Abstract
We have used molecular dynamics simulations to study the structure and dynamics of a range of DNA duplexes containing the 2,6-diamino-4-hydroxy-5-formamidopyrimidine (FapydG) lesion that can result from oxidative damage at guanine. Compared to the corresponding undamaged DNA duplexes, FapydG-containing duplexes show little gross structural changes—the damaged base remains stacked in to the DNA double helix and retains hydrogen bonds to its cytosine partner. However, the experimentally observed reduction in DNA stability that accompanies lesion formation can be explained by a careful energetic analysis of the simulation data. Irrespective of the nature of the base pairs on either side of the lesion site, conversion of a guanine to a FapydG base results in increased dynamical flexibility in the base (but not in the DNA as a whole) that significantly weakens its hydrogen-bonding interactions. Surprisingly, the stacking interactions with its neighbours are not greatly altered. The formamido group adopts a non-planar conformation that can interact significantly and in a sequence-dependent manner with its 3′-neighbour. We conclude that the recognition of FapydG lesions by the repair protein formamidopyrimidine-DNA glycosylase probably does not involve the protein capturing an already-extrahelical FapydG base, but rather it relies on detecting alterations to the DNA structure and flexibility created by the lesion site.
Keywords: DNA; 2,6-diamino-4-hydroxy-5-formamidopyrimidine; formamidopyrimidine; molecular dynamics; simulation; molecular mechanics/generalized Born surface area
1. Introduction
DNA suffers oxidative damage when exposed to reactive oxygen species (ROS), which include the highly reactive hydroxyl radical (·OH), superoxide radical (O2·−) and non-radical hydrogen peroxide (H2O2). Cell metabolism, UV radiation and chemicals can all increase the level of ROS in cells leading to the creation of several oxidation products such as 5-hydroxycytosine, thymine glycol, 8-oxopurine and formamidopyrimidine lesions. Of these lesions, oxidative guanine damage is the most extensively studied because it is easily measured (Marnett 2000) and guanine is the most easily oxidized among the DNA bases owing to its lowest oxidation potential (Steenken & Jovanovic 1997; Burrows & Muller 1998).
2,6-Diamino-4-hydroxy-5-formamidopyrimidine (FapydG) is one of the most prevalent guanine-derived lesions formed under O2-deficient conditions by ionizing radiation and other agents that produce ROS (Douki et al. 1997; Pouget et al. 2000; Crespo-Hernandez & Arce 2004). This ring-opened lesion has received less attention than the well-documented 8-oxo-7,8-dihydroguanine (8OG) lesion, which is often regarded as the chief guanine oxidation product developed under aerobic conditions. However, a number of studies have revealed that FapydG is detected at equal or greater yields than 8OG after oxidative stress (Dizdaroglu et al. 1993; Mori et al. 1993; Kasprzak et al. 1997; Pouget et al. 2000; Hu et al. 2005). It has been documented that FapydG induces misincorporation of A opposite it in vitro resulting in G:C→T:A transversion mutations (Wiederholt & Greenberg 2002) and might also block the progression of DNA polymerases (Tudek 2003). A recent study of the relationship between the frequency of occurrence and the mutagenic potential of some important DNA lesions has demonstrated that FapydG appears to be the most ubiquitous lesion associated with high mutagenicity in DNA (Wilson & Bohr 2007).
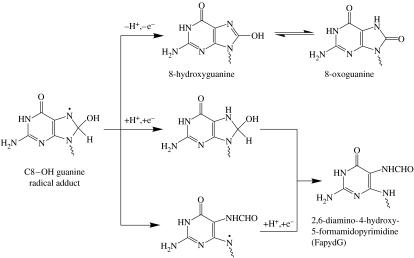
FapydG is usually generated through an addition of a hydroxyl radical (·OH) at the C8 position of guanine giving the C8–OH adduct radical (Evans et al. 2004), as illustrated in figure 1. Subsequent cleavage of the C8–N9 bond of the imidazole ring and one-electron reduction gives rise to FapydG whereas an oxidation of the adduct yields 8OG. The C8–OH adduct radical may also undergo the reduction prior to ring cleavage leading to the formation of 7,8-dihydro-8-hydroxyguanine, which is then transformed to yield FapydG.
Figure 1.
Formation of FapydG and 8OG through the C8–OH adduct radical of guanine in the absence of oxygen. Adapted from Evans et al. (2004).
Although the formation of both FapydG and 8OG lesions is through the same intermediate as shown in figure 1, the remarkable changes in the structure of FapydG from the original guanine base give rise to greatly increased conformational flexibility of the N-glycosidic bond and formamide group, generating fascinating questions concerning the effects of this lesion on DNA structure, and on lesion recognition by repair systems. Unfortunately, the first problem in studying FapydG experimentally is the difficulty in producing pure oligonucleotides containing a β-FapydG lesion, which is assumed to be the naturally occurring anomer in the DNA duplex (Patro et al. 2004). Chemically synthesized β-FapydG tends to rapidly anomerize to give α-FapydG under conditions required for DNA synthesis (Haraguchi & Greenberg 2001). Thus, there has been relatively few experimental studies on FapydG itself; however, two FapydG analogues have been produced, which can be inserted site specifically into DNA as the pure β-anomer.
The first analogue is a non-hydrolysable FapydG with a C-glycosidic bond (β-C-FapydG) (Delaney & Greenberg 2002). This β-C-nucleoside analogue is effectively an inhibitor of formamidopyrimidine-DNA glycosylase (Fpg) since Fpg can tightly bind to DNA containing β-C-FapydG pairing to C in vitro but is unable to excise the damage (Wiederholt et al. 2003). Another analogue is a carbocyclic FapydG (cFapydG), which contains a cyclopentane ring instead of the 2′-deoxyribose sugar (Ober et al. 2003). The crystal structure of a bacterial Fpg enzyme, LlFpg from Lactococcus lactis, bound to a cFapydG-containing duplex has been solved by Coste et al. (2004). This noteworthy study provides us with a structural basis for recognition of FapydG by Fpg, which is a prime step in the base excision repair. The structure is deposited in the Protein Data Bank (PDB) as two PDB entries, 1TDZ (solved at 1.80 Å resolution with a missing loop from residues 220–224) and the complete model with 1.95 Å resolution (PDB code 1XC8, released 1 year after 1TDZ). A key feature of the structure of the complex is that the lesion base is flipped out of the DNA double helix into the active site of Fpg. This obviously raises the question as to whether the protein captures an already-extrahelical lesion, or whether the ‘flipping’ event is an integral part of the recognition process.
Measuring the effects of DNA damage on its structure and stability is clearly of vital importance if we are to understand issues such as mutagenic potential and lesion detection and repair. Molecular modelling provides a variety of methods to do this, of varying complexity, speed and accuracy. Molecular dynamics (MD) simulations are a particularly valuable approach to the study of DNA structure and dynamics (Cheatham & Kollman 2000; Cozmuta & Mehrez 2007), but by themselves allow a limited exploration of energetic factors. The MM/GB(PB)SA method combines a molecular mechanics (MM) analysis of solute enthalpy with either a generalized Born (GB) or a Poisson–Boltzmann (PB) continuum model for solvation free energy, and a solvent accessibility (SA) correction for non-polar solvent–solute interactions (Srinivasan et al. 1998; Kollman et al. 2000). Because solvent-associated terms are handled using continuum (implicit) approaches, it is a particularly convenient method to use for the estimation of binding free energies. The MM/GB(PB)SA method has been used to study relative stabilities of A- and B-form helices of nucleic acids (Srinivasan et al. 1998) and widely applied to estimate the binding affinities of small molecules binding to biomolecules (Kollman et al. 2000; Massova & Kollman 2000; Wang & Laughton 2007), also in protein–protein (Gohlke & Case 2004) and protein–DNA interactions (Song et al. 2006). The PB model is theoretically more exact than the GB model but is considerably more computationally expensive; in practice, a well-parametrized GB model appears to perform as well (Wang & Laughton 2007).
In this study, the aim was to identify the initial molecular perturbation to the normal structure of duplex DNA brought about by the presence of FapydG, which may trigger the damage recognition by Fpg. The influence of FapydG on DNA stability compared to its normal equivalent was studied using an MD simulation technique. Prior to performing MD simulations, a potential structure of an intrahelical FapydG with an appropriate force-field parameter set was first developed based on both X-ray crystal and NMR structures available to date plus theoretical methods (given in the electronic supplementary material). Unrestrained MD simulations were performed on a series of dodecamer DNA duplexes containing an anti-FapydG or G residue paired with cytosine in the Watson–Crick-like hydrogen-bonding pattern (figure 2). The simulations were analysed to study the stability of the duplexes using the molecular mechanics/generalized Born surface area (MM/GBSA) approach, and whether the FapydG lesion might potentially reveal itself to repair processes through local alterations in DNA structure or dynamics.
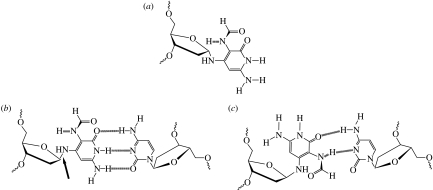
Figure 2.
(a) The α-anomer of FapydG; (b) base pairing of β-anti-FapydG with C in a Watson–Crick-like hydrogen-bonding pattern (the anti-glycosidic bond is arrowed); (c) alternative hydrogen-bonding scheme for a syn-FapydG:C base pair.
2. Methods
2.1 Simulation methods
All simulations were performed using the Amber 8 suite of programs (Case et al. 2004). An initial 12-mer B-DNA duplex d(CTTTTGCAAAAG)2, termed TGC, was generated using the nucgen module. The dynamics of this duplex have been extensively studied in previous work (Harris et al. 2001). It was shown that the sequence remained stable over the 5 ns simulations and its time-averaged structure was in excellent agreement with the NMR-derived structures. Parametrization of FapydG was based on extensive quantum mechanical calculations at the HF/6-31G* level, paying particular attention to the energy barriers for rotations of the formamido group. The parametrized FapydG was then incorporated into the TGC sequence to give d(CTTTTFCAAAAG)·d(CTTTTGCAAAAG), termed TFC, where F was FapydG; an anti-conformation (240°) of FapydG was employed based on the NMR structure of the formamidopyrimidine adduct of aflatoxin B1 (Mao et al. 1998). In preliminary experiments, an alternative, syn, conformation of the base (Ober et al. 2005) was also investigated using the MMGBSA method (see below) but found to be approximately 10 kcal mol−1 less stable than the anti-form, into which it spontaneously converted after 500 ps of MD (results not shown). To check if the conformational preferences and stability of the FapydG base were influenced by its neighbouring bases, the original model systems, featuring a TXC sequence (X is G or FapydG), were modified (using insight II) to give AXA-, AXC-, TXA-, AXG- and TXT-containing sequences. These sequences were chosen to provide at least one example each of the lesion base being flanked by two purines, two pyrimidines, or both combinations of one purine and one pyrimidine.
All models were then solvated with an explicit TIP3P water model (Jorgensen et al. 1983) in a truncated octahedral box with a minimum 10 Å distance between any solute atom and a box edge. Periodic boundary conditions (PBC) were applied. The systems were neutralized with a minimum number of potassium ions. In this study, potassium was preferred to sodium since it may be regarded as the more relevant ion within the environment of the nucleus and there are some reports that in long MD simulations sodium ions can congregate in the DNA minor groove and distort it, even at ionic strengths well below those that under experimental conditions are observed to promote DNA condensation (Várnai & Zakrzewska 2004).
Initially constructed systems were energy minimized in two stages (first just the solvent and ions, then all atoms). Equilibration was performed using our multi-step protocol for gentle heating and harmonic restraint reduction over a period of 90 ps (Shields et al. 1998), and a 100 ps unrestrained equilibration was subsequently done. A production phase of the unrestrained MD was carried out at constant temperature (300 K) and pressure (1 atm) using the Berendsen algorithm (Berendsen et al. 1984) to control the simulation temperature by readjusting the atomic velocities. An integration time step of 2 fs was used and all bond lengths involving hydrogen were constrained using Shake (Ryckaert et al. 1977). The particle mesh Ewald (PME) approach (Darden et al. 1993) was used to estimate long-range electrostatic interactions with a direct-space non-bonded cut-off of 9 Å. Trajectories, which contain coordinates and velocities of each atom with respect to time, were saved every 2 ps over the course of 5 ns production MD for further analyses. All simulations were carried out using the Sander module.
2.2 Energetic analysis
2.2.1 Relative binding energies
To investigate how conversion of a guanine base to FapydG affects its interactions with the surrounding DNA structure, we treat the system as a ligand/receptor complex, in which the targeted nucleotide is the ligand and the rest of the solute is the receptor. In this way, it is amenable to analysis using the MM/GBSA approach to calculate the free energies of molecules (Gx) in solution (see equation (2.1); Srinivasan et al. 1998; Kollman et al. 2000). The molecular mechanical energies, the van der Waals interaction (EvdW); electrostatic interaction (Ees); and internal energy (Ein), as well as the free energy of GB solvated systems (Gpol) are calculated with the Sander program. For Gpol we use the GB solvent model developed by Onufriev, Basford and Case (GBOBC; Onufriev et al. 2000). The hydrophobic contribution to the solvation free energy (Gnonpol) is estimated using the solvent accessible surface area of the solute (Sitkoff et al. 1994) and equation (2.2) (Still et al. 1990),
| (2.1) |
The binding energies of guanine and FapydG to their ‘receptors’ are calculated in the usual way (equation (2.3)), averaging over snapshots extracted from the simulations every 50 ps. However, individually these values are artefactual since they include an internal energy term associated with the cleavage of the bonds between the ‘ligand’ base and those to its 5′- and 3′-sides. However, the relative free energies of binding are meaningful since this term will cancel out (equation (2.4)),
| (2.2) |
| (2.3) |
and
| (2.4) |
2.2.2 Intra- and inter-strand interactions
Intra- and inter-strand interactions were calculated over each 5 ns simulation using the Amber anal module to calculate stacking energies and base pair hydrogen-bonding energies. For these calculations the DNA backbones were ignored, the charge on C1′ was therefore adjusted to maintain a total neutral charge in each base, and van der Waals' parameters were applied for all atoms except C1′. The stacking energy calculations take into consideration interactions with the flanking bases on the same strand, but not cross-strand stacking interactions, which are very minor.
3. Results and discussion
3.1 General MD results
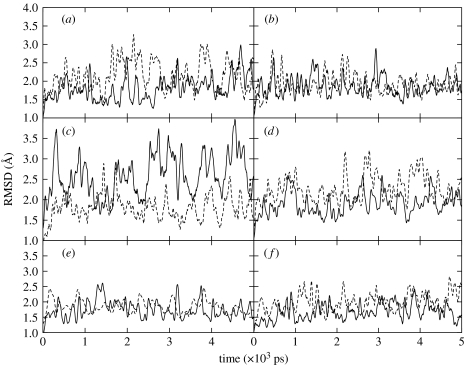
Nucleobases of FapydG or guanine paired with cytosine in Watson–Crick geometries were incorporated in the central AXA, AXC, AXG, TXA, TXC and TXT sequences. The RMSD time series of each duplex, which provides a measure of conformational changes with respect to the initial conformation, are all stable, but seem to fluctuate more broadly in the presence of FapydG, as depicted in figure 3. The average RMSD for each sequence is shown in table 1. DNA structures containing FapydG lesions tend to deviate slightly more from the canonical B-form starting structure than their normal (G) counterparts, but this is not always the case, and visual inspection of the structures does not reveal any consistent type of structural change accompanying FapydG incorporation. We discuss the significance of this in more detail below. No tendency for base opening was detected in any of the 5 ns simulations.
Figure 3.
RMSD plots of damaged (X=F, dashed curve) and undamaged (X=G, solid curve) sequences compared to the starting structure as a function of simulation time. (a) AXA, (b) AXC, (c) AXG, (d) TXA, (e) TXC and (f) TXT.
Table 1.
Average RMSD values of damaged and undamaged sequences compared to the starting structure over its 5 ns simulation.
| sequence | average RMSD (Å) | sequence | average RMSD (Å) |
|---|---|---|---|
| AGA | 1.81 | TGA | 1.90 |
| AFA | 2.16 | TFA | 2.26 |
| AGC | 1.86 | TGC | 1.84 |
| AFC | 1.91 | TFC | 1.75 |
| AGG | 2.58 | TGT | 1.67 |
| AFG | 1.80 | TFT | 1.99 |
3.2 Energetic properties and DNA stability
3.2.1 Relative binding energies
DNA duplexes in which G:C base pairs have been replaced with cFapydG:C show a considerable reduction in melting temperature (Ober et al. 2003). This suggests that FapydG has a major deleterious effect on duplex stability. The MM/GBSA approach was used to estimate relative binding free energies of the FapydG base compared to its normal counterpart in a variety of sequence contexts. As shown in table 2, in all the cases ΔGbinding of damaged duplexes are less than the normal duplexes of approximately 5–10 kcal mol−1, mainly destabilized by the electrostatic interaction (ΔEes) and the polar part of the solvation free energy (ΔGpol). The destabilization of DNA-containing FapydG may be explained by two factors: (i) the weaker hydrogen bonding of the FapydG:C base pair due to the high degree of freedom of the glycosidic bond and (ii) unfavourable electrostatic interactions between the non-planar formamide group and the 3′-flanking base.
Table 2.
Estimated relative binding free energy of damaged and undamaged DNA duplexes from MM/GBSA approach. (In each case, F is base paired to C in the anti-conformation (figure 2).)
| sequence | ΔΔEvdW (kcal mol−1) | ΔΔEes (kcal mol−1) | ΔΔEin (kcal mol−1) | ΔΔGpol (kcal mol−1) | ΔΔGnonpol (kcal mol−1) | ΔΔGbinding (kcal mol−1) |
|---|---|---|---|---|---|---|
| AFA-AGA | 0.4±3.0 | 5.5±5.8 | −2.4±1.7 | 3.1±9.4 | −0.3±0.1 | 6.4±8.7 |
| AFC-AGC | 1.2±3.3 | −0.1±5.2 | −2.9±1.6 | 7.0±9.9 | −0.2±0.1 | 5.1±9.4 |
| AFG-AGG | 0.5±3.0 | 7.3±6.4 | −3.1±1.5 | 4.6±9.2 | −0.2±0.1 | 9.1±7.8 |
| TFA-TGA | −0.6±3.2 | 8.5±5.8 | −3.0±1.9 | −1.4±10.2 | −0.3±0.1 | 5.1±10.0 |
| TFC-TGC | 0.3±3.3 | 4.4±5.6 | −2.4±1.8 | 5.1±9.3 | −0.4±0.1 | 7.0±8.2 |
| TFT-TGT | −0.3±3.0 | 8.8±5.6 | −2.7±1.6 | 4.5±10.5 | −0.3±0.1 | 10.0±8.9 |
3.2.2 Hydrogen bonding and stacking interactions
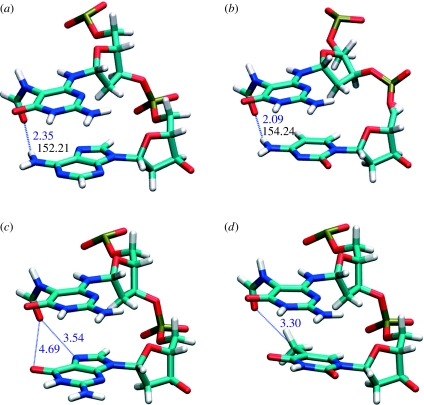
To further substantiate the above hypothesis regarding the origins of DNA destabilization by FapydG, non-bonded interactions—both the inter-strand (hydrogen bonding) and intra-strand (stacking interaction)—of the FapydG:C and G:C base pair in each sequence context were calculated. The reliability of the Amber force field for this type of analysis is well established (Hobza et al. 1997). The resulting interactions are shown in table 3. The most noticeable feature is the consistently poorer hydrogen bonding of FapydG to its cytosine partner: approximately 8 kcal mol−1 lower than for normal G:C base pairs. Although the FapydG:C base pair exhibits a G:C-like hydrogen-bonding pattern, the high mobility of the linkage between the sugar and the base (through two single bonds, rather than the normal one) results in the instability of those hydrogen bonds. Stacking energy calculations for FapydG or guanine flanked by various 5′- and 3′-neighbouring bases resulted unexpectedly in the observation that the non-planar FapydG has the tendency to generate more stable base stacking in B-DNA than normal guanine. Such preferences occur when the nucleobase to the 3′-side of FapydG is adenine, cytosine or guanine, whereas FapydG displays unfavourable interactions with 3′-thymine. Careful examination of structures from the MD trajectories reveals that the non-planar formamido group can generate direct hydrogen bonding with an amino group in adenine and cytosine, while favourable dipole–dipole interactions may enhance interactions with 3′-guanine. In addition, though this must be an effect additional to the stacking energy term calculated here, the relative orientations of the formamido carbonyl group and N7 and O6 of the guanine base are ideal to stabilize bridging water molecules (figure 4). The unfavourable interactions between the FapydG and 3′-thymine base step could be explained by a steric effect between the out-of-plane formamide group of FapydG and the thymine methyl group.
Table 3.
Inter-strand (hydrogen bonding) and intra-strand (stacking interaction) interactions of a FapydG:C or G:C base pair in six different sequence contexts.
| sequence | hydrogen bond energy (kcal mol−1) | stacking energy (kcal mol−1) |
|---|---|---|
| AFA-AGA | 7.8±2.4 | −1.5±2.5 |
| AFC-AGC | 7.8±2.1 | −4.9±2.5 |
| AFG-AGG | 8.0±2.3 | −0.7±2.6 |
| TFA-TGA | 8.0±2.3 | 0.3±2.7 |
| TFC-TGC | 8.3±2.2 | −2.8±2.6 |
| TFT-TGT | 8.4±3.0 | 1.2±3.0 |
Figure 4.
Proposed interactions between a formyl functional group of FapydG and 3′-neighbouring nucleobases (schematic generated from their time-averaged structures). (a) FapydG/A and (b) FapydG/C base steps show hydrogen bond lengths at 2.35 and 2.09 Å, and the angles at 152 and 154°, respectively. (c) FapydG/G shows opportunities for bridging water molecules while (d) FapydG/T indicates unfavourable steric interactions between the formamido group and the methyl group of thymine.
4. Conclusions
The overall stability and reproducibility of the simulations, and general agreement with the available experimental data, give us confidence in the parametrization of FapydG undertaken for this study, and in the general correctness of the FapydG:C base pair geometry. In particular, they support the adoption of the anti-glycosidic conformation by the FapydG base, and suggest a non-planar orientation of the formamido group. By performing a number of simulations on FapydG-containing duplexes and their normal counterparts, we have been able to identify structural and energetic consequences of the lesion that are distinguishable above the ‘noise’ associated with sequence-dependent properties of DNA. Energetic analysis shows that duplex destabilization by FapydG is primarily due to the weaker hydrogen bonding with its cytosine partner. Despite the destruction of the large planar purine ring system, stacking interactions are not greatly perturbed. This explains the observation above that the introduction of a FapydG lesion is not always reflected in a more globally distorted DNA structure, as measured by RMSD. The formamido group may modulate the structure and stability of lesion sites through its sequence-dependent interactions with the 3′-base. Interestingly, these simulations suggest that a 3′-thymine is particularly destabilizing, and so that lesions in this context might be particularly easy to be repaired by Fpg. However, other investigations underway suggest that this sequence is harder to bend in the direction required for protein–DNA recognition, and so repair may be inhibited. Unfortunately, there is currently no experimental evidence available regarding this, either way. Despite the weaker hydrogen bonding in the FapydG:C base pair, it is still strong enough to maintain a stacked-in conformation of the base, at least on the MD time scale. However, time scale is an issue: it could be argued that if the kinetic barrier to flipping is high, it might be that in fact the flipped-out state is the stable one for FapydG, and these simulations were just not long enough to see this. Our argument against this is related to the fact that the difference in free energy between a normal G:C base pair in the flipped-in and flipped-out state is estimated to be in the region of 10–15 kcal mol−1 (Varnai & Lavery 2002); therefore, even if FapydG lesions are 6–10 kcal mol−1 less stable, the equilibrium is still significantly on the side of the flipped-in state. This suggests that the base flipping required to insert the damaged base into the pocket of the repair enzyme Fpg can only occur with the assistance of the protein; the enzyme does not recognize and capture a ‘pre-flipped’ state. Further studies are underway to examine this issue in more detail, and also to examine the effects of the FapydG lesion on the local solvation pattern, which could both influence intrinsic stability and relate to lesion recognition by the repair protein.
Acknowledgments
S.J. was supported by a Royal Thai Government Scholarship.
Footnotes
One contribution of 9 to a Theme Supplement ‘Biomolecular simulation’.
Supplementary Material
Atom types and atomic charges of the FapydG lesion and the modified Amber force field for the nonplanar formamido functional group are provided.
References
- Berendsen H.J.C., Postma J.P.M., van Gunsteren W.F., DiNola A., Haak J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984;81:3684–3690. doi: 10.1063/1.448118. [DOI] [Google Scholar]
- Burrows C.J., Muller J.G. Oxidative nucleobase modifications leading to strand scission. Chem. Rev. 1998;98:1109–1152. doi: 10.1021/cr960421s. [DOI] [PubMed] [Google Scholar]
- Case D.A., et al. University of California; San Francisco, CA: 2004. Amber 8. [Google Scholar]
- Cheatham T.E., Kollman P.A. Molecular dynamics simulation of nucleic acids. Annu. Rev. Phys. Chem. 2000;51:435–471. doi: 10.1146/annurev.physchem.51.1.435. [DOI] [PubMed] [Google Scholar]
- Coste F., Ober M., Carell T., Boiteux S., Zelwer C., Castaing B. Structural basis for the recognition of the Fapy·dG lesion (2,6-diamino-4-hydroxy-5-formamidopyrimidine) by formamidopyrimidine-DNA glycosylase. J. Biol. Chem. 2004;279:44 074–44 083. doi: 10.1074/jbc.M405928200. [DOI] [PubMed] [Google Scholar]
- Cozmuta I., Mehrez H. DNA modelling within ab initio and empirical methods. J. Comp. Theor. Nanosci. 2007;4:349–383. [Google Scholar]
- Crespo-Hernandez C.E., Arce R. Formamidopyrimidines as major products in the low- and high-intensity UV irradiation of guanine derivatives. J. Photochem. Photobiol. B: Biol. 2004;73:167–175. doi: 10.1016/j.jphotobiol.2003.11.006. [DOI] [PubMed] [Google Scholar]
- Darden T., York D., Pedersen L. Particle mesh Ewald: an N log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10 089–10 092. doi: 10.1063/1.464397. [DOI] [Google Scholar]
- Delaney M.O., Greenberg M.M. Synthesis of oligonucleotides and thermal stability of duplexes containing the β-C-nucleoside analogue of Fapy·dG. Chem. Res. Tox. 2002;15:1460–1465. doi: 10.1021/tx025588x. [DOI] [PubMed] [Google Scholar]
- Dizdaroglu M., Olinski R., Doroshow J.H., Akman S.A. Modification of DNA bases in chromatin of intact target human cells by activated human polymorphonuclear leukocytes. Cancer Res. 1993;53:1269–1272. [PubMed] [Google Scholar]
- Douki T., Martini R., Ravanat J.L., Turesky R.J., Cadet J. Measurement of 2,6-diamino-4-hydroxy-5-formamidopyrimidine and 8-oxo-7,8-dihydroguanine in isolated DNA exposed to gamma radiation in aqueous solution. Carcinogenesis. 1997;18:2385–2391. doi: 10.1093/carcin/18.12.2385. [DOI] [PubMed] [Google Scholar]
- Evans M.D., Dizdaroglu M., Cooke M.S. Oxidative DNA damage and disease: induction, repair and significance. Mut. Res. 2004;567:1–61. doi: 10.1016/j.mrrev.2003.11.001. [DOI] [PubMed] [Google Scholar]
- Gohlke H., Case D.A. Converging free energy estimates: MM-PB(GB)SA studies on the protein–protein complex Ras–Raf. J. Comput. Chem. 2004;25:238–250. doi: 10.1002/jcc.10379. [DOI] [PubMed] [Google Scholar]
- Haraguchi K., Greenberg M.M. Synthesis of oligonucleotides containing Fapy·dG (N6-(2-deoxy-α, β-d-erythro-pentofuranosyl)-2,6-diamino-4-hydroxy-5-formamidopyrimidine) J. Am. Chem. Soc. 2001;123:8636–8637. doi: 10.1021/ja0160952. [DOI] [PubMed] [Google Scholar]
- Harris S.A., Gavathiotis E., Searle M.S., Orozco M., Laughton C.A. Cooperativity in drug-DNA recognition: a molecular dynamics study. J. Am. Chem. Soc. 2001;123:12 658–12 663. doi: 10.1021/ja016233n. [DOI] [PubMed] [Google Scholar]
- Hobza P., Kabelac M., Sponer J., Mejzlik P., Vondrasek J. Performance of empirical potentials (Amber, CFF95, CVFF, CHARMM, OPLS, POLTV), semiempirical quantum chemical methods (AM1, MNDO/M, PM3) and ab initio Hartree–Fock method for interaction of DNA bases: comparison with nonempirical beyond Hartree–Fock results. J. Comput. Chem. 1997;18:1136–1150. doi: 10.1002/(SICI)1096-987X(19970715)18:9%3C1136::AID-JCC3%3E3.0.CO;2-S. [DOI] [Google Scholar]
- Hu J., de Souza-Pinto N.C., Haraguchi K., Hogue B.A., Jaruga P., Greenberg M.M., Dizdaroglu M., Bohr V.A. Repair of formamidopyrimidines in DNA involves different glycosylases: role of the OGG1, NTH1, and NEIL1 enzymes. J. Biol. Chem. 2005;280:40 544–40 551. doi: 10.1074/jbc.M508772200. [DOI] [PubMed] [Google Scholar]
- Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. doi: 10.1063/1.445869. [DOI] [Google Scholar]
- Kasprzak K.S., Jaruga P., Zastawny T.H., North S.L., Riggs C.W., Olinski R., Dizdaroglu M. Oxidative DNA base damage and its repair in kidneys and livers of nickel(II)-treated male F344 rats. Carcinogenesis. 1997;18:271–277. doi: 10.1093/carcin/18.2.271. [DOI] [PubMed] [Google Scholar]
- Kollman P.A., et al. Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models. Acc. Chem. Res. 2000;33:889–897. doi: 10.1021/ar000033j. [DOI] [PubMed] [Google Scholar]
- Mao H., Deng Z., Wang F., Harris T.M., Stone M.P. An intercalated and thermally stable FAPY adduct of aflatoxin B1 in a DNA duplex: structural refinement from 1H NMR. Biochemistry. 1998;37:4374–4387. doi: 10.1021/bi9718292. [DOI] [PubMed] [Google Scholar]
- Marnett L.J. Oxyradicals and DNA damage. Carcinogenesis. 2000;21:361–370. doi: 10.1093/carcin/21.3.361. [DOI] [PubMed] [Google Scholar]
- Massova I., Kollman P.A. Combined molecular mechanical and continuum solvent approach (MM-PBSA/GBSA) to predict ligand binding. Pers. Drug Disc. Des. 2000;18:113–135. doi: 10.1023/A:1008763014207. [DOI] [Google Scholar]
- Mori T., Hori Y., Dizdaroglu M. DNA base damage generated in vivo in hepatic chromatin of mice upon whole body gamma-irradiation. Int. J. Radiat. Biol. 1993;64:645–650. doi: 10.1080/09553009314551881. [DOI] [PubMed] [Google Scholar]
- Ober M., Linne U., Gierlich J., Carell T. The two main DNA lesions 8-oxo-7,8-dihydroguanine and 2,6-diamino-5-formamido-4-hydroxypyrimidine exhibit strongly different pairing properties. Angew. Chem. 2003;115:5097–5101. doi: 10.1002/ange.200351287. [DOI] [PubMed] [Google Scholar]
- Ober M., Mller H., Pieck C., Gierlich J., Carell T. Base pairing and replicative processing of the formamidopyrimidine-dG DNA lesion. J. Am. Chem. Soc. 2005;127:18 143–18 149. doi: 10.1021/ja0549188. [DOI] [PubMed] [Google Scholar]
- Onufriev A., Bashford D., Case D.A. Modification of the generalized Born model suitable for macromolecules. J. Phys. Chem. B. 2000;104:3712–3720. doi: 10.1021/jp994072s. [DOI] [Google Scholar]
- Patro J.N., Haraguchi K., Delaney M.O., Greenberg M.M. Probing the configurations of formamidopyrimidine lesions Fapy·dA and Fapy·dG in DNA using endonuclease IV. Biochemistry. 2004;43:13 397–13 403. doi: 10.1021/bi049035s. [DOI] [PubMed] [Google Scholar]
- Pouget J.P., Douki T., Richard M.J., Cadet J. DNA damage induced in cells by gamma and UVA radiation as measured by HPLC/GC-MS and HPLC-EC and Comet assay. Chem. Res. Tox. 2000;13:541–549. doi: 10.1021/tx000020e. [DOI] [PubMed] [Google Scholar]
- Ryckaert J.P., Ciccotti G., Berendsen H.J.C. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 1977;23:327–341. doi: 10.1016/0021-9991(77)90098-5. [DOI] [Google Scholar]
- Shields G.C., Laughton C.A., Orozco M. Molecular dynamics simulation of a PNA·DNA·PNA triple helix in aqueous solution. J. Am. Chem. Soc. 1998;120:5895–5904. doi: 10.1021/ja9723444. [DOI] [Google Scholar]
- Sitkoff D., Sharp K.A., Honig B. Accurate calculation of hydration free energies using macroscopic solvent models. J. Phys. Chem. 1994;98:1978–1988. doi: 10.1021/j100058a043. [DOI] [Google Scholar]
- Song K., Hornak V., de los Santos C., Grollman A.P., Simmerling C. Computational analysis of the mode of binding of 8-oxoguanine to formamidopyrimidine-DNA glycosylase. Biochemistry. 2006;45:10 886–10 894. doi: 10.1021/bi060380m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan J., Cheatham T.E., Cieplak P., Kollman P.A., Case D.A. Continuum solvent studies of the stability of DNA, RNA, and phosphoramidate-DNA helices. J. Am. Chem. Soc. 1998;120:9401–9409. doi: 10.1021/ja981844+. [DOI] [Google Scholar]
- Steenken S., Jovanovic S.V. How easily oxidizable is DNA? One-electron reduction potentials of adenosine and guanosine radicals in aqueous solution. J. Am. Chem. Soc. 1997;119:617–618. doi: 10.1021/ja962255b. [DOI] [Google Scholar]
- Still W.C., Tempczyk A., Hawley R.C., Hendrickson T. Semianalytical treatment of solvation for molecular mechanics and dynamics. J. Am. Chem. Soc. 1990;112:6127–6129. doi: 10.1021/ja00172a038. [DOI] [Google Scholar]
- Tudek B. Imidazole ring-opened DNA purines and their biological significance. J. Biochem. Mol. Biol. 2003;31:12–19. doi: 10.5483/bmbrep.2003.36.1.012. [DOI] [PubMed] [Google Scholar]
- Varnai P., Lavery R. Base flipping in DNA: pathways and energetics studied with molecular dynamics simulations. J. Am. Chem. Soc. 2002;124:7272–7273. doi: 10.1021/ja025980x. [DOI] [PubMed] [Google Scholar]
- Várnai P., Zakrzewska K. DNA and its counterions: a molecular dynamics study. Nucleic Acids Res. 2004;32:4269–4280. doi: 10.1093/nar/gkh765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Laughton C.A. Molecular modelling methods for prediction of sequence-selectivity in DNA recognition. Methods. 2007;42:196–203. doi: 10.1016/j.ymeth.2006.09.002. [DOI] [PubMed] [Google Scholar]
- Wiederholt C.J., Greenberg M.M. Fapy·dG instructs Klenow exo(-) to misincorporate deoxyadenosine. J. Am. Chem. Soc. 2002;124:7278–7279. doi: 10.1021/ja026522r. [DOI] [PubMed] [Google Scholar]
- Wiederholt C.J., Delaney M.O., Pope M.A., David S.S., Greenberg M.M. Repair of DNA containing Fapy·dG and its β-C-nucleoside analogue by formamidopyrimidine DNA glycosylase and MutY. Biochemistry. 2003;42:9755–9760. doi: 10.1021/bi034844h. [DOI] [PubMed] [Google Scholar]
- Wilson D.M., III, Bohr V.A. The mechanics of base excision repair, and its relationship to aging and disease. DNA Repair. 2007;6:544–559. doi: 10.1016/j.dnarep.2006.10.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Atom types and atomic charges of the FapydG lesion and the modified Amber force field for the nonplanar formamido functional group are provided.