Abstract
To find inherited causes of autism-spectrum disorders, we studied families in which parents share ancestors, enhancing the role of inherited factors. We mapped several loci, some containing large, inherited, homozygous deletions that are likely mutations. The largest deletions implicated genes, including PCDH10 (protocadherin 10) and DIA1 (deleted in autism1, or c3orf58), whose level of expression changes in response to neuronal activity, a marker of genes involved in synaptic changes that underlie learning. A subset of genes, including NHE9 (Na+/H+ exchanger 9), showed additional potential mutations in patients with unrelated parents. Our findings highlight the utility of “homozygosity mapping” in heterogeneous disorders like autism but also suggest that defective regulation of gene expression after neural activity may be a mechanism common to seemingly diverse autism mutations.
Autism is a severe neuropsychiatric disorder characterized by impaired social interaction and communication and by repetitive and stereotyped interests and behavior. Autism includes mental retardation in up to 70% (1) and seizures in 20 to 25% of cases (2). Although autism spectrum disorders (ASDs) are highly heritable, they exhibit wide clinical variability and heterogeneous genetic architecture, which have hindered gene identification (3, 4).
The great majority of identified ASD genes have high rates of de novo mutation. Large, de novo, microscopically evident chromosomal anomalies have been reported in 1 to 2% of cases of autism (5), and recent work has identified submicroscopic deletions and duplications, collectively called copy number variants (CNVs), affecting many loci, in 10% or more of sporadic cases (4, 6–9). Mutations in FMR1, TSC1, TSC2, NF1, UBE3A, and MECP2 that cause mono-genic neurological disorders also cause syndromic autism and are all associated with high rates of de novo or recent mutation. The extreme genetic heterogeneity of autism, and the high de novo mutation rate, have hindered linkage studies of inherited autism susceptibility loci (3, 4). The accumulating number of distinct, individually rare genetic causes in autism (5, 10, 11) suggests that the genetic architecture of autism resembles that of mental retardation and epilepsy, with many syndromes, each individually rare, as well as other cases potentially reflecting complex interactions between inherited changes (12).
“Homozygosity mapping” (13, 14) in pedigrees with shared ancestry has been a successful methodology to discover autosomal recessive disease genes for many genetically heterogeneous neurodevelopmental conditions, such as brain malformations and mental retardation (15–17). Because of the large amount of genetic information that can be obtained from pedigrees in which parents share a recent common ancestor, the need to pool information from multiple families is reduced. Homozygosity mapping has been suggested as potentially useful for mapping complex traits as well (18), but this hypothesis has not been tested, other than one study of patent ductus arteriosus (generally considered to be a multifactorial condition) (19). Although segregation analyses have supported a role for autosomal recessive genes in ASD (20), homozygosity mapping has not been applied to autism to date. Here, we show that homozygosity mapping can be useful for identifying loci and genes in ASDs in consanguineous populations.
Ascertainment of pedigrees with autism and recent shared ancestry
The Homozygosity Mapping Collaborative for Autism (HMCA) (21) has recruited 104 families (79 simplex and 25 multiplex) from the Arabic Middle East, Turkey, and Pakistan (table S1 and fig. S1), of which 88 pedigrees (69 simplex and 19 multiplex) have cousin marriages (i.e., parental consanguinity). To establish thorough research diagnoses, international participating clinicians received training in accepted autism research scales. When research scales were not available in the language of their country, these clinicians enrolled patients and family members based on DSM-IV-TR diagnoses that were informed by these clinicians’ experience with validated research scales. Additional direct assessments of patients were conducted by clinical members of the Boston team, which included developmental psychologists (J.W., E.L., R.M.J.), pediatric neurologists (G.M., A.P.), a clinical geneticist (W.H.T.), and a neuropsychiatrist (E.M.M.). Reliability between clinician assessments was high; a description of clinical methods is available in (22).
Marriage between first cousins increases the prevalence of neurological birth defects by about 100%, with this excess attributable to increased autosomal recessive causes (23, 24), and with de novo chromosome anomalies representing a correspondingly reduced portion of the total (24). Although comparable epidemiological data for autism are not available, we reasoned that a prominent involvement of autosomal recessive genes in autism would be signaled by differences in the male-to-female (M/F) ratio of affected children in consanguineous (related) versus nonconsanguineous marriages (although recessive causes of autism may still retain some gender-specific difference in penetrance). Across the HMCA, the M/F ratio of affected individuals was typical, at 4.8:1 (115 males: 24 females). However, in consanguineous, multiplex pedigrees, the M/F ratio was 2.6:1 (34 males: 13 females) (fig. S1), compared to 7.4:1 (81 males: 11 females) for the other categories of families (i.e., nonconsanguineous and consanguineous simplex) (chi-square = 5.37, df = 1, P = 0.02). The M/F ratio of 2.6:1 is close to what would be predicted if the prevalence of autism were doubled in these families, with the excess attributable to recessive causes (23, 24).
An increased role for inherited factors in autism families with shared ancestry was also suggested by a low rate of de novo CNVs that segregated with disease, despite the use of two sensitive methods for detecting them: the Affymetrix Gene-Chip Human Mapping 500K single-nucleotide polymorphism (SNP) array, as well as bacterial artificial chromosome (BAC) comparative genomic hybridization (CGH) microarrays [principally an extensively validated, commercial microarray from Signature Genomics (see 22)]. Whereas rates of inherited CNVs (some potentially causative) were high in both the SNP and BAC arrays, ranging in size from 1.4 kb to 3.9 Mb (tables S2 and S3), overall rates of de novo CNVs that segregated with ASD were 0% in consanguineous multiplex (0 of 42 patients) and 1.9% in consanguineous simplex families (1 of 52 patients), which were considerably lower than reported for non-consanguineous families: 1.28% in the HMCA overall versus 7.1% using representational oligonucleotide microarray analysis in autism (6) (chi-square = 4.438, df = 1, P < 0.02), or versus 27.5% (7) (chi-square = 17.733, df = 1, P < 0.01) using another BAC array in syndromic autism. A large study, using identical BAC arrays run in the same lab as our study, found 5.6% (84 of 1500) of patients referred to Signature Genomics with de novo or pathogenic CNVs (chi-square = 3.052, df = 1, P < 0.05) (25). The HMCA rate of de novo CNVs was similar to previously reported rates in multiplex pedigrees with autism [1.28% in the HMCA versus 2.6%, or 2 of 77, in multiplex autism (6), chi-square = 0.557, df = 1, P = 0.22] and in controls [1.28% HMCA versus 1.0%, or 2 of 196, in control subjects (6), chi-square = 0.001, df = 1, P = 0.49], despite the fact that the 500K platform used here has significantly higher coverage. The single large de novo CNV discovered was a 3-Mb deletion at 22q11.21, the velocardiofacial syndrome (VCFS) locus (encompassing all SNPs between rs432770 and rs1014626), which has been previously reported in autistic patients (26). The relatively reduced M/F ratio of affected children and the reduced rate of linked de novo CNVs in the consanguineous sample (not significantly different from rates in control) both suggest that consanguineous pedigrees with autism are enriched for autosomal recessive causes similar to other congenital neurological disorders in consanguineous populations (23, 24).
Homozygosity mapping implicates heterogeneous loci and genes
Homozygosity mapping in consanguineous autism pedigrees suggested considerable genetic heterogeneity, implicating several genetic loci, with limited overlap between pedigrees. Using the Affymetrix Gene-Chip Human Mapping 500K SNP arrays, a locus-exclusionary approach was taken assuming a model of autosomal recessive inheritance and high penetrance. Several single families showed one or two loci with strong support for linkage [multipoint logarithm of the odds ratio for linkage (lod) scores ranging from 2.4 to 2.96] (table S4), corroborated by microsatellite analysis. Potentially linked loci were generally nonoverlapping between families, consistent with genetic heterogeneity, although two families shared linkage to an overlapping region of chromosome 2q (AU-4500, lod = 2.41, and AU-4200, lod = 1.81) that has been previously implicated in other autism linkage studies (27). The higher lod scores from single families, although not achieving genome-wide significance, are comparable to the highest lod scores achieved by pooling hundreds of nonconsanguineous pedigrees (3, 4, 27).
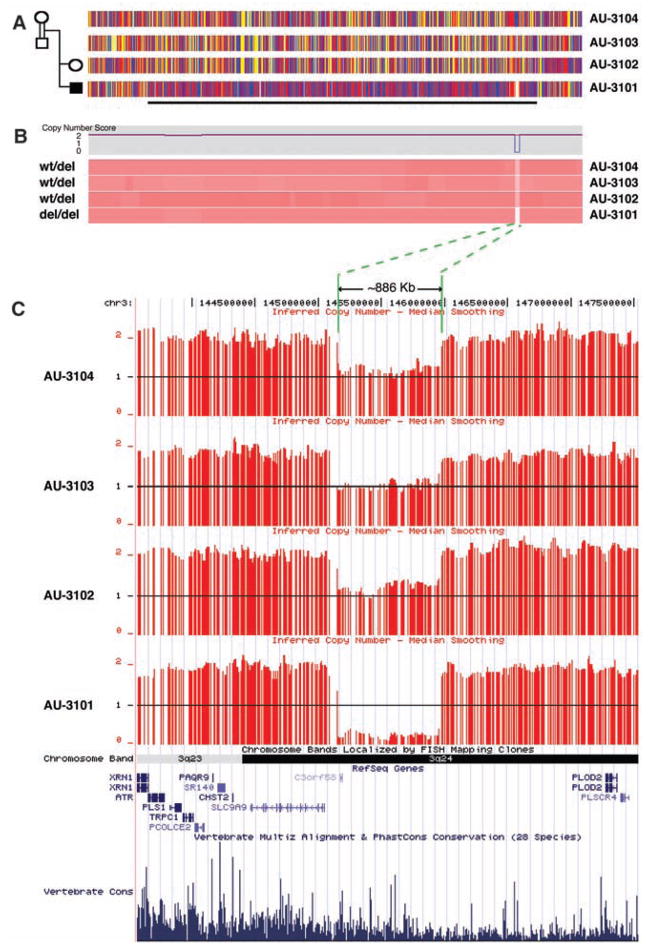
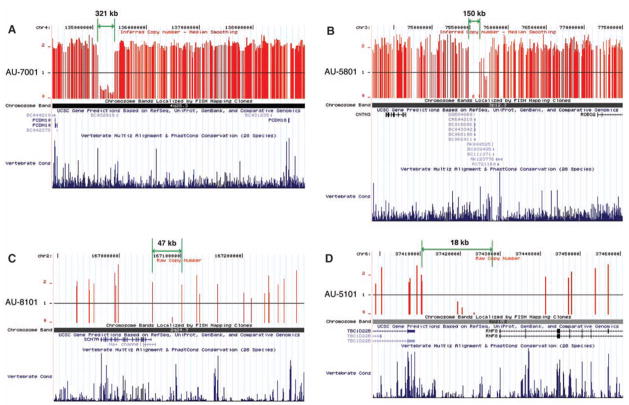
Although the large size of linked loci precluded systematic gene sequencing in most cases, we were surprised to see that several consanguineous pedigrees showed large, rare, inherited homozygous deletions within linked regions, some of which are very likely causative mutations (Figs. 1 and 2 and table S5). Such deletions were present in 5 of 78 consanguineous pedigrees (6.4%) and ranged in size from 18 thousand base pairs (kbp) to > 880 kbp. For example, patient AU-3101, a boy diagnosed with autistic disorder and seizures, demonstrated a 74-cM segment of identity by descent (IBD) on chromosome 3q (lod score = 1.45) (Fig. 1), with an ~886-kbp homozygous deletion within 3q24. The deletion is hemizygous in both parents and an unaffected sibling (hence inherited from a common grandparent or more distant ancestor) but was not present in any of the other 393 samples from our autism pedigrees, nor in 184 Middle Eastern control chromosomes, nor in 2200 samples from the Autism Genetic Resource Exchange (AGRE) repository (www.agre.org). This deletion was confirmed using Agilent oligo arrays (fig. S2) and polymerase chain reaction (PCR) (fig. S3). The deletion completely removes c3orf58, which encodes an uncharacterized protein with a signal peptide that localizes to the Golgi (28). Moreover, the deletion is near the 5′ region of NHE9, such that only 60 to 85 kbp upstream of the transcription initiation site is spared. NHE9 (also known as SLC9A9) encodes a (Na+, K+)/H+ exchanger previously reported to have been disrupted in a pedigree with a developmental neuropsychiatric disorder and mild mental retardation (29). Of note, SNPs within NHE9 and less than 100 kb from c3orf58 were among the top 21 regions (P < 0.00001) in the human genome showing adaptive selection (i.e., evidence for recent evolutionary selection) in a recent genome-wide analysis (30). A second >300 kbp, linked, homozygous deletion (again not present in >2000 individuals other than this family) is closest to PCDH10 on 4q28 (Fig. 2 and table S5), which encodes a cadherin superfamily protein essential for normal forebrain axon outgrowth (31). Smaller deletions (also unique to the individual family) (table S5) were closest to CNTN3, encoding BIG-1, an immunogloglobulin superfamily protein that stimulates axon outgrowth (32); RNF8, encoding a RING finger protein that acts as a ubiquitin ligase and transcriptional co-activator (33); and SCN7A (amid a cluster of voltage-gated sodium channels that also includes SCN1A, SCN2A, SCN3A, and SCN9A) on 2q. Homozygous deletions were confirmed by PCR (22). Of note, all of the implicated genes have high levels of expression in brain. Although without further data it is not known that all of these “private” homozygous deletions are causative, some are very likely to be, with larger deletions also more commonly pathogenic than smaller ones (4, 6).
Fig. 1.
Homozygosity mapping in pedigree AU-3100 reveals an ~886-kb inherited homozygous deletion at 3q24 within a 74 cM block of IBD in patient AU-3101, who has autism with seizures. (A) SNP genotypes for each subject in the pedigree using the 500K SNP microarray along chromosome 3q. The four horizontal tracks represent SNP genotyping data along 3q from centromere to telomere moving left to right, aligned with each individual in the pedigree. Red and blue vertical hatches represent homozygous SNPs, and yellow hatches indicate heterozygosity. The horizontal black line demarcates the 74-cM region of IBD in patient 3101 that is not found in an unaffected sibling or parents. (B) Copy number data using the 500K SNP microarray and dCHIP (45) hidden Markov model inferred methodology aligned with the genotyping SNPs from (A). The top panel indicates copy number (CN) score for AU-3101, and the lower panel shows pink tracks corresponding to CN data for all corresponding SNPs along 3q24 above. Dark pink shade indicates CN = 2 for the majority of this region. A white area in AU-3101 represents the homozygous deletion. The light pink equivalent region in AU-3102, AU-3103, and AU-3104 represents CN = 1 or carrier status of the wild-type deletion (wt/del). (C) Mapping of inferred CN data SNP-by-SNP on the University of California Santa Cruz (UCSC) genome browser demonstrates the deletion of c3orf58 and an extensive genomic (likely regulatory) region 5′ to the transcriptional start of SLC9A9 (NHE9). Horizontal red lines indicate each SNP with copy number of 0, 1, or 2. Green lines and arrows demarcate the extent of the deletion. Alignment of annotated genes in the National Center for Biotechnology Information RefSeq database are shown, as well as a representation of vertebrate conservation using multiz and related tools in the UCSC/Penn State Bioinformatics comparative genomic alignment pipeline.
Fig. 2.
Homozygous deletions within regions of IBD that segregate with disease were identified using the Affymetrix 500K microarray and are represented as schematic diagrams using the UCSC genome browser. Vertical red lines indicate each SNP with copy number of 0, 1, or 2. The green lines and arrows indicate the distance between the two SNPs with copy number equal to or greater than 1 flanking each deletion. Chromosomal bands containing deletions, genes in the vicinity of deletions, and vertebrate conservation using multiz and related tools in the UCSC/Penn State Bioinformatics comparative genomic alignment pipeline are also shown. A second large deletion: (A) Homozygous deletion in AU-7001 within a protocadherin cluster proximal to PCDH10. Smaller deletions: (B) Homozygous deletion in AU-5801 encompasses 5′ noncoding region of CNTN3. (C) Homozygous deletion in AU-8101 contains 5′ noncoding regions of SCN7A and a related sodium channel isoform. (D) Homozygous deletion in AU-5101 removes 5′ region of RNF8 and 3′ noncoding region of TBC1D228, an un-characterized Rab guanosine triphosphatase. This deletion was fine-mapped using PCR to demonstrate that the deletion excludes the first exon of RNF8. (See also table S5.)
Autism-associated genes are regulated by neuronal activity
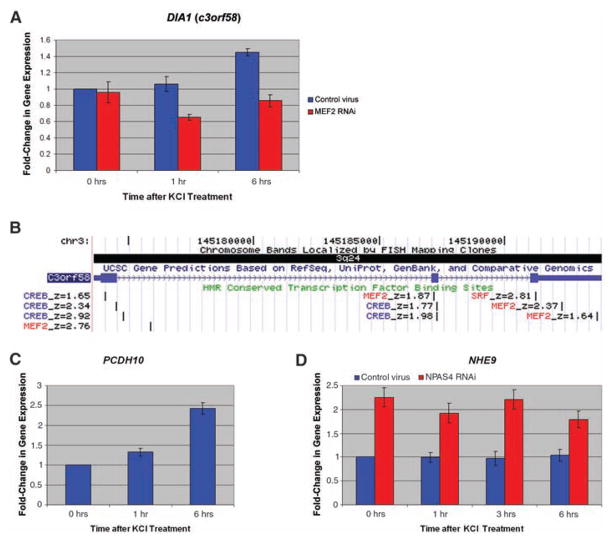
Unexpectedly, the three genes within or closest to the two largest deletions (c3orf58, NHE9, and PCDH10) were all independently identified by unbiased screens looking for genes regulated by neuronal activity or for targets of transcription factors induced by activity. Neuronal activity induces a set of transcription factors (including MEF2, NPAS4, CREB, EGR, SRF, and others) with time courses of minutes to hours, and these transcription factors induce or repress specific target genes that mediate synaptic development and plasticity (34). This activity-based gene expression results in protein changes that selectively enhance or repress synapses, likely forming a part of learning paradigms (35). In microarray screens using cultured rat hippocampal neurons (performed blind to the genetic study), 1005 complementary DNAs (cDNAs) (of 22,407 nonredundant genes tested, i.e., ≈5% of the transcriptome) were identified as altered in expression after neuronal membrane depolarization by elevated KCl (21). Among these “neural activity–regulated” genes, c3orf58 (deleted in patient AU-3101) was robustly increased within 6 hours of membrane depolarization (Fig. 3A). c3orf58 contained several evolutionarily conserved binding sites for MEF2, CREB, and SRF (Fig. 3B), and depolarization-dependent transcription of c3orf58 was strongly inhibited by RNA interference (RNAi) knock-down of the MEF2 transcription factor (Fig. 3A), which suggests that c3orf58 may be a direct or indirect MEF2 target. We propose renaming this gene DIA1 (deleted in autism-1). In the same forward screen, transcription of PCDH10 (the gene closest to the second-largest, >300-kbp, homozygous deletion in patient AU-7001) (Fig. 2A) was strongly up-regulated in hippocampal neurons in response to membrane depolarization (Fig. 3C). Although PCDH10 was not greatly affected by MEF2 RNAi, PCDH10 was robustly induced by a MEF2-VP16 fusion protein, reaching 1.31 ± 0.09 fold-induction of transcription and 1.94 ± 0.23 fold-induction at 1 hour and 2.5 hours, respectively. This transcriptional activation also suggests strongly that PCDH10 is a transcriptional target of MEF2. Because the KCl depolarization assay identified fewer than 5% of the transcriptome as altered in expression, the identification in this assay of two of three genes associated with the two largest deletions is quite unlikely by chance alone (binomial test, P < 0.006 for two or more genes in the “altered expression” category). Enrichment of genes induced by membrane depolarization remains significant even when genes closest to all of the five homozygous deletions are considered (two of six transcripts found on the array, binomial test, P < 0.027). Moreover, a separate screen using RNAi knock-down of NPAS4, a transcription factor activated in response to depolarization, showed that NHE9 was one of 292 out of 22,407 cDNAs (1.3%) whose transcription was significantly altered (in this case, increased) (Fig. 3D) (22), although NHE9 expression was not detectably altered by membrane depolarization alone. These transcriptional effects suggest that the homozygous deletions in autism patients may preferentially involve activity-regulated genes, either mutating their coding sequence (e.g., DIA1) or potentially affecting conserved DNA sequences (NHE9, PCDH10) that may be critical for proper transcriptional regulation.
Fig. 3.
Genes within or juxtaposed to homozygous deletions show activity-dependent gene regulation or are targets of transcription factors regulated by neuronal activity. (A) Activation of DIA1 (c3orf58) gene expression in rat hippocampal cultures (0, 1, and 6 hours) after membrane depolarization with KCl. Control lentivirus shown in blue, and cultures transduced with MEF2A and MEF2D RNAi lentivirus shown in red. (B) The genomic structure of DIA1 (c3orf58), also showing highly conserved transcription factor binding sites based on meeting computation thresholds of conservation in human/mouse/rat alignment with the Transfac Matrix Database (v7.0) (www.gene-regulation.com). Prominent activity-regulated transcription factor sites, namely MEF2/SRF (red) and CREB (blue), are shown. Z-scores of evolutionary conservation are also shown, z-score > 1.64 corresponding to P < 0.05, and z-score > 2.33 corresponding to P < 0.01. (C) Activation of PCDH10 gene expression in rat hippocampal cultures (0, 1, and 6 hours) after membrane depolarization. (D) Activation of NHE9 gene expression in hippocampal cultures (0, 1, 3, and 6 hours) after membrane depolarization with KCl in control lentivirus cultures (blue) and NPAS4 RNAi lentivirus (red).
A subset of genes identified in HMCA samples shows potential mutations in nonconsanguineous pedigrees
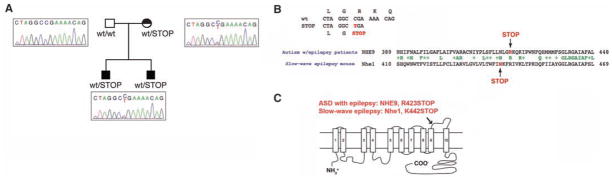
Further analysis of NHE9 demonstrated deleterious sequence variants associated with similiar autistic phenotypes in patients whose parents were not related. Because the pro-band AU-3101 with the deletion juxtaposed to NHE9 showed autism as well as epilepsy, we sequenced NHE9 in other patients with autism and epilepsy. A heterozygous CGA to TGA transition, changing arginine 423 residue to a stop codon, was found, and it creates a predicted protein truncation in the final extracellular loop of this multispanning transmembrane protein (Fig. 4, A to C). This nonsense change occurs within two amino acids of a similar nonsense mutation in Nhe1 that causes slow-wave epilepsy in mice (36) (Fig. 4B). The swe mouse mutation results in a gene dosage-dependent reduction of protein levels and loss of function in brain (36). A similar nonsense mutation in the final extracellular loop has recently been found in the related NHE6 gene in a patient with an Angelman-like syndrome, which involves both autism symptoms and epilepsy (37). The NHE9 nonsense change here is carried by two male siblings with autistic disorder as well as their mother, who was reported to have had childhood language delay based on a parental language questionnaire (blind to genotype). One autistic son has electroencephalogram-confirmed epilepsy, and the second autistic son had two probable seizures but does not have known epilepsy. This nonsense change was not found in greater than 3800 control chromosomes. Complete resequencing of all exons and exon-intron boundaries in a greater than fivefold excess (480) of controls revealed no nonsense changes (table S6). Rare, nonconservative coding changes were more common in patients with autism with epilepsy compared with control subjects (5.95% versus 0.63%, Fisher’s exact test, P = 0.005 for total changes), although autistic patients without seizures did not differ significantly from controls in the rate of nonconservative changes (1.14% autism without seizures versus 0.63% controls). The heterozygous changes, in particular the nonsense mutation, likely affect gene dosage and suggest that the study of consanguineous pedigrees may identify genes of importance in nonconsanguineous populations as well.
Fig. 4.
Mutational analysis of NHE9 in nonconsanguineous pedigrees with comorbid autism and epilepsy. (A) Mutational analysis of NHE9 in an AGRE pedigree reveals a nonsense change highly similar to the nonsense mutation in Nhe1 in the slow-wave epilepsy mouse. The AGRE pedigree structure includes two sons with autistic disorder. Patient 1 has comorbid epilepsy, and patient 2 had potential seizures at a younger age but does not currently carry the diagnosis of epilepsy. The mother does not have autism but is reported to have had a speech delay as a child (represented by a half-shading). Both patients and the mother carry the nonsense change. Sequence traces demonstrate the heterozygous C→T transition in Exon 11. This transition occurs at a CpG position consistent with a mutation at a methylated CpG. (B) Sequence trace indicating the position and consequences of the C→T transition. The CGA→TGA transition results in a change from an arginine residue at position 423 to a stop codon. The lower trace demonstrates that the position of this nonsense change in NHE9 occurs in a similar position as the causative, null missense mutation in Nhe1 in the slow-wave epilepsy mouse. (C) Nonsense mutations in the last extracellular loop of NHE proteins: NHE9 in patients with comorbid autism and epilepsy, and Nhe1 in the slow-wave epilepsy mouse. The human nonsense change was not found in greater than 3800 control chromosomes. Complete resequencing of all exons and exon-intron boundaries in a fivefold excess (480) of controls revealed no nonsense changes (table S6).
Discussion
Our copy number analysis, linkage, and resequencing together support other recent studies (3, 4, 6) suggesting that autism is highly heterogeneous genetically, but our data further suggest that homozygosity mapping provides an important approach to dissect this heterogeneity. We show that individuals with related parents are more likely to have inherited causes of disease, likely autosomal and recessive, and that these pedigrees allow mapping of loci from small numbers of families. Such families can also provide linkage evidence to support the identification of mutations, both coding and noncoding. Genes that act in a recessive manner may make good candidates for future analysis in association or resequencing studies, because they may show interactions with other nonlinked mutations (38). Our data implicating noncoding elements in patients with shared ancestry, as well as the heterozygous nonsense changes in patients without shared ancestry, suggest that loss of proper regulation of gene dosage may be an important genetic mechanism in autism. This possibility is also strongly supported by the numerous heterozygous CNVs found in patients from noncon-sanguineous pedigrees (4, 6).
Our data add to accumulating evidence for numerous individually rare loci in autism. These include FMR1, MECP2, NLGN3, NLGN4 (9), SHANK3 (10), CNTNAP2 (39–41), A2BP1 (41), NRXN1 (4) (also implicated by inherited CNVs in our study, table S2), and now candidate genes such as PCDH10, DIA1 (c3orf58), NHE9, CNTN3, SCN7A, and RNF8, in addition to chromosomal and CNV anomalies (4, 6–8, 26). Whereas genes involved in glutamatergic transmission seem to be important in autism (4), data from our study and others (6, 42) implicate other biological mechanisms as well. Potential disease mechanisms include failures in neuronal cell adhesion molecules such as NLGN3, NLGN4, and NRXN1; PCDH10 and CNTN3, identified as potential candidates here, may have similar roles. Endosomal trafficking and protein turnover is another potential mechanism implicated by NHE9, which itself is localized to endosomes (43). Further, mutations in NHE6 (which encodes a protein highly related to that encoded by NHE9) were found in a series of patients with an Angelman syndrome–like phenotype, with epilepsy and autism-like symptoms in some patients (37). DIA1 (c3orf58) appears to encode a protein localized to the Golgi apparatus (28), and so may also relate to protein trafficking.
The regulation of expression of some autism candidate genes by neuronal membrane depolarization suggests the appealing hypothesis that neural activity–dependent regulation of synapse development may be a mechanism common to several autism mutations. Early brain development is driven largely by intrinsic patterns of gene expression that do not depend on experience-driven synaptic activity (44). Mutations in the genes active in early development can lead to brain malformations or severe mental retardation. In contrast, postnatal brain development requires input from the environment that triggers the release of neurotransmitter and promotes critical aspects of synaptic maturation. During this process, neural activity alters the expression of hundreds of genes, each with a defined temporal course that may be particularly vulnerable to gene dosage changes. The connection between experience-dependent neural activity and gene expression in the postnatal period forms the basis of learning and memory, and autism symptoms typically emerge during these later stages of development. Our finding that deletions of genes regulated by neuronal activity or regions potentially involved in regulation of gene expression in autism suggests that defects in activity-dependent gene expression may be a cause of cognitive deficits in patients with autism. Therefore, disruption of activity-regulated synaptic development may be one mechanism common to at least a subset of seemingly heterogeneous autism-associated mutations.
Supplementary Material
www.sciencemag.org/cgi/content/full/321/5886/218/DC1
Methods
Figs. S1 to S3
Tables S1 to S7
References
References and Notes
- 1.Fombonne E. J Autism Dev Disord. 2003;33:365. doi: 10.1023/a:1025054610557. [DOI] [PubMed] [Google Scholar]
- 2.Canitano R. Eur Child Adolesc Psychiatry. 2006;16:61. doi: 10.1007/s00787-006-0563-2. [DOI] [PubMed] [Google Scholar]
- 3.Risch N, et al. Am J Hum Genet. 1999;65:493. doi: 10.1086/302497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Szatmari P, et al. Nat Genet. 2007;39:319. doi: 10.1038/ng1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vorstman JA, et al. Mol Psychiatry. 2006;11:1. doi: 10.1038/sj.mp.4001781. [DOI] [PubMed] [Google Scholar]
- 6.Sebat J, et al. Science. 2007;316:445. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jacquemont ML, et al. J Med Genet. 2006;43:843. doi: 10.1136/jmg.2006.043166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weiss LA, et al. N Engl J Med. 2008;358:667. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- 9.Kumar RA, et al. Hum Mol Genet. 2008;17:628. doi: 10.1093/hmg/ddm376. [DOI] [PubMed] [Google Scholar]
- 10.Jamain S, et al. Nat Genet. 2003;34:27. doi: 10.1038/ng1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Durand CM, et al. Nat Genet. 2007;39:25. doi: 10.1038/ng1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang YH, et al. Am J Med Genet. 2004;131:1. doi: 10.1002/ajmg.a.30297. [DOI] [PubMed] [Google Scholar]
- 13.Lander ES, Botstein D. Science. 1987;236:1567. doi: 10.1126/science.2884728. [DOI] [PubMed] [Google Scholar]
- 14.Smith CAB. J R Stat Soc B. 1953;15:153. [Google Scholar]
- 15.Bond J, et al. Nat Genet. 2002;32:316. doi: 10.1038/ng995. [DOI] [PubMed] [Google Scholar]
- 16.Piao X, et al. Science. 2004;303:2033. doi: 10.1126/science.1092780. [DOI] [PubMed] [Google Scholar]
- 17.Dixon-Salazar T, et al. Am J Hum Genet. 2004;75:979. doi: 10.1086/425985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rudan I, Campbell H, Carothers AD, Hastie ND, Wright AF. Nat Genet. 2006;38:1224. doi: 10.1038/ng1106-1224. [DOI] [PubMed] [Google Scholar]
- 19.Mani A, et al. Proc Natl Acad Sci USA. 2002;99:15054. doi: 10.1073/pnas.192582999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ritvo ER, et al. Am J Psychiatry. 1985;142:187. doi: 10.1176/ajp.142.2.187. [DOI] [PubMed] [Google Scholar]
- 21.The Homozygosity Mapping Collaborative for Autism: Amman, Jordan: University of Jordan, Pediatrics, Najwa Khuri-Bulos, Amira Masri. Irbid, Jordan: King Abdullah University Hospital, Pediatrics and Child Neurology, Azhar Daoud. Jeddah, KSA: King Faisal Specialist Hospital and Research Centre, Pediatric Neurology, G.G.; Child Development and Pediatrics, S.B. Riyadh, KSA: King Faisal Specialist Hospital and Research Center, Genetics, Brian Meyer. Kuwait City, Kuwait: Kuwait Medical Genetics Centre, Sadika Al-Awadi; Kuwait Centre for Autism, S.A.-S. Muscat, Oman: Royal Hospital, Genetics Unit, Anna Rajab. Lahore, Pakistan: University of Health Sciences, A.H., in addition to Pakistan Psychiatric Research Centre, Haroon Rashid Chaudhry and Saima Niaz. Doha, Qatar: Weill Cornell Medical College in Qatar, Genetics and Pediatrics, Ahmad Teebi. Istanbul, Turkey: Istanbul University, Child Psychiatry, N.M.M., in addition to Sabri Herguner. Ankara, Turkey: SB Diskapi Children’s Hospital, Ozgur Oner. Al Ain, UAE: United Arab Emirates University, Lihadh Al-Gazali, Valsamma Eapen. Dubai, UAE: Al Noor Centre for Children with Special Needs; Dubai Autism Center; Dubai Harvard Foundation for Medical Research, Muna Al Saffar. Sharjah, UAE: Manzil Centre for Challenged Individuals; Sharjah Autism Centre. Boston, MA, USA: C.A.W., E.M.M., S.Y.Y., R.S.H., J.N.P., B.B., R.M.J., J.W., in addition to Children’s Hospital Boston, Developmental Medicine Center, Leonard Rappaport, Ramzi Nasir, Elaine LeClair, Ellen Hanson; Neurology, Annapurna Poduri; Clinical Genetics, Wen-Hann Tan; Massachusetts General Hospital, Pediatric Neurology, Ganeshwaran H. Mochida.
- 22.Materials and methods are available as supporting material on Science Online.
- 23.Hamamy HA, Masri AT, Al-Hadidy AM, Ajlouni KM. Saudi Med J. 2007;28:1015. [PubMed] [Google Scholar]
- 24.Hoodfar E, Teebi AS. J Med Genet. 1996;33:212. doi: 10.1136/jmg.33.3.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shaffer LG, et al. J Pediatr. 2006;149:98. doi: 10.1016/j.jpeds.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 26.Vorstman JA, et al. J Am Acad Child Adolesc Psychiatry. 2006;45:1104. doi: 10.1097/01.chi.0000228131.56956.c1. [DOI] [PubMed] [Google Scholar]
- 27.International Molecular Genetic Study of Autism Consortium. Am J Hum Genet. 2001;69:570. [Google Scholar]
- 28.Takatalo MS, Kouvonen P, Corthals G, Nyman TA, Ronnholm RH. Proteomics. 2006;6:3502. doi: 10.1002/pmic.200500516. [DOI] [PubMed] [Google Scholar]
- 29.de Silva MG, et al. J Med Genet. 2003;40:733. doi: 10.1136/jmg.40.10.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Williamson SH, et al. PLoS Genet. 2007;3:e90. doi: 10.1371/journal.pgen.0030090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Uemura M, Nakao S, Suzuki ST, Takeichi M, Hirano S. Nat Neurosci. 2007;10:1151. doi: 10.1038/nn1960. [DOI] [PubMed] [Google Scholar]
- 32.Yoshihara Y, et al. Neuron. 1994;13:415. doi: 10.1016/0896-6273(94)90357-3. [DOI] [PubMed] [Google Scholar]
- 33.Takano Y, et al. J Biol Chem. 2004;279:18926. doi: 10.1074/jbc.M309148200. [DOI] [PubMed] [Google Scholar]
- 34.West AE, Griffith EC, Greenberg ME. Nat Rev Neurosci. 2002;3:921. doi: 10.1038/nrn987. [DOI] [PubMed] [Google Scholar]
- 35.Flavell SW, et al. Science. 2006;311:1008. doi: 10.1126/science.1122511. [DOI] [PubMed] [Google Scholar]
- 36.Cox GA, et al. Cell. 1997;91:139. doi: 10.1016/s0092-8674(01)80016-7. [DOI] [PubMed] [Google Scholar]
- 37.Gilfillan GD, et al. Am J Hum Genet. 2008;82:1003. doi: 10.1016/j.ajhg.2008.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ji W, et al. Nat Genet. 2008;40:592. doi: 10.1038/ng.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alarcon M, et al. Am J Hum Genet. 2008;82:150. doi: 10.1016/j.ajhg.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arking DE, et al. Am J Hum Genet. 2008;82:160. doi: 10.1016/j.ajhg.2007.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bakkaloglu B, et al. Am J Hum Genet. 2008;82:165. doi: 10.1016/j.ajhg.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martin CL, et al. Am J Med Genet B Neuropsychiatr Genet. 2007;144:869. doi: 10.1002/ajmg.b.30530. [DOI] [PubMed] [Google Scholar]
- 43.Park M, et al. Neuron. 2006;52:817. doi: 10.1016/j.neuron.2006.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sur M, Rubenstein JL. Science. 2005;310:805. doi: 10.1126/science.1112070. [DOI] [PubMed] [Google Scholar]
- 45.Li C, Wong WH. Genome Biol. 2001;2:8. doi: 10.1186/gb-2001-2-8-research0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.We are grateful to the families who participated in this study. We also thank C. Lord for advice on phenotyping; D. Altshuler, M. Daly, and C. Lee for helpful discussions; C. Seidman, J. Seidman, and D. Housman for sharing control DNA; K. Allen, S. Tzakas, and C. Austin for technical assistance; and members of the Program in Medical and Populations Genetics at the Broad Institute, the Autism Consortium, and the Walsh laboratory for helpful discussions. We gratefully acknowledge the National Center for Research Resources Broad Institute Center for Genotyping and Analysis for expert design and execution of the SNP genotyping reported herein and the resources provided by the AGRE Consortium. AGRE is a program of Cure Autism Now (CAN) and is supported, in part, by grant MH64547 from the National Institute of Mental Health to Daniel H. Geschwind (PI). Genotyping services were also provided by the Center for Inherited Disease Research (CIDR). CIDR is fully funded through a federal contract from the National Institutes of Health to The Johns Hopkins University, contract number N01-HG-65403. We are grateful for support from CAN, the Nancy Lurie Marks Family Foundation, the Simons Foundation, the Harvard Kuwait Project, the Developmental Disabilities Research Center of Children’s Hospital Boston (5P30HD018655-26), the Clinical Investigator Training Program of Harvard and Massachusetts Institute of Technology in collaboration with Pfizer Inc. and Merck & Co., the Anne and Paul Marcus Foundation, the Charles H. Hood Foundation, and NIH (1K23MH080954-01 to E.M.M, 1R01 MH083565 to C.A.W., 1K01MH71801 to R.J.F., and 5R01NS048276-05 to M.E.G.). E.M.M. holds a Career Award for Medical Scientists from the Burroughs Wellcome Fund and is also grateful for support from the Rappaport Research Scholarship in Neuroscience at Massachusetts General Hospital. S.-Y.Y. is a postdoctoral fellow of the National Alliance for Autism Research. M.E.G. is grateful for support from the F. M. Kirby Foundation. C.A.W. is an Investigator of the Howard Hughes Medical Institute.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
www.sciencemag.org/cgi/content/full/321/5886/218/DC1
Methods
Figs. S1 to S3
Tables S1 to S7
References