Summary
The mammalian secondary palate exhibits morphological, pathological, and molecular heterogeneity along the anterior-posterior axis. Although the cell proliferation rates are similar in the anterior and posterior regions during palatal outgrowth, previous studies have identified several signaling pathways and transcription factors that specifically regulate the growth of the anterior palate. In contrast, no factor has been shown to preferentially regulate posterior palatal growth. Here we show that mice lacking the transcription factor Mn1 have defects in posterior but not anterior palatal growth. We show that Mn1 mRNA exhibits differential expression along the anterior-posterior axis of the developing secondary palate, with preferential expression in the middle and posterior regions during palatal outgrowth. Extensive analyses of palatal gene expression in wild-type and Mn1-/- mutant mice identified Tbx22, the mouse homolog of the human X-linked cleft palate gene, as a putative downstream target of Mn1 transcriptional activation. Tbx22 exhibits a similar pattern of expression with that of Mn1 along the anterior-posterior axis of the developing palatal shelves and its expression is specifically down-regulated in Mn1-/- mutants. Moreover, we show that Mn1 activated reporter gene expression driven by either the human or mouse Tbx22 gene promoters in co-transfected NIH3T3 cells. Overexpression of Mn1 in NIH3T3 cells also increased endogenous Tbx22 mRNA expression in a dose-dependent manner. These data indicate that Mn1 and Tbx22 function in a novel molecular pathway regulating mammalian palate development.
Keywords: cleft palate, Mn1, palate development, anterior-posterior patterning, Tbx22
Introduction
The meningioma-1 (MN1) gene was first identified as the gene disrupted by a balanced chromosomal translocation that caused meningioma, a benign brain tumor (Lekanne Deprez et al., 1995). MN1 encodes a protein of 1,319 amino acids, with no homology to any known functional domains, but the gene is evolutionarily conserved from Drosophila to human (Lekanne Deprez et al., 1995). While its relation to meningioma remains unclear since no mutations or deletions of the MN1 gene have been found in other meningioma patients, the MN1 gene has been shown to play important roles in acute myeloid leukemia (AML) pathogenesis (reviewed by Grosveld, 2007). MN1 is the target of a recurrent chromosomal translocation, t(12;22)(p13;q12), associated with human AML (Buijs et al., 1995). The translocation fuses the MN1 gene with the TEL gene that encodes an ETS family DNA-binding transcription factor. Both in vitro and in vivo studies showed that the MN1-TEL fusion protein is oncogenic and that the transforming activity of the fusion protein depended on the N-terminal 500 amino acid residues of the MN1 protein (Buijs et al., 2000; Kawagoe and Grosveld, 2005a; Kawagoe and Grosveld, 2005b; Carella et al., 2006). In addition, MN1 is overexpressed in many AML patients associated with other chromosomal abnormalities and in some AML patients without karyotype abnormalities (Ross et al., 2004; Valk et al., 2004; Du et al., 2005; Heuser et al., 2006; reviewed by Grosveld, 2007). Moreover, overexpression of MN1 in the bone marrow caused malignant myeloid disease in mice (Carella et al., 2007). While the molecular mechanisms involving MN1 in AML pathogenesis remains to be elucidated, several biochemical studies have showed that MN1 can function as a transcriptional coactivator of the nuclear hormone receptors for retinoic acid or vitamin D (van Wely et al., 2003; Sutton et al., 2005). MN1 has also been shown to interact with the transcriptional coactivators p300/CBP and RAC3 and to mediate transcriptional activation via CACCC-rich DNA sequences (van Wely et al., 2003; Meester-Smoor et al., 2007). Thus, in addition to involvement in AML, MN1 may interact with other transcription factors to regulate cell proliferation and cell differentiation during mammalian development.
To further investigate the roles of MN1 in oncogenesis and development, Meester-Smoor et al. (2005) generated mice with a targeted deletion in the orthologous Mn1 gene. Although the mutant mice did not exhibit any increased incidence of tumor formation, all Mn1-/- homozygous mutant mice died shortly after birth and exhibited severe craniofacial developmental defects, including cleft palate, and some Mn1+/- heterozygous mutant mice also had cleft palate (Meester-Smoor et al., 2005). In mice, as in humans, the secondary palate develops from bilateral outgrowth on the oral side of the developing maxillary processes. The palatal processes initially grow vertically flanking the developing tongue. At a specific developmental time the bilateral palatal shelves reorient to the horizontal position above the tongue, grow toward and fuse with each other at the midline to form the intact roof of the oral cavity (Ferguson, 1988). Cleft palate may result from disturbances in the growth, elevation, or fusion of the palatal shelves. Gene inactivation studies in mice have demonstrated that many genes play essential roles in palatal shelf growth, including Bmp4, Bmpr1a, Fgf10, Fgfr2b, Msx1, Osr2, Shox2, and Tgfbr2, indicating that multiple molecular pathways interact to regulate palate development (Zhang et al., 2002; Han et al., 2003; Ito et al., 2003; Lan et al., 2004; Rice et al., 2004; Alappat et al., 2005; Liu et al. 2005; Yu et al., 2005). Moreover, since palate development occurs concurrently with significant growth and morphogenesis of the craniofacial complex, gross defects in structures outside of the palatal shelves may sometime hinder palatal shelf elevation or contact, resulting in cleft palate (reviewed by Chai and Maxson, 2006). To understand the roles of Mn1 in palate development, we have characterized its expression patterns during normal palate development and have identified a primary role for Mn1 in differential regulation of palatal growth along the anterior-posterior axis.
The mammalian secondary palate is divided anatomically into the anterior bony region (hard palate) and the posterior muscular region (soft palate) (Sperber, 2002). Cleft palate defects affecting the entire palate (complete cleft of the secondary palate) or either the anterior or posterior regions (incomplete cleft of the secondary palate) have been well documented (reviewed by Hilliard et al., 2005). Consistent with the morphological and pathological differences in the anterior and posterior palate, recent studies have clearly demonstrated that there is molecular heterogeneity along the anterior-posterior axis of the developing secondary palate (reviewed by Hilliard et al., 2005; Li and Ding, 2007). During early palate development, expression of several critical signaling molecules and transcription factors, including Bmp4, Fgf10, Msx1 and Shox2, is highly restricted along the anterior-posterior axis. Expression of Bmp4 and Msx1 is restricted to the most anterior 25% whereas Fgf10 and Shox2 are expressed in the anterior half of the developing palatal shelves, up to the level of the first molar tooth buds, prior to palatal fusion (Zhang et al., 2002; Alappat et al., 2005; Yu et al., 2005; Hilliard et al., 2005; Li and Ding, 2007). Fgf10 signals through the Fgfr2b receptor to regulate palatal epithelial cell proliferation and survival (Rice et al., 2004; Alappat et al., 2005). Bmp4 and Msx1 appeared to function in a positive feedback loop to regulate mesenchymal proliferation in the anterior palate (Zhang et al., 2002). Interestingly, exogenous Bmp4 induced Msx1 expression and cell proliferation in the anterior but not in the posterior palatal mesenchyme in explant culture assays (Zhang et al., 2002; Hilliard et al., 2005). Shox2 is also required for growth of the anterior palate and mice lacking Shox2 exhibited incomplete cleft within the anterior palate while the mutant posterior palate fused normally (Yu et al., 2005). Although the anterior and posterior palatal regions exhibit similar growth rates during palatal outgrowth (Zhang et al., 2002; Li and Ding, 2007), however, no factor has been reported to preferentially regulate the growth of the posterior palate. The Meox2 homeobox gene has been reported as being expressed in the posterior but not in the anterior palatal shelves in certain strains of mice (Jin and Ding, 2006; Li and Ding, 2007). Some, but not all, Meox2 mutant mice exhibited cleft palate, but they did not have defects in palatal shelf growth and their cleft palate defect appeared to result from postfusion rupture (Jin and Ding, 2006). In this report, we show that Mn1 preferentially regulates the growth of the posterior palate in mice. In addition, we show that palatal expression of Tbx22, in which mutations cause X-linked cleft palate and ankyloglossia (CPX) in humans (Braybrook et al., 2001; 2002), is specifically regulated by Mn1. These data provide novel insights into the molecular mechanisms regulating the regional growth and patterning of the secondary palate.
Materials and methods
Mice
Mice carrying the targeted mutation in Mn1 have been described previously (Meester-Smoor et al., 2005). Mn1+/- mice were maintained in the FVB congenic background and were intercrossed to generate homozygous mutant embryos for experimental analysis. Wild-type C57BL/6J and CD-1 mice were also used for in situ hybridization analysis of Mn1 and Tbx22 mRNA expression during palate development.
In situ hybridization and histological analyses
Embryos at different stages were dissected, fixed in 4% paraformaldehyde (PFA) in PBS overnight at 4°C. Whole mount in situ hybridization was performed as described previously (Lan et al., 2001). For section in situ hybridization, PFA-fixed embryos were dehydrated through graded alcohols and embedded in paraffin, sectioned at 7 μm thickness, followed by prehybridization processing and by hybridization with digoxigenin-labeled cRNA probes as described previously (Zhang et al., 1999).
For histology, embryos were collected at predetermined stages, fixed in either Bouin’s fixative or 4% PFA overnight, dehydrated through graded ethanol, embedded in paraffin wax and sectioned at 7 μm thickness, followed by staining with haematoxylin and eosin.
Analyses of cell proliferation and cell apoptosis
Cell proliferation was measured by BrdU incorporation assays or detection of Ki67. For BrdU incorporation assays, timed mating was set up between Mn1+/- heterozygous mice and pregnant female mice were injected intraperitoneally with BrdU (5-bromo-2-deoxy-uridine, Roche) labeling reagent at gestational day 12 or 13, with a dosage of 15 μl/g body weight. One hour after injection, embryos were dissected, fixed in Carnoy’s fixative, dehydrated through graded ethanol, embedded in paraffin wax and sectioned at 5 μm thickness. Sections from anterior, middle or posterior regions of the developing palatal shelves were selected for detection of BrdU-labeled cells by using the BrdU labeling and detection kit (Roche) following the manufacturer’s protocol. Following BrdU detection, sections were counterstained with nuclear fast red (Vector Laboratories, Inc.) to label all cellular nuclei. The total number of cells and the number of BrdU-positive cells in the palatal epithelium and mesenchyme on each of five consecutive sections were counted. Cell proliferation index was calculated as the percentage of the total cells being BrdU-positive. ANOVA was applied for statistical analyses and a P value less than 0.01 was considered statistically significant.
For detection of Ki67, Paraffin sections from selected palatal regions of staged mouse embryos were stained with an antibody against Ki67 as described previously (Casey et al., 2006). Cell apoptosis was detected by TUNEL assays. Paraffin sections from selected palatal regions of staged mouse embryos were analyzed by using the DeadEnd™ Fluorometric TUNEL System (Promega) following the manufacturer’s instructions.
Expression vectors and promoter-luciferase constructs
The Mn1 expression vector was constructed by subcloning an Mn1 cDNA fragment containing the full-length protein-coding region into the pcDNA3TOPO (Invitrogen) expression vector. The human TBX22 promoter-luciferase reporter vectors (pGL3-TBX22 hP0 and pGL3-TBX22 hP1) were described previously (Andreou et al., 2007). The mouse Tbx22 promoter-luciferase reporter vectors were similarly constructed by PCR amplifying the mouse Tbx22 promoter regions (Fig. S3) followed by subcloning into the pGL3-basic vector (Promega). All subcloned fragments were sequence verified.
Cell culture, transfection, and luciferase reporter assays
NIH3T3 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin. For luciferase reporter assays, cells were plated in 24-well tissue culture plates (Corning) and co-transfected with 0.05 μg of a luciferase reporter vector, 0.05 μg of the pRL-Renilla luciferase expression vector (Promega), and increasing amounts of the Mn1 expression vector. Transfections were performed by using the Lipofectamine reagents (Invitrogen) in accordance with the manufacture’s instructions. Cells were cultured for 48 hours after transfection and then assayed using the Dual-Luciferase Assay Kit (Promega). Firefly luciferase activity was normalized to Renilla luciferase activity. All transfection experiments were carried out in triplicates and data were summarized from three repeat experiments.
Real-time RT-PCR
For detection of the effects of Mn1 on endogenous Tbx22 gene expression, NIH3T3 cells were plated in 6-well tissue culture plates and transfected with increasing amounts of the Mn1 expression vector. Cells were cultured for 48 hours after transfection and total RNA was extracted using Trizol reagents (Invitrogen). First strand cDNA was synthesized using SuperScript First-Strand Synthesis System (Invitrogen). Quantitative PCR amplifications were performed in an iCycler real-time PCR machine (Bio-Rad) using the SYBR GreenER™ qPCR Supermix (Invitrogen). Mn1 gene-specific PCR primers are 5′-AGATCCAGCTGCAGAGACAA-3′ and 5′-TACTCATGGCGCTCTTGACT-3′. Tbx22 gene-specific PCR primers are 5′-GACCTGTCCCTGATTGAGTCC-3′ and 5′-GCTGGTTTTGGTAAGCTGTCA-3′. Hprt gene-specific primers are 5’-TGCTGGTGAAAAGGACCTCTCG-3’ and 5’-CTGGCAACATCAACAGGACTCC-3’. For each sample, the relative levels of Mn1 and Tbx22 mRNAs were normalized to that of HPRT using the standard curve method.
Results
Expression of Mn1 mRNA during early mouse embryogenesis and in the developing palate
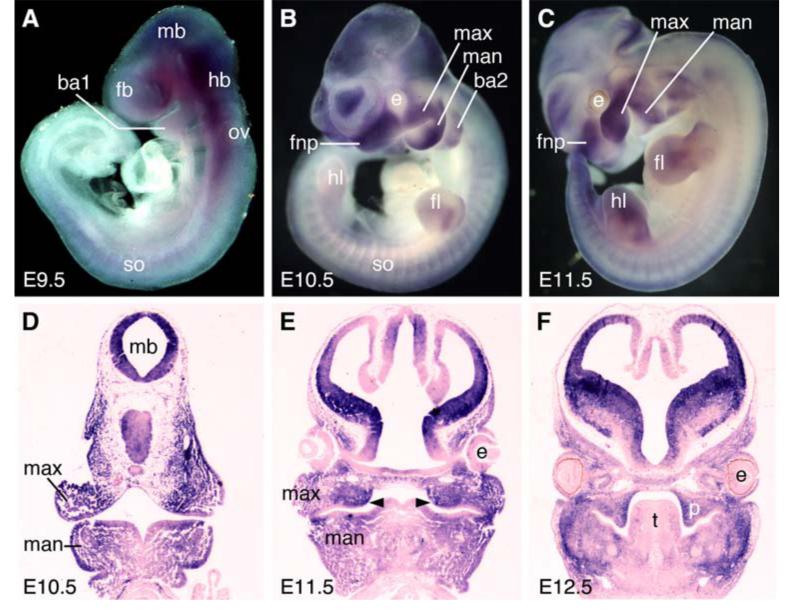
By whole mount in situ hybridization, Mn1 mRNA expression was first detected at embryonic day E9.5 (Fig. 1A). At this stage, Mn1 mRNA was strongly expressed in the midbrain and hindbrain tissues and in the craniofacial mesenchyme. By E10.5, Mn1 mRNA was highly expressed in all of the prominences of the developing face, including frontonasal processes, maxillary processes, mandibular processes, and in the second branchial arch (Fig. 1, B and D). Mn1 mRNA was also detected in a subset of mesenchymal cells in the developing somites and limb buds. This pattern of Mn1 mRNA expression persists at E11.5 (Fig. 1, C and E). At this stage, palatal outgrowth had initiated on the oral sides of the maxillary processes and Mn1 mRNA was strongly expressed in the palatal primordia (Fig. 1E). In situ hybridization of sections of E12.5 embryos showed that Mn1 mRNA was highly expressed in the developing palatal mesenchyme and in the preossification mesenchymal cells in the mandible (Fig. 1F). Strong Mn1 mRNA expression persists in the brain, in particular in the ventricular zone (Fig. 1F).
Fig. 1.
Expression pattern of Mn1 mRNA in developing mouse embryos. mRNA signals were detected by whole mount in situ hybridization (A-C) or section in situ hybridization (D-F). (A) Mn1 mRNA expression was first detected in the developing brain tissues and in the craniofacial mesenchyme at E9.5. (B-E) Strong Mn1 mRNA expression was observed in the developing brain, frontonasal processes, maxillary processes, mandibular processes, the second branchial arch, the developing somites and limb buds at E10.5 (B, D) and E11.5 (C, E). (F) Section in situ hybridization showing strong Mn1 mRNA expression in the developing palatal mesenchyme and in the preossification mesenchymal cells in the mandible. ba1, first branchial arch; ba2, second branchial arch; e, eye; fb, forebrain; fl, forelimb; fnp, frontonasal process; hb, hindbrain; hl, hindlimb; man, mandibular process; max, maxillary process; mb, midbrain; ov, otic vesicle; p, palatal shelf; so, somite; t, tongue.
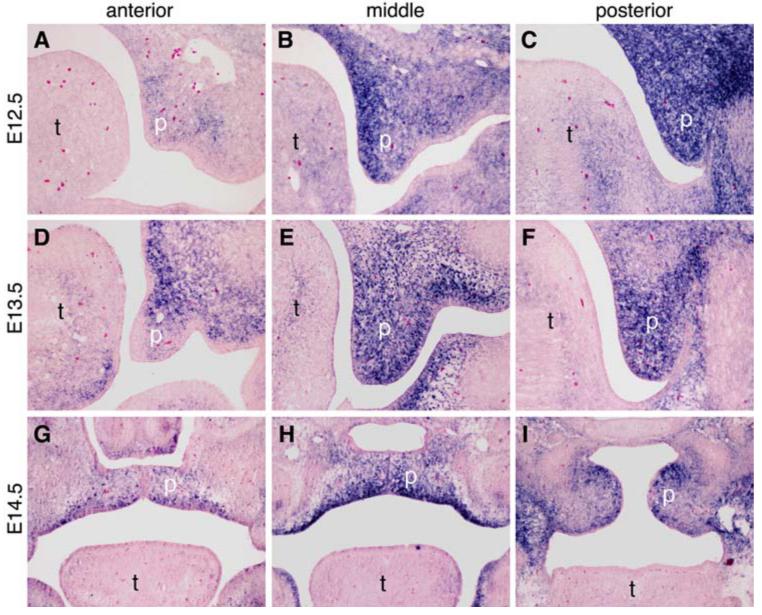
The reported cleft palate phenotype of the Mn1 mutant mice prompted us to carry out a detailed analysis of Mn1 mRNA expression during palate development. Recent studies have demonstrated that several genes exhibit differential expression along the anterior-posterior axis of the developing palatal shelves in mice (Zhang et al., 2002; Alappat et al., 2005; Yu et al., 2005; Hilliard et al., 2005; Li and Ding, 2007). The distinct gene expression patterns appear to divide the developing palatal shelves into three regions along the anterior-posterior axis: (1) anterior palate that expresses high levels of Msx1 and Shox2 mRNAs (Zhang et al., 2002; Hilliard et al., 2005; Yu et al., 2005); (2) middle palate (roughly corresponding to the region flanked by the upper first molar tooth germs, Alappat et al., 2005) that lacks Msx1 expression and exhibits anterior-to-posterior expansion of Shox2 mRNA expression from E12.5 to E14.5 (Li and Ding, 2007); and (3) posterior palate that expresses high levels of Meox2 mRNA but lacks Msx1 and Shox2 mRNA expression (Li and Ding, 2007). In situ hybridization of serial sections of the developing palatal shelves showed that Mn1 mRNA is differentially expressed along the anterior-posterior axis of the developing palatal shelves, with high levels of expression in both mesenchyme and epithelium in the middle and posterior regions and very low levels in the anterior region of the palatal shelves during the vertical growth period from E12.5 to E13.5 as well as after palatal shelf elevation at E14.5 (Fig. 2).
Fig. 2.
The expression pattern of Mn1 mRNA along the anterior-posterior axis of the developing palate. Middle palate corresponds to the palatal region flanked by the maxillary first molar tooth germs. (A-C) At E12.5, Mn1 mRNA expression in the developing secondary palate exhibits an anterior-posterior gradient, with low levels in the anterior palate (A), moderate levels in the middle palate (B), and highest levels in the posterior palate (C). (D-F) At E13.5, Mn1 mRNA expression is still high in the middle (E) and posterior (F) regions and much weaker in the anterior region (D) of the developing palate. (G-I) At E14.5, the bilateral palatal shelves have elevated to the horizontal position above the tongue and have initiated contact and fusion in the anterior (G) and middle (H) regions. Mn1 mRNA expression remains strong in the middle and posterior palatal regions and relatively weak in the anterior palate. p, palatal shelf; t, tongue.
Mn1-/- mutant mice exhibit palatal retardation and failure of palatal shelf elevation
To investigate which palatal developmental steps require Mn1 function, we carried out detailed histological analyses of Mn1-/- mouse embryos throughout palate development. Mn1-/- mutant embryos displayed normal palatal shelf outgrowth at E12.5 (data not shown). At E13.5, the palatal shelves of Mn1-/- mutant embryos exhibited similar size and shape with those of wild-type embryos (Fig. 3A, B). By E14.5, the palatal shelves of wild-type embryos had already elevated to the horizontal position and initiated fusion by forming the midline epithelial seam (Fig. 3C), which was disintegrated by E15.5 to form the fused secondary palate (Fig. 3E). In contrast, the bilateral palatal shelves of Mn1-/- mutant embryos failed to elevate and remained at the vertical position at E14.5 and at E15.5 (Fig. 3, D and F).
Fig. 3.
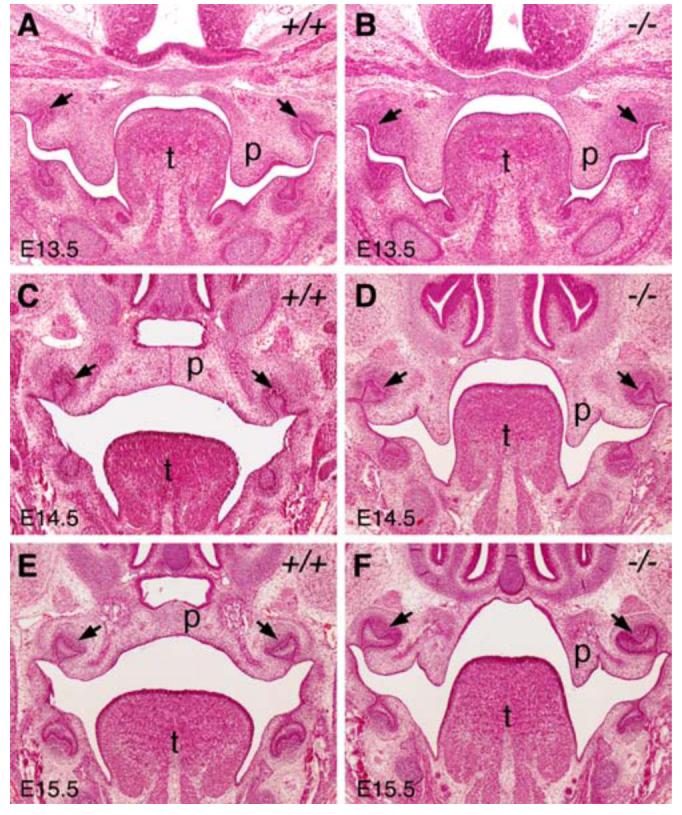
Mn1-/- mutant mice exhibit palatal retardation and failure of palatal shelf elevation. All panels shown are from middle palate regions. (A, B) At E13.5, wild-type (A) and Mn1-/- homozygous mutant (B) embryos exhibited similar palatal shelf size and shape. (C, D) At E14.5, the wild-type (C) palatal shelves had elevated to the horizontal position above the tongue and initiated fusion by forming the midline epithelial seam, while the Mn1-/- homozygous mutant (D) palatal shelves were still vertically oriented. (E, F) At E15.5, the wild-type (E) palatal shelves had completed fusion, but the Mn1-/- homozygous mutant (F) palatal shelves remained vertically oriented. Arrows point to the first molar tooth germs. p, palatal shelf; t, tongue.
Histological analyses of Mn1-/- mutant embryos at later stages revealed different fates of the palatal shelves along the anterior-posterior axis. At E16.5, the middle and posterior palatal shelves in Mn1-/- mutant embryos were still vertically oriented and appeared greatly reduced in size (Fig. 4B). By E18.5, the posterior palatal shelves were further retarded and retracted to the maxillary processes (Fig. 4D), while the anterior palatal shelves were still clearly visible and often elevated on one or both sides (Fig. 4C).
Fig. 4.
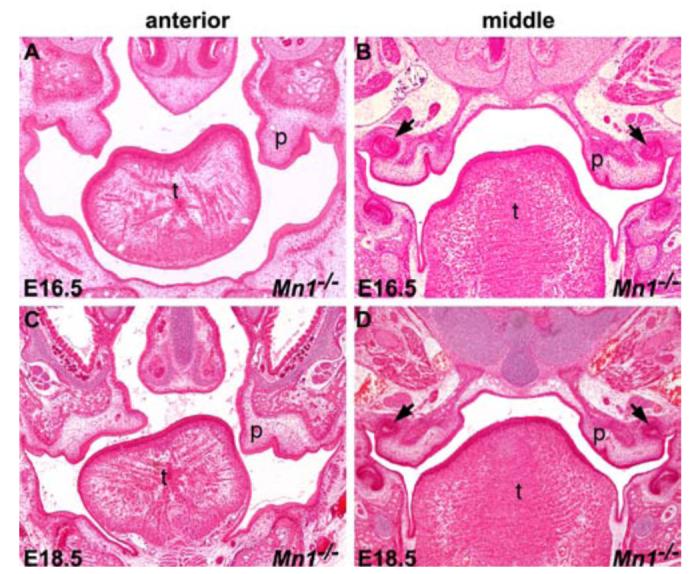
Histological analyses of palate development in Mn1-/- mutants at late stages. (A, B) At E16.5, the palatal shelves in Mn1-/- mutant embryos were still vertically oriented. In addition, the palatal shelves appeared to be significantly diminished in size in the middle to posterior regions (B). (C, D) At E18.5, whereas the palatal shelves in the anterior region in the Mn1-/- mutant mice were partially elevated (C), the middle and posterior regions of the palatal shelves were further retarded and retracted into the maxillary processes (D). Arrows in B and D point to the upper first molar tooth germs. p, palatal shelf; t, tongue.
Mn1 is required for proper palatal shelf growth
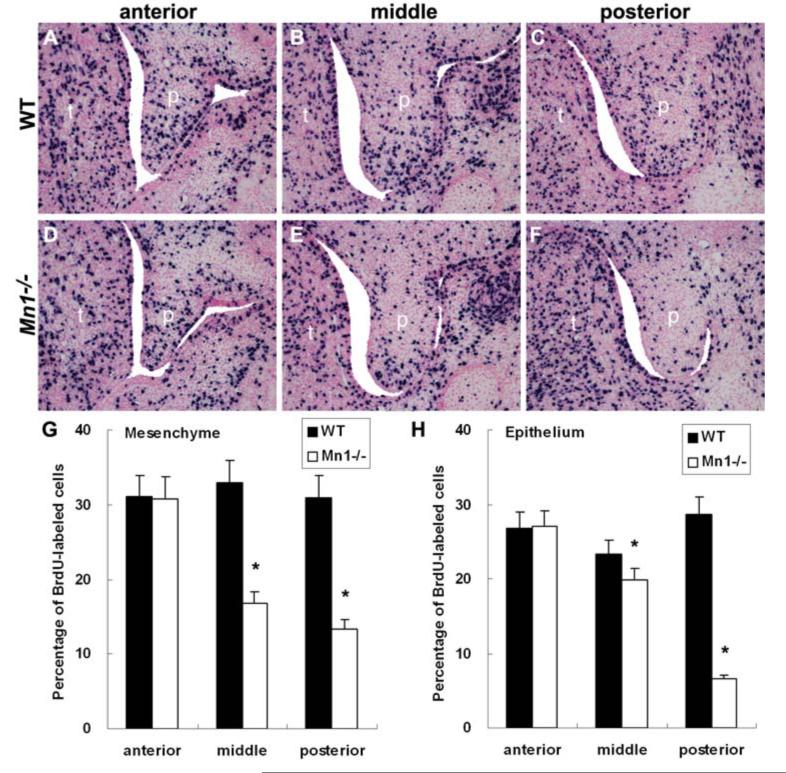
Impairment of palatal shelf elevation is often accompanied by and partially due to retarded palatal shelf growth, as has been reported in the Osr2-/- mutant mice (Lan et al., 2004). To investigate the cellular mechanisms of palatal shelf retardation and elevation failure in Mn1-/- mutant mice, we examined whether there are alterations in cell proliferation and cell survival during palate development in Mn1-/- mutant mice. No differences in cell apoptosis were found in the palatal shelves between wild-type and Mn1-/- mutant embryos at E12.5 and at E13.5 (data not shown). No significant alterations in cell proliferation were observed in the palatal shelves in Mn1-/- mutant embryos at E12.5 (data not shown). At E13.5, we detected a 57% reduction (P<0.01) in the posterior and a 49% reduction (P<0.01) in the middle regions of the palatal mesenchyme in Mn1-/- mutant embryos in comparison with their wild-type littermates (Fig. 5G). In contrast, the cell proliferation index was not significantly different in the anterior palatal mesenchyme in the same Mn1-/- mutant and wild-type embryos (Fig 5G). Similarly, palatal epithelial cell proliferation was also significantly reduced in the middle and posterior palate but not in the anterior palate in Mn1-/- mutant embryos, in comparison with the wild-type littermates (Fig. 5H). The selective reduction in palatal cell proliferation in the middle and posterior regions of the palatal shelves in Mn1-/- mutant mice correlates with the differential expression of Mn1 mRNA along the anterior-posterior axis during normal palate development.
Fig. 5.
Analyses of cell proliferation during palate development in wild-type and Mn1-/- mutant embryos. In comparison with the wild-type littermates (A-C), the number of BrdU-labeled cells in the developing palatal shelves was greatly reduced in the middle (E) and posterior (F) palatal shelves, but not in the anterior (D) palatal mesenchyme in the Mn1-/- mutant embryos by E13.5. p, palatal shelf; t, tongue. (G, H) Comparison of the percentage of BrdU-labeled cells in the palatal mesenchyme (G) and epithelium (H), respectively, in the anterior, middle, and posterior palatal regions in E13.5 wild-type (WT) and Mn1-/- mutant embryos. Error bars represent standard deviation and asterisk denotes a significant difference (P<0.01) between the wild-type and Mn1-/- mutant samples.
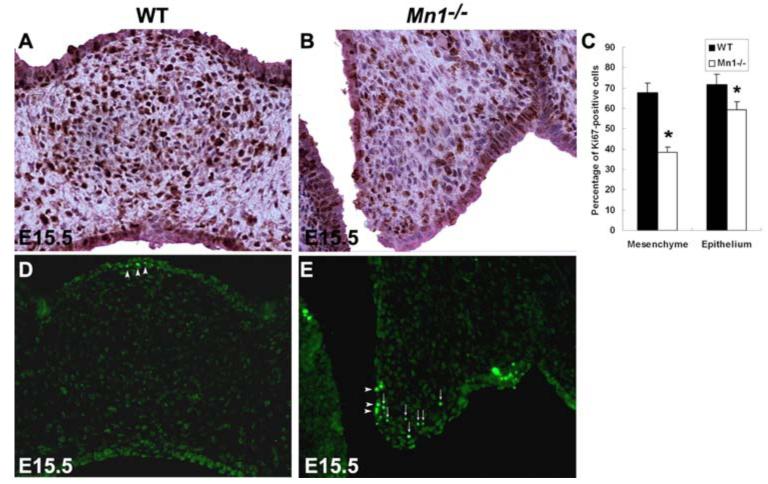
To understand the dramatic retardation of the palatal shelves at later stages in Mn1-/- mutant embryos, we also examined cell proliferation and cell apoptosis in E14.5 and E15.5 embryos. The decrease in cell proliferation rate in the posterior and middle regions of the palatal shelves in the Mn1-/- mutant embryos continued through E15.5 (Fig. 6 A-C and data not shown), indicating that Mn1 is an important regulator of palatal shelf growth before and after palatal shelf elevation. Moreover, by TUNEL assays, we detected increased apoptosis at E15.5 in the posterior regions of the palatal mesenchyme in Mn1-/- mutant embryos (Fig. 6D, E). These data indicate that the dramatic degeneration of the posterior palatal shelves observed by E18.5 in Mn1-/- mutant embryos resulted from decreased proliferation and increased apoptosis in the posterior palatal shelves, in combination with retraction of the freely projecting palatal shelves into the maxillary processes due to the morphogenetic expansion of the craniofacial width.
Fig. 6.
Analyses of cell proliferation and cell apoptosis in wild-type (WT) and Mn1-/- mutant embryos at E15.5. (A-C) Cell proliferation in palatal mesenchyme detected using immunohistochemical staining of the Ki67 protein. In comparison with wild-type embryos (A), cell proliferation in the middle palatal region in Mn1-/- mutant embryos (B) was greatly reduced. Error bars in (C) represent standard deviation and asterisk denotes a significant difference (P<0.01) between the wild-type and mutant samples. (D-E) TUNEL assays mid-palatal sections of E15.5 palatal shelves of wild-type (D) and Mn1-/- mutant (E) embryos showed increased cell apoptosis in the palatal mesenchyme in the mutant. Small arrows point to highly TUNEL-positive palatal mesenchymal cells and arrowheads point to highly TUNEL-positive palatal epithelial cells.
Palatal shelf growth defect in Mn1-/- mutants is accompanied by a region-specific decrease in CyclinD2 expression
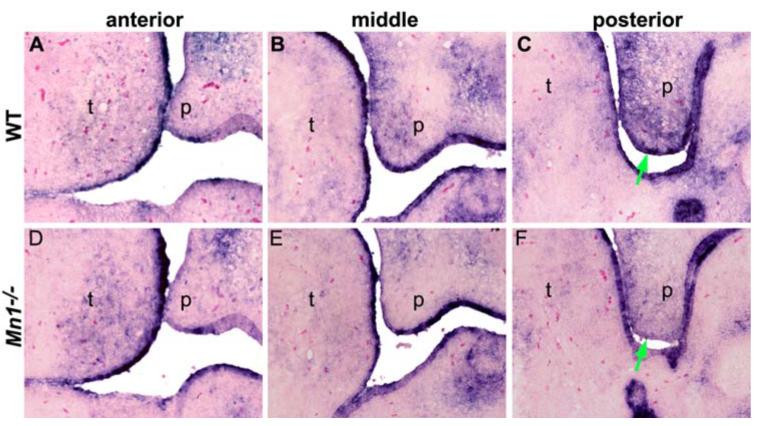
Several molecular pathways have been shown to regulate cell proliferation during palate development by regulating the expression of D-type cyclins. D-type cyclins are important cell cycle regulators, which bind CDK4 or CDK6 to control cell cycle progression through the G1 phase (Matsushime et al., 1991; Xiong et al., 1991; Motokura et al., 1991; Morgan et al., 1997). Conditional inactivation of Tgfβr2 in cranial neural crest (CNC) cells down-regulated CyclinD1 expression and reduced CNC cell proliferation in the palatal mesenchyme (Ito et al., 2003). Msx1 regulates neural crest cell proliferation and differentiation also through maintaining CyclinD1 expression (Hu et al., 2001). To understand the cellular mechanism by which Mn1 regulates palatal growth in the middle and posterior palatal shelves, we investigated possible alterations in the expression levels of D-type cyclins in Mn1-/- mutant embryos. No change in the levels of CyclinD1 expression was detected in either the epithelium or mesenchyme of the developing palate in Mn1-/- embryos in comparison with wild-type littermates (data not shown). In contrast, the expression of CyclinD2 is reduced in the middle and posterior regions of the palatal mesenchyme as well as in the posterior palatal epithelium in Mn1-/- embryos at E13.5 (Fig. 7). These data suggest that Mn1 regulates palatal shelf growth, at least in part, through maintaining CyclinD2 expression in the middle and posterior palatal shelves.
Fig. 7.
Expression of CyclinD2 was down-regulated in Mn1-/- mutant palatal shelves at E13.5. (A,D) In the anterior region of the developing palate, expression of CyclinD2 was similarly weak in wild-type (A) and Mn1-/- mutant (D) embryos. (B,E) In the middle palate, CyclinD2 is highly expressed in both the epithelium and mesenchyme in the wild-type embryo (B) but its expression is much reduced in the Mn1-/- palatal mesenchyme (E). (C) In the posterior palate region, strong CyclinD2 expression was detected in both the epithelium and mesenchyme in the wild-type palate. (F) In comparison to the wild-type littermate, CyclinD2 expression is much reduced in both the palatal epithelium and mesenchyme in the posterior palatal region in the Mn1-/- mutant embryo. Green arrows in C and F point to the medial edge epithelium of the palatal shelves.
Effects of Mn1 deficiency on palatal gene expression
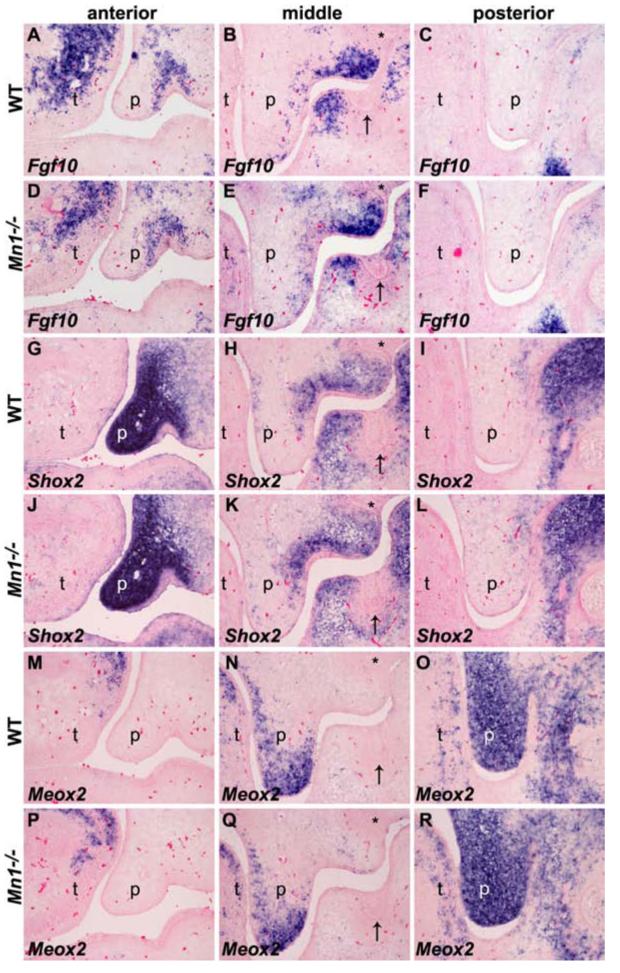
To investigate the molecular mechanisms involving Mn1 in palate development, we examined the expression patterns of other genes known to play important role in palate development, including Fgf10, Fgfr2, Osr2, Shh, Patch1, Pax9, Shox2, and Tgfβ3. No obvious differences in either levels or patterns of expression of these genes were found in the developing palatal shelves in wild-type and Mn1-/- mutant embryos (Fig. 8 and Supplemental Fig. 1). In particular, the differential expression patterns of Fgf10, Shox2, and Meox2 mRNAs along the anterior-posterior axis of the developing palatal shelves are maintained in the Mn1-/- mutant embryos (Fig. 8), indicating that there is no gross anterior-posterior patterning defects in the developing palate in Mn1-/- mutant embryos.
Fig. 8.
The anterior-posterior pattern of the developing palatal shelves is not disrupted in the Mn1-/- mutant embryos. (A-F) Expression of Fgf10 along the anterior-posterior axis of the developing palatal shelves in wild-type (A-C) and Mn1-/- mutant (D-F) embryos at E13.5. Fgf10 showed similar restricted expression pattern in the anterior and middle palatal regions in both wild-type and Mn1-/- mutant embryos. (G-L) Expression of Shox2 along the anterior-posterior axis of the developing palatal shelves in wild-type (G-I) and Mn1-/- mutant (J-L) embryos at E13.5. Shox2 mRNA is strongly expressed in the anterior and absent in the posterior palatal regions in wild-type embryos (G-I). In the middle palate, Shox2 mRNA expression is restricted in the proximolateral region (H). The expression pattern and levels of Shox2 mRNA are not altered in Mn1-/- mutant (J-L) embryos. (M-O) Meox2 mRNA expression is restricted to the middle and posterior regions and absent from the anterior region of the developing palate in wild-type embryos at E13.5. (P-R) The levels and anterior-posterior pattern of Meox2 mRNA expression pattern in E13.5 Mn1-/- mutant embryo are comparable to that of the wild-type littermate (M-O). p, palatal shelf; t, tongue. Arrows point to the mandibular first molar tooth germs and asterisks mark the maxillary first molar tooth germs in the mid-palatal sections.
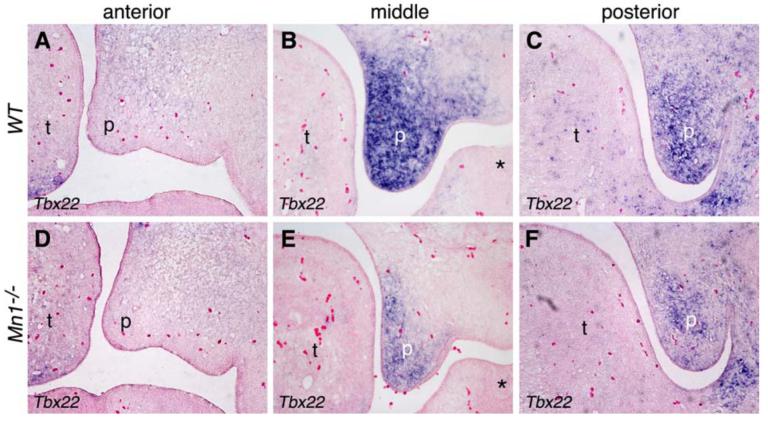
Extensive expression analyses of other genes implicated in palate development showed that expression of Tbx22, homologous to the gene associated with CPX in humans, is specifically reduced in the developing palatal shelves in Mn1-/- mutant embryos (Fig. 9). Interestingly, expression of Tbx22 mRNA also exhibits a posterior preference in the developing palatal shelves, similar to the expression pattern of Mn1, during normal palate development in mice (Fig. 9A-C). Compared to wild-type embryos, the expression levels of Tbx22 are dramatically reduced in the middle and posterior palatal shelves of Mn1-/- mutant embryos (Fig. 9D-F), indicating that Mn1 and Tbx22 function in the same molecular pathway to regulate mammalian palate development.
Fig. 9.
Tbx22 is specifically down-regulated in Mn1-/- mutant palatal shelves at E13.5. (A-C) Tbx22 mRNA is strongly expressed in the middle (B) and posterior (C) palatal mesenchyme, but barely expressed in the anterior (A) palatal mesenchyme in wild-type embryos. (D-F) Tbx22 expression is much reduced in the middle (E) and posterior (F) palatal mesenchyme in the Mn1-/- mutant littermates. Asterisk in B and E marks the mandibular first molar tooth germ. p, palatal shelf; t, tongue.
Mn1 is a transcriptional activator of Tbx22 gene expression
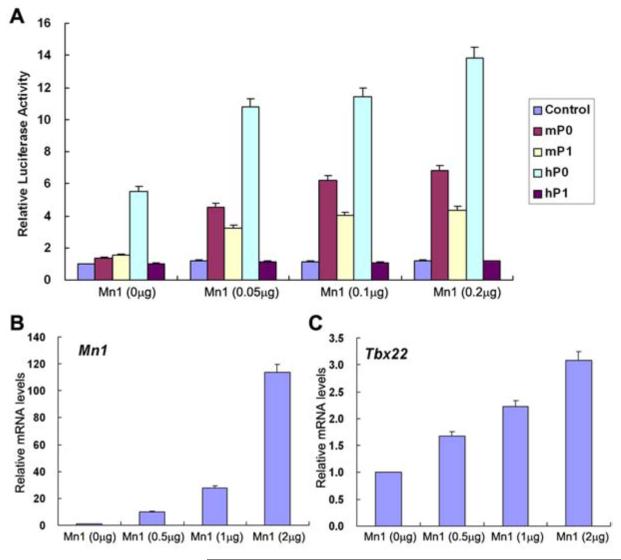
The similar expression patterns of Mn1 and Tbx22 mRNAs in the developing wild-type palatal shelves and the specific down-regulation of Tbx22 mRNA in Mn1-/- mutant embryos suggest that Mn1 may directly regulate Tbx22 gene expression during palate development. We previously showed that the human TBX22 gene is transcribed from two different promoters that are approximately 10 Kb apart (Andreou et al., 2007). Quantitative RT-PCR assays of human embryonic RNA samples indicated that embryonic TBX22 mRNA expression was predominantly generated by the hP0 promoter (data not shown). Analyses of 5’ RACE (rapid amplification of cDNA ends) products from E12.5 mouse embryonic craniofacial RNA templates revealed that the mouse Tbx22 gene is also transcribed from at least two distinct promoters, a distal promoter corresponding to the human TBX22 hP0 and a proximal promoter corresponding to the human TBX22 hP1 (Fig. S2). Interestingly, the previously determined core binding DNA sequence for the Mn1 transcription factor, CACCC, is present in various locations within 2 Kb of the transcription start site for each of the mouse and human TBX22 promoters (Figure S3). To investigate whether Mn1 can activate transcription from the mouse and human TBX22 gene promoters, we constructed Tbx22 promoter-luciferase reporter plasmids (see Materials and Methods) and tested the effects of Mn1 co-transfection on luciferase reporter expression in NIH3T3 cells. Transfection of each of the promoter-luciferase plasmids showed that the mouse mP0 and mP1 as well as human hP0 promoters each had promoter activity in NIH3T3 cells, with the hP0 promoter being three-fold stronger than the mP0 or mP1 promoter (Fig. 10A). In contrast, the hP1-luciferase construct did not show promoter activity in NIH3T3 and three other cell lines tested (Fig. 10A) (Andreou et al., 2007, and data not shown). Co-transfection with the Mn1 expression vector resulted in a dose-dependent increase in luciferase expression from each of the mP0-, mP1-, and hP0-luciferase constructs but not the hP1 promoter-luciferase construct (Fig. 10A). Moreover, transfection of NIH3T3 cells with the Mn1 expression vector resulted in a dose-dependent increase in endogenous Tbx22 mRNA expression (Fig. 10B,C). These data indicate that Mn1 functions as a transcriptional activator of Tbx22 gene expression.
Fig. 10.
Transcriptional regulation of Tbx22 by Mn1. (A) Tbx22 promoter-luciferase reporter activity in transfected NIH3T3 cells in the absence or presence of increasing amounts of co-transfected Mn1 expression vector. Luciferase activity was normalized against the control (promoter-less pGL3-basic) vector transfected cells. Error bar represents standard deviation. (B,C) Effect of Mn1 overexpression on endogenous Tbx22 mRNA expression in NIH3T3 cells. Expression levels of Mn1 (B) and Tbx22 (C) mRNAs were quantified by real time RT-PCR and normalized against mock-transfected cells. Error bar represents standard deviation.
Discussion
Cleft palate is a common birth defect in humans, occurring in approximately 1 in 1000 live births (Vanderas, 1987; Schutte and Murray, 1999; Gorlin et al., 2001). Approximately 50% of cleft palate cases are non-syndromic, although cleft palate has been associated with more than 300 syndromic developmental disorders. Several genes underlying syndromic forms of cleft palate have been identified, including IRF6 (Van der Woude and popliteal pterygium syndromes) (Kondo et al., 2002), MSX1 (cleft lip or palate with hypodontia) (van den Boogaard et al., 2000), P63 (ectodermal dysplasia and cleft lip or palate) (Celli et al., 1999; Ianakiev et al., 2000), PVRL1 (autosomal recessive cleft lip/palate with ectodermal dysplasia) (Suzuki et al., 2000), and TBX22 (CPX) (Braybrook et al., 2001). In addition, genetic studies in mice have revealed that mutations in more than sixty different genes each resulted in cleft palate and have uncovered specific molecular and cellular mechanisms controlling palate development (reviewed by Gritli-Linde, 2007). Interestingly, recent studies have demonstrated that palatal shelf growth is differentially regulated along the anterior-posterior axis (reviewed by Hilliard et al., 2005). Zhang et al. (2002) first demonstrated that Msx1 and Bmp4 are both only expressed in and required for the growth of the anterior region of the developing palatal shelves. In palatal explant culture assays, exogenous Bmp4 induced Msx1 mRNA expression and increased cell proliferation in the anterior but not in posterior palatal mesenchyme (Zhang et al., 2002; Hilliard et al., 2005). The Shox2 transcription factor is also specifically expressed in the anterior palate and mice lacking Shox2 exhibited growth defects in the anterior but not posterior regions of the developing palatal shelves (Yu et al., 2005). However, previous studies have also showed that the anterior and posterior palatal regions have similar cell proliferation rates during palatal outgrowth (Zhang et al., 2002; Li and Ding, 2007), suggesting that there must be factors that preferentially regulate posterior palatal growth. Our finding that Mn1 is differentially expressed along the anterior-posterior axis and preferentially regulates middle and posterior palatal cell proliferation fills a longstanding gap in the understanding of the molecular mechanisms of palate development.
Mn1 specifically regulates Tbx22 gene expression during palate development
Mutations in the human TBX22 gene, including nonsense, frameshift, splice-site and missense changes, have been associated with CPX (Braybrook et al., 2001; 2002; Chaabouni et al., 2005). In addition, mutation-screening studies of patients with isolated cleft palate showed that mutations in TBX22 represent the most common single cause of cleft palate known (Marcano et al., 2004; Suphapeetiporn et al., 2007). These data suggest that TBX22 plays primary and critical roles in palate development. Consistent with this hypothesis, Braybrook et al. (2002) showed that TBX22 mRNA is strongly expressed in the developing human palatal shelves prior to palatal elevation and fusion. TBX22 mRNA expression was also detected in the base of the developing tongue, which correlated well with the ankyloglossia phenotype in CPX patients. Several laboratories independently isolated the mouse Tbx22 gene and reported Tbx22 mRNA expression in the developing palatal shelves and at the base of the developing tongue (Braybrook et al., 2002; Bush et al., 2002; Herr et al., 2003). Although the exact roles of Tbx22 in palate development remain to be elucidated, these studies showed that palatal expression of Tbx22 mRNA was high during vertical palatal growth and declined after palatal shelf elevation in both human and mice, suggesting that Tbx22 may play a conserved role in palatal shelf growth. Welsh et al. (2007) showed, by whole mount in situ hybridization, that Tbx22 mRNA is preferentially expressed in the posterior palate in E13.5 and E14.5 mouse embryos. We confirmed, by using section in situ hybridization, the differential expression of Tbx22 mRNA along the anterior-posterior axis of the developing secondary palate (Fig. 9). Interestingly, Tbx22 mRNA expression was dramatically down-regulated in the Mn1-/- mutant palatal shelves. The fact that palatal expression of many genes, including that of Meox2, which is also preferentially expressed in the posterior palate during palatal outgrowth (Li and Ding, 2007, and Fig. 8), is not affected by Mn1 deficiency indicates that Tbx22 is a specific downstream target of Mn1. Our data that Mn1 activated, in a dose-dependent manner, expression of endogenous Tbx22 mRNA and the luciferase reporter gene driven by either the human or mouse Tbx22 promoter sequences in transfected NIH3T3 cells provide further support that Mn1 activates transcription of the Tbx22 gene. These data link two spatially co-expressed transcription factors in a novel molecular pathway for the regulation of posterior palate development.
Primary and secondary effects of Mn1 deficiency on palate development
It was previously suggested that the cleft palate defect in the Mn1-/- mutant mice might be secondary to the severe cranial skeletal defects (Meester-Smoor et al., 2005). We found that Mn1 is differentially expressed along the anterior-posterior axis of the developing palatal shelves and required for proper palatal growth in the middle and posterior regions but not in the anterior region. The specific down-regulation of Tbx22 mRNA expression in the Mn1-/- mutant palatal shelves also identifies a primary role for Mn1 in palate development. However, Mn1-/- mutant mice exhibit complete cleft of the secondary palate, although no significant growth deficiency was found in the anterior palate at any stage of palate development. In addition, the palatal shelves failed to elevate, in particular in the middle and posterior regions, in the Mn1-/- mutant mice. Impairment of palatal shelf elevation is often caused by mechanical hindrance, such as aberrant palatal-tongue and palatal-mandible fusions in the Jag2-/- and Fgf10-/- mutant mice, respectively (Casey et al., 2006; Alappat et al., 2005). Defects in mandibular development or deformation of the tongue have also been suggested as causes of failure of palatal shelf elevation in several mutant mouse strains, including Ryk-/- and Foxf2-/- mutant mice (Halford et al., 2000; Wang et al., 2003). In addition, Pax9-/- mutant mice exhibited impairment of palatal shelf elevation attributable to deformation of the palatal shelves themselves (Peters et al., 1998). Mn1-/- mutant mice exhibited normal mandibular development (Meester-Smoor et al., 2005). However, the developing tongue did not properly descend to the floor of the mouth during the time of palatal shelf elevation in the Mn1-/- mutant mice, in comparison with that in the wild-type littermates (Fig. 3). Whereas Mn1 mRNA is strongly expressed in the middle and posterior palatal mesenchyme, little Mn1 expression was observed in the developing tongue (Fig. 2). Thus, the palatal elevation defect in the Mn1-/- mutant mice may be due to an intrinsic defect in the palatal shelves or secondary to as yet unidentified defects in a mandibular component involved in tongue movement. Taken together, the cleft palate phenotype in Mn1-/- mutant mice likely results from a combination of the primary defects in palatal growth and secondary effects of other craniofacial abnormalities.
Supplementary Material
Acknowledgement
This work was supported by the NIH/NIDCR grants R01DE013681 and R01DE015207 to RJ.
References
- Alappat SR, Zhang Z, Suzuki K, Zhang X, Liu H, Jiang R, Yamada G, Chen Y. The cellular and molecular etiology of the cleft secondary palate in Fgf10 mutant mice. Dev. Biol. 2005;277:102–113. doi: 10.1016/j.ydbio.2004.09.010. [DOI] [PubMed] [Google Scholar]
- Andreou AM, Pauws E, Jones MC, Singh MK, Bussen M, Doudney K, Moore GE, Kispert A, Brosens JJ, Stanier P. TBX22 missense mutations found in patients with X-linked cleft palate affect DNA binding, sumoylation, and transcriptional repression. Am. J. Hum. Genet. 2007;81:700–712. doi: 10.1086/521033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braybrook C, Doudney K, Marçano AC, Arnason A, Bjornsson A, Patton MA, Goodfellow PJ, Moore GE, Stanier P. The T-box transcription factor gene TBX22 is mutated in X-linked cleft palate and ankyloglossia. Nat. Genet. 2001;29:179–183. doi: 10.1038/ng730. [DOI] [PubMed] [Google Scholar]
- Braybrook C, Lisgo S, Doudney K, Henderson D, Marçano AC, Strachan T, Patton MA, Villard L, Moore GE, Stanier P, Lindsay S. Craniofacial expression of human and murine TBX22 correlates with the cleft palate and ankyloglossia phenotype observed in CPX patients. Hum. Mol. Genet. 2002;11:2793–804. doi: 10.1093/hmg/11.22.2793. [DOI] [PubMed] [Google Scholar]
- Buijs A, Sherr S, van Baal S, van Bezouw S, van der Plas D, van Kessel A. Geurts, Riegman P, Deprez R. Lekanne, Zwarthoff E, Hagemeijer A. Translocation (12;22) (p13;q11) in myeloproliferative disorders results in fusion of the ETS-like TEL gene on 12p13 to the MN1 gene on 22q11. Oncogene. 1995;10:1511–1519. [PubMed] [Google Scholar]
- Buijs A, van Rompaey L, Molijn AC, Davis JN, Vertegaal AC, Potter MD, Adams C, van Baal S, Zwarthoff EC, Roussel MF, Grosveld GC. The MN1-TEL fusion protein., encoded by the translocation (12;22)(p13;q11) in myeloid leukemia, is a transcription factor with transforming activity. Mol. Cell. Biol. 2000;20:9281–93. doi: 10.1128/mcb.20.24.9281-9293.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush JO, Lan Y, Maltby KM, Jiang R. Isolation and developmental expression analysis of Tbx22, the mouse homolog of the human X-linked cleft palate gene. Dev. Dyn. 2002;225:322–6. doi: 10.1002/dvdy.10154. [DOI] [PubMed] [Google Scholar]
- Carella C, Bonten J, Rehg J, Grosveld GC. MN1-TEL, the product of the t(12;22) in human myeloid leukemia, immortalizes murine myeloid cells and causes myeloid malignancy in mice. Leukemia. 2006;20:1582–92. doi: 10.1038/sj.leu.2404298. [DOI] [PubMed] [Google Scholar]
- Carella C, Bonten J, Sirma S, Kranenburg TA, Terranova S, Klein-Geltink R, Shurtleff S, Downing JR, Zwarthoff EC, Liu PP, Grosveld GC. MN1 overexpression is an important step in the development of inv(16) AML. Leukemia. 2007;21:1679–90. doi: 10.1038/sj.leu.2404778. [DOI] [PubMed] [Google Scholar]
- Casey LM, Lan Y, Cho ES, Maltby KM, Gridley T, Jiang R. Jag2-Notch1 signaling regulates oral epithelial differentiation and palate development. Dev. Dyn. 2006;235:1830–44. doi: 10.1002/dvdy.20821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celli J, Duijf P, Hamel BC, Bamshad M, Kramer B, Smits AP, Newbury-Ecob R, Hennekam RC, Van Buggenhout G, van Haeringen A, Woods CG, van Essen AJ, de Waal R, Vriend G, Haber DA, Yang A, McKeon F, Brunner HG, van Bokhoven H. Heterozygous germline mutations in the p53 homolog p63 are the cause of EEC syndrome. Cell. 1999;99:143–53. doi: 10.1016/s0092-8674(00)81646-3. [DOI] [PubMed] [Google Scholar]
- Chai Y, Maxson RE., Jr. Recent advances in craniofacial morphogenesis. Dev. Dyn. 2006;235:2353–75. doi: 10.1002/dvdy.20833. [DOI] [PubMed] [Google Scholar]
- Chaabouni M, Smaoui N, Benneji N, M’rd R, Jemaa LB, Hachicha S, Chaabouni H. Mutation analysis of TBX22 reveals new mutation in Tunisian CPX family. Clin. Dysmorphol. 2005;14:23–25. [PubMed] [Google Scholar]
- Du Y, Jenkins NA, Copeland NG. Insertional mutagenesis identifies genes that promote the immortalization of primary bone marrow progenitor cells. Blood. 2005;106:3932–9. doi: 10.1182/blood-2005-03-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson MW. Palate development. Development Suppl. 1988;103:41–60. doi: 10.1242/dev.103.Supplement.41. [DOI] [PubMed] [Google Scholar]
- Grosveld GC. MN1, a novel player in human AML. Blood. Cells. Mol. Dis. 2007;39:336–9. doi: 10.1016/j.bcmd.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlin RJ, Cohen MM, Jr., Hennekam RCM. Oxford University Press; New York: 2001. Syndromes of the head and neck. [Google Scholar]
- Gritli-Linde A. Molecular control of secondary palate development. Dev. Biol. 2007;301:309–26. doi: 10.1016/j.ydbio.2006.07.042. [DOI] [PubMed] [Google Scholar]
- Halford MM, Armes J, Buchert M, Meskenaite V, Grail D, Hibbs ML, Wilks AF, Farlie PG, Newgreen DF, Hovens CM, Stacker SA. Ryk-deficient mice exhibit craniofacial defects associated with perturbed Eph receptor crosstalk. Nat. Genet. 2000;25:414–418. doi: 10.1038/78099. [DOI] [PubMed] [Google Scholar]
- Han J, Ito Y, Yeo JY, Sucov HM, Maas R, Chai Y. Cranial neural crest-derived mesenchymal proliferation is regulated by Msx1-mediated p19(INK4d) expression during odontogenesis. Dev. Biol. 2003;261:183–196. doi: 10.1016/s0012-1606(03)00300-2. [DOI] [PubMed] [Google Scholar]
- Herr A, Meunier D, Müller I, Rump A, Fundele R, Ropers HH, Nuber UA. Expression of mouse Tbx22 supports its role in palatogenesis and glossogenesis. Dev. Dyn. 2003;226:579–586. doi: 10.1002/dvdy.10260. [DOI] [PubMed] [Google Scholar]
- Heuser M, Beutel G, Krauter J, Döhner K, von Neuhoff N, Schlegelberger B, Ganser A. High meningioma 1 (MN1) expression as a predictor for poor outcome in acute myeloid leukemia with normal cytogenetics. Blood. 2006;108:3898–905. doi: 10.1182/blood-2006-04-014845. [DOI] [PubMed] [Google Scholar]
- Hilliard SA, Yu L, Gu S, Zhang Z, Chen YP. Regional regulation of palatal growth and patterning along the anterior-posterior axis in mice. J. Anat. 2005;207:655–667. doi: 10.1111/j.1469-7580.2005.00474.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Lee H, Price SM, Shen MM, Abate-Shen C. Msx homeobox genes inhibit differentiation through upregulation of cyclin D1. Development. 2001;128:2373–2384. doi: 10.1242/dev.128.12.2373. [DOI] [PubMed] [Google Scholar]
- Ianakiev P, Kilpatrick MW, Toudjarska I, Basel D, Beighton P, Tsipouras P. Split-hand/split-foot malformation is caused by mutations in the p63 gene on 3q27. Am. J. Hum. Genet. 2000;67:59–66. doi: 10.1086/302972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y, Yeo JY, Chytil A, Han J, Bringas P, Jr., Nakajima A, Shuler CF, Moses HL, Chai Y. Conditional inactivation of Tgfbr2 in cranial neural crest causes cleft palate and calvaria defects. Development. 2003;130:5269–5280. doi: 10.1242/dev.00708. [DOI] [PubMed] [Google Scholar]
- Jin JZ, Ding J. Analysis of Meox2 mutant mice reveals a novel postfusion-based cleft palate. Dev. Dyn. 2006;235:539–546. doi: 10.1002/dvdy.20641. [DOI] [PubMed] [Google Scholar]
- Kawagoe H, Grosveld GC. Conditional MN1-TEL knock-in mice develop acute myeloid leukemia in conjunction with overexpression of HOXA9. Blood. 2005a;106:4269–77. doi: 10.1182/blood-2005-04-1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawagoe H, Grosveld GC. MN1-TEL myeloid oncoprotein expressed in multipotent progenitors perturbs both myeloid and lymphoid growth and causes T-lymphoid tumors in mice. Blood. 2005b;106:4278–86. doi: 10.1182/blood-2005-04-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo S, Schutte BC, Richardson RJ, Bjork BC, Knight AS, Watanabe Y, Howard E, de Lima RL, Daack-Hirsch S, Sander A, McDonald-McGinn DM, Zackai EH, Lammer EJ, Aylsworth AS, Ardinger HH, Lidral AC, Pober BR, Moreno L, Arcos-Burgos M, Valencia C, Houdayer C, Bahuau M, Moretti-Ferreira D, Richieri-Costa A, Dixon MJ, Murray JC. Mutations in IRF6 cause Van der Woude and popliteal pterygium syndromes. Nat Genet. 2002;32:285–9. doi: 10.1038/ng985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan Y, Kingsley PD, Cho ES, Jiang R. Osr2, a new mouse gene related to Drosophila odd-skipped, exhibits dynamic expression patterns during craniofacial, limb, and kidney development. Mech. Dev. 2001;107:175–179. doi: 10.1016/s0925-4773(01)00457-9. [DOI] [PubMed] [Google Scholar]
- Lan Y, Ovitt CE, Cho ES, Maltby KM, Wang Q, Jiang R. Odd-skipped related 2 (Osr2) encodes a key intrinsic regulator of secondary palate growth and morphogenesis. Development. 2004;131:3207–3216. doi: 10.1242/dev.01175. [DOI] [PubMed] [Google Scholar]
- Deprez R. H. Lekanne, Riegman PH, Groen NA, Warringa UL, van Biezen NA, Molijn AC, Bootsma D, de Jong PJ, Menon AG, Kley NA, et al. Cloning and characterization of MN1, a gene from chromosome 22q11, which is disrupted by a balanced translocation in a meningioma. Oncogene. 1995;10:1521–1528. [PubMed] [Google Scholar]
- Li Q, Ding J. Gene expression analysis reveals that formation of the mouse anterior secondary palate involves recruitment of cells from the posterior side. Int. J. Dev. Biol. 2007;51:167–172. doi: 10.1387/ijdb.062212ql. [DOI] [PubMed] [Google Scholar]
- Liu W, Sun X, Braut A, Mishina Y, Behringer RR, Mina M, Martin JF. Distinct functions for Bmp signaling in lip and palate fusion in mice. Development. 2005;132:1453–61. doi: 10.1242/dev.01676. [DOI] [PubMed] [Google Scholar]
- Marcano ACB, Doudney K, Braybrook C, Squires R, Patton MA, Lees M, Richieri-Costa A, Lidral AC, Murray JC, Moore GE, et al. TBX22 mutations are a frequent cause of cleft palate. J. Med. Genet. 2004;41:68–74. doi: 10.1136/jmg.2003.010868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushime H, Roussel MF, Ashmun RA, Sherr CJ. Colony-stimulating factor 1 regulates novel cyclins during the G1 phase of the cell cycle. Cell. 1991;65:701–713. doi: 10.1016/0092-8674(91)90101-4. [DOI] [PubMed] [Google Scholar]
- Meester-Smoor MA, Molijn AC, Zhao Y, Groen NA, Groffen CA, Boogaard M, van Dalsum-Verbiest D, Grosveld GC, Zwarthoff EC. The MN1 oncoprotein activates transcription of the IGFBP5 promoter through a CACCC-rich consensus sequence. J. Mol. Endocrinol. 2007;38:113–25. doi: 10.1677/jme.1.02110. [DOI] [PubMed] [Google Scholar]
- Meester-Smoor MA, Vermeij M, van Helmond MJ, Molijn AC, van Wely KH, Hekman AC, Vermey-Keers C, Riegman PH, Zwarthoff EC. Targeted disruption of the Mn1 oncogene results in severe defects in development of membranous bones of the cranial skeleton. Mol. Cell. Biol. 2005;25:4229–4236. doi: 10.1128/MCB.25.10.4229-4236.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan DO. Cyclin-dependent kinases: engines, clocks, and microprocessors. Annu. Rev. Cell. Dev. Biol. 1997;13:261–291. doi: 10.1146/annurev.cellbio.13.1.261. [DOI] [PubMed] [Google Scholar]
- Motokura T, Bloom T, Kim HG, Jüppner H, Ruderman JV, Kronenberg HM, Arnold A. A novel cyclin encoded by a bcl1-linked candidate oncogene. Nature. 1991;350:512–515. doi: 10.1038/350512a0. [DOI] [PubMed] [Google Scholar]
- Peters H, Neubüser A, Kratochwil K, Balling R. Pax9-deficient mice lack pharyngeal pouch derivatives and teeth and exhibit craniofacial and limb abnormalities. Genes. Dev. 1998;12:2735–2747. doi: 10.1101/gad.12.17.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice R, Spencer-Dene B, Connor EC, Gritli-Linde A, McMahon AP, Dickson C, Thesleff I, Rice DPC. Disruption of Fgf10/Fgfr2b-coordinated epithelial-mesenchymal interactions causes cleft palate. J. Clin. Invest. 2004;113:1692–1700. doi: 10.1172/JCI20384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross ME, Mahfouz R, Onciu M, Liu HC, Zhou X, Song G, Shurtleff SA, Pounds S, Cheng C, Ma J, Ribeiro RC, Rubnitz JE, Girtman K, Williams WK, Raimondi SC, Liang DC, Shih LY, Pui CH, Downing JR. Gene expression profiling of pediatric acute myelogenous leukemia. Blood. 2004;104:3679–87. doi: 10.1182/blood-2004-03-1154. [DOI] [PubMed] [Google Scholar]
- Schutte BC, Murray JC. The many faces and factors of orofacial clefts. Hum. Mol. Genet. 1999;8:1853–1859. doi: 10.1093/hmg/8.10.1853. [DOI] [PubMed] [Google Scholar]
- Sperber GH. Palatogenesis: closure of the secondary palate. In: Wyszynski DF, editor. Cleft Lip & Palate, From Origin To Treatment. Oxford University Press; 2002. pp. 14–24. [Google Scholar]
- Suphapeetiporn K, Tongkobpetch S, Siriwan P, Shotelersuk V. TBX22 mutations are a frequent cause of non-syndromic cleft palate in the Thai population. Clin. Genet. 2007;72:478–483. doi: 10.1111/j.1399-0004.2007.00891.x. [DOI] [PubMed] [Google Scholar]
- Sutton AL, Zhang X, Ellison TI, Macdonald PN. The 1,25(OH)2D3-regulated transcription factor MN1 stimulates vitamin D receptor-mediated transcription and inhibits osteoblastic cell proliferation. Mol. Endocrinol. 2005;19:2234–44. doi: 10.1210/me.2005-0081. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Hu D, Bustos T, Zlotogora J, Richieri-Costa A, Helm s J. A., Spritz RA. Mutations of PVRL1, encoding a cell-cell adhesion molecule/herpesvirus receptor, in cleft lip/palate-ectodermal dysplasia. Nat. Genet. 2000;25:427–30. doi: 10.1038/78119. [DOI] [PubMed] [Google Scholar]
- Valk PJ, Delwel R, Löwenberg B. Gene expression profiling in acute myeloid leukemia. Curr. Opin. Hematol. 2004;12:76–81. doi: 10.1097/01.moh.0000149610.14438.9a. [DOI] [PubMed] [Google Scholar]
- van den Boogaard MJ, Dorland M, Beemer FA, van Amstel HK. MSX1 mutation is associated with orofacial clefting and tooth agenesis in humans. Nat. Genet. 2000;24:342–3. doi: 10.1038/74155. [DOI] [PubMed] [Google Scholar]
- van Wely KH, Molijn AC, Buijs A, Meester-Smoor MA, Aarnoudse AJ, Hellemons A, den Besten P, Grosveld GC, Zwarthoff EC. The MN1 oncoprotein synergizes with coactivators RAC3 and p300 in RAR-RXR-mediated transcription. Oncogene. 2003;22:699–709. doi: 10.1038/sj.onc.1206124. [DOI] [PubMed] [Google Scholar]
- Vanderas AP. Incidence of cleft lip, cleft palate, and cleft lip and palate among races: a review. Cleft. Palate. J. 1987;24:216–225. [PubMed] [Google Scholar]
- Wang T, Tamakoshi T, Uezato T, Shu F, Kanzaki-Kato N, Fu Y, Koseki H, Yoshida N, Sugiyama T, Miura N. Forkhead transcription factor Foxf2 (LUN)-deficient mice exhibit abnormal development of secondary palate. Dev. Biol. 2003;259:83–94. doi: 10.1016/s0012-1606(03)00176-3. [DOI] [PubMed] [Google Scholar]
- Welsh IC, Hagge-Greenberg A, O’Brien TP. A dosage-dependent role for Spry2 in growth and patterning during palate development. Mech. Dev. 2007;124:746–761. doi: 10.1016/j.mod.2007.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Connolly T, Futcher B, Beach D. Human D-type cyclin. Cell. 1991;65:691–699. doi: 10.1016/0092-8674(91)90100-d. [DOI] [PubMed] [Google Scholar]
- Yu L, Gu S, Alappat S, Song Y, Yan M, Zhang X, Zhang G, Jiang Y, Zhang Z, Zhang Y, Chen Y. Shox2-deficient mice exhibit a rare type of incomplete clefting of the secondary palate. Development. 2005;132:4397–4406. doi: 10.1242/dev.02013. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zhao X, Hu Y, ***St Amand T, Zhang M, Ramamurthy R, Qiu M, Chen Y. Msx1 is required for the induction of Patched by Sonic hedgehog in the mammalian tooth germ. Dev. Dyn. 1999;215:45–53. doi: 10.1002/(SICI)1097-0177(199905)215:1<45::AID-DVDY5>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Song Y, Zhao X, Zhang X, Fermin C, Chen Y. Rescue of cleft palate in Msx1-deficient mice by transgenic Bmp4 reveals a network of BMP and Shh signaling in the regulation of mammalian palatogenesis. Development. 2002;129:4135–4146. doi: 10.1242/dev.129.17.4135. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.