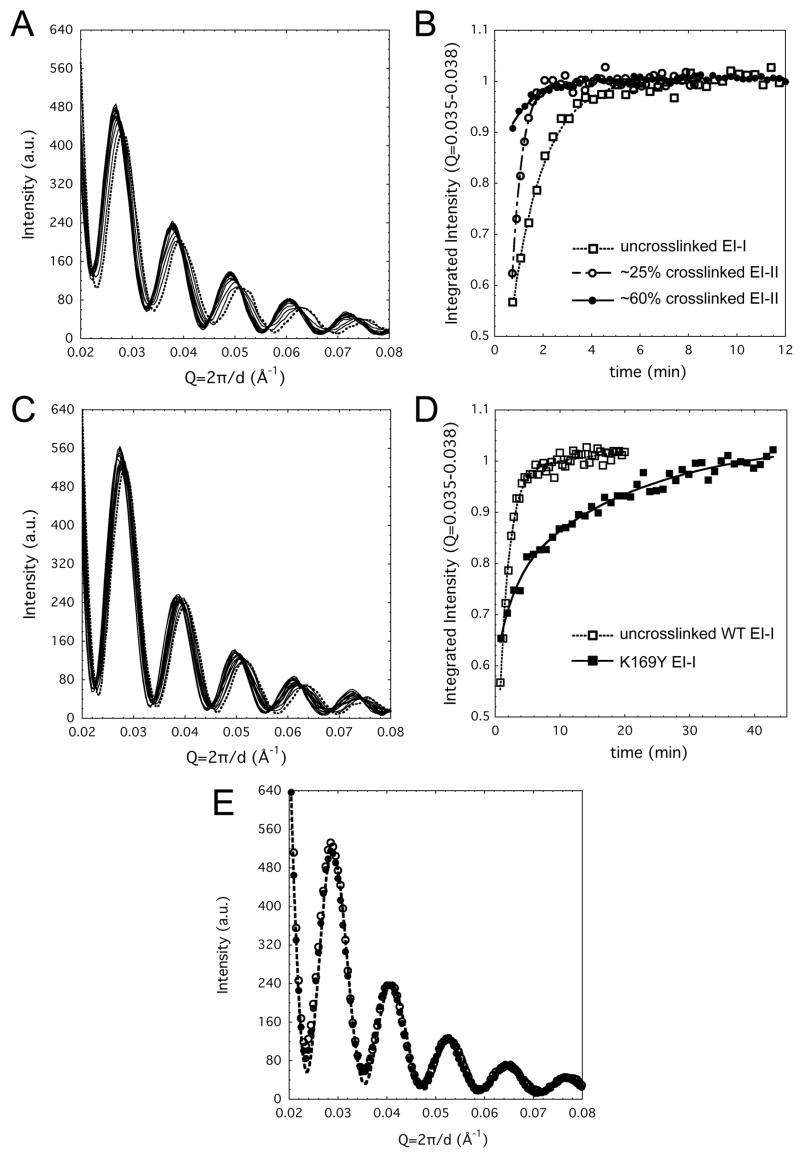
Figure 3. Time-resolved SAXS following reneutralization-induced conversion of EI-I/II to Balloon particles.
(A) Un-crosslinked WT EI-I particles convert to Balloon in a two-state fashion following reneutralization (dashed line indicates the first pattern measured); curves shown are at 30 s intervals. At later timepoints, Balloon converts to Head-II; these two particle forms are not distinguishabe by SAXS. (B) The kinetics of WT maturation (open squares) show that the majority of particles convert from EI-I to Balloon in the 1 to 4 min. time frame, and all have converted by ~5 min. If the initial population is composed of EI-II particles, similar in quaternary structure to EI-I but with 25% (open circles) or 60% (solid circles) of total crosslinks, maturation to Balloon is accelerated. Lines through the datapoints are drawn to guide the eye and do not represent fits to specific kinetic models. (C) The crosslink-defective K169Y mutant exhibits a similar transition to what is observed with WT; curves shown are at 60 s intervals. (D) The kinetics of the K169Y transition are several times slower (solid squares) than for WT particles (open squares); a slight difference in integrated amplitude at the earliest time points is due to the presence of ~5–10% residual P-II particles in the WT case. (E) SAXS patterns for the acidified then reneutralized trpWT particles at 1 h (solid circles) or >10 h (open circles) post-reneutralization are superimposable with that measured for untrypsinized WT EI-I/II particles at pH 4 (dashed line).
EI-I/II particles were also incubated at pH 3.9 for 2 h and for more than 72 h allowing them to accumulate greater amounts of crosslinking corresponding to ~25% (~100/420) and ~60% (~250/420) of total possible crosslinks, respectively. When these particles were exposed to neutral pH and monitored by SAXS, we observed a clear correlation of faster maturation kinetics with greater degree of initial crosslinking (Figure 3B). The transitions remained two-state, however.
The kinetics of conversion of EI-I to Balloon as monitored by SAXS for the crosslink-deficient K169Y mutant were dramatically slowed relative to WT, as shown in Figure 3C, D. The isoscattering points that were observed in the K169Y experiments were not as sharp as in the WT case, possibly indicating that the transition occurs with a lower degree of cooperativity among subunits in the absence of crosslinking.
In both WT and K169Y cases, the kinetics could not be described purely as first-order, possibly reflecting the presence of underlying mixtures of populations that are not distinguishable by SAXS; for example the number of captured E-loops in each particle may vary throughout the population.
Trypsinized WT and ΔEloop are trapped in the EI-I particle state
Two additional crosslink-defective variants, trypsinized WT (trpWT) and the ΔEloop mutant, which has a truncated E-loop, were also examined. Trypsinization of WT P-II cleaves the capsid protein at a single site, after K166 on the E-loop, leaving the K169 crosslink residue intact (Duda et al., 1995a). We observed that the trpWT and ΔEloop variants undergo the first maturation stage in identical fashion to WT P-II (Supplementary Fig. 1), confirming that crosslinking is not a factor in the P-II to EI-I transition (Lee et al., 2005). However, they do not progress further. When acidified trpWT particles were reneutralized under identical conditions to unproteolyzed WT, they were found to persist in the EI-I form even after several hours (Figure 3). After 72 h, a minor population converted to the larger Balloon form, but the overwhelming majority remained in the compact EI-I state. The ΔEloop mutant, in which the flexible tip of the E-loops including K169, is replaced by an abbreviated turn, also exhibited inhibition of the EI-I to Balloon transition. Maturation of these particles beyond EI-I was not detected even days after re-neutralization (data not shown).
Expansion Intermediate pseudoatomic model
In order to gain insight into the structural basis for the significant effect crosslink formation has upon conformational switching, a pseudoatomic model of the EI-I/II particle was composed. The 3.4 Å resolution crystal structures of mature H-II subunits were docked into a 14 Å resolution cryo-EM reconstruction of minimally crosslinked EI-I/II particles (Figure 4) (Helgstrand et al., 2003; Ross et al., 2005). The 7 quasiequivalent subunits were fitted independently into the EM electron density map using rigid body modeling of the subunit core (A and P domains, treated as a unit). Overall, the subunit cores fit well into the cryo-EM density envelope, consistent with previous indications that relatively minor structural changes occur in the A- and P-domain core during maturation (Conway et al., 2001; Wikoff et al., 2006). We estimate the accuracy of positioning for the core domain residue Cα coordinates to be ~2–3 Å (Baker and Johnson, 1996; Rossmann, 2000).
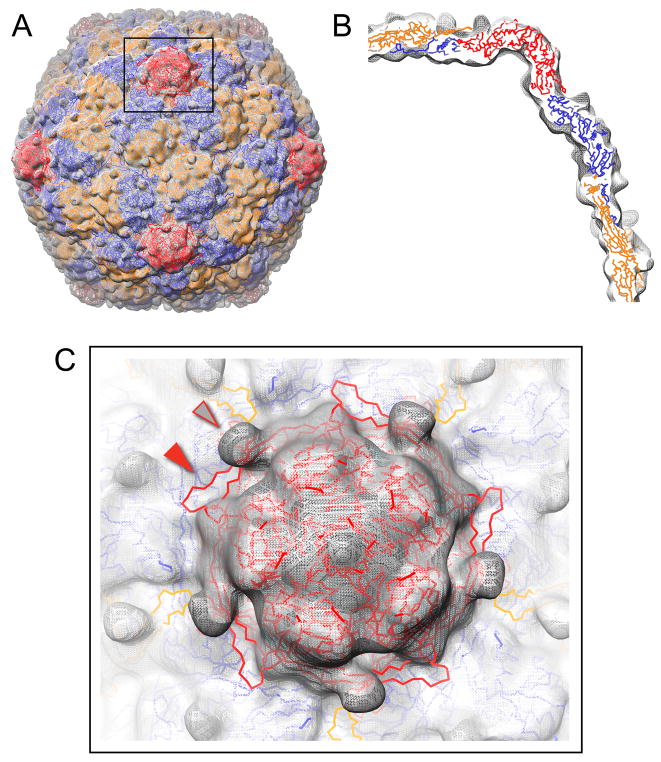
Figure 4. Pseudoatomic model of the EI-I/II particle.
(A) 3.4 Å Head-II subunit structures (backbone trace shown; A-C subunits orange, D-F subunits blue, G subunits red) were modeled as rigid bodies into a 14 Å cryo-EM reconstruction of minimally-crosslinked, WT EI-I particles (grey mesh). (B) Cross-sectional view of the pseudoatomic model fit into the EM density map. (C) Exterior view down a 5-fold axis of symmetry. In the model shown, the E-loops are in the crosslinked, down conformation found in Head-II (solid arrow), whereas the EM density for the un-crosslinked EI-I particles shows nubs of density that project outward from the surface for the E-loops in the uncrosslinked, up conformation (open arrow).
Compared to a previously reported model for the EI-I particle form that was based upon a 25 Å cryo-EM reconstruction, the current model exhibits a significantly better fit to the higher resolution 14 Å EM map, with real-space correlation coefficient of 0.88 versus 0.57 (Wikoff et al., 2006). This improvement is attributable to the increase in structural detail provided by the higher resolution EM reconstruction, which constrained the modeling and provided better “fiducials” for accurately determining subunit positions.
The SAXS pattern and the P(r) electron distance distribution plot for the EI-I/II pseudoatomic model were computed and found to be superimposable with the SAXS pattern measured for EI-I/II particles at pH 3.9 (Supplementary Fig. 2). Likewise, ab initio 3-D shape reconstruction based upon the measured SAXS data for EI-I/II particles showed good agreement with the pseudoatomic model for the EI-I/II particles. These results confirm the consistency of the cryo-EM model and the SAXS data.
As shown in Figure 5, the pseudoatomic model reveals a notable feature, centered at the 3-fold and quasi-3-fold vertices of the capsid. A suite of conserved interactions involving loops from the P-domains at this nexus has been identified in P-II and H-II particles (Gertsman et al., in preparation). Based upon the EI-I/II model, this set of interactions is maintained in the EI-I/II state, while the rest of the subunits surrounding the P-domain loops have rotated into conformations that are intermediate between P-II and H-II (Figure 5, Table 1). This suggests that the intersection of loops from the three P-domains acts as a hinge during maturation. The dihedral angles formed by the hinge base (first plane defined by the 3 N356 residues) and Glu-292 at the distal-most point of the subunits (second plane defined by Glu-292 and its two proximal-most N356 residues) has an average value of 151° ±7° among the 7 quasiequivalent subunits in P-II (Table 1 and Figure 5). In the EI-I/II state, the subunits are still angled, but somewhat less-so than in P-II, and less variation is observed among the quasiequivalent subunits with an average angle of 163°±4°. By contrast, the hinges in H-II are nearly planar, averaging 175°±4°.
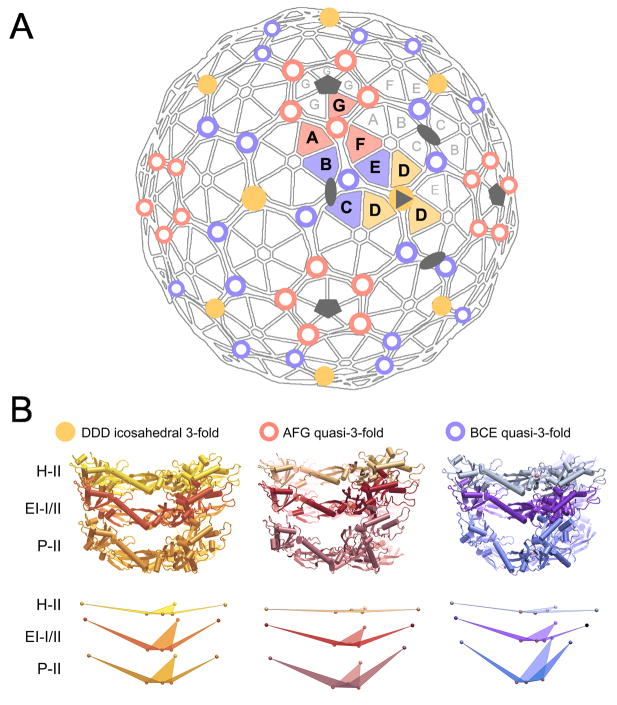
Figure 5. Hinges in HK97 capsid lattice.
(A) Distribution of the 3 quasiequivalent varieties of P-domain hinges in the T=7l capsid lattice (DDD yellow dot; AFG red circle; BCE blue circle). (B) In EI-I/II, the hinges are in conformations that are intermediate in structure between those observed in P-II and H-II. The crosslink residue, N356, is situated at the crux of the P-domain hinge. As depicted in the bottom row of (B), upon conversion from P-II to EI-I/II, the subunits hinge by up to 21°, and in converting from EI-I/II to H-II, subunits hinge by a further 10–16°, measured as the dihedral angle between the pivot base plane defined by 3 N356 residues and the plane defined by residue Glu-292 at the distal end of each subunit and the two proximal N356 residues.
Table 1. Subunit hinging at icosahedral 3-fold and quasi-3-fold axes in HK97 Prohead-II, Expansion Intermediate-I/II, and Head-II particles.
Dihedral angles measured between the hinge’s base plane defined by 3 N356 residues and the plane defined by residue Glu-292 at the distal end of each subunit and the subunit’s two proximal N356 residues.
| Hinge | Subunit | Subunit tilt relative to hinge base in P-II (EI-I/II vs P-II difference) | Subunit tilt relative to hinge base in EI-I/II (H-II vs EI-I/II difference) | Subunit tilt relative to hinge base in H-II |
|---|---|---|---|---|
| DDD | D | 158 (−1) | 157 (16) | 173 |
| BCE q3 | B | 148 (18) | 166 (12) | 178 |
| C | 139 (21) | 160 (10) | 170 | |
| E | 150 (16) | 166 (12) | 178 | |
| AFG q3 | A | 160 (7) | 167 (11) | 176 |
| F | 154 (13) | 167 (13) | 180 | |
| G | 149 (12) | 161 (12) | 173 | |
| Average | — | 151±7° | 163±4° | 175±4° |
Crosslink classes
The pseudoatomic EI-I/II model allows us to estimate the distances between the crosslinking residues (K169 and N356 Cα atom positions) in the capsid and possibly to infer which crosslinks form most readily in EI-I/II (Table 2). There are 7 quasiequivalent sets of crosslink sites in a T=7 structure, each set having 60 members distributed over 60 equivalent sites on the icosahedral capsid. The distances between Cα atoms for K169 and N356 at the 7 quasiequivalent positions fall into two discrete groups: for the A→A and E→D the distances are 4–5 Å; for the other 5 positions the distances are more than twice as large. We do not expect these values to be highly accurate, because the modeling was done using the E-loop conformations observed in H-II. However this is the most appropriate available structure for estimating residue positions when the E-loop is in the “down” crosslinked conformation. We believe that the two distance groupings of crosslink residues we see in the EI-II model may represent two broad classes of crosslink sites.
Table 2. Relative residue positions for the crosslink residues in the EI-I/II pseudoatomic model and in Head-II.
Estimated accuracy of coordinate positions ~2–3Å.
| K169 subunit | N356 subunit | Cα-Cα (Å) In EI-I/II model | Cα-Cα (Å) In H-II | |
|---|---|---|---|---|
| DDD 3-fold | E | D | 5 | 8 |
| BCE q3-fold | D | B | 12 | 8 |
| F | C | 10 | 8 | |
| C | E | 12 | 8 | |
| AFG q3-fold | A | A | 4 | 8 |
| G | F | 13 | 8 | |
| B | G | 12 | 8 |
Since ~60% of crosslinks have formed in the EI-II particles that exhibit the ladder pattern, we surmise that the majority of the crosslinks in the first class involving A→A and E→D (K169→N356 subunits) pairs, which have closer residue spacings most likely have formed in these particles. The A subunits are the subunits of hexons that immediately surround the pentons in the structure, and they crosslink with each other to form the 5-circles of subunits. Because the 5-circles are uniquely formed from the A subunits and the A→A crosslinks, biochemical detection of 5-circles serves as an indication that A→A crosslinking is nearing completion. In fact we find that 5-circles appear relatively early during in vitro crosslinking (Figure 2). These results strengthen the assertion that close spacing of the crosslink residues in the pseudoatomic model corresponds to the crosslink classes that form most readily. The E→D crosslinks, the other group that appears from the model to be likely to crosslink, surround the icosahedral 3-fold axis in the middle of the icosahedral face. The A→A and E→D crosslinks are not sufficient by themselves to give rise to the ladder of gel bands that we observe for the EI-II particles; there must also be some bonds formed in the other quasiequivalent sets of crosslinking sites. The larger spacing seen in the model for the other five quasiequivalent types of sites means that there is a low, but not necessarily zero, probability of close spacing and crosslink formation that is only rarely sampled in the EI-I/II state by the mobile surface E-loops.
Discussion
Capsids of dsDNA viruses such as bacteriophage HK97 assemble first as a compact, relatively fragile prohead. Maturation converts prohead into a more robust structure termed head with twice the volume, sufficient to contain the tightly packaged chromosome. During maturation, HK97 capsids form intersubunit covalent bonds that connect capsid proteins into closed, interlocking rings of 5 and 6 subunits and that make Head-II significantly more stable than Prohead-II (Ross et al., 2005). These intersubunit crosslinks begin to form only after particles have converted from P-II into the maturation intermediate, EI-I/II. This conversion is most likely a relaxation of the capsid lattice that, in vivo, occurs in response to packaging of the initial portions of the phage’s chromosome. As DNA packaging proceeds, the shell must expand further and strengthen to withstand the increasing internal pressure exerted by the DNA. The experiments presented here illuminate the relationships between conformational maturation of the capsid and formation of crosslinks, and they provide insight into the way the process is driven forward.
The HK97 variants, trpWT and ΔEloop, reveal that EI-I is the resting conformational state for the capsid lattice when the crosslinking machinery is not functional (Figure 3). If the crosslinking machinery is intact and functional, at neutral pH EI-I/II particles convert readily to Balloon and Head-II. The crosslink-defective K169Y mutant is still capable of maturing beyond EI-I; we argue that this is due to numerous noncovalent bonds that form between the E-loop and the capsid substrate including a putative hydrogen bond between Y169 and N356 that conserves the crosslinking connectivity observed between the two residue positions in WT (Gan et al., 2006). These noncovalent bonds preserve some degree of influence of the E-loop position on capsid maturation, albeit less effectively than the covalent crosslinking bond. In trypsinized WT by contrast, the E-loops have been clipped and are frayed, making it difficult for the arms of the E-loop to form the interactions that would lash them into the down position. The ΔEloop mutant that has 9 residues at the tip of the E-loops deleted, including K169 and others involved in interactions between the E-loop and capsid surface, shows complete inhibition of the EI-I to Balloon conversion. Thus, more severe modification of the E-loop-related crosslinking machinery correlates with more severe inhibition of the EI-I/II to Balloon conversion
Our findings are consistent with previous EM studies, in which WT particle populations showed substantial conversion to Balloon while the population of crosslink defective K169Y particles was heavily weighted towards the less mature EI-I/II morphology (Ross et al., 2005). It was suggested that crosslink formation in HK97 capsids biases the transition towards Balloon. Indeed our findings demonstrate that while particle structure does not change measurably in conjunction with crosslink accumulation up to formation of the ladder pattern (Figure 2), the greater the number of crosslinks that form in EI-I/II prior to the quaternary structural change, the more rapidly the population of particles convert to Balloon when exposed to neutral pH (Figure 3). The bimodal shift in crosslink patterns is simultaneous with the two-state quaternary change observed by SAXS, consistent with the hypothesis that the accumulation of crosslinks— up to the tipping point embodied in the ladder pattern— greatly facilitates the transition. The crosslinks therefore modulate the propensity of the capsid to undergo its structural reorganization by shifting the bias of the switch (Figure 6).
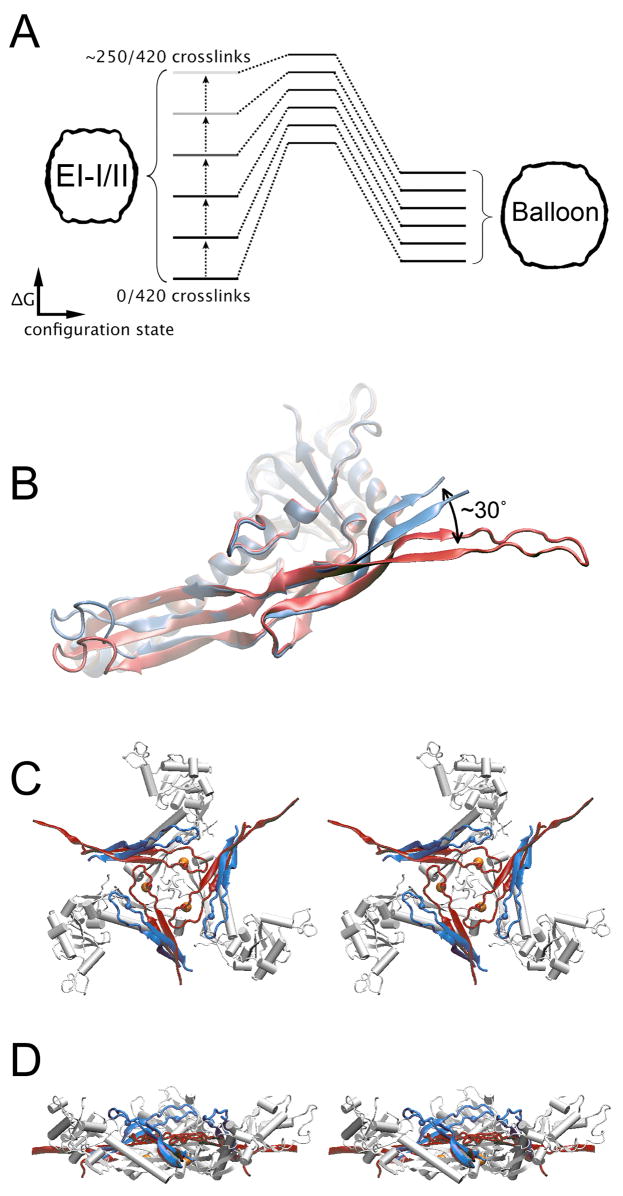
Figure 6. Crosslink-dependent E-loop bending spring-loads the 3-fold and quasi-3-fold hinges and switches the conformational balance towards Balloon.
(A) In the absence of crosslinks the EI-I/II quaternary organization appears to be the low energy, resting state for the HK97 capsid. Covalent crosslink formation shifts the switch bias towards the Balloon state. (B) E-loops are in an “up” position in the absence of crosslinks (P-II A subunit, blue). Crosslinking deflects the loops downward by ~30° (H-II A subunit, red). (C) Top view (stereo) of DDD hinge in the EI-I/II model. E-loops from adjacent E subunits whose K169 residues (red and blue spheres) will crosslink with the D-subunit N356 (yellow spheres) are shown in blue (un-crosslinked) and in red (crosslinked). Crosslinking deflects the E-loop from its resting position and the resulting strain may be transferred to the P-domain hinge via N356. A side view of the hinge is shown in (D).
Crosslinking and the capture of E-loops in the “down” conformation may potentiate the change in the forward direction by destabilizing EI-I/II particles. EM reconstructions of P-II and minimally crosslinked EI-I/II capsids indicate that the E-loops are highly mobile and vary in orientation about a preferred “up” position in which the β-strands make an oblique angle to the capsid shell (Conway et al., 2001; Ross et al., 2005). As each mobile E-loop is captured by crosslinking, it is deflected from its preferred equilibrium position to the “down” position in which the β-strands are held parallel to the capsid surface. We posit that capture of the mobile E-loops likely introduces tension into the capsid, raising the EI-I/II particle to a higher energy relative to the Balloon state (Figure 6). As crosslinks accumulate, the conformational balance is tipped to favor the more mature Balloon structure. In this scheme, the capture of each E-loop does not affect local structure, but it “ratchets” forward the overall tension in the capsid lattice.
The pseudoatomic model of EI-I/II (Figures 4, 5) supports the hypothesis that the tightly interacting P-domain loops involving AFG, DDD, and BCE subunit trimers are hinges about which subunits pivot as they transition from Prohead-II to the more mature Balloon and Head-II particles (Gertsman et al., in preparation). Notably, the covalent crosslinks between E-loop K169 and N356 residues form directly over the trigonal P-domain hinges (Figure 6). Hence, once a crosslink forms, the strain introduced by capturing the E-loop in the “down” position would be transferred directly to the hinge. Moreover, within each subunit, E-loops are directly linked to P-domains by the outgoing β-strand of the E-loop, which goes on to form one of 3 strands in the P-domain β-sheet (Figure 1A). The P-domain hinges are thus not only “spring-loaded” by the crosslink formation, but mechanically linked to one another as well. This linkage likely facilitates capsid expansion by coupling the hinges into an integrated network.
The 3-fold and quasi-3-fold vertices in other icosahedral capsids may also serve as hinges that facilitate capsid remodeling during maturation. As a result of their flexible nature, however, these hinges may be weak points in the lattice that must be reinforced, before the full pressure of the packaged DNA can be contained. In phage λ, the trimers of the gpD protein bind to positions on the T=7 lattice that are directly analogous to the crosslink sites in HK97 (Dokland and Murialdo, 1993; Lander et al., 2008). In T4 phage, a trimer of Soc proteins stabilizes the capsid by binding at the trigonal vertices where three gp23* hexons meet (Iwasaki et al., 2000). In herpes simplex viruses the triplex complex is situated at 3-fold vertices between hexons and plays an important role in mediating assembly and maturation (Heymann et al., 2003; Trus et al., 1996). Hence, while these structurally related viruses may lack covalent crosslinks, they employ accessory proteins to achieve a similar effect in bolstering the mature capsids.
EXPERIMENTAL PROCEDURES
Production of HK97 particles
All particles used in this study were initially purified as Prohead-II particles, following previously reported procedures (Duda et al., 1995b; Lee et al., 2004). P-II particles were stored in 20 mM Tris pH 7.5, 40 mM NaCl (Buffer A).
In the ΔEloop truncation mutant, the amino acid sequence NADVVAEKALKPE in the E-loop of gp5 was replaced with APGD, a sequence that has a propensity to form reverse turns (Dyson et al., 1998). The mutant was constructed by PCR amplifications using the strategy of splicing by overlap extension (Horton et al., 1989). PCR conditions are provided in Supplementary Information.
Trypsinized WT (trpWT) P-II particles were produced by starting with an aliquot of WT P-II particles. The particle solution was diluted to 1 mg/ml in Buffer A and trypsin was added to a final concentration of 0.02 mg/ml. The digestion proceeded for 1 h at 37 °C and was terminated by addition of PMSF (to 1 mM). The proteolyzed P-II particles were purified from the trypsin digestion mixture by anion exchange chromatography in the presence of 1 mM PMSF. The trpWT P-II fractions were then concentrated and buffer exchanged by high-speed pelleting twice at 148,000× g for 2 h at 4 °C. Pelleted particles were resuspended in Buffer A. Trypsinization of P-II particles yields cleavage of a single site, following K166 on the E-loop (Duda et al., 1995a). The efficiency of proteolysis in our trypsinization reaction was assessed by SDS-PAGE.
Solution small-angle x-ray scattering
Solution scattering data were recorded at the SSRL Beam Line 4–2. A bent cylinder mirror focused the beam onto the detector position, and a pair of Si(111) crystals was used as a monochromator. X-ray wavelength was 1.38 Å in these experiments. SPEAR operating parameters, beamline configuration, and detector instrumentation and calibration methods were identical to those used in previously reported experiments (Lee et al., 2005) except where noted. The sample-to-detector distance was 2.5 m. Radial averaging, intensity scaling and background subtraction were performed by MarParse (Smolsky et al., 2007), and the small level of non-uniformity of detector response was corrected using an in-house program.
A polycarbonate sample cell with mica windows was used for both static and time-resolved measurements. Sample temperature was held at 20 °C. Static SAXS measurements were taken for P-II, EI-II particles that had been acidified for more than 72 h, EI-IV Balloon and H-II; matching blank solutions were Buffer A for P-II and H-II, and 250mM KCl, 50 mM Na-citrate pH 3.9 acidification buffer for EI-II and EI-IV particles. Particle concentrations were 2–10 mg/ml. No concentration dependent effects were observed in the SAXS measurements.
Reneutralization experiments were performed by diluting concentrated stock solutions of P-II at least 10× into acidification buffer, and then adding one-sixth volume of 1 M Tris-HCl, pH 8.3 such that the final solution contained ~5 mg/ml particles and had a pH of 7.5. WT particles were acidified for 40 min, 2 h, or more than 72 h, in order to generate EI-I/II particles with minimal crosslinks, ~25%, and ~60% of the total complement of crosslinks, respectively. Each of these samples was then reneutralized in identical fashion. The same method of acidification followed by reneutralization was performed with K169Y, trpWT, and ΔEloop particles. Following reneutralization, a series of 5 s exposures at intervals ranging between 30 and 60 s were gathered for WT experiments. Due to the slower maturation kinetics, K169Y measurements were taken at 30–600 s intervals. A 60 s dead time preceded the first measurements due to the time required to mix the sample, load the sample cell, and reset the beamline hutch interlock.
Singular Value Decomposition of time-resolved SAXS data was carried out as described previously (Lee et al., 2005).
Time course of crosslink formation assayed by SDS PAGE
Crosslink formation was assayed under conditions that paralleled those present in the time-resolved SAXS experiments, with the exception of lower protein concentration in the SDS-PAGE assays. A solution of 77 mg/ml WT P-II was diluted 350-fold into 250 mM KCl, 50mM citrate, pH 3.9 buffer and incubated for 40 min at room temperature to convert the population of particles to EI-I. At 40 min, the entire EI-I solution volume was reneutralized by addition of one-sixth volume of 1 M Tris-HCl, pH 8.3. Under these conditions, the population takes tens of minutes to fully convert to H-II. Aliquots of the reneutralized particle solution were taken at 0.25–60 minute intervals. At each timepoint, an aliquot was TCA-precipitated, washed with acetone, and the sample pellet was stored at −20 °C. Once all of the timepoints were collected, each pellet was resuspended in SDS load buffer and analyzed by SDS-PAGE using a low-crosslink acrylamide gel (Duda, 1998). P-II, EI-IV and H-II standards were processed similarly.
Pseudoatomic model refinement
Previous studies in which HK97 crystal structures were docked into moderate-resolution cryo-EM maps found that the most physically reasonable model was obtained if the subunit coordinates were first converted to poly-alanine coordinates; this in effect accounts for the repacking of interfacial side chains (Gan et al., 2006). Thus, in composing the pseudoatomic model for the EI-I/II particle the refined H-II subunit coordinates (Helgstrand et al., 2003) were converted to poly-alanine coordinates and manually fitted into a 14 Å resolution cryo-EM map of minimally crosslinked EI-I (Ross et al., 2005) using the program “O” (Jones et al., 1991). Structure factors were then calculated from the map and the phases were used as the experimental target with the program CNS. Rigid body refinement of the subunit core (A and P domains treated as a single entity) positions was done using data up to 25 Å resolution and then extended to 14 Å resolution. The E-loops (residues 155–175) and N-arms (residues 104–125) were not included in the first round of rigid body refinement because these subdomains exhibit conformational variability during maturation (Benevides et al., 2004; Lee et al., 2004; Szymczyna et al., 2007).
The resulting model and experimental map had a real-space correlation coefficient (RSCC) of 0.80. The model was used to mask out just the capsid electron density from the experimental map, which has noise in the solvent regions. This masked map was used to calculate new structure factors to 14Å resolution and subjected to refinement as above. The resulting model exhibited somewhat disjointed interfacial packing with subunits offset from each other, particularly at the 3-fold and quasi-3-fold axes of symmetry. The refinement procedure was repeated over a range of magnifications of the EM map, and an optimal fit was found for a 1.3% magnification. This magnitude of magnification uncertainty is not uncommon in image reconstructed cryo-EM density maps (Rossmann, 2000). The resulting model and experimental map had an RSCC of 0.85. The SAXS pattern calculated from this model also exhibited better agreement with the measured SAXS for EI-I/II particles (Supplementary Fig. 2).
In a final round of refinement, the N-arms (residues 104–125) were manually repositioned into knobs of density on the internal surface of the capsid and then treated as independent entities during rigid-body refinement. This did not result in gross changes to the core domain positions but improved the model’s fit to the EM map, with an RSCC of 0.88.
When the E-loops (residues 155–175) were fit as independent entities during refinement, they partially occupied the nubs of density in the EM map, which project outward from the surface and which have been attributed to the two-stranded β-sheet stems of uncrosslinked E-loops, whose outer tips are highly mobile (Ross et al., 2005). These nubbins of density do not, however, allow E-loop orientations to be uniquely determined.
Supplementary Material
Acknowledgments
We thank Ilya Gertsman (TSRI) for providing the Prohead-II PDB coordinates used in our analysis. This work was supported by National Institutes of Health funding R01AI040101 (JEJ) R01GM047795 (RWH), F32GM65013 (KKL), and supported in part (ACS) by the Intramural Research Program of NIAMS. SAXS experiments were carried out at SSRL BL4-2, a national facility operated by Stanford University on behalf of the U.S. Dept. of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the DoE, Office of Biological and Environmental Research, and by the NIH, National Center for Research Resources, Biomedical Technology Program. The content is solely the responsibility of the authors and does not necessarily reflect the official views of NCRR or NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agirrezabala X, Velazquez-Muriel JA, Gomez-Puertas P, Scheres SH, Carazo JM, Carrascosa JL. Quasi-atomic model of bacteriophage t7 procapsid shell: insights into the structure and evolution of a basic fold. Structure. 2007;15:461–472. doi: 10.1016/j.str.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Baker ML, Jiang W, Rixon FJ, Chiu W. Common ancestry of herpesviruses and tailed DNA bacteriophages. J Virol. 2005;79:14967–14970. doi: 10.1128/JVI.79.23.14967-14970.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker TS, Johnson JE. Low resolution meets high: towards a resolution continuum from cells to atoms. Current opinion in structural biology. 1996;6:585–594. doi: 10.1016/s0959-440x(96)80023-6. [DOI] [PubMed] [Google Scholar]
- Belnap DM, Filman DJ, Trus BL, Cheng N, Booy FP, Conway JF, Curry S, Hiremath CN, Tsang SK, Steven AC, Hogle JM. Molecular tectonic model of virus structural transitions: the putative cell entry states of poliovirus. J Virol. 2000;74:1342–1354. doi: 10.1128/jvi.74.3.1342-1354.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benevides JM, Bondre P, Duda RL, Hendrix RW, Thomas GJ., Jr Domain structures and roles in bacteriophage HK97 capsid assembly and maturation. Biochemistry. 2004;43:5428–5436. doi: 10.1021/bi0302494. [DOI] [PubMed] [Google Scholar]
- Buck CB, Thompson CD, Pang YY, Lowy DR, Schiller JT. Maturation of papillomavirus capsids. J Virol. 2005;79:2839–2846. doi: 10.1128/JVI.79.5.2839-2846.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canady MA, Tihova M, Hanzlik TN, Johnson JE, Yeager M. Large conformational changes in the maturation of a simple RNA virus, nudaurelia capensis omega virus (NomegaV) Journal of molecular biology. 2000;299:573–584. doi: 10.1006/jmbi.2000.3723. [DOI] [PubMed] [Google Scholar]
- Conway JF, Duda RL, Cheng N, Hendrix RW, Steven AC. Proteolytic and conformational control of virus capsid maturation: the bacteriophage HK97 system. Journal of molecular biology. 1995;253:86–99. doi: 10.1006/jmbi.1995.0538. [DOI] [PubMed] [Google Scholar]
- Conway JF, Wikoff WR, Cheng N, Duda RL, Hendrix RW, Johnson JE, Steven AC. Virus maturation involving large subunit rotations and local refolding. Science. 2001;292:744–748. doi: 10.1126/science.1058069. [DOI] [PubMed] [Google Scholar]
- Dokland T, Murialdo H. Structural transitions during maturation of bacteriophage lambda capsids. Journal of molecular biology. 1993;233:682–694. doi: 10.1006/jmbi.1993.1545. [DOI] [PubMed] [Google Scholar]
- Duda RL. Protein chainmail: catenated protein in viral capsids. Cell. 1998;94:55–60. doi: 10.1016/s0092-8674(00)81221-0. [DOI] [PubMed] [Google Scholar]
- Duda RL, Hempel J, Michel H, Shabanowitz J, Hunt D, Hendrix RW. Structural transitions during bacteriophage HK97 head assembly. Journal of molecular biology. 1995a;247:618–635. doi: 10.1006/jmbi.1995.0168. [DOI] [PubMed] [Google Scholar]
- Duda RL, Martincic K, Xie Z, Hendrix RW. Bacteriophage HK97 head assembly. FEMS Microbiol Rev. 1995b;17:41–46. doi: 10.1111/j.1574-6976.1995.tb00186.x. [DOI] [PubMed] [Google Scholar]
- Dyson HJ, Bolinger L, Feher VA, Osterhout JJ, Jr, Yao J, Wright PE. Sequence requirements for stabilization of a peptide reverse turn in water solution--proline is not essential for stability. European journal of biochemistry/FEBS. 1998;255:462–471. doi: 10.1046/j.1432-1327.1998.2550462.x. [DOI] [PubMed] [Google Scholar]
- Effantin G, Boulanger P, Neumann E, Letellier L, Conway JF. Bacteriophage T5 structure reveals similarities with HK97 and T4 suggesting evolutionary relationships. Journal of molecular biology. 2006;361:993–1002. doi: 10.1016/j.jmb.2006.06.081. [DOI] [PubMed] [Google Scholar]
- Fokine A, Leiman PG, Shneider MM, Ahvazi B, Boeshans KM, Steven AC, Black LW, Mesyanzhinov VV, Rossmann MG. Structural and functional similarities between the capsid proteins of bacteriophages T4 and HK97 point to a common ancestry. Proc Natl Acad Sci U S A. 2005;102:7163–7168. doi: 10.1073/pnas.0502164102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan L, Conway JF, Firek BA, Cheng N, Hendrix RW, Steven AC, Johnson JE, Duda RL. Control of crosslinking by quaternary structure changes during bacteriophage HK97 maturation. Mol Cell. 2004;14:559–569. doi: 10.1016/j.molcel.2004.05.015. [DOI] [PubMed] [Google Scholar]
- Gan L, Speir JA, Conway JF, Lander G, Cheng N, Firek BA, Hendrix RW, Duda RL, Liljas L, Johnson JE. Capsid conformational sampling in HK97 maturation visualized by X-ray crystallography and cryo-EM. Structure. 2006;14:1655–1665. doi: 10.1016/j.str.2006.09.006. [DOI] [PubMed] [Google Scholar]
- Helgstrand C, Wikoff WR, Duda RL, Hendrix RW, Johnson JE, Liljas L. The refined structure of a protein catenane: the HK97 bacteriophage capsid at 3.44 A resolution. Journal of molecular biology. 2003;334:885–899. doi: 10.1016/j.jmb.2003.09.035. [DOI] [PubMed] [Google Scholar]
- Hendrix RW. Bacteriophage HK97: assembly of the capsid and evolutionary connections. Adv Virus Res. 2005;64:1–14. doi: 10.1016/S0065-3527(05)64001-8. [DOI] [PubMed] [Google Scholar]
- Hewat EA, Blaas D. Cryoelectron microscopy analysis of the structural changes associated with human rhinovirus type 14 uncoating. J Virol. 2004;78:2935–2942. doi: 10.1128/JVI.78.6.2935-2942.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heymann JB, Cheng N, Newcomb WW, Trus BL, Brown JC, Steven AC. Dynamics of herpes simplex virus capsid maturation visualized by time-lapse cryo-electron microscopy. Nat Struct Biol. 2003;10:334–341. doi: 10.1038/nsb922. [DOI] [PubMed] [Google Scholar]
- Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene. 1989;77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- Iwasaki K, Trus BL, Wingfield PT, Cheng N, Campusano G, Rao VB, Steven AC. Molecular architecture of bacteriophage T4 capsid: vertex structure and bimodal binding of the stabilizing accessory protein, Soc. Virology. 2000;271:321–333. doi: 10.1006/viro.2000.0321. [DOI] [PubMed] [Google Scholar]
- Jardine PJ, Coombs DH. Capsid expansion follows the initiation of DNA packaging in bacteriophage T4. Journal of molecular biology. 1998;284:661–672. doi: 10.1006/jmbi.1998.2179. [DOI] [PubMed] [Google Scholar]
- Jiang W, Chang J, Jakana J, Weigele P, King J, Chiu W. Structure of epsilon15 bacteriophage reveals genome organization and DNA packaging/injection apparatus. Nature. 2006;439:612–616. doi: 10.1038/nature04487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Li Z, Zhang Z, Baker ML, Prevelige PE, Jr, Chiu W. Coat protein fold and maturation transition of bacteriophage P22 seen at subnanometer resolutions. Nat Struct Biol. 2003;10:131–135. doi: 10.1038/nsb891. [DOI] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Lander G, Evilevitch A, Jeembaeva M, Potter C, Carragher B, Johnson JE. Bacteriophage lambda stabilization by auxiliary protein gpD: timing, location, and mechanism of attachment determined by cryoEM. Structure. 2008 doi: 10.1016/j.str.2008.05.016. accepted for publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lata R, Conway JF, Cheng N, Duda RL, Hendrix RW, Wikoff WR, Johnson JE, Tsuruta H, Steven AC. Maturation dynamics of a viral capsid: visualization of transitional intermediate states. Cell. 2000;100:253–263. doi: 10.1016/s0092-8674(00)81563-9. [DOI] [PubMed] [Google Scholar]
- Lee KK, Gan L, Tsuruta H, Hendrix RW, Duda RL, Johnson JE. Evidence that a local refolding event triggers maturation of HK97 bacteriophage capsid. Journal of molecular biology. 2004;340:419–433. doi: 10.1016/j.jmb.2004.05.008. [DOI] [PubMed] [Google Scholar]
- Lee KK, Tsuruta H, Hendrix RW, Duda RL, Johnson JE. Cooperative reorganization of a 420 subunit virus capsid. Journal of molecular biology. 2005;352:723–735. doi: 10.1016/j.jmb.2005.07.024. [DOI] [PubMed] [Google Scholar]
- Morais MC, Choi KH, Koti JS, Chipman PR, Anderson DL, Rossmann MG. Conservation of the capsid structure in tailed dsDNA bacteriophages: the pseudoatomic structure of phi29. Mol Cell. 2005;18:149–159. doi: 10.1016/j.molcel.2005.03.013. [DOI] [PubMed] [Google Scholar]
- Ross PD, Cheng N, Conway JF, Firek BA, Hendrix RW, Duda RL, Steven AC. Crosslinking renders bacteriophage HK97 capsid maturation irreversible and effects an essential stabilization. Embo J. 2005;24:1352–1363. doi: 10.1038/sj.emboj.7600613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossmann MG. Fitting atomic models into electron-microscopy maps. Acta crystallographica. 2000;56:1341–1349. doi: 10.1107/s0907444900009562. [DOI] [PubMed] [Google Scholar]
- Smolsky IL, Liu P, Niebuhr M, Ito K, Weiss TM, Tsuruta H. Biological small-angle X-ray scattering facility at the Stanford Synchrotron Radiation Laboratory. J Appl Cryst. 2007;40:s453–s458. [Google Scholar]
- Steven AC, Heymann JB, Cheng N, Trus BL, Conway JF. Virus maturation: dynamics and mechanism of a stabilizing structural transition that leads to infectivity. Current opinion in structural biology. 2005;15:227–236. doi: 10.1016/j.sbi.2005.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymczyna BR, Gan L, Johnson JE, Williamson JR. Solution NMR studies of the maturation intermediates of a 13 MDa viral capsid. Journal of the American Chemical Society. 2007;129:7867–7876. doi: 10.1021/ja071118j. [DOI] [PubMed] [Google Scholar]
- Trus BL, Booy FP, Newcomb WW, Brown JC, Homa FL, Thomsen DR, Steven AC. The herpes simplex virus procapsid: structure, conformational changes upon maturation, and roles of the triplex proteins VP19c and VP23 in assembly. Journal of molecular biology. 1996;263:447–462. doi: 10.1016/s0022-2836(96)80018-0. [DOI] [PubMed] [Google Scholar]
- Turner BG, Summers MF. Structural biology of HIV. Journal of molecular biology. 1999;285:1–32. doi: 10.1006/jmbi.1998.2354. [DOI] [PubMed] [Google Scholar]
- Wikoff WR, Conway JF, Tang J, Lee KK, Gan L, Cheng N, Duda RL, Hendrix RW, Steven AC, Johnson JE. Time-resolved molecular dynamics of bacteriophage HK97 capsid maturation interpreted by electron cryo-microscopy and X-ray crystallography. J Struct Biol. 2006;153:300–306. doi: 10.1016/j.jsb.2005.11.009. [DOI] [PubMed] [Google Scholar]
- Wikoff WR, Liljas L, Duda RL, Tsuruta H, Hendrix RW, Johnson JE. Topologically linked protein rings in the bacteriophage HK97 capsid. Science. 2000;289:2129–2133. doi: 10.1126/science.289.5487.2129. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.