Abstract
Replication of the single-stranded linear DNA genome of parvovirus minute virus of mice (MVM) starts with complementary strand synthesis from the 3′-terminal snap-back telomere, which serves as a primer for the formation of double-stranded replicative form (RF) DNA. This DNA elongation reaction, designated conversion, is exclusively dependent on cellular factors. In cell extracts, we found that complementary strand synthesis was inhibited by the cyclin-dependent kinase inhibitor p21WAF1/CIP1 and rescued by the addition of proliferating cell nuclear antigen, arguing for the involvement of DNA polymerase (Pol) δ in the conversion reaction. In vivo time course analyses using synchronized MVM-infected A9 cells allowed initial detection of MVM RF DNA at the G1/S phase transition, coinciding with the onset of cyclin A expression and cyclin A-associated kinase activity. Under in vitro conditions, formation of RF DNA was efficiently supported by A9 S cell extracts, but only marginally by G1 cell extracts. Addition of recombinant cyclin A stimulated DNA conversion in G1 cell extracts, and correlated with a concomitant increase in cyclin A-associated kinase activity. Conversely, a specific antibody neutralizing cyclin A-dependent kinase activity, abolished the capacity of S cell extracts for DNA conversion. We found no evidence for the involvement of cyclin E in the regulation of the conversion reaction. We conclude that cyclin A is necessary for activation of complementary strand synthesis, which we propose as a model reaction to study the cell cycle regulation of the Pol δ-dependent elongation machinery.
Eukaryotic DNA replication is restricted to the S phase of the cell cycle. Cell cycle progression is regulated through the action of cyclin-dependent kinases (cdks) that are sequentially activated on association with different cyclins (1). In vertebrate cells, the transition from G1 to S phase is controlled by the activities of cyclin E- and cyclin A-dependent kinases (2). There is accumulating evidence for the involvement of cyclin A and cyclin E in the regulation of cellular DNA replication. Cyclin A is localized at nuclear replication foci in mammalian cells (3, 4), and both cyclin A and cdk2 have been found to be associated with replicating DNA in the simian virus 40 (SV40) in vitro replication system (5). Addition of recombinant cyclin A/cdk2 and cyclin E/cdk2 to G1 phase nuclei has been shown to trigger the initiation of DNA replication in a human cell-free system (6). Similarly, the inability of human G1 cell extracts to replicate SV40 origin-containing DNA in vitro could be overcome by the addition of cyclin A or active cdc2 kinase (7, 8). Conversely, immunodepletion of cyclin A from S cell extracts partially inhibited SV40 origin-driven plasmid replication (9), whereas a significant decrease in the ability of Xenopus egg extracts to replicate sperm DNA was observed after depletion of cyclin E or cdk2 (10, 11). However, it has been difficult to identify replication factors that are targets for cdks in vivo and to associate the above mentioned cyclin A- and cyclin E-dependent effects with distinct DNA replication steps.
Parvoviruses are unique among viruses in having a single-stranded (ss) linear DNA genome. The termini of this 5-kb genome contain palindromic sequences that fold into stable hairpin structures allowing self-primed DNA synthesis from the 3′-OH group (12). Parvovirus DNA replication starts with the synthesis of a complementary strand, converting the ss virion DNA into double-stranded monomer replicative form (mRF) DNA. This so-called conversion reaction relies only on cellular factors (12, 13). Subsequent amplification of RF DNA proceeds through the formation of multimeric intermediates by a unidirectional leading strand synthesis mechanism, and requires the nonstructural protein NS1, the major replicative protein of the autonomous parvoviruses (12). The unidirectional replication mode sets parvoviruses apart from viruses such as SV40 that replicate their genomes bidirectionally through leading and lagging strand synthesis (14, 15). Both parvoviruses and SV40 are strictly dependent on cellular DNA polymerases (Pols) for their replication. However, whereas SV40 DNA replication involves Pol α/primase-driven synthesis and initial extension of RNA primers followed by a switch to Pol δ-dependent elongation, parvovirus DNA replication appears to require only one Pol for elongation of in-built primers (16).
Parvovirus DNA replication is known to be S phase-dependent (12, 17), but the distinct replicative events tethered to S phase remained to be investigated. Furthermore, the identity of factor(s) mediating the cell cycle dependence of parvovirus DNA replication has been unknown. In this study, we show by means of an in vitro system that complementary strand synthesis from the genome of minute virus of mice (MVM), an autonomous parvovirus, is coupled with S phase and activated by cyclin A. In agreement with this finding, duplex RF DNA first became detectable in MVM-infected A9 cells at the time of appearance of cyclin A and associated kinase activity at the G1/S phase transition. The requirement for proliferating cell nuclear antigen (PCNA) in in vitro conversion reactions strongly argues for the involvement of Pol δ in MVM DNA replication. Given its independence of viral factors, parvoviral DNA conversion constitutes a model reaction to analyze the cell cycle regulation of the Pol δ-dependent elongation machinery.
Materials and Methods
Time Course Analysis of MVM DNA Replication After Infection of Synchronized Cells.
Mouse A9 fibroblasts (18) were grown in suspension culture as described (13). Cells were synchronized by serum starvation through incubation in MEM containing 0.5% FCS for 72 h. G0/G1-synchronized cells were infected with the prototype strain of MVM (MVMp) at a multiplicity of infection (MOI) of 10 plaque-forming units (pfu) per cell. At 4 h postinfection (p.i.) cells were treated with neuraminidase (100 μg/ml, Sigma), further incubated under low serum conditions for 8 h, and then released into S phase by addition of serum (20% final concentration). Cell cycle distribution was monitored by fluorescence-activated cell sorting (FACS) analysis. Whole-cell extracts from MVM-infected cells were prepared as described (19). Viral DNA was extracted according to the method of Hirt (20) and subjected to Southern blot analysis (21).
Preparation of Viral ss DNA and Cytosolic Extracts.
The preparation of MVM ss DNA and cytosolic extracts was described elsewhere (13). Cells were synchronized in G1 phase by serum starvation for 72 h and in S phase by subsequent addition of serum (20% final concentration) and further incubation for 16 h at 37°C.
In Vitro Replication Assay.
In vitro DNA replication was performed as described (13). Briefly, replication mixtures (50 μl) were supplemented with nucleotides, [α-32P]dCTP (3,000 Ci/mmol), an ATP-regenerating system, and 60–80 μg of cytosolic extract. The reaction was started by addition of MVM ss DNA (20 ng), allowed to proceed for 2 h at 37°C, and terminated by SDS/Proteinase K treatment. Reaction products were analyzed by neutral agarose gel electrophoresis (0.8%). Human p21WAF1/CIP1, bacterially expressed human PCNA (provided by U. Hübscher, University of Zürich, Switzerland), and anti-cyclin A mAbs E23 (22) and E72 (Neomarkers, Fremont, CA) were added to in vitro replication reactions as indicated in the figure legends.
Production of Recombinant Proteins.
Recombinant baculovirus vectors were used for the expression of glutathione S-transferase (GST)-cyclin A, hemagglutinin (HA)-cdk2 fusion proteins, and cyclin E protein in Sf9 insect cells. Baculovirus infection and preparation of whole-cell extracts from infected Sf9 cells were performed as described (13). GST-cyclin A was purified by adsorption to glutathione Sepharose 4B (Amersham Pharmacia) and eluted with 10 mM glutathione in 50 mM Tris⋅HCl (pH 8.0). HA-cdk2/cyclin E were copurified by adsorption to a HA-Ab (Babco, Richmond, CA) column and eluted with 1 mg/ml 12CA5 peptide in 50 mM Hepes (pH 7.6)/0.1 mM EDTA/50 mM KCl/1 mM DTT. Recombinant proteins were dialyzed against 40 mM Hepes (pH 7.6)/8 mM MgCl2/0.5 mM DTT, before being added to in vitro replication reactions in the amounts noted in the figure legends.
Protein Analyses.
The levels of viral and cellular proteins were determined by immunoblotting, using polyclonal Abs against NS1 (Sp8) (23), cyclin A (provided by M. Pagano, New York University), cyclin E (M20, Santa Cruz Biotechnology), and an mAb against PCNA (PC10, Upstate Biotechnology).
Histone H1 kinase assays were performed essentially as described (24). Briefly, cyclin/cdk complexes were immunoprecipitated from 60 or 80 μg cell extract by incubation with 2 μl of polyclonal Abs against cyclin A or cyclin E, respectively, or 2 μl of mAb against cyclin B (GNS1, Santa Cruz Biotechnology) for 2 h at 4°C. The precipitated complexes were incubated with histone H1 (1 μg per assay) and [γ-32P]ATP (5,000 Ci/mmol) for 30 min at 30°C in a 30-μl phosphorylation reaction. After electrophoresis through 15% SDS-polyacrylamide gels, phosphorylated histone H1 was detected by autoradiography.
Results
The Onset of MVM DNA Replication Coincides with Activation of Cyclin A-Dependent Kinase in Vivo.
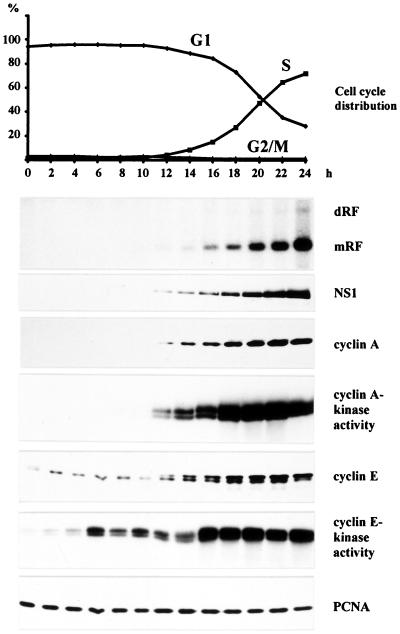
To investigate the S phase-dependence of MVM DNA replication, we initially conducted in vivo time course experiments using synchronized A9 cell cultures. G0/G1-blocked cells were infected with MVM, released from the block 12 h p.i., and subsequently analyzed for cell cycle progression and MVM DNA replication over a 24-h period. Given the involvement of cyclin E and cyclin A in the temporal control of cellular DNA synthesis (2), the expression of both cyclins and their associated kinase activities were monitored. In addition, expression of the parvoviral replication protein NS1 and the cellular replication factor PCNA was measured in the course of time. As shown in Fig. 1, formation of viral double-stranded mRF DNA was first detected at the onset of S phase, 12 h after release from the G1 block. The production of mRF DNA correlated with the expression of NS1 protein, which was consistent with the dependence of parvoviral gene transcription on both a double-stranded DNA template and S phase (25). Strikingly, the appearance of mRF DNA coincided with the induction of cyclin A protein expression as well as cyclin A-associated kinase activity. In contrast, both cyclin E protein and cyclin E-dependent kinase activity were already detectable at earlier time points and did not correlate with the onset of MVM DNA replication. PCNA was present throughout the observation time period, reflecting its constitutive expression in G1 and S phase cells (26). These data pointed to cyclin A and its associated kinase as a candidate cell cycle factor involved in the control of MVM DNA replication.
Figure 1.
Time course analysis of MVM DNA replication in synchronized A9 cells. Suspension cultures were synchronized in G0/G1 by serum starvation and infected with MVM (MOI = 10 pfu/cell). At 12 h p.i., cells were released from the G1 block by addition of FCS (20% final concentration). Culture samples were taken at 2-h intervals and monitored for cell cycle distribution, production of MVM DNA replication intermediates, and viral and cellular protein expression. Cyclin A- and E-associated kinase activities were determined as described in Materials and Methods. mRF, monomer replicative form DNA; dRF, dimer replicative form DNA.
Complementary Strand Synthesis in Vitro Is Cell Cycle-Dependent.
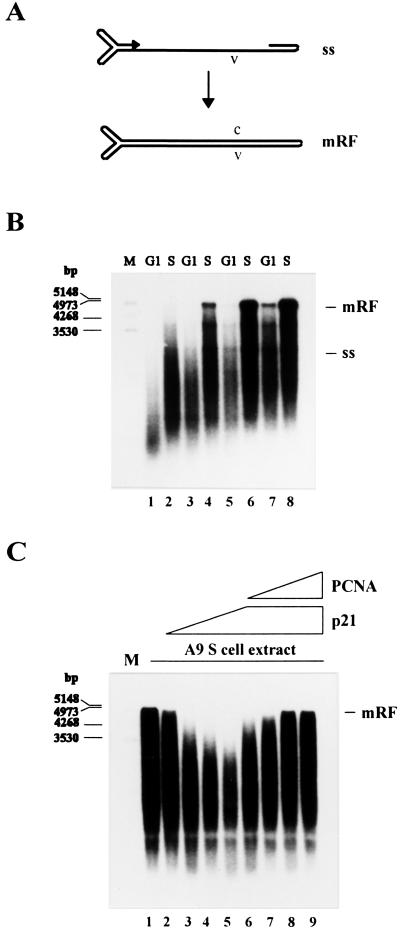
Our failure to detect mRF DNA in G1 cells suggested that complementary strand synthesis (Fig. 2A) may be coupled with S phase. Yet, the in vivo data did not rule out that this replicative step occurred before S phase, but at an undetectable level in the absence of further RF DNA amplification. To directly determine the cell cycle dependence of the conversion reaction, we prepared extracts from uninfected A9 cell cultures synchronized in G1 or S phase. These extracts were used for in vitro replication analyses according to ref. 13. In the absence of parvoviral NS1 proteins, this in vitro system supports the conversion of viral ss DNA into the covalently closed subform of mRF DNA (cRF), mimicking cRF formation in vivo (27), but fails to further process and amplify this mRF species.
Figure 2.
Conversion of MVM ss DNA into RF DNA in extracts from A9 cells synchronized in G1 and S phase. (A) Schematic representation of the conversion reaction. (B) MVM ss DNA (20 ng) was incubated in G1 or S cell extracts corresponding to 40 μg (lanes 1 and 2), 60 μg (lanes 3 and 4), 80 μg (lanes 5 and 6), or 100 μg (lanes 7 and 8) of cellular proteins. (C) MVM ss DNA (20 ng) was incubated in S cell extract alone (lane 1) or supplemented with 0.4 μg (lane 2), 0.8 μg (lane 3), 1.2 μg (lane 4), or 1.6 μg (lanes 5 to 9) of recombinant p21WAF1/CIP1 and additionally 0.4 μg (lane 6), 0.8 μg (lane 7), 1.2 μg (lane 8), or 1.6 μg (lane 9) of recombinant PCNA. Replication products were analyzed by neutral agarose gel electrophoresis (0.8%). ss, single-stranded virion DNA; mRF, monomer replicative form DNA; v, viral strand; c, complementary strand; M, DNA size markers in bp.
Increasing amounts of G1 or S cell extracts were tested for their ability to drive the conversion reaction. The efficiency of complementary strand synthesis was found to be consistently higher in S cell extracts as compared with G1 cell extracts (Fig. 2B). This argued for the regulation of conversion by an S phase-specific activator(s) and/or a G1 phase-specific inhibitor(s). To differentiate between these two possibilities, in vitro replication reactions were carried out using mixtures of G1 and S cell extracts in various ratios. The conversion yield proved to be unaffected by the addition of increasing amounts of G1 cell extract to a given S cell extract, whereas it increased proportionally to the amount of S cell extract added to a given G1 cell extract (data not shown). We concluded that the inefficiency of G1 cell extracts to support complementary strand synthesis was because of the absence of an S phase-specific activator(s). The radioactively labeled material running ahead of the mRF product is likely to result from incomplete conversion and/or DNA synthesis primed on partially degraded ss template DNA.
A p21-Induced Block of the Conversion Reaction Is Abrogated by PCNA.
Complementary strand synthesis from MVM ss DNA can be achieved with a variety of purified Pols in vitro (28, 29). To identify the Pol that is responsible for conversion in cell extracts, we supplemented S cell extracts with increasing amounts of the recombinant protein p21WAF1/CIP1. Besides its interaction with cdks, p21WAF1/CIP1 binds to the Pol δ cofactor PCNA, resulting in the inhibition of Pol δ-dependent DNA synthesis (30, 31). As is apparent from Fig. 2C (lanes 2–5), the addition of p21WAF1/CIP1 led to a dose-dependent reduction in the capacity of S cell extracts for MVM DNA conversion. p21-mediated inhibition of Pol δ activity has been reported to be reversible on addition of excess PCNA (30, 31). By supplementing S cell extracts with increasing amounts of PCNA in the presence of a constant level of p21WAF1/CIP1, we were indeed able to relieve the p21-mediated inhibition of conversion (Fig. 2C, lanes 6–9). From the sensitivity of MVM complementary strand synthesis to p21WAF1/CIP1 and its rescue by PCNA, it was concluded that the conversion reaction is specifically driven by the cellular Pol δ.
Cyclin A Activates Complementary Strand Synthesis in Vitro.
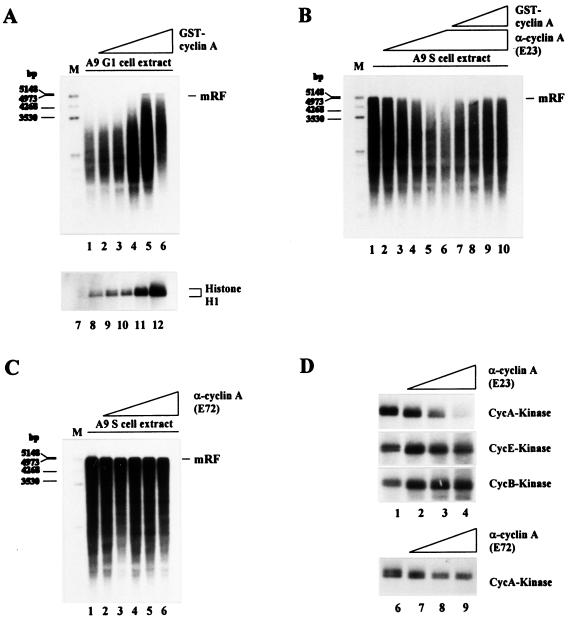
The correlation between onset of MVM DNA replication and induction of cyclin A and associated cdk activity in vivo (Fig. 1) prompted us to determine whether the different capacities of G1 and S cell extracts for MVM DNA conversion could be traced back to their respective cyclin A contents. As shown in Fig. 3A, the inability of G1 cell extracts to convert MVM ss DNA into mRF in vitro was overcome by the addition of GST-cyclin A in a dose-dependent manner. In contrast, addition of recombinant GST alone in up to a 3-fold molar excess had no effect (data not shown). The cyclin A-induced stimulation of conversion in G1 cell extracts correlated with an increase in cyclin A-associated kinase activity (Fig. 3A), suggesting that cyclin A may stimulate MVM complementary strand synthesis by means of phosphorylation of replication factors.
Figure 3.
Effect of cyclin A on the ability of A9 cell extracts to convert MVM ss DNA into RF. (A) MVM ss DNA (20 ng) was incubated in G1 cell extract alone (80 μg) (lane 1) or in the presence of 0.1 μg (lane 2), 0.2 μg (lane 3), 0.4 μg (lane 4), 0.6 μg (lane 5), or 0.8 μg (lane 6) of GST-cyclin A. Cyclin A-dependent kinase activity was determined by phosphorylation of histone H1 after addition of increasing amounts of GST-cyclin A (0.0 μg, lane 7; 0.1 μg, lane 8; 0.2 μg, lane 9; 0.4 μg, lane 10; 0.8 μg, lane 11; 1.6 μg, lane 12) to G1 cell extract samples (80 μg protein) and cyclin A/cdk complex immunoprecipitation. (B) MVM ss DNA (20 ng) was incubated in S cell extract alone (lane 1) or in the presence of 0.4 μg (lane 2), 0.8 μg (lane 3), 1.2 μg (lane 4), 1.6 μg (lane 5), or 2.0 μg (lanes 6 to 10) of the cyclin A-specific neutralizing mAb E23 and additionally 0.2 μg (lane 7), 0.4 μg (lane 8), 0.8 μg (lane 9), or 1.2 μg (lane 10) of GST-cyclin A. (C) MVM ss DNA (20 ng) was incubated in S cell extract alone (lane 1) or in the presence of 0.4 μg (lane 2), 0.8 μg (lane 3), 1.2 μg (lane 4), 1.6 μg (lane 5), or 2.0 μg (lane 6) of the cyclin A-specific nonneutralizing mAb E72. (D) Samples (80 μg protein) from S cell extract were supplemented with increasing amounts of the cyclin A-specific Abs E23 or E72 (no Ab, lanes 1 and 6; 0.6 μg, lanes 2 and 7; 1.2 μg, lanes 3 and 8; 1.8 μg, lanes 4 and 9), and kinase activities were determined as described in Materials and Methods. mRF, monomer replicative form DNA; M, size markers in bp.
To further substantiate our results, we preincubated S cell extracts with increasing amounts of the cyclin A-specific neutralizing Ab E23, which inhibits cyclin A-dependent kinase activity (Fig. 3D). Addition of this Ab to S cell extracts inhibited conversion in a dose-dependent manner (Fig. 3B, lanes 2–6). This inhibition was specific because the conversion capacity of neutralizing Ab-treated S cell extracts could be progressively rescued by supplementing them with increasing amounts of recombinant cyclin A (Fig. 3B, lanes 7–10). In contrast, complementary strand synthesis was not impaired after preincubation of S cell extracts with the nonneutralizing cyclin A-specific Ab E72 (Fig. 3C). Correspondingly, cyclin A-associated phosphorylation of histone H1 could be fully inhibited by the neutralizing Ab, whereas the same amount of nonneutralizing Ab only slightly reduced cyclin A-dependent kinase activity (Fig. 3D). No inhibitory effect of the neutralizing Ab on cyclin E- and cyclin B-dependent kinase activities was detected. Altogether, our results show that cyclin A is required for Pol δ-dependent complementary strand synthesis in vitro and that cyclin E or cyclin B cannot substitute for cyclin A in this function. The qualitative correlation observed between the relative capacities of different anti-cyclin A Abs for inhibiting MVM DNA conversion and cyclin A/cdk-activity corroborates the idea that cyclin A may modulate conversion by means of its associated cdk.
Cyclin E Is Unable to Rescue MVM DNA Conversion in G1 Phase.
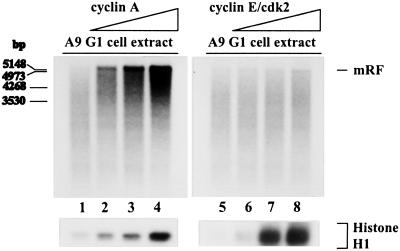
Some in vitro studies have suggested a role for cyclin E or cyclin E-associated kinase in the regulation of cellular DNA replication (6, 10). This prompted us to investigate whether cyclin E contributed to the differential capacity of S vs. G1 cell extracts for MVM DNA conversion. In vitro replication reactions were carried out using G1 cell extracts that had been supplemented with increasing amounts of recombinant cyclin E/cdk2. Parallel samples were treated with recombinant cyclin A, resulting in the above mentioned dose-dependent increase in the ability of G1 cell extracts to support complementary strand synthesis (Fig. 4, lanes 1–4). In contrast, the efficiency of the conversion reaction could not be enhanced by supplying G1 cell extracts with exogenous cyclin E/cdk2 (Fig. 4, lanes 5–8), although a comparable or even higher increase in cdk activity was exhibited by cyclin E- vs. cyclin A-treated samples (Fig. 4 Bottom). This result argued against the involvement of cyclin E/cdk2 in the activation of MVM DNA conversion at the G1/S transition.
Figure 4.
Effect of cyclins A and E on the capacity of A9 G1 cell extracts for conversion of MVM ss DNA into RF. MVM ss DNA (20 ng) was incubated in G1 cell extract (60 μg) alone (lanes 1 and 5) or in the presence of increasing amounts of cyclin A (80 ng, lane 2; 160 ng, lane 3; 320 ng, lane 4) or cyclin E/cdk2 (50 ng, lane 6; 100 ng, lane 7; 200 ng, lane 8). The yields of conversion are shown on the top, whereas corresponding cyclin A- and cyclin E-associated kinase activities are illustrated on the bottom.
Discussion
Parvovirus DNA replication is coupled with the S phase of the cell cycle (12, 17). Unlike some other DNA viruses, parvoviruses are unable to induce quiescent cells to initiate DNA synthesis; hence, parvoviral DNA replication is delayed until host cells reach S phase of their own volition (32). Besides cellular replication factors, the parvoviral regulatory protein NS1 is essential for the replication of parvoviral DNA (12). Transcription of the NS gene has recently been reported to be S phase-dependent (25) and may thus contribute to the coupling of viral DNA amplification with cellular DNA synthesis. Yet, NS1 expression is thought to require a prior replicative event, namely the conversion of the ss parvoviral genome into a double-stranded intermediate (mRF) that can serve as a transcription template. The present (Fig. 1) and previous (17) results indicate that mRF can only be detected after cells have entered S phase, suggesting that conversion may be the primary S phase-associated event of the parvoviral life cycle. However, these in vivo data do not rule out that the S phase-dependence relates to an intracellular step in the viral life cycle preceding conversion or the amplification of parental mRF to a detectable level.
The present study provides direct evidence for the coupling of conversion with S phase by means of an in vitro replication system that is competent for the formation of parental mRF molecules from virion ss DNA (13). Thus, S and G1 cell extracts could be distinguished by the much greater capacity of the former to support self-primed complementary strand synthesis. This S phase-dependence could be traced back to the fact that G1 cells lack an S phase-specific activator, which was identified as being cyclin A. Indeed, the incompetence of G1 cell extracts and capacity of S cell extracts for MVM DNA conversion could be reversed through the addition or neutralization of cyclin A, respectively. This effect was specific for cyclin A, because cyclin E/cdk2 failed to rescue the conversion in G1 cell extracts, although this complex is involved in the G1/S transition. Stimulation of conversion by cyclin A in vitro and onset of RF DNA synthesis in vivo correlated with an increase in cyclin A-associated kinase activity. Furthermore, conversion in S cell extracts could be inhibited by means of an anti-cyclin A Ab that was able to interfere with cyclin A/cdk activity. Altogether, these results suggest that cyclin A may stimulate complementary strand synthesis through the activation of its associated kinase, although other effects of cyclin A, such as a direct interaction with the replication machinery, would also be consistent with our data.
In vitro conversion of virion ss DNA into RF DNA can be achieved using a number of partially purified eukaryotic and prokaryotic Pols (28, 29). In contrast, our results indicate that, in cell extracts, this reaction is specifically driven by Pol δ. Indeed, the MVM DNA conversion reaction could be fully inhibited by treating competent A9 cell extracts with p21WAF1/CIP1. This block was overcome by using an excess of PCNA, pointing to the involvement of Pol δ, which absolutely requires PCNA as a cofactor for processive DNA synthesis (33). Binding of p21WAF1/CIP1 to PCNA is independent of binding to cdk (30, 31), yet we do not exclude that concomitant cdk inactivation may contribute to the inhibitory effect of p21 on DNA replication. However, p21 mutants that could bind cyclin/cdk, but not PCNA, were shown to only achieve an incomplete inhibition of SV40 DNA replication in S cell extracts (9). In support of our conclusion, there is in vitro evidence for the participation of Pol δ in late parvovirus DNA replication steps, which, like conversion, involve a continuous unidirectional strand elongation process (16, 34). Furthermore, we failed to inhibit the conversion reaction by adding anti-Pol α neutralizing Abs in amounts that suppressed in vitro SV40 DNA replication (data not shown). It is worth noting that the parvoviral genome folds back at the 3′-end, thereby providing a primer-template structure that can be readily used for complementary strand elongation, obviating the need for Pol α-associated primase activity (12). In addition, PCNA in conjunction with ATP and replication factor C (RFC) has been shown to inhibit Pol α function at primer-template junctions (35, 36).
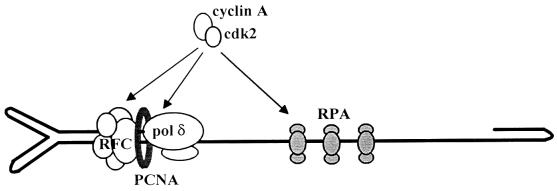
As far as its structure is concerned, the parvoviral genome is reminiscent of primed ss DNA templates, which were used to show that Pol δ and the accessory cofactors PCNA, RFC, and replication protein A (RPA) are sufficient for the formation and functioning of a DNA strand elongation complex (35, 36). RFC is needed to load PCNA onto DNA, thus allowing the recruitment of Pol δ, whereas RPA binds to and stabilizes ss DNA regions ahead of the nascent strand. Interestingly, PCNA and RPA were shown to be required for rolling circle replication initiated from a parvoviral origin (16). Hence, Pol δ, PCNA, RFC, and RPA can be assumed to represent the minimal elements necessary for parvovirus DNA conversion and are candidates for mediating the cyclin A-induced activation of complementary strand synthesis, as depicted in Fig. 5. All these factors are potential targets for cdk-driven phosphorylation, as they contain putative cdk phosphorylation sites. Pol δ and the p32 subunit of RPA (RPA32) have been shown to be phosphorylated in a cell cycle-dependent manner in vivo (8, 37). However, the functional relevance of RPA phosphorylation remains unclear, because mutations affecting both cdk phosphorylation sites of RPA32 did not impair SV40 DNA replication in vitro (38). Similarly, the large subunit of Pol δ (p125) was found to be phosphorylated by several cyclin-cdk complexes in vitro, but no concomitant functional change was observed (39). It should be stated that cyclin A may also modulate parvovirus DNA conversion irrespective of its associated cdk activity. Indeed, cyclin A was found at sites of cellular DNA replication (3) and shown to be associated with replicating SV40 origin-containing DNA (5), suggesting that cyclin A is a constituent of the elongation machinery in which it may play a structural role.
Figure 5.
Schematic representation of the cellular factors assembled at the MVM virion DNA primer-template junction and thought to be involved in complementary strand elongation. Cyclin A or the cyclin A/cdk2 complex may regulate the conversion reaction by forming part of the elongation machinery and/or phosphorylating some of its constituents.
Cyclin A also regulates the replication of SV40 origin-containing DNA under in vitro conditions (7, 9). In this system, cyclin A was shown to stimulate a replication step that occurs downstream of SV40 T antigen-induced unwinding of the origin but before ongoing elongation (9). It remained to be determined whether this step was Pol α- and/or Pol δ-dependent. Our results argue for the latter, as cyclin A could be shown to stimulate Pol δ-dependent DNA synthesis. Furthermore, it was recently reported that Pol α-dependent initiation of SV40 DNA replication in vitro was stimulated by cyclin E/cdk2, but inhibited by cyclin A/cdk2 (40). Thus, by concomitantly inactivating Pol α-associated functions and activating Pol δ and/or some of its cofactors, cyclin A may contribute to the switch from Pol α-dependent initiation to Pol δ-driven strand elongation.
Parvovirus DNA conversion is unique in that it starts from an in-built terminal primer and relies solely on a presumably limited number of cellular proteins. The relative simplicity of this reaction together with its hereby demonstrated cell cycle dependence, allow parvoviral complementary strand synthesis to be considered as a model for studying the regulation of the Pol δ-associated elongation machinery at the molecular level.
Acknowledgments
We thank U. Hübscher for providing recombinant PCNA and p21WAF1/CIP1, M. Pagano for supplying a polyclonal Ab against cyclin A, and H. Piwnica-Worms and J. M. Roberts for providing recombinant baculoviruses for the production of cyclin A and cyclin E/cdk2.
Abbreviations
- MVM
minute virus of mice
- mRF
monomer replicative form
- PCNA
proliferating cell nuclear antigen
- Pol
DNA polymerase
- cdk
cyclin-dependent kinase
- SV40
simian virus 40
- NS
nonstructural protein
- ss
single-stranded
- GST
glutathione S-transferase
- RPA
replication protein A
- MOI
multiplicity of infection
- p.i.
postinfection
- pfu
plaque-forming unit
- HA
hemagglutinin
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.090485297.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.090485297
References
- 1.Reed S I. Annu Rev Cell Biol. 1992;8:529–561. doi: 10.1146/annurev.cb.08.110192.002525. [DOI] [PubMed] [Google Scholar]
- 2.Nigg E. BioEssays. 1995;17:471–480. doi: 10.1002/bies.950170603. [DOI] [PubMed] [Google Scholar]
- 3.Cardoso M C, Leonhardt H, Nadal-Ginard B. Cell. 1993;74:979–992. doi: 10.1016/0092-8674(93)90721-2. [DOI] [PubMed] [Google Scholar]
- 4.Sobczak-Thepot J, Harper F, Florentin Y, Zindy F, Brechot C, Puvion E. Exp Cell Res. 1993;206:43–48. doi: 10.1006/excr.1993.1118. [DOI] [PubMed] [Google Scholar]
- 5.Fotedar R, Roberts J M. Cold Spring Harbor Symp Quant Biol. 1991;56:325–333. doi: 10.1101/sqb.1991.056.01.039. [DOI] [PubMed] [Google Scholar]
- 6.Krude T, Jackman M, Pines J, Laskey R A. Cell. 1997;88:109–119. doi: 10.1016/s0092-8674(00)81863-2. [DOI] [PubMed] [Google Scholar]
- 7.D'Urso G, Marraccino R L, Marshak D R, Roberts J M. Science. 1990;250:786–791. doi: 10.1126/science.2173140. [DOI] [PubMed] [Google Scholar]
- 8.Dutta A, Stillman B. EMBO J. 1992;11:2189–2199. doi: 10.1002/j.1460-2075.1992.tb05278.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fotedar A, Cannella D, Fitzgerald P, Rousselle T, Gupta S, Dorée M, Fotedar R. J Biol Chem. 1996;271:31627–31637. doi: 10.1074/jbc.271.49.31627. [DOI] [PubMed] [Google Scholar]
- 10.Jackson P K, Chevalier S, Philippe M, Kirschner M W. J Cell Biol. 1995;130:755–769. doi: 10.1083/jcb.130.4.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fang F, Newport J W. Cell. 1991;66:731–742. doi: 10.1016/0092-8674(91)90117-h. [DOI] [PubMed] [Google Scholar]
- 12.Cotmore S F, Tattersall P. Adv Virus Res. 1987;33:91–174. doi: 10.1016/s0065-3527(08)60317-6. [DOI] [PubMed] [Google Scholar]
- 13.Baldauf A Q, Willwand K, Mumtsidu E, Nüesch J P F, Rommelaere J. J Virol. 1997;71:971–980. doi: 10.1128/jvi.71.2.971-980.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kelly T J. J Biol Chem. 1988;263:17889–17892. [PubMed] [Google Scholar]
- 15.Stillman B. Annu Rev Cell Biol. 1989;5:197–245. doi: 10.1146/annurev.cb.05.110189.001213. [DOI] [PubMed] [Google Scholar]
- 16.Christensen J, Cotmore S F, Tattersall P. J Virol. 1997;71:1405–1416. doi: 10.1128/jvi.71.2.1405-1416.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wolter S, Richards R, Armentrout R W. Biochim Biophys Acta. 1980;607:420–431. doi: 10.1016/0005-2787(80)90152-5. [DOI] [PubMed] [Google Scholar]
- 18.Littlefield J W. Nature (London) 1964;203:1142–1144. doi: 10.1038/2031142a0. [DOI] [PubMed] [Google Scholar]
- 19.Kumar V, Chambon P. Cell. 1988;55:145–156. doi: 10.1016/0092-8674(88)90017-7. [DOI] [PubMed] [Google Scholar]
- 20.Hirt B. J Mol Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 21.Southern E M. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 22.Adamczewski J P, Gannon J V, Hunt T. J Virol. 1993;67:6551–6557. doi: 10.1128/jvi.67.11.6551-6557.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Faisst S, Faisst S R, Dupressoir T, Plaza S, Pujol A, Jauniaux J C, Rhode S L, Rommelaere J. J Virol. 1995;69:4538–4543. doi: 10.1128/jvi.69.7.4538-4543.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsushime H, Quelle D E, Shurtleff S A, Shibuya M, Sherr C J, Kato J Y. Mol Cell Biol. 1994;14:2066–2076. doi: 10.1128/mcb.14.3.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deleu L, Fuks F, Spitkovsky D, Hörlein R, Faisst S, Rommelaere J. Mol Cell Biol. 1998;18:409–419. doi: 10.1128/mcb.18.1.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kelman Z. Oncogene. 1997;14:629–640. doi: 10.1038/sj.onc.1200886. [DOI] [PubMed] [Google Scholar]
- 27.Cotmore S F, Gunther M, Tattersall P. J Virol. 1989;63:1002–1006. doi: 10.1128/jvi.63.2.1002-1006.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Faust E A, Rankin C D. Nucleic Acids Res. 1982;10:4181–4201. doi: 10.1093/nar/10.14.4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bourguignon G J, Tattersall P J, Ward D C. J Virol. 1976;20:290–306. doi: 10.1128/jvi.20.1.290-306.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Waga S, Hannon G J, Beach D, Stillman B. Nature (London) 1994;369:574–578. doi: 10.1038/369574a0. [DOI] [PubMed] [Google Scholar]
- 31.Flores-Rozas H, Kelman Z, Dean F B, Pan Z Q, Harper J W, Elledge S J, O'Donnell M, Hurwitz J. Proc Natl Acad Sci USA. 1994;91:8655–8659. doi: 10.1073/pnas.91.18.8655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tattersall P, Bratton J. J Virol. 1983;46:944–955. doi: 10.1128/jvi.46.3.944-955.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tan C K, Castillo C, So A G, Downey K M. J Biol Chem. 1986;261:12310–12316. [PubMed] [Google Scholar]
- 34.Cossons N, Faust E A, Zannis-Hadjopoulos M. Virology. 1996;216:258–264. doi: 10.1006/viro.1996.0058. [DOI] [PubMed] [Google Scholar]
- 35.Tsurimoto T, Stillman B. J Biol Chem. 1991;266:1950–1960. [PubMed] [Google Scholar]
- 36.Maga G, Hübscher U. Biochemistry. 1996;35:5764–5777. doi: 10.1021/bi952455k. [DOI] [PubMed] [Google Scholar]
- 37.Zeng X R, Hao H, Jiang Y, Lee M Y W T. J Biol Chem. 1994;269:24027–24033. [PubMed] [Google Scholar]
- 38.Henricksen L A, Wold M S. J Biol Chem. 1994;269:24203–24208. [PubMed] [Google Scholar]
- 39.Wu S M, Zhang P, Zeng X R, Zhang S J, Mo J, Li B Q, Lee M Y W T. J Biol Chem. 1998;273:9561–9569. doi: 10.1074/jbc.273.16.9561. [DOI] [PubMed] [Google Scholar]
- 40.Voitenleitner C, Rehfuess C, Hilmes M, O'Rear L, Liao P C, Gage D A, Ott R, Nasheuer H P, Fanning E. Mol Cell Biol. 1999;19:646–656. doi: 10.1128/mcb.19.1.646. [DOI] [PMC free article] [PubMed] [Google Scholar]