Abstract
Three new galloyl arbutins, hyemalosides A–C (1–3), along with nine known compounds were isolated from the evergreen tree Eugenia hyemalis. The structures of compounds 1–3 were determined by analysis of NMR and MS data. Compounds 1–3 inhibited HIV-1 RNase H in vitro with IC50 values of 1.46, >18, and 1.19 μM, respectively. However, in a XTT-based cell viability assay using the human T-cell line CEM-SS infected with HIV-1RT, none of the compounds inhibited the cytopathic effect of HIV-1 infection at the highest dose tested (20 μg/mL).
As part of a molecularly targeted screening program to discover drug leads from natural products, we investigated the chemistry of the evergreen tree Eugenia hyemalis L. Cambess (Myrtaceae). The organic extract of the plant showed inhibition of ribonuclease H (RNase H) enzymatic activity in a high-throughput screening assay.1 RNase H is a second, distinct enzymatic activity of the HIV-1 reverse transcriptase protein. It is a nonspecific nuclease that is responsible for hydrolyzing the RNA strand of a RNA/DNA heteroduplex and allowing the incorporation of viral genetic information into the host cell genome. Because RNase H activity is required for viral infectivity, it is a viable target for screening for potential anti-HIV drugs, and a high-throughput fluorescence resonance energy transfer (FRET) assay1 was developed for this purpose.
Bioassay-guided fractionation of the extract led to the isolation of three new compounds, hyemalosides A–C (1–3), along with nine known compounds. The structures of the nine known compounds were determined on the basis of spectroscopic evidence and comparison with literature values. These were characterized as 2-O-galloylarbutin,2 4-O-galloylarbutin, 2,6-di-O-galloylarbutin,3 2,4,6-tri-O-galloylarbutin,4 1,2,6-tri-O-galloyl-β-d-glucose,5 kaempferol-3-O-(6-O-galloyl-β-d-glucopyranoside),6 afzelin 2″-O-gallate,7 afzelin 3″-O-gallate,7 and quercitrin.7
HREIMS of compound 1 showed a peak at m/z 727.1191 [M – H]− consistent with a molecular formula of C33H28O19. The 1H NMR spectrum showed a proton doublet at δ 5.13 (J = 7.9 Hz) along with six resonances from δ 3.88 to 5.49, which were linked by COSY data and indicated a hexose moiety. Large coupling constants for the sugar ring protons (J = 7.9–9.5 Hz) indicated a series of trans diaxial couplings characteristic of glucose and also revealed that the anomeric oxygen was β-oriented. 13C NMR resonances at δ 152.1, 116.8 (2C), 119.9 (2C), and 154.4 along with proton doublets at δ 6.58 (2H, J = 8.5 Hz) and 6.80 (2H, J = 8.7 Hz) were consistent with a para-substituted benzene ring. An HMBC correlation from the anomeric H-1 (δ 5.13) to C-7 (δ 152.1) established that the para-substituted ring was attached to glucose via an ether link between C-1 (δ 102.3) and C-7 (δ 152.1). A literature search and comparison of spectroscopic data identified this fragment as an arbutin moiety (4).8 The remaining 21 13C peaks were clustered into three groups each containing 13C NMR resonances around δ 121, 110, 146, 140, and 168, which, along with three 2-H singlets at δ 6.98, 7.02, and 7.15, were characteristic of the presence of three galloyl groups. The position of the galloyl groups was determined by HMBC correlations. Correlations from H-2, H-3, and H-6a and -6b to ester carbonyls at position 7 on the galloyl groups established the structure of hyemaloside A (1) as 2,3,6-tri-O-galloylarbutin (0.05% yield).
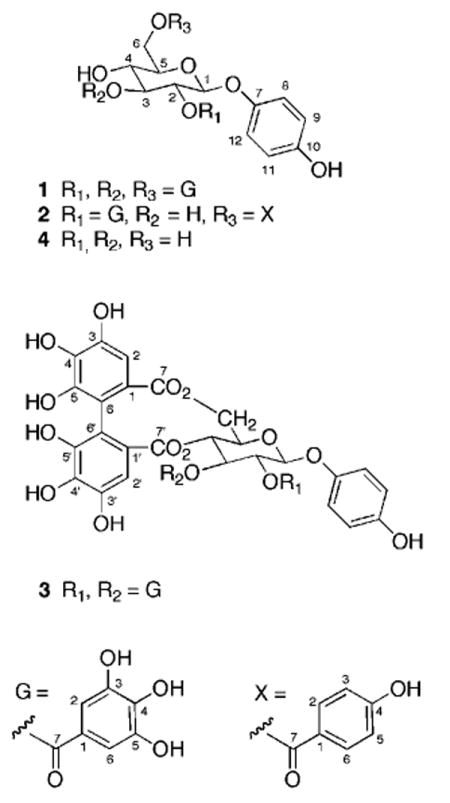
Compound 2 (0.06% yield) also contained a substituted arbutin moiety, with proton signals from δ 3.55 to 5.13 attributed to a hexose and large coupling constants (J = 8.1–9.5 Hz) consistent with a β-linked glucose moiety. HREIMS data (m/z 567.1105 [M + Na]+) indicated its molecular formula to be C26H24O13. 13C NMR resonances at δ 152.2, 116.6 (2C), 119.9 (2C), and 154.2 along with proton doublets at δ 6.54 (2H, J = 8.8 Hz) and 6.79 (2H, J = 8.9 Hz) revealed a para-substituted benzene ring that was attached to the glucose moiety at C-1 (δ 102.5) by an HMBC correlation from H-1 (δ 4.98) to C-7 (δ 152.2). An additional set of doublets in the 1H spectrum at δ 6.86 (2H, J = 8.9) and 7.92 (2H, J = 8.8 Hz) was attributed to a para-substituted benzoic acid moiety and a galloyl group was indicated by a 2-H singlet at δ 7.10. Characteristic 13C resonances supported the presence of these structural fragments. HMBC correlations from H-2 (δ 5.13) to an ester carbonyl at C-7 (δ 167.7) on the galloyl group established the position of the galloyl group. The location of the esterified benzoic acid subunit was revealed by an HMBC correlation from H-6a (δ 4.41) and H-6b (δ 4.71) to an ester carbonyl (C-7, δ 167.91), which completed the structural assignment of hyemaloside B (2).
Compound 3 (0.02%) showed a negative molecular ion at m/z 877.1129 [M – H]− in the HREIMS spectrum, indicative of a molecular formula of C40H30O23 and consistent with 15 double-bond equivalents. The 1H NMR spectrum had, in addition to the arbutin skeleton, two 2-H singlets at δ 7.00 and 6.91 and 14 13C resonances (δ 120.5, 120.7, 110.4, 110.5, 146.2, 146.5, 140.2, 140.0, 167.0, 167.6), which were attributed to two galloyl groups. Two additional aromatic singlets equivalent to one proton each at δ 6.46 and 6.63 were assigned to a hexahydroxydiphenoyl (HHDP) group. HMBC correlations from the glucose methylene (H-6a, δ 5.36 and H-6b, δ 3.94) and from H-2 (δ 6.63) of the HHDP to the C-7 (δ 169.6) HHDP ester carbonyl established the linkage of one end of the HHDP group. The other attachment was established by HMBC correlations from the glucose H-4 (δ 5.20) and from H-2′ (δ 6.46) of the HHDP to C-7′ (δ 169.1).
The remaining two galloyl groups were therefore attached to C-2 (δ 73.4) and C-3 (δ 74.1) of the glucose moiety. Comparison with literature values showed that hyemaloside C differs from the known compound tellimagrandin II9 by the substitution of a 4-hydroxyphenyl group at the anomeric carbon of 3 instead of a 3,4,5-hydroxyphenyl group.
All of the compounds were tested for activity against a small panel consisting of RNase H enzymes from E. coli, HIV-1, HIV-2, and humans. None of the compounds inhibited E. coli RNase H at 10 μg/mL (data not shown). All of the compounds tested were active against human and HIV-2 RNase H except for 2-O-galloylarbutin and 4-O-galloylarbutin (Table 1). The compounds were also tested for anti-HIV activity in a XTT-based cell viability assay using the human T-cell line CEM-SS infected with HIV-1RT.10 Surprisingly, even the compounds that were active against HIV-1 RNase H were not protective in the cell-based cytopathicity screen at 20 μg/mL (data not shown), and therefore, additional follow-up studies on the compounds were not completed, including testing for reverse transcriptase activity. Potent activity in enzymatic assays but a lack of activity in the cell-based cytopathicity assay is an emerging theme for gallate esters in this assay.11, 12 That these compounds do not have activity in the cell-based screen is very different from the many small phenolic compounds with HIV-1 activity we have isolated in our laboratory over the years.13–15 We do not believe that this is a case of general nonspecific binding since the extracts are run in the presence of 1% bovine serum albumin (BSA) and active extracts from plants were subjected to a polyamide protocol to eliminate those whose activity is due solely to tannins. The observation remains that these types of phenolic compounds have been seen often enough with this screen to be considered nuisance compounds.
Table 1.
Biological Activity of Compounds against RNase H Enzymes
| IC50, μM | ||||
|---|---|---|---|---|
| compound | high conc, μM | HIV-1 | HIV-2 | human |
| hyemaloside A (1) | 13.7 | 1.46 ± 0.01 | 0.58 ± 0.14 | 0.18 ± 0.01 |
| hyemaloside B (2) | 18.3 | >18.3 | 3.32 ± 0.13 | 2.44 ± 0.25 |
| hyemaloside C (3) | 11.3 | 1.19 ± 0.05 | 0.84 ± 0.16 | 0.07 ± 0.01 |
| 2-O-galloylarbutin | 23.5 | >23.5 | >23.5 | >23.5 |
| 4-O-galloylarbutin | 23.5 | >23.5 | >23.5 | >23.5 |
| 2,6-di-O-galloylarbutin | 17.7 | 5.05 ± 0.10 | 0.28 ± 0.10 | 0.55 ± 0.09 |
| 2,4,6-tri-O-galloylarbutin | 13.7 | 0.68 ± 0.01 | 0.10 ± 0.05 | 0.04 ± 0.003 |
| 1,2,6-tri-O-galloyl-β-d-glucose | 13.7 | 10.21 ± 0.01 | 0.28 ± 0.10 | 0.55 ± 0.09 |
| kaempferol-3-O-(6-O-galloyl-β-d-glucopyranoside) | 16.6 | >16.6 | 5.19 ± 1.08 | 9.50 ± 0.36 |
| afzelin 2″-O-gallate | 17.1 | > 17.1 | 3.38 ± 0.32 | 3.03 ± 0.15 |
| afzelin 3″-O-gallate | 17.1 | > 17.1 | 9.12 ± 0.76 | 7.25 ± 0.13 |
| quercitrin | 22.3 | >22.3 | 21.27 ± 0.49 | 12.46 ± 0.05 |
Experimental Section
General Experimental Procedures
Optical rotations were measured on a Perkin-Elmer 241 polarimeter. UV spectra were recorded on a Varian Cary 50 UV–vis spectrophotometer. NMR spectra were recorded on a Varian Unity Inova 500 in CD3OD at 500 MHz for 1H and 125 MHz for 13C. MS spectra were measured with an Agilent Technologies 6510 Q-TOF LC/MS and an Applied Biosystems, Inc. QSTAR XL hybrid triple-quad time-of-flight (QqTOF) mass spectrometer. Column chromatography was performed using Sephadex LH-20 (Amersham Biosciences).
Plant Material
The plant material was collected in Nueva Colombia, Cordillera, Paraguay (May 1, 1990), by N. Trushell of the New York Botanical Gardens under contract to the National Cancer Institute. A voucher specimen (collection number Q65V1530) is maintained at the New York Botanical Gardens.
Extraction and Isolation
The whole plant, minus roots (0.55 kg), was extracted successively with CH2Cl2–MeOH (1:1) and MeOH. The combined extracts were reduced to dryness in vacuo to give 41.40 g of crude extract. A portion of this extract (5.00 g) was subjected to a solvent–solvent partitioning scheme16 that concentrated the activity in the ethyl acetate (2.48 g)-soluble fraction. Size exclusion chromatography of the active EtOAc material on Sephadex LH-20 (3 × 135 cm) with CH2Cl2–MeOH (1:1) provided 10 fractions (A–J). Fractions H and J were purified by HPLC on Rainin Dynamax C18 (1 × 25 cm) reversed-phase HPLC eluting with a gradient of 0–60% acetonitrile in 0.05% aqueous TFA in 40 min at a flow rate of 3 mL/min to provide hyemaloside A (1) (2.5 mg) and hyemaloside C (3) (1.1 mg). The MeOtBu fraction from the solvent–solvent partitioning scheme was also subjected to size exclusion chromatography on Sephadex LH-20 with CH2Cl2–MeOH (1:1), which provided 10 fractions (A–J). Fraction J was purified by HPLC as above to provide hyemaloside B (2) (3.0 mg).
Biological Testing
Details of the RNase H FRET assay have been previously described.1 The HIV cytopathicity assay was done as previously reported using CEM-SS cells infected with strain RF of HIV-1.10
Hyemaloside A (1)
brown, amorphous powder; [α]25 +58.3 (c 0.18, MeOH); UV (MeOH) λmax (log ε) 221 (5.02), 278 (4.67) nm; 1H NMR (CD3OD, 500 MHz) δ 7.15 (2H, s, H-2, H-6 of R3), 7.02 (2H, s, H-2, H-6 of R2), 6.98 (2H, s, H-2, H-6 of R1), 6.80 (2H, d, J = 8.7 Hz, H-9, H-11), 6.58 (2H, d, J = 8.5 Hz, H-8, H-12), 5.49 (1H, t, J = 9.3 Hz, H-3), 5.34 (1H, t, J = 8.3 Hz, H-2), 5.13 (1H, d, J = 7.9 Hz, H-1), 4.63 (1H, dd, J = 11.1, 8.0 Hz, H-6b), 4.53 (1H, dd, J = 11.7, 6.4 Hz, H-6a), 3.95 (1H, m, H-5), 3.88 (1H, t, J = 9.5 Hz, H-4); 13C NMR (CD3OD, 125 MHz) δ 168.14 (C, C-7 of R3), 167.79 (C, C-7 of R2), 167.25 (C, C-7 of R1), 154.36 (C, C-10), 152.08 (C, C-7), 146.61 (C, C-3, C-5 of R2), 146.41 (C, C-3, C-5 of R1), 146.35 (C, C-3, C-5 of R3), 140.11 (C, C-4 of R2), 139.99 (C, C-4 of R3), 139.94 (C, C-4 of R1), 121.33 (C, C-1 of R3), 121.04 (C, C-1 of R2), 120.78 (C, C-1 of R1), 119.86 (CH, C-9, C-11), 116.78 (CH, C-8, C-12), 110.39 (CH, C-2, C-6 of R2), 110.32 (CH, C-2, C-6 of R1), 110.26 (CH, C-2, C-6 of R3), 102.34 (CH, C-1), 76.67 (CH, C-3), 75.72 (CH, C-5), 73.31 (CH, C-2), 70.20 (CH, C-4), 64.44 (CH2, C-6); HREIMS m/z 727.1191 [M – H]− (calcd for C33H27O19, 727.1146).
Hyemaloside B (2)
brown, amorphous powder; [α]25 −36.6 (c 0.18, MeOH); UV (MeOH) λmax (log ε) 271 (3.95) nm; 1H NMR (CD3OD, 500 MHz) δ 7.92 (2H, d, J = 8.8 Hz, H-2, H-6 of R3), 7.10 (2H, s, H-2, H-6 of R1), 6.86 (2H, d, J = 8.9 Hz, H-3, H-5 of R3), 6.79 (2H, d, J = 8.9 Hz, H-9, H-11), 6.54 (2H, d, J = 8.8 Hz, H-8, H-12), 5.13 (1H, dd, J = 9.4, 9.2 Hz, H-2), 4.98 (1H, d, J = 8.1 Hz, H-1), 4.71 (1H, dd, J = 12.0, 1.7 Hz, H-6b), 4.41 (1H, dd, J = 11.9, 7.4 Hz, H-6a), 3.80 (1H, m, H-5), 3.74 (1H, t, J = 9.5 Hz, H-3), 3.55 (1H, t, J = 9.5 Hz, H-4); 13C NMR (CD3OD, 125 MHz) δ 167.91 (C, C-7 of R3), 167.67 (C, C-7 of R1), 163.66 (C, C-4 of R3), 154.22 (C, C-10), 152.15 (C, C-7), 146.49 (C, C-3, C-5 of R1), 139.93 (C, C-4 of R1), 132.94 (CH, C-2, C-6 of R3), 122.16 (C, C-1 of R3), 121.48 (C, C-1 of R1), 119.87 (CH, C-9, C-11), 116.63 (CH, C-8, C-12), 116.25 (CH, C-3, C-5 of R3), 110.34 (CH, C-2, C-6 of R1), 102.46 (CH, C-1), 76.20 (CH, C-3), 75.73 (CH, C-5), 75.32 (CH, C-2), 72.20 (CH, C-4), 64.90 (CH2, C-6); HREIMS m/z 567.1105 [M + Na]+ (calcd for C26H24O13Na, 567.1114).
Hyemaloside C (3)
brown, amorphous powder; [α]25 +22.2 (c 0.12, MeOH); UV (MeOH) λmax (log ε) 221 (4.17), 284 (4.20) nm; 1H NMR (CD3OD, 500 MHz) δ 7.00 (2H, s, H-2, H-6 of R1), 6.91 (2H, s, H-2, H-6 of R2), 6.63 (2H, s, H-2′, H-6′), 6.46 (2H, s, H-2, H-6 of HDDP), 6.85 (2H, d, J = 9.3 Hz, H-9, H-11), 6.66 (2H, d, J = 8.9 Hz, H-8, H-12), 5.66 (1H, t, J = 9.6 Hz, H-3), 5.43 (1H, dd, J = 9.5, 8.0 Hz, H-2), 5.36 (1H, dd, J = 13.7, 6.7 Hz, H-6b), 5.24 (1H, d, J = 8.1 Hz, H-1), 5.20 (1H, t, J = 10.4 Hz, H-4), 4.32 (1H, m, H-5), 3.94 (1H, dd, J = 13.0, 1.0 Hz, H-6a); 13C NMR (CD3OD, 125 MHz) δ 169.60 (C, C-7′), 169.14 (C, C-7 of HDDP), 167.60 (C, C-7 of R2), 166.95 (C, C-7 of R1), 154.62 (C, C-10), 151.82 (C, C-7), 146.46 (C, C-3, C-5 of R2), 146.22 (C, C-3, C-5 of R1), 145.92 (C, C-3, C-3′ of HDDP), 144.82 (C, C-5, C-5′ of HDDP), 140.19 (C, C-4 of R1), 140.02 (C, C-4 of R2), 137.65 (C, C-4, C-4′ of HDDP), 126.28 (C, C-1′), 125.82 (C, C-1 of HDDP), 120.67 (C, C-1 of R2), 120.49 (C, C-1 of R1), 120.08 (C, C-9, C-11), 116.78 (CH, C-8, C-12), 110.52 (CH, C-2, C-6 of R2), 110.37 (CH, C-2, C-6 of R1), 108.64 (CH, C-2′, C-6′), 108.25 (CH, C-2, C-6 of HDDP), 102.73 (CH, C-1), 74.12 (CH, C-3), 73.44 (CH, C-2), 72.83 (CH, C-5), 71.42 (CH, C-4), 63.97 (CH2, C-6); HREIMS m/z 877.1129 [M – H]− (calcd for C40H29O23, 877.1099).
Acknowledgments
This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract N01-CO-12400. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. We thank N. Trushell (NYBG) and G. Cragg (NPB) for the contract collection, T. McCloud for extractions, and J. Wilson for the HIV cytopathicity assay.
Footnotes
Supporting Information Available: This material is available free of charge via the Internet at http://pubs.acs.org.
References and Notes
- 1.Parniak MA, Min KL, Budihas SR, Le Grice SF, Beutler JA. Anal Biochem. 2003;322:33–39. doi: 10.1016/j.ab.2003.06.001. [DOI] [PubMed] [Google Scholar]
- 2.Britton G, Haslam E. J Chem Soc. 1965:7312–7319. [Google Scholar]
- 3.Thuong PT, Kang HJ, Na M, Jin W, Youn UJ, Seong YH, Song KS, Min BS, Bae K. Phytochemistry. 2007;68:2432–2438. doi: 10.1016/j.phytochem.2007.05.031. [DOI] [PubMed] [Google Scholar]
- 4.Fukushima M, Kanbe N. Japan Patent JP 62215512 A. 1987
- 5.Nawwar MAM, Hussein SAM, Merfort I. Phytochemistry. 1994;36:793–798. [Google Scholar]
- 6.Masuda T, Iritani K, Yonemori S, Oyama Y, Takeda Y. Biosci Biotechnol Biochem. 2001;65:1302–1309. doi: 10.1271/bbb.65.1302. [DOI] [PubMed] [Google Scholar]
- 7.Moharram FA, Marzouk MS, Ibrahim MT, Mabry TJ. Nat Prod Res. 2006;20:927–934. doi: 10.1080/14786410500378494. [DOI] [PubMed] [Google Scholar]
- 8.Wiedenfeld H, Zych M, Buchwald W, Furmanowa M. Sci Pharm. 2007;75:29–34. [Google Scholar]
- 9.Wilkins CK, Bohm BA. Phytochemistry. 1976;15:211–214. [Google Scholar]
- 10.Gulakowski RJ, McMahon JB, Staley PG, Moran RA, Boyd MR. J Virol Methods. 1991;33:87–100. doi: 10.1016/0166-0934(91)90010-w. [DOI] [PubMed] [Google Scholar]
- 11.Takada K, Bermingham A, O'Keefe BR, Wamiru A, Beutler JA, Le Grice SF, Lloyd J, Gustafson KR, McMahon JB. J Nat Prod. 2007;70:1647–1649. doi: 10.1021/np0702279. [DOI] [PubMed] [Google Scholar]
- 12.Dat NT, Bae K, Wamiru A, McMahon JB, LeGrice SFJ, Bona M, Beutler JA, Kim YH. J Nat Prod. 2007;70:839–841. doi: 10.1021/np060359m. [DOI] [PubMed] [Google Scholar]
- 13.Cardellina JH, II, Munro MHG, Fuller RW, Manfredi KP, McKee TC, Tischler M, Bokesch HR, Gustafson KR, Beutler JA, Boyd MR. J Nat Prod. 1993;56:1123–1129. doi: 10.1021/np50097a016. [DOI] [PubMed] [Google Scholar]
- 14.Bokesch HR, McKee TC, Cardellina JH, II, Boyd MR. Nat Prod Lett. 1994;4:155–157. [Google Scholar]
- 15.Bokesch HR, McKee TC, Currens MJ, Gulakowski RJ, McMahon JB, Cardellina JH, II, Boyd MR. Nat Prod Lett. 1996;8:133–136. [Google Scholar]
- 16.Meragelman KM, McKee TC, Boyd MR. J Nat Prod. 2000;63:427–428. doi: 10.1021/np990570g. [DOI] [PubMed] [Google Scholar]


