Abstract
Sirtuins are NAD+-dependent deacetylases that mediate cellular processes such as lifespan extension and metabolic regulation. Sirtuins form a unique metabolite, 2′-O-acetyl-ADP-ribose (OAADPr), shown to block oocyte maturation, bind to chromatin-related proteins, and activate ion channels. Given the various sirtuin phenotypes, the potential of OAADPr as a signaling molecule is extensive. However, exploring of the biological roles of OAADPr has been hindered by the lack of in vivo evidence and a reliable method for quantification. Here, we provide the first direct evidence and quantification of cellular OAADPr. Compared to endogenous OAADPr levels (0.56 ± 0.13 μM) in wildtype S. cerevisiae, deletion of all five yeast sirtuins (Sir2, Hst1-4) yielded essentially no detectable OAADPr. The single deletion of Hst2 yielded 0.37 ± 0.12 μM OAADPr. Deletion of an enzyme, Ysa1, previously shown in vitro to hydrolyze OAADPr resulted in a significant increase (0.85 ± 0.24 μM) in OAADPr. Together, these data provide evidence that cellular levels of OAADPr are controlled by the action of sirtuins and can be modulated by the Nudix hydrolase Ysa1. Our methodology consisting of internal standard 13C-OAADPr and LC-MS/MS analysis, displays excellent sensitivity and a linear dynamic range from 0.2 to 500 pmol. Moreover, extraction efficiencies were >75 %. This methodology is an essential tool in probing the biological roles of OAADPr, especially under conditions in which sirtuin phenotypes are well established.
Keywords: 2′-O-acetyl-ADP-ribose, OAADPr, sirtuin, Sir2, Hst2, Ysa1
Introduction
Sirtuins, class III histone deacetylases (HDACs), are found in all kingdoms of life and have been implicated in a growing number of diverse cellular processes such as gene silencing [1; 2; 3], lifespan extension [4; 5; 6; 7], and direct metabolic regulation [8; 9; 10]. In contrast to class I and II HDACs, which generate free acetate [11], sirtuins require NAD+ and subsequently generate a unique metabolite, 2′-O-acetyl-ADP-ribose (OAADPr) [12; 13; 14; 15; 16; 17]. Though Sir2 homologs from yeast, Drosophilia, and humans have been shown to produce OAADPr in vitro [18], there is no experimental evidence of its existence or roles in vivo. To elucidate the biological roles of OAADPr and sirtuins, a method to identify and quantify endogenous OAADPr is crucial.
In yeast there are five sirtuins which may contribute to endogenous OAADPr, the founding member yeast silent information regulator 2 (ySir2) and Hst1 - 4 [2; 3; 13; 19]. HST2 is of particular interest because it has been reported to be the most active yeast sirtuin [13] and has been extensively characterized [13; 20; 21; 22; 23; 24; 25]. Additionally, endogenous OAADPr may be degraded by a variety of enzymes such as Nudix hydrolases or esterases [18; 26; 27]. Nudix hydrolase YSA1 not only hydrolyzes OAADPr but also degrades its related metabolite ADPr [26].
Though many sirtuins have been well characterized with respect to their acetylated protein targets (e.g. histones, transcription factors, α-tubulin and metabolic enzymes), little is known about the sirtuin metabolite OAADPr. Initial studies by Borra et al. showed that microinjection of OAADPr can block oocyte maturation in starfish [18]. Additional in vitro studies demonstrated that OAADPr binds targets such as histone macroH2A1.1 [28], the SIR complex [29], and cation channel TRPM2 [30]. These in vitro studies suggest OAADPr may function as a signaling molecule or second messenger; however, there is a paucity of information that directly links cellular OAADPr with sirtuin functions. To explore the roles of sirtuins at the molecular level, it will be essential to establish a robust method to identify and quantify endogenous OAADPr.
Here we have developed the first method to detect and quantify endogenous OAADPr in biological extracts. First, the isotopically labeled internal standard, 13C-OAADPr, was synthesized enzymatically using yeast sirtuin HST2 and a histone H3 peptide substrate. Second, a method for detection and quantification of OAADPr was developed using LC-MS/MS with HILIC chromatography coupled to Multiple Reaction Monitoring (MRM) MS/MS. This methodology has a lower limit of detection of 0.2 pmol and displays a linear response over three orders of magnitude from 0.2 to 500 pmol. The utility of this method is evidenced by the first direct verification of OAADPr in yeast. Furthermore, OAADPr was quantified in wild-type S. cerevisiae and compared to a strain lacking all sirtuins and to a strain lacking an OAADPr-consuming enzyme. .
Materials and Methods
Materials
Materials were purchased from Sigma-Aldrich (St. Louis, MO) or Fisher Scientific (Pittsburgh, PA) and were of the highest quality available, unless otherwise indicated. 13C- and 12C-acetyl-H3 peptides (TGGK(ac)APR), corresponding to histone H3 tail residues 11-17, were synthesized at the UW Madison Biotechnology Peptide Synthesis Facility using acetic acid-1-13C (99 atom %) or acetic acid. NADase was obtained from Sigma-Aldrich. Yeast strains, S. cerevisiae BY4742, were obtained from Open Biosystems (Huntsville, AL).
Synthesis of 13C-OAADPr and Native OAADPr
13C-OAADPr and OAADPr (Figure 1) were synthesized enzymatically using yeast sirtuin HST2, following procedures previously described for OAADPr [18]. Briefly, the reaction consisted of 40 μM HST2, 0.8 mM NAD+, 1.6 mM 13C- or 12C- acetyl-H3 peptide (TGGK(ac)APR) in 50 mM Tris, pH 8 and was purified over a small-pore Vydac C18 column using an acetonitrile gradient (A: 0.05 % (v/v) trifluoroacetic acid (TFA) in water, B: 0.02 % (v/v) TFA in acetonitrile). Identity and purity were confirmed by mass spectrometry at the UW Madison Human Proteomics Program, supported by the Wisconsin Partnership Fund for a Healthy Future.
Figure 1.
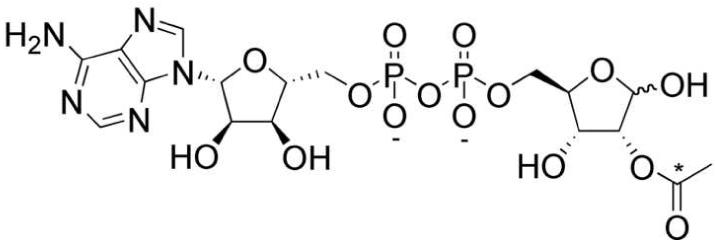
Structure of Novel Sirtuin Metabolite OAADPr, 2′-O-acetyl-ADP-ribose. The 13C carbon in internal standard 13C-OAADPr is indicated by an asterisk.
Yeast samples
Wild-type, Δhst2, and Δysa1 BY4742 S. cerevisiae yeast were obtained from Open Biosystems (Huntsville, AL). The complete sirtuin deletion strain (Δsir2Δhst1Δhst2Δhst3Δhst4) was a generous gift from Brian Kennedy at University of Washington. This strain also harbored Δfob1. Yeast were grown in YPD media overnight to OD600 of 0.6. Cells were harvested by centrifugation (1500 g, 10 min, 4 °C), weighed, and 2 nmol of 13C-OAADPr standard was added to each extract. Cell pellets were lysed by vortexing with glass-beads in 10 % (v/v) TFA, at maximum speed for 10 min at 4 °C [26]. Using this method, over 90 % of the yeast were lysed, as confirmed by microscopy. Next, cell debris was pelleted by centrifugation (17000 g, 60 min, 4 °C) and the supernatant immediately analyzed by LC-MS/MS, corresponding to approximately 4 × 107 cells per injection.
LC-MS/MS Analysis
Metabolites were analyzed at the UW Madison Biotechnology Center using an Agilent 1100 HPLC coupled to an ABI 3200 Q-trap mass spectrometer. Chromatography consisted of a HILIC column (Nest Group, 300Å 5μm polyhydroxyethyl A, 4.6 × 200 mm) eluted with a gradient of A: acetonitrile versus B: 10 mM ammonium acetate at 0.2 mL/min. The HPLC gradient was held at 20 % B for 15 min before ramping to 80 % B over 20 min. The gradient then ramped to 100 % B over 20 min where it was held for 5 min before ramping back to 20 % B over 5 min. The system was then equilibrated in 20 % B for 40 min before the next run.
The Q-trap mass spectrometer parameters were optimized using a direct infusion of 0.1 μM OAADPr in 50 % acetonitrile using a flow of 80 μL/min. The parameters were set as follows: curtain gas 20, probe temperature 350 °C, nebulizer gas 20, auxillary gas 5, interface heater on, collision gas medium, ion spray -4500V, declustering potential -55V, entrance potential -10V, collision energy -35V, and collision exit potential -4V.
The mass spectrometer was operated in negative MRM (multiple reaction monitoring) mode and monitored the following MRM transitions, 600.3/346.0 and 601.3/346.0, for OAADPr and 13C-OAADPr, respectively.
Data Analysis
Data were manually analyzed using the ABI Analyst Software 1.4.1. All data were quantified and normalized by comparison to the 13C-OAADPr internal standard.
Results
Synthesis of Internal Standard 13C-OAADPr and Native OAADPr
To begin investigating the biological roles of OAADPr, we developed a specific, sensitive method to identify and quantify OAADPr from biological samples. The first step was to generate an internal standard, 13C-OAADPr (Fig. 1). Using methodologies previously developed for OAADPr [18], 13C-OAADPr and native OAADPr (Fig. 1) were synthesized enzymatically and purified by reverse-phase chromatography. Briefly, yeast sirtuin HST2 was used to deacetylate 13C- or 12C-acetyl-H3 peptide (TGGK(ac)APR) in the presence of NAD+, generating the corresponding OAADPr standards. 13C-OAADPr and OAADPr were then individually purified over a small-pore Vydac C18 column [18]. Purity and identity were confirmed by analytical HPLC (data not shown) and MS. Mass spectra demonstrating the isotopic purities of OAADPr and 13C-OAADPr are shown in Figure 2.
Figure 2.
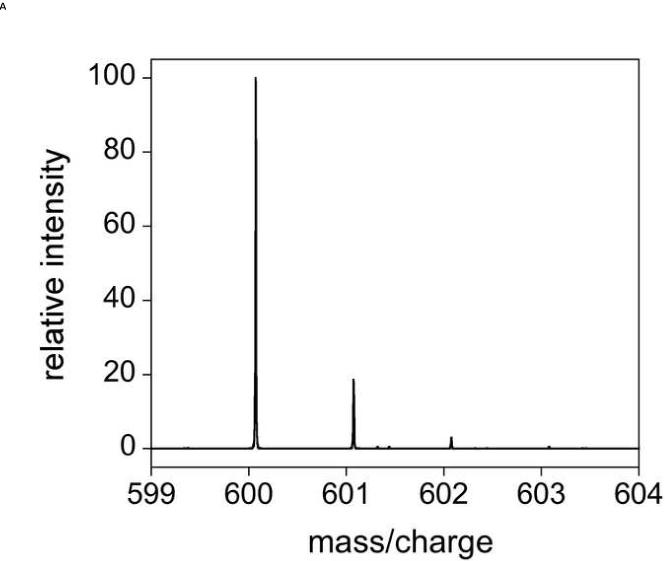

Mass spectra of sirtuin metabolite OAADPr and internal standard 13C-OAADPr. Mass spectra were acquired using 10 μM of A) OAADPr (m/z 600.072) and B) 13C-OAADPr (m/z 601.075) in 50 % acetonitrile, using a TriVersa NanoMate coupled to a Thermo Scientific LTQ-FT operated in negative mode.
Development of LC-MS/MS Method
Next, the internal standard 13C-OAADPr and native OAADPr were used to develop the LC-MS/MS methodology. Mass spectrometer parameters were optimized by directly infusing OAADPr as described in the Methods section. Under these conditions, OAADPr, 13C-OAADPr, or related metabolite ADPr, all sharing the same major fragment ion of 346.0, had similar MRM intensities (data not shown). Using the internal standard 13C-OAADPr, our methodology was shown to have excellent sensitivity, with a lower limit of detection of 0.2 pmol (Fig. 3). Additionally, the linear dynamic range extends over three orders of magnitude from 0.2 to 500 pmol (Fig. 3).
Figure 3.
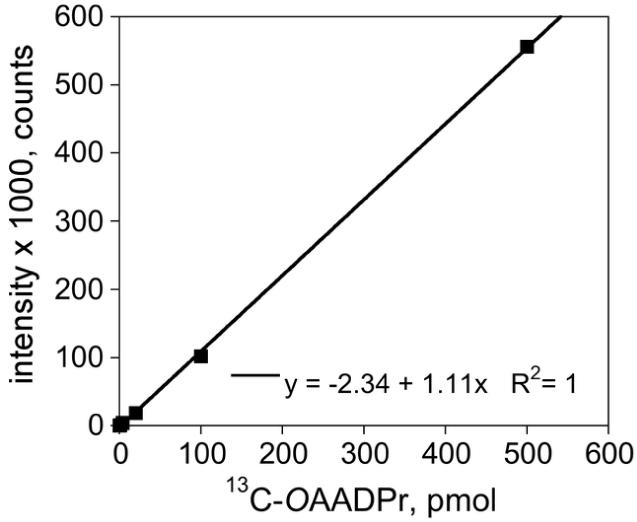
Detection Limits of OAADPr LC-MS/MS Methodology. MRM intensity of internal standard 13C-OAADPr is linear over 3 orders of magnitude from 0.2 to 500 pmol, using the LC-MS/MS methodology. The equation of the standard curve is shown inset.
Direct Evidence of Endogenous OAADPr
Here we provide the first direct evidence of endogenous OAADPr (Fig. 4). As described in the Methods section, 13C-OAADPr was spiked into yeast cell pellets prior to cell lysis and LC-MS/MS analysis. Figure 4 shows example MRM chromatograms with intensity versus time for OAADPr (solid line) within wild-type yeast (Fig. 4). There is a distinct signal from OAADPr (solid line, Fig. 4) that co-elutes exactly with the internal standard 13C-OAADPr (dashed line, Fig. 4) in all samples types tested (Fig. 4 and data not shown). Control extracts without the internal standard 13C-OAADPr display significant intensities from native OAADPr only (data not shown). The split peaks seen in Figure 4 are the 2′ and 3′ isomers of both OAADPr and 13C-OAADPr. Though sirtuins enzymatically generate the 2′ isomer, this is rapidly transesterified to form an ∼1:1 mixture of 2′: 3′ isomers [31; 32].
Figure 4.
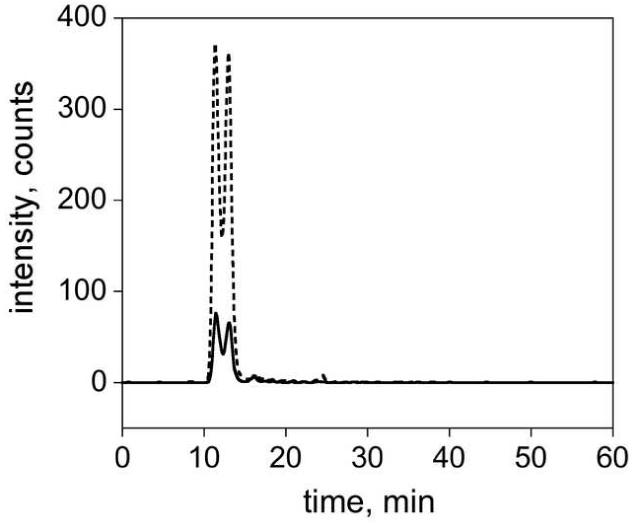
Direct Evidence of Endogenous OAADPr. Example MRM chromatograms show intensity versus time for native OAADPr (solid line) and 13C-OAADPr standard (dashed line) and in wild-type S. cerevisiae extracts.
Calculation of Extraction Efficiency
To calculate exact OAADPr concentrations, 13C-OAADPr intensities were used to calculate extraction efficiencies and to normalize between biological replicates (Fig. 5). At least three separate yeast cultures (biological replicates) were analyzed for each experimental group. Figure 5 shows example MRM chromatograms for two biological replicates from wild-type yeast (Fig. 5). Not only do the chromatograms in Figure 5 display the same retention times and split peaks as shown in Figure 4, it is apparent that the extraction efficiency was similar for all of the biological replicates. The average extraction efficiencies of the 13C-OAADPr standard were 77 ± 8 % for wild-type, 75 ± 16 % for Δhst2, and 130 ± 19 % for Δsir2Δhst1Δhst2Δhst3Δhst4, and 77 ± 4 % for Δysa1samples, with n = 3 - 6 and errors given as one standard deviation. Additionally, we have shown that the ratio of 12C/13C MRM intensities is reproducible across technical replicates, with an average variability of less than 3 % (data not shown).
Figure 5.

Direct Comparison of Biological Extracts using Internal Standard 13C-OAADPr. Example MRM chromatograms show intensity versus time for 13C-OAADPr internal standard within two different biological replicates in wild-type S. cerevisiae extracts.
Rigorous Quantification of OAADPr
Here we report the first cellular concentrations for the sirtuin metabolite OAADPr. OAADPr concentrations were determined by comparison to the 13C-OAADPr standard curve (Fig. 3) and the samples were normalized for extraction efficiency using the internal standard 13C-OAADPr.
It is important to address two minor points regarding the isotopic distributions between OAADPr and 13C-OAADPr. The natural abundance 13C-isotope within endogenous OAADPr contributes to the intensity of the 13C-OAADPr standard. This contribution was theoretically and experimentally determined to be 19 % of the OAADPr intensity (Fig. 2A). Additionally, the small amount of OAADPr present in the 13C-OAADPr standard was determined to be 2 % of the 13C-OAADPr intensity (Fig. 2B). Under our conditions of approximately 5-fold more internal standard than endogenous OAADPr, the small amount of OAADPr arising from the 13C-OAADPr standard was subtracted from the total OAADPr present in the sample.
To calculate the OAADPr concentration in yeast cells, the number of cells were estimated by OD600 [33]. All results are from 3 - 6 biological replicates, with errors given as one standard deviation. The OAADPr concentration was 0.56 ± 0.13 μM in wild-type yeast (Fig. 6). In the sirtuin deletion strain (Δhst2), the OAADPr concentration was lower at 0.37 ± 0.12 μM (Fig. 6). Comparing wildtype to Δhst2 strains, an unpaired t test yielded a p value of 0.064. Deletion of all five yeast sirtuins (Δsir2Δhst1Δhst2Δhst3Δhst4) resulted in no detectable OAADPr levels (-0.14 ± 0.33 μM, Fig. 6). Essentially, the levels were within the detection limit of the method. The p value of this data set as compared to the wild-type data is 0.0006. Deletion of YSA1, one of the enzymes implicated in OAADPr hydrolysis, resulted in a significant increase in OAADPr concentration (0.85 ± 0.24 μM). Comparing these samples to the wild-type control yielded a p value of 0.0456.
Figure 6.

Endogenous OAADPr Concentrations in BY4742 S. cerevisiae. OAADPr concentrations in wild-type (0.56 ± 0.13 μM), Δhst2 (0.37 ± 0.12 μM), complete sirtuin deletion strain Δsir2Δhst1Δhst2Δhst3Δhst4 (-0.14 ± 0.33 μM), and Nudix hydrolase deletion strain Δysa1 (0.85 ± 0.24 μM) were determined as described in the text. All values are averages from 3 - 6 biological replicates, with errors given as one standard deviation.
Discussion
Though NAD+ and its related metabolite ADPr have been studied by a variety of methods, such as enzyme cycling assays [34; 35; 36], HPLC [37; 38; 39; 40], MS [41], and LC-MS/MS [42], there has been a lack of information about the cellular levels of novel sirtuin metabolite OAADPr. Accordingly, we have developed an LC-MS/MS method to identify and quantify endogenous OAADPr.
In yeast, we found cellular levels of OAADPr to be 0.4 - 0.8 μM. For comparison, NAD+ levels in yeast are estimated to be 1 - 3 mM (summarized in [43], also [44]). In mammalian cells, NAD+ has been estimated to be ∼370 μM, both in red blood cells and tissue culture cells [41; 42]. Moreover, ADP-ribose, one of the major products that may be formed from degradation of OAADPr or directly from NAD+ hydrolysis, has been calculated to be 44 - 73 μM in tissue culture cells [40]. If indeed OAADPr functions as a second messenger or signaling molecule, levels in the low micromolar range are not unexpected. Steady-state levels of OAADPr in vivo will depend on the rate of synthesis by sirtuins, its slow degradation by spontaneous hydrolysis [18], and on the rates of utilization by a variety of endogenous OAADPr-metabolizing enzymes [18; 26; 27].
Significantly, we show sirtuin deletion (partial in Δhst2, and complete in Δsir2Δhst1Δhst2Δhst3Δhst4) or Nudix hydrolase deletion (Δysa1) affect OAADPr levels (Fig. 6). Hst2, a well-characterized yeast sirtuin [13; 20; 21; 22; 23; 24; 25] has been implicated in transcriptional regulation [20; 25]. Yeast Δhst2 samples show 30 % less OAADPr than the control samples (0.37 ± 0.12 μM versus 0.56 ± 0.13 μM, p = 0.064), under the conditions tested. Dramatically, deletion of all five yeast sirtuins results in OAADPr levels within the limits of detection (-0.14 ± 0.33 μM, Fig. 6). Comparison of the complete sirtuin deletion strain to wild-type yields a p-value of 0.0006, indicating a highly significant difference between the data sets.
Previously, we have described three different enzymatic activities in yeast extracts that are capable of utilizing OAADPr as a substrate. These enzyme activities include (1) an esterase that removes the acetyl-group to form ADPr, (2) a Nudix hydrolase that cleaves the pyrophosphate bond to yield AMP and acetylated phospho-ribose, and (3) an acetyltransferase that transfers the acetate to an unknown acceptor [18; 26; 27]. Interestingly, these same OAADPr-utilizing activities have been observed in mammalian tissue culture cells ([18; 26; 27]). Here we have shown that deletion of one of these OAADPr-utilizing enzymes, Nudix hydrolase Ysa1, results in a significant increase (p value of 0.0456 ) in OAADPr concentration (0.85 ± 0.24 μM versus 0.56 ± 0.13 μM). These data provide direct evidence that Ysa1 can modulate the cellular levels of OAADPr.
As pointed out in the Results section, there is minor isotopic spillover between the endogenous OAADPr and the 13C-OAADPr standard. This is easily taken into account to correct the observed levels from biological samples, however, future use of an isotopic standard labeled at multiple positions, such as 13Cx, 15Nx-OAADPr would alleviate this necessary correction.
In summary, we have developed the first methodology to detect and quantify the unique sirtuin metabolite OAADPr, using LC-MS/MS and internal standard 13C-OAADPr. This system results in rigorous quantification by correlating chromatographic retention times, parent masses, and fragment masses for both OAADPr and the internal standard 13C-OAADPr. Additionally, 13C-OAADPr was used to normalize for extraction efficiency and to quantify OAADPr levels. The MRM methodology provides higher confidence in metabolite identification versus traditional enzyme cycling or UV-based HPLC methods, which are affected by contaminants or co-eluting species [42]. We have demonstrated that 13C-OAADPr exactly co-elutes with native OAADPr with a lower limit of detection of 0.2 pmol and a linear range over 3 orders of magnitude from 0.2 to 500 pmol. Using our methodology, average extraction efficiencies were ≥ 80 %.
Most importantly, we have provided the first direct evidence of endogenous OAADPr in biological extracts. We also demonstrate that genetic manipulation of OAADPr producing and degrading enzymes regulate the cellular levels of OAADPr. Decreased OAADPr levels were observed in yeast sirtuin deletions strains, Δhst2 and Δsir2Δhst1Δhst2Δhst3Δhst4, while increased OAADPr was observed with the Δysa1 strain. We anticipate this methodology will be essential in determining the biological roles of OAADPr, studying its roles as a possible secondary messenger, and establishing the regulatory pathways that affect the production and degradation of OAADPr.
Acknowledgements
We thank Jim Brown at the UW Madison Biotechnology Center for assistance with the mass spectrometry. We also thank Brian Kennedy at the University of Washington for the complete sirtuin deletion strain.
Abbreviations
- OAADPr
2′-O-acetyl-ADP-ribose
- LC-MS/MS
liquid chromatography coupled to tandem mass spectrometry
- NAD+
nicotinamide adenine dinucleotide
- TFA
trifluoroacetic acid
- MRM
multiple reaction monitoring
- YPD
yeast extract peptone dextrose
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Tanny JC, Dowd GJ, Huang J, Hilz H, Moazed D. An enzymatic activity in the yeast Sir2 protein that is essential for gene silencing. Cell. 1999;99:735–745. doi: 10.1016/s0092-8674(00)81671-2. [DOI] [PubMed] [Google Scholar]
- [2].Gasser SM, Cockell MM. The molecular biology of the SIR proteins. Gene. 2001;279:1–16. doi: 10.1016/s0378-1119(01)00741-7. [DOI] [PubMed] [Google Scholar]
- [3].Rusche LN, Kirchmaier AL, Rine J. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu Rev Biochem. 2003;72:481–516. doi: 10.1146/annurev.biochem.72.121801.161547. [DOI] [PubMed] [Google Scholar]
- [4].Tissenbaum HA, Guarente L. Increased dosage of a Sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410:227–30. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- [5].Rogina B, Helfand SL. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc Natl Acad Sci U S A. 2004;101:15998–16003. doi: 10.1073/pnas.0404184101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Picard F, Kurtev M, Chung N, Topark-Ngarm A, Senawong T, Machado De Oliveira R, Leid M, McBurney MW, Guarente L. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature. 2004;429:771–6. doi: 10.1038/nature02583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Cohen HY, Miller C, Bitterman KJ, Wall NR, Hekking B, Kessler B, Howitz KT, Gorospe M, de Cabo R, Sinclair DA. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science. 2004;305:390–2. doi: 10.1126/science.1099196. [DOI] [PubMed] [Google Scholar]
- [8].Starai VJ, Celic I, Cole RN, Boeke JD, Escalante-Semerena JC. Sir2-dependent activation of acetyl-CoA synthetase by deacetylation of active lysine. Science. 2002;298:2390–2392. doi: 10.1126/science.1077650. [DOI] [PubMed] [Google Scholar]
- [9].Hallows WC, Lee S, Denu JM. Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. PNAS. 2006;103:10230–10235. doi: 10.1073/pnas.0604392103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Schwer B, Bunkenborg J, Verdin RO, Andersen JS, Verdin E. Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2. PNAS. 2006;103:10224–10229. doi: 10.1073/pnas.0603968103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Grozinger CM, Schreiber SL. Deacetylase enzymes: biological functions and the use of small-molecule inhibitors. Chem Biol. 2002;9:3–16. doi: 10.1016/s1074-5521(02)00092-3. [DOI] [PubMed] [Google Scholar]
- [12].Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- [13].Smith JS, Brachmann CB, Celic I, Kenna MA, Muhammad S, Starai VJ, Avalos JL, Escalante-Semerena JC, Grubmeyer C, Wolberger C, Boeke JD. A phylogenetically conserved NAD(+)-dependent protein deacetylase activity in the Sir2 protein family. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:6658–6663. doi: 10.1073/pnas.97.12.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Landry J, Sutton A, Tafrov ST, Heller RC, Stebbins J, Pillus L, Sternglanz R. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc Natl Acad Sci U S A. 2000;97:5807–11. doi: 10.1073/pnas.110148297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Tanner KG, Landry J, Sternglanz R, Denu JM. Silent information regulator 2 family of NAD-dependent histone/protein deacetylases generates a unique product, 1-O-acetyl-ADP-ribose. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:14178–14182. doi: 10.1073/pnas.250422697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Tanny JC, Moazed D. Coupling of histone deacetylation to NAD breakdown by the yeast silencing protein Sir2: Evidence for acetyl transfer from substrate to an NAD breakdown product. Proc Natl Acad Sci U S A. 2001;98:415–20. doi: 10.1073/pnas.031563798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Sauve AA, Celic I, Avalos J, Deng H, Boeke JD, Schramm VL. Chemistry of gene silencing: the mechanism of NAD+-dependent deacetylation reactions. Biochemistry. 2001;40:15456–63. doi: 10.1021/bi011858j. [DOI] [PubMed] [Google Scholar]
- [18].Borra MT, O’Neill FJ, Jackson MD, Marshall B, Verdin E, Foltz KR, Denu JM. Conserved enzymatic production and biological effect of O-acetyl-ADP-ribose by silent information regulator 2-like NAD(+)-dependent deacetylases. Journal of Biological Chemistry. 2002;277:12632–12641. doi: 10.1074/jbc.M111830200. [DOI] [PubMed] [Google Scholar]
- [19].Frye RA. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem Biophys Res Commun. 2000;273:793–8. doi: 10.1006/bbrc.2000.3000. [DOI] [PubMed] [Google Scholar]
- [20].Perrod S, Cockell MM, Laroche T, Renauld H, Ducrest AL, Bonnard C, Gasser SM. A cytosolic NAD-dependent deacetylase, Hst2p, can modulate nucleolar and telomeric silencing in yeast. Embo J. 2001;20:197–209. doi: 10.1093/emboj/20.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Zhao K, Chai X, Clements A, Marmorstein R. Structure and autoregulation of the yeast Hst2 homolog of Sir2. Nat Struct Biol. 2003;10:864–71. doi: 10.1038/nsb978. [DOI] [PubMed] [Google Scholar]
- [22].Zhao K, Chai X, Marmorstein R. Structure of the yeast Hst2 protein deacetylase in ternary complex with 2′-O-acetyl ADP ribose and histone peptide. Structure. 2003;11:1403–11. doi: 10.1016/j.str.2003.09.016. [DOI] [PubMed] [Google Scholar]
- [23].Lamming DW, Latorre-Esteves M, Medvedik O, Wong SN, Tsang FA, Wang C, Lin SJ, Sinclair DA. HST2 mediates SIR2-independent life-span extension by calorie restriction. Science. 2005;309:1861–4. doi: 10.1126/science.1113611. [DOI] [PubMed] [Google Scholar]
- [24].Smith BC, Denu JM. Sir2 Protein Deacetylases: Evidence for Chemical Intermediates and Functions of a Conserved Histidine. Biochemistry. 2006;45:272–282. doi: 10.1021/bi052014t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Durand-Dubief M, Sinha I, Fagerstroem-Billai F, Bonilla C, Wright A, Grunstein M, Ekwall K. Specific functions for the fission yeast Sirtuins Hst2 and Hst4 in gene regulation and retrotransposon silencing. Embo Journal. 2007;26:2477–2488. doi: 10.1038/sj.emboj.7601690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Rafty LA, Schmidt MT, Perraud AL, Scharenberg AM, Denu JM. Analysis of O-acetyl-ADP-ribose as a target for Nudix ADP-ribose hydrolases. Journal of Biological Chemistry. 2002;277:47114–47122. doi: 10.1074/jbc.M208997200. [DOI] [PubMed] [Google Scholar]
- [27].Ono T, Kasamatsu A, Oka S, Moss J. The 39-kDa poly(ADP-ribose) glycohydrolase ARH3 hydrolyzes O-acetyl-ADP-ribose, a product of the Sir2 family of acetyl-histone deacetylases. PNAS. 2006 doi: 10.1073/pnas.0607911103. 0607911103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Kustatscher G, Hothorn M, Pugieux C, Scheffzek K, Ladurner AG. Splicing regulates NAD metabolite binding to histone macroH2A. Nature Structural & Molecular Biology. 2005;12:624–625. doi: 10.1038/nsmb956. [DOI] [PubMed] [Google Scholar]
- [29].Liou G-G, Tanny JC, Kruger RG, Walz T, Moazed D. Assembly of the SIR Complex and Its Regulation by O-Acetyl-ADP-Ribose, a Product of NAD-Dependent Histone Deacetylation. Cell. 2005;121:515–527. doi: 10.1016/j.cell.2005.03.035. [DOI] [PubMed] [Google Scholar]
- [30].Grubisha O, Rafty LA, Takanishi CL, Xu X, Tong L, Perraud A-L, Scharenberg AM, Denu JM. Metabolite of SIR2 Reaction Modulates TRPM2 Ion Channel. J. Biol. Chem. 2006;281:14057–14065. doi: 10.1074/jbc.M513741200. [DOI] [PubMed] [Google Scholar]
- [31].Sauve AA, Celic I, Avalos J, Deng HT, Boeke JD, Schramm VL. Chemistry of gene silencing: The mechanism of NAD(+)-dependent deacetylation reactions. Biochemistry. 2001;40:15456–15463. doi: 10.1021/bi011858j. [DOI] [PubMed] [Google Scholar]
- [32].Jackson AD, Denu JM. Structural identification of 2 ′- and 3 ′-O-acetyl-ADP-ribose as novel metabolites derived from the Sir2 family of beta-NAD(+)-dependent histone/protein deacetylases. Journal of Biological Chemistry. 2002;277:18535–18544. doi: 10.1074/jbc.M200671200. [DOI] [PubMed] [Google Scholar]
- [33].Burke D, Dawson D, Stearns T. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Manual. Cold Spring Harbor Laboratory Press; 2000. [Google Scholar]
- [34].Bernofsky C, Swan M. An improved cycling assay for nicotinamide adenine dinucleotide. Analytical Biochemistry. 1973;53:452–458. doi: 10.1016/0003-2697(73)90094-8. [DOI] [PubMed] [Google Scholar]
- [35].Jacobson EL, Jacobson MK. Pyridine nucleotide levels as a function of growth in normal and transformed 3T3 cells. Archives of Biochemistry and Biophysics. 1976;175:627–634. doi: 10.1016/0003-9861(76)90553-1. [DOI] [PubMed] [Google Scholar]
- [36].Lin SS, Manchester JK, Gordon JI. Enhanced Gluconeogenesis and Increased Energy Storage as Hallmarks of Aging in Saccharomyces cerevisiae. J. Biol. Chem. 2001;276:36000–36007. doi: 10.1074/jbc.M103509200. [DOI] [PubMed] [Google Scholar]
- [37].Jones DP. Determination of pyridine dinucleotides in cell extracts by high-performance liquid chromatography. Journal of Chromatography B: Biomedical Sciences and Applications. 1981;225:446–449. doi: 10.1016/s0378-4347(00)80293-5. [DOI] [PubMed] [Google Scholar]
- [38].Litt MR, Potter JJ, Mezey E, Mitchell MC. Analysis of pyridine dinucleotides in cultured rat hepatocytes by high-performance liquid chromatography. Analytical Biochemistry. 1989;179:34–36. doi: 10.1016/0003-2697(89)90196-6. [DOI] [PubMed] [Google Scholar]
- [39].Ryll T, Wagner R. Improved ion-pair high-performance liquid chromatographic method for the quantification of a wide variety of nucleotides and sugar--nucleotides in animal cells. Journal of Chromatography: Biomedical Applications. 1991;570:77–88. doi: 10.1016/0378-4347(91)80202-n. [DOI] [PubMed] [Google Scholar]
- [40].Gasser A, Guse AH. Determination of intracellular concentrations of the TRPM2 agonist ADP-ribose by reversed-phase HPLC. Journal of Chromatography B-Analytical Technologies in the Biomedical and Life Sciences. 2005;821:181–187. doi: 10.1016/j.jchromb.2005.05.002. [DOI] [PubMed] [Google Scholar]
- [41].Yang H, Yang T, Baur J, Perez E, Matsui T, Carmona J, Lamming D, Souza-Pinto N, Bohr V, Rosenzweig A, de Cabo R, Sauve A, Sinclair D. Nutrient-Sensitive Mitochondrial NAD(+) Levels Dictate Cell Survival. Cell. 2007;130:1095–107. doi: 10.1016/j.cell.2007.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Yamada K, Hara N, Shibata T, Osago H, Tsuchiya M. The simultaneous measurement of nicotinamide adenine dinucleotide and related compounds by liquid chromatography/electrospray ionization tandem mass spectrometry. Analytical Biochemistry. 2006;352:282–285. doi: 10.1016/j.ab.2006.02.017. [DOI] [PubMed] [Google Scholar]
- [43].Lin S-J, Ford E, Haigis M, Liszt G, Guarente L. Calorie restriction extends yeast life span by lowering the level of NADH. Genes Dev. 2004;18:12–16. doi: 10.1101/gad.1164804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Belenky P, Racette FG, Bogan KL, McClure JM, Smith JS, Brenner C. Nicotinamide Riboside Promotes Sir2 Silencing and Extends Lifespan via Nrk and Urh1/Pnp1/Meu1 Pathways to NAD+ Cell. 2007;129:473–484. doi: 10.1016/j.cell.2007.03.024. [DOI] [PubMed] [Google Scholar]


