Abstract
The Neisseria gonorrhoeae type IV secretion system secretes chromosomal DNA that acts in natural transformation. To examine the mechanism of DNA processing for secretion, we made mutations in the putative relaxase gene traI and used nucleases to characterize the secreted DNA. The nuclease experiments demonstrated that the secreted DNA is single-stranded and blocked at the 5′ end. Mutation of traI identified Tyr93 as required for DNA secretion, while substitution of Tyr201 resulted in intermediate levels of DNA secretion. TraI exhibits features of relaxases, but also has features that are absent in previously characterized relaxases, including an HD phosphohydrolase domain and an N-terminal hydrophobic region. The HD domain residue Asp120 was required for wild-type levels of DNA secretion. Subcellular localization studies demonstrated that the TraI N-terminal region promotes membrane interaction. We propose that Tyr93 initiates DNA processing and Tyr201 is required for termination or acts in DNA binding. Disruption of an inverted-repeat sequence eliminated DNA secretion, suggesting that this sequence may serve as the origin of transfer for chromosomal DNA secretion. The TraI domain architecture, although not previously described, is present in fifty-three uncharacterized proteins, suggesting that the mechanism of TraI function is a widespread process for DNA donation.
Introduction
Neisseria gonorrhoeae encodes an unusual type IV secretion system (T4SS). T4SSs include conjugation systems (Lawley et al., 2003), which transport DNA from one bacterium directly into another, and effector translocator systems, which secrete proteins into host cells that exert virulence functions (Christie, 2001). One of the best-characterized T4SSs is that of Agrobacterium tumefaciens, which transports proteins and plasmid DNA directly into plant cells (Christie et al., 2005). The gonococcal T4SS secretes chromosomal DNA into the environment, where the secreted DNA transforms other gonococci in the population (Dillard and Seifert, 2001; Hamilton et al., 2001). This unique, non-lytic mechanism for DNA donation may partly explain the high frequency of genetic exchange in N. gonorrhoeae and its panmictic population structure (Maynard Smith et al., 1993). The high rate of transformation results in the rapid spread of antibiotic resistance genes and increased variation of surface molecules, allowing evasion of the human immune response (Reviewed in Hamilton and Dillard, 2006). Mutations in gonococcal T4SS genes result in decreased DNA in culture supernatants and in decreased DNA donation for natural transformation (Dillard and Seifert, 2001; Hamilton et al., 2005).
The N. gonorrhoeae T4SS genes are encoded in the gonococcal genetic island (GGI) (Dillard and Seifert, 2001; Hamilton et al., 2005). The GGI is a 57-kb horizontally-acquired genomic region present in ∼80% of N. gonorrhoeae strains, and a few N. meningitidis strains, and may contribute to the virulence of these bacteria (Dillard and Seifert, 2001; Snyder et al., 2005). The GGI-encoded T4SS is similar to the F-plasmid family of conjugation T4SSs in terms of gene arrangement and limited sequence similarity. Mutational analysis demonstrated that DNA secretion requires several T4SS genes predicted to encode components of the secretion apparatus, as well as a relaxase, two peptidoglycanases, a disulphide bond isomerase, and a chromosome partitioning protein (Dillard and Seifert, 2001; Hamilton et al., 2001; Hamilton et al., 2005; Kohler et al., 2007). Because of the similarity to F-plasmid and the requirement for T4SS homologues, we predict that chromosomal DNA donation in N. gonorrhoeae may occur by a mechanism similar to DNA transfer from Escherichia coli Hfr strains, but with the DNA secreted into the surrounding milieu.
Although it was shown that gonococci secrete chromosomal DNA during growth, the characteristics of the secreted DNA have not been determined. The ability of the secreted DNA to transform other gonococci may be influenced by whether the DNA is single-stranded or double-stranded and whether it has protein bound to it or not. Previous studies using exogenously added DNA suggested that double-stranded DNA was the preferred substrate for natural transformation of N. gonorrhoeae (Biswas and Sparling, 1981), while one report suggested that single-stranded and double-stranded DNA transform with similar efficiency (Stein, 1991). Transformation experiments comparing DNA donation by wild-type versus a T4SS-mutant donor revealed that wild-type donors produce more transformants, even when the cultures were allowed to proceed to autolysis (Dillard and Seifert, 2001). These results suggest that the secreted DNA is more efficient at transforming recipients than DNA released by autolysis. However, the donation of DNA via a T4SS suggests that the secreted DNA is single-stranded, as is the case for conjugated DNA and DNA transferred by the A. tumefaciens T4SS (Llosa et al., 2002). If gonococci secrete DNA by a mechanism similar to conjugation, then the DNA should be cut by a nicking enzyme or relaxase and the enzyme may remain bound to the DNA (Gomis-Ruth et al., 2004).
Relaxases bind and nick supercoiled DNA in a sequence- and strand-specific manner at the origin of transfer (oriT) (Grinter, 1981). The tyrosine-mediated nicking reaction results in the covalent binding of the relaxase to the 5′ end of the cut DNA (Grinter, 1981; Pansegrau et al., 1990). The nucleoprotein complex is recognized by the coupling protein for transport via the type IV secretion apparatus (Llosa and O’Callaghan, 2004). Therefore, relaxases function as pilot proteins for DNA transport (Grinter, 1981; Pansegrau et al., 1990). Relaxases are characterized by the presence of three conserved amino acid sequence motifs (Pansegrau and Lanka, 1991). Motif I contains one or two catalytic tyrosines (Pansegrau et al., 1990; Pansegrau et al., 1993); motif II is thought to be involved in facilitating DNA-protein interactions at the 3′ side of the cleavage site (Pansegrau et al., 1994); and motif III, also known as the three-histidine (3H) motif, has been historically used as the hallmark of relaxases and is involved in metal ion coordination (Francia et al., 2004). N. gonorrhoeae encodes a putative relaxase, TraI, required for type IV secretion (Hamilton et al., 2005). TraI exhibits features of relaxases, but also exhibits features that are absent in previously characterized relaxases, suggesting that DNA processing for secretion in N. gonorrhoeae may have important differences compared to DNA processing by known T4SSs. To characterize DNA secretion by N. gonorrhoeae and gain a better understanding of DNA donation for natural transformation, we examined the mechanism of DNA processing focusing on the putative relaxase TraI and the sensitivities of the secreted DNA to specific nucleases.
Here we show that the putative relaxase TraI has features not found in known relaxases, including an N-terminal hydrophobic region and an HD (histidine and aspartate) phosphohydrolase domain. Site-directed mutagenesis of traI identified two tyrosines necessary for DNA secretion, with one predicted to act in the initial cleavage of chromosomal DNA for secretion. Also a mutation in motif I of the HD domain significantly decreased DNA secretion, as did mutations affecting the unique hydrophobic region found at the N-terminus. Remarkably, this study identified fifty-three proteins with the same domain architecture as TraI. These proteins conserve an N-terminal putative relaxase domain fused to the HD domain of phosphohydrolases, all within the first three-hundred amino acids. These proteins may represent uncharacterized conjugation or DNA donation proteins. Nuclease treatment of gonococcal culture supernatants and co-culture transformation in the presence of specific nucleases revealed that the secreted DNA is only degraded by nucleases that have single-strand 3′→5′ exonuclease activity or endonuclease activity, suggesting that gonococci secrete single-stranded DNA protected at the 5′ end. An insertion disrupting an inverted repeat in a non-coding region of the GGI caused loss of DNA secretion, suggesting that this region may contain an oriT. Altogether, this work reveals important properties of the secreted DNA and the mechanism of DNA processing for type IV secretion in N. gonorrhoeae.
Results
Site-directed mutagenesis replacing Tyr93 and Tyr201 reveals their requirement for TraI function in DNA secretion
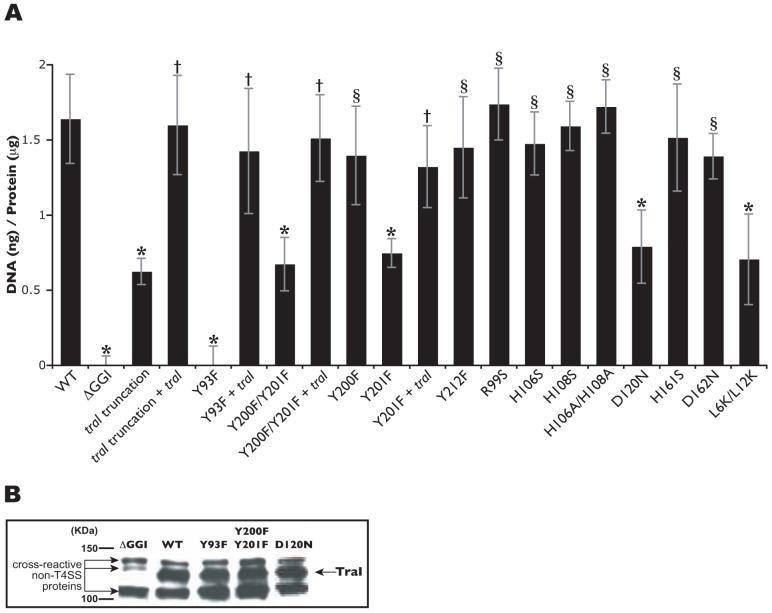
Previously we found that a strain carrying a traI insertion (truncating TraI at amino acid 200) is deficient in DNA secretion, indicating a requirement for TraI in type IV secretion (Hamilton et al., 2005). Complementation of this strain with the wild-type copy of traI at a distant location on the chromosome restored DNA secretion to wild-type levels, confirming that the DNA secretion deficiency was not due to a polar effect on downstream genes (Fig. 1A). These results provide further evidence that DNA secretion in N. gonorrhoeae is dependent on the putative relaxase TraI. An alignment of TraI and the Xyllela fastidiosa putative relaxase revealed conservation of Tyr93, Tyr201 and Tyr212 among the proteins (Fig. 2). These conserved tyrosines are potential catalytic amino acid residues. The F-family of relaxases conserves pairs of tyrosines reported to play different roles in DNA processing (Grandoso et al., 2000). Thus, Tyr200 was also of interest since it was located next to Tyr201 forming a tyrosine pair. To determine the functional relevance of Tyr93, Tyr200, Tyr201 and Tyr212, we used site-directed mutagenesis to replace these conserved tyrosines with phenylalanines. DNA secretion was eliminated in the strain carrying the Y93F mutation, while the Y212F mutation had no effect on DNA secretion (Fig.1A). Interestingly, the strain expressing a double tyrosine substitution (Y200F/Y201F) showed intermediate levels of DNA secretion (Fig. 1A). Individual tyrosine substitutions were made to determine if the intermediate DNA secretion phenotype observed in the strain carrying the Y200F/Y201F mutation could be ascribed to a particular tyrosine. The Y200F mutant showed wild-type levels of DNA secretion, whereas DNA secretion by the strain carrying the Y201F mutation resulted in the intermediate DNA secretion phenotype observed with the Y200F/Y201F substitutions (Fig. 1A). Introduction of wild-type traI at a distant site on the chromosome restored wild-type levels of DNA secretion in the strains carrying traI Y93F, Y200F/Y201F, or Y201F mutations (Fig. 1A). Analysis of native expression of TraIY93F and TraIY200F/Y201F by Western blot revealed that the proteins are expressed at levels similar to that of the wild-type protein (Fig.1B). These results provide evidence that Tyr93 and Tyr201 are required for the function of TraI in DNA secretion. Furthermore, the different effects of the Y93F and Y201F mutations on DNA secretion suggest non-redundant roles of these tyrosines in the activity of TraI. Tyr93 may be involved in the initiation of DNA processing, while Tyr201 may be involved either in a second cleavage for termination of DNA processing or in the stability of the DNA-protein interaction.
Fig. 1.
A. Fluorometric detection of secreted DNA. Piliated gonococcal strains were grown for 2.5h in liquid culture. Cell-free culture supernatants were collected, and DNA was detected with the fluorescent DNA-binding dye PicoGreen and normalized to total cell protein. MS11 was used as the wild-type (WT) strain and ND500 (ΔGGI in MS11) as the negative control. The results are an average of at least four independent experiments. *p-value<0.003 when compared to wild-type. †p-value<0.003 when compared to the respective mutant. §Not significantly different than wild type. B. Detection of TraI by Western blot. TraI was produced by wild-type (wt) and mutant strains at approximately the same levels. TraI is not detected in strain ND500, a strain in which the GGI was deleted.
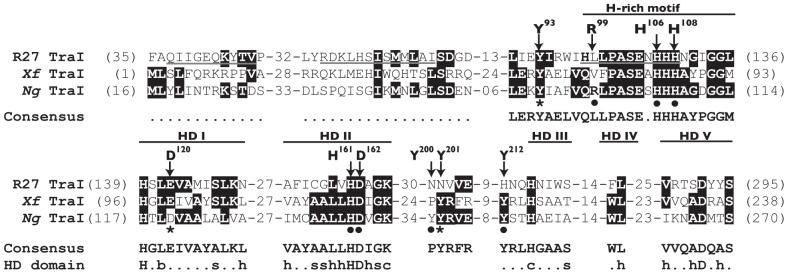
Fig. 2.
Amino acid sequence alignment of predicted relaxases from plasmid R27 (R27 TraI), X. fastidiosa (Xf TraI), and N. gonorrhoeae (Ng TraI). Identical amino acids are highlighted. Underlined sequences in R27 TraI were reported to correspond to the three motifs of classical relaxases. However, the first two motifs are not conserved in the Ng TraI family. The hallmarks of the Ng TraI family are a conserved tyrosine (Tyr93 in Ng TraI) closely followed by a histidine (H)-rich motif, and the HD phosphohydrolase domain. The signature sequence of the H-rich motif is h(Q/H)xhPASExHHHx3GG(L/M)h, where h is a hydrophobic residue and x is any residue. The HD domain motifs are labeled HD I to V (where HD II is the signature motif). The consensus amino acid sequence for the fifty-four proteins in the Ng TraI family of predicted relaxases is shown (the dotted lines represent no consensus). The consensus amino acid sequence for the HD domain is also shown [(b) big, (s) small, (h) hydrophobic, (c) charged, and the capital letters represent invariant amino acids]. The arrows indicate the amino acids in Ng TraI that were the targets for site-directed mutagenesis in this study. Mutations that decrease DNA secretion are indicated with a star. Mutations that have no effect on DNA secretion are indicated with a solid circle.
Single amino acid substitutions within the histidine-rich motif result in wild-type levels of DNA secretion
A histidine (H)-rich motif is found five amino acid residues from Tyr93 (Fig. 2). This sequence aligns well with a region from the relaxase (TraI) of Salmonella typhi plasmid R27, where it was referred to as the 3H motif of relaxases (Lawley et al., 2002). The gonococcal TraI H-rich motif conserves Arg99, His106, and His108 (Fig. 2). Arg99 was of interest because it aligns well with Arg150 of the F-plasmid family 3H motif (data not shown), which was shown to contribute to relaxation of DNA in vitro (Harley et al., 2002). To determine the functional relevance of Arg99, His106, and His108 in DNA secretion, we constructed strains carrying traI R99S, H106S, or H108S mutations. Surprisingly, these strains exhibit wild-type levels of DNA secretion (Fig. 1A). A possible explanation for this result is that single histidine substitutions or the substitution to serine may be insufficient to eliminate a possible function in metal coordination. To address this possibility, we constructed a strain expressing a double histidine substitution (H106A/H108A), where the histidines were replaced by alanines. The strain carrying traIH106A/H108A also showed wild-type levels of DNA secretion (Fig. 1A). These results indicate that Arg99, His106, and His108 are not required for TraI function in DNA secretion.
N. gonorrhoeae TraI has features unlike any characterized relaxase
TraI was identified as a putative relaxase based on N-terminal amino acid sequence homology to the putative nickase from Pseudomonas resinovorans and the putative relaxase from X. fastidiosa (Hamilton et al., 2005). Strikingly, TraI conserves the five motifs characteristic of the HD domain of the metal-dependent phosphohydrolase superfamily, all within the first three-hundred amino acids (Fig. 2) (Aravind and Koonin, 1998). To determine if the presence of the HD domain was unique to N. gonorrhoeae (Ng) TraI among relaxases, we performed a database search using full-length TraI as the query. The search resulted in the identification of fifty-three proteins with a high degree of amino acid conservation to TraI (Table 1, Fig. S1). Some of the proteins in this group are annotated as putative relaxases or hydrolases, but most of them are hypothetical proteins (Table 1). These proteins come from a number of γ- and β-proteobacteria, and for a large number of them, the sequences have just recently become available. A phylogenetic tree obtained from an amino acid alignment of the full-length proteins resulted in the clustering of TraI with putative relaxases from other pathogenic bacterial species including Vibrio cholerae, avian pathogenic Escherichia coli, Salmonella typhi, Proteous vulgaris, and Yersinia pestis (Fig. 3). Interestingly, the large majority of these proteins are chromosomally encoded, and of these, some can be found in mobile genomic islands (e.g. integrated pKLC102) or conjugative/integrative elements (e.g. pICEhin1056). Only six of the proteins in this family are plasmid-encoded (Fig. 3).
Table 1.
Proteins with similar domain architecture as N. gonorrhoeae TraI
| Protein origin | Predicted Function | Length (bp) | Identify/Range (aa) | Acc. Number |
|---|---|---|---|---|
| Neisseria gonorrhoeae str. MS11 | Relaxase | 850 | 100%/850 | AAW83058.1 |
| Acidovorax avenae ssp. citrulli str. AAC00-1 | Phosphohydrolase | 861 | 35%/26 | YP_971114.1 |
| Acidovorax avenae ssp. citrulli str. AAC00-1 | Relaxase | 623 | 47%/86 | YP_968947.1 |
| Acidovorax sp. JS42 | Relaxase | 627 | 36%/115 | YP_985545.1 |
| Acidovorax sp. JS42 | Relaxase | 614 | 36%/100 | ABM42340.1 |
| Acidovorax sp. JS42 | Relaxase | 610 | 26%/284 | YP_985678.1 |
| Methylibium petroleiphilum str. PM1 | Hypothetical | 612 | 36%/100 | YP_001021595.1 |
| Burkholderia xenovorans str. LB400 | Hypothetical | 615 | 39%/101 | YP_559832.1 |
| Ralstonia metallidurans str. CH34 | Relaxase | 680 | 37%/115 | YP_584510.1 |
| Azoarcus sp. EbN1 | Hypothetical | 711 | 54%/42 | YP_157225.1 |
| Azoarcus sp. EbN1 | Hypothetical | 623 | 46%/88 | YP_158431.1 |
| Azoarcus sp. EbN1 | Hypothetical | 604 | 46%/88 | YP_158356.1 |
| Nitrosomonas eutropha str. C91 | Relaxase | 621 | 37%/100 | ABI58357.1 |
| Haemophilus somnus str. 129PT | Hypothetical | 640 | 37%/94 | YP_718205.1 |
| Haemophilus influenzae str. 86-028NP | Hypothetical | 638 | 46%/77 | YP_247808.1 |
| Haemophilus influenzae plasmid pICEhin1056 | Hypothetical | 635 | 27%/393 | CAF29070.1 |
| Haemophilus influenzae str. R2866 | HD hydrolase | 603 | 27%/393 | ZP_00157643.1 |
| Haemophilus ducreyi str. 35000HP | Hypothetical | 217 | 41%/86 | NP_873395.1 |
| Xanthomonas campestris pv. campestris ATCC33913 | Hypothetical | 340 | 37%/103 | NP_638468.1 |
| Xanthomonas campestris pv. campestris str. 8004 | Hypothetical | 642 | 44%/92 | YP_243101.1 |
| Xanthomonas campestris pv. vesicatoria str. 85-10 | Hypothetical | 610 | 35%/100 | YP_364087.1 |
| Xanthomonas axonopodis pv. citri str. 306 | Hypothetical | 633 | 38%/107 | NP_642513.1 |
| Xylella fastidiosa str. 9a5c | Hypothetical | 616 | 34%/101 | NP_299042.1 |
| Pseudomonas syringae str. BR2R | Hypothetical | 638 | 35%/103 | AAL99262.1 |
| Pseudomonas syringae pv. syringae str. B728a | Relaxase | 318 | 35%/103 | YP_233835.1 |
| Pseudomonas syringae pv. syringae str. B728a | Relaxase | 634 | 35%/103 | YP_234511.1 |
| Pseudomonas syringae pv. phaseolicola | HD hydrolase | 578 | 35%/103 | CAI36140.1 |
| Pseudomonas syringae pv. tomato str. DC3000 | Hypothetical | 642 | 76%/17 | NP_790676.1 |
| Pseudomonas aeruginosa str. 6077 | Hypothetical | 643 | 40%/100 | ABD94612.1 |
| Pseudomonas aeruginosa clone C plasmid pKLC102 | Hypothetical | 639 | 41%/100 | AAP22591.1 |
| Pseudomonas aeruginosa clone C | Hypothetical | 608 | 40%/100 | AAP94681.1 |
| Pseudomonas aeruginosa str. PA14 | Pathogenesis-related | 639 | 40%100 | AAP84130.1 |
| Pseudomonas aeruginosa str. SG17M | Hypothetical | 600 | 37%/100 | AAN62266.1 |
| Pseudomonas aeruginosa clone C | Hypothetical | 630 | 37%/115 | AAN62129.1 |
| Pseudomonas aeruginosa str. 2192 | HD hydrolase | 312 | 40%/100 | ZP_00971291.1 |
| Pseudomonas aeruginosa str. 2192 | HD hydrolase | 602 | 37%/115 | ZP_00974810.1 |
| Pseudomonas aeruginosa str. PACS2 | Hypothetical | 614 | 38%/101 | ZP_01366009.1 |
| Pseudomonas resinovorans str. CA10 plasmid pCAR1 | Nickase | 900 | 70%/27 | NP_758664.1 |
| Pseudomonas fluorescence str. PfO-1 | Hypothetical | 575 | 47%/76 | YP_348763.1 |
| Pseudomonas fluorescence str. Pf-5 | Hypothetical | 628 | 39%/100 | YP_261833.1 |
| Pseudomonas putida str. F1 | Hypothetical | 595 | 44%/97 | ZP_00898124.1 |
| Azotobacter vinelandii str. AvOP | Relaxase | 627 | 40%/108 | ZP_00419624.1 |
| Yersinia pestis bv. orientalis str. IP275 | HD hydrolase | 992 | 66%/36 | ZP_01175343.1 |
| Providencia rettgeri str. R391 | Relaxase | 716 | 42%/87 | AAM08003.1 |
| Proteus vulgaris str. UR-75 plasmid Rts 1 | DNA helicase | 892 | 69%/26 | NP_640161.1 |
| Salmonella enterica ssp. enterica sv. typhi plasmid pHCM1 | Hypothetical | 1011 | 44%/68 | NP_569454.1 |
| Salmonella typhi plasmid R27 | Relaxase | 1011 | 44%/68 | NP_058333.1 |
| Escherichia coli plasmid pAPEC-01-R sv. 01:K1 | Relaxase | 1050 | 66%/27 | ABF67837.1 |
| Serratia mercescens plasmid R478 | Hypothetical | 1050 | 66%/27 | NP_941280.1 |
| Marinobacter aquaeoli str. VT8 | Relaxase | 624 | 48%/80 | YP_957144.1 |
| Shewanella sp. W3-18-1 plasmid 1 | Phosphohydrolase | 716 | 42%/87 | YP_962502.1 |
| Shewanella sp. ANA-3 | Relaxase | 941 | 29%/164 | YP_863780.1 |
| Vibrio cholerae | Relaxase | 716 | 42%/87 | ABA87024.1 |
| Vibrio cholerae str. MO10 | Relaxase | 716 | 42%/87 | EAZ47260.1 |
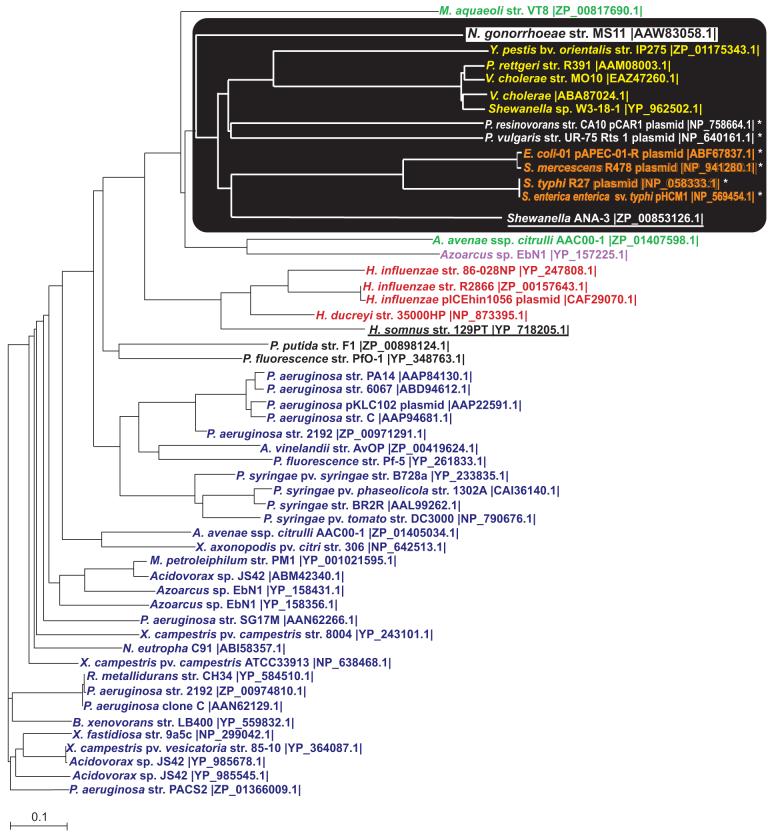
Fig. 3.
Phylogenetic tree of proteins with the same domain architecture as Ng TraI. The phylogenetic tree was generated from the CLUSTAL W alignment of the full-length proteins. A black box highlights the cluster containing Ng TraI. The different font colors represent proteins with similar predicted amphipathic α helices at the N-terminus proximal region: yellow, orange, red, and navy blue. Black font and white font indicate proteins with unique amphipathic α helices. Ng TraI, black font (white background), contains an N-terminal amphipathic α helix. Azoarcus sp. EbN1 (purple font) contains a predicted N-terminal transmembrane domain in addition to the N-terminus proximal amphipathic α helix. Green font indicates proteins that contain two predicted N-terminus proximal amphipathic α helices. Underlined proteins do not contain predicted amphipathic α helices. H. somnus contains a charged reside on the hydrophobic side, and the Shewanella ANA-3 sequence available is truncated. Orange font indicates proteins that conserve motif I and II of relaxases. *Plasmid-encoded proteins.
The only other putative relaxase from the Ng TraI family shown to be required for type IV secretion is TraI from the S. typhi R27 plasmid. R27 TraI contains the three signature motifs of relaxases (Lawley et al., 2002). However, alignment of the fifty-four proteins in the Ng TraI family revealed that the R27-plasmid TraI motifs I and II (Fig. 2) are only conserved in pAPEC-01-R, R478 plasmid, and pHCM1 (Fig. 3 and Fig. S1). Amino acid conservation within the Ng TraI family starts with the motif containing Ng TraI Tyr93, conserved in forty-three of the proteins, followed by the H-rich motif of signature h(Q/H)xhPASExHHHx3GG(L/M)h (where x is any residue and h is a hydrophobic residue), present in all the proteins. Moreover, all the proteins conserve Ng TraI Tyr201 and the HD phosphohydrolase domain (Fig. S1, consensus sequence shown in Fig. 2). Finally, almost all the proteins conserve a domain of unknown function (DUF1528) near the C-terminus. This information suggests that TraI belongs to a newly discovered family of relaxases or relaxase-like proteins, and unlike any of the characterized relaxases, the domain architecture includes the HD domain of phosphohydrolases.
The HD domain motif I mutation D120N reduces DNA secretion
The HD phosphohydrolase domain highly conserves histidines and aspartates (Fig. 2). Therefore, it has been proposed that the primary function of this domain is in the coordination of divalent cations (Aravind and Koonin, 1998). The HD domain motifs I, II, and V were reported to be conserved in the entire HD domain superfamily (Aravind and Koonin, 1998). Therefore, we focused on His161 and Asp162, which are the signature amino acids of the HD domain (present in motif II), and Asp120, present in motif I (Fig. 2) (Aravind and Koonin, 1998). Asp120 was of interest because secondary-structure-based threading (LOOPP) placed it in close proximity to Tyr93. There is evidence that interaction of an aspartate with an active site tyrosine is required for the activity of relaxases (Guasch et al., 2003). Thus, to determine the contribution of these amino acids in the function of TraI in DNA secretion, we constructed strains carrying traI H161S, D162N, or D120N mutations. The N. gonorrhoeae strains carrying the H161S or the D162N mutation showed wild-type levels of DNA secretion (Fig. 1A). However, the strain carrying the D120N mutation exhibited significantly reduced levels of DNA in culture supernatants (Fig. 1A). Analysis of native expression of TraID120N by Western blot revealed that the protein is expressed at levels similar to that of the wild-type protein (Fig.1B). The precise role of the HD domain in DNA processing is not clear. Nevertheless, these results indicate that the HD domain contributes to the function of TraI in DNA secretion.
Unique N-terminal region in N. gonorrhoeae TraI
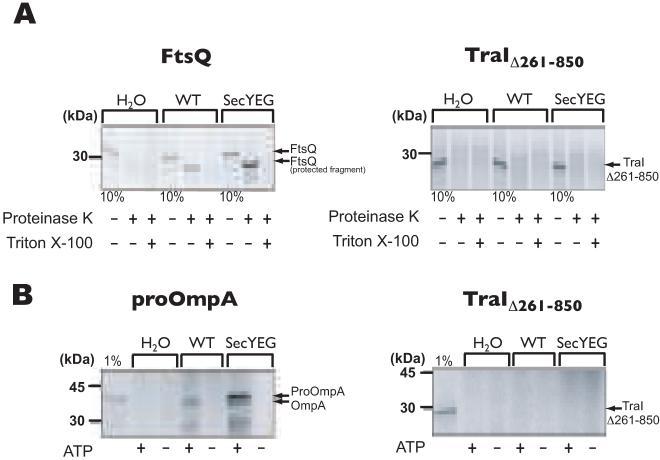
A feature that makes TraI unique among the members of its family and all characterized relaxases is the presence of an N-terminal region that is identified (by SignalP algorithm analysis) as a signal sequence (Fig. 4A). If this region were to function as a signal sequence, TraI would be transported into the periplasm via the sec-translocase prior to export by the T4SS. To address the functionality of the putative signal peptide in targeting TraI to the membrane and transporting it via the sec-translocase, we used the well-established in vitro translocation assay with isolated inverted inner membrane vesicles of E. coli (Cunningham and Wickner, 1989). 35S-labeled TraIΔ261-850 was used in a co-translational targeting/transport assay in which the translocation into inverted inner membrane vesicles was monitored by the accessibility of TraIΔ261-850 to the externally added proteinase K. If TraI contains a functional signal sequence, the protein would be transported into the vesicles where it would be protected from degradation by proteinase K, and therefore, detectable by autoradiography following electrophoresis. However, if TraI’s N-terminal region does not function as a sec-dependent signal sequence, it would remain accessible to proteinase K and be degraded. Protease-protected fragments of TraIΔ261-850 were not observed in the co-translational targeting/transport assay, unlike the protease-protected fragments detected in the positive control FtsQ (Fig. 5A). A post-translational transport assay of 35S-labeled TraIΔ261-850 also resulted in no detection of protease-protected fragments, unlike the ones detected for the positive control proOmpA (Fig. 5B). These results indicate that the N-terminal region does not function as a signal peptide to target TraI for transport into the periplasm via the Sec system.
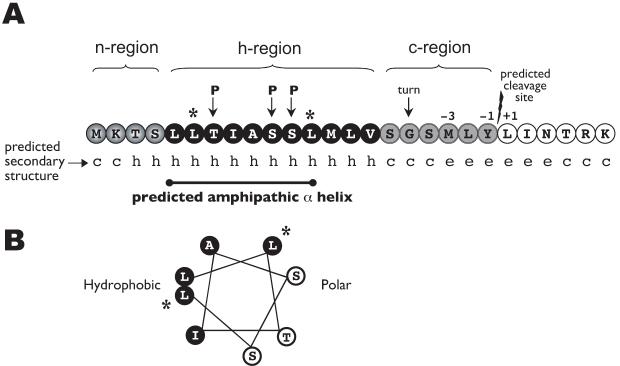
Fig. 4.
Map of the TraI N-terminal region.
A. A SignalP 3.0 algorithm analysis of the Ng TraI amino acid sequence predicted the presence of a signal sequence (0.923 probability) with a cleavage site between Tyr21 and Leu22 (0.5 probability). The hydrophobic (h) region is interrupted by polar residues (P), which results in the secondary structure prediction of an amphipathic α helix (c, coil; h, helix; e, sheet).
B. Helical wheel of amino acids 5 to 12. Black font, white background represents amino acids in the polar side of the helix. White font, black background represents amino acids in the hydrophobic side. *Charged residues were introduced into the hydrophobic side of the predicted amphipathic α helix at positions 6 and 12 (L6K/L12K).
Fig. 5.
In vitro translocation assay with isolated inverted inner membrane vesicles of E. coli.
A. FtsQ and TraIΔ261-850 were used in a co-translational targeting/transport assay in which the translocation into inverted inner membrane vesicles was monitored by the accessibility of the transported protein to externally added proteinase K, in the presence or absence of Triton X-100. To detect even small amounts of transported protein, inverted membrane vesicles derived from a strain overexpressing the SecYEG translocase were also used. Ten (10) % of the reaction mixture before addition of proteinase K was loaded as a synthesis control. FtsQ is a membrane protein whose insertion into the vesicles is mediated by Sec proteins. Only a small domain of FtsQ is exposed on the outside of the vesicles and is accessible to degradation by proteinase K.
B. proOmpA and TraIΔ261-850 were used in a post-translation protein transport assay in the presence or absence of inner membrane vesicles, as described above, with or without ATP. One (1) % of synthesized protein before addition of proteinase K was loaded as a control. proOmpA contains a signal peptide for transport into the periplasm and is processed by leader peptidase.
The N-terminal region of N. gonorrhoeae TraI mediates membrane association and is necessary for wild-type levels of DNA secretion
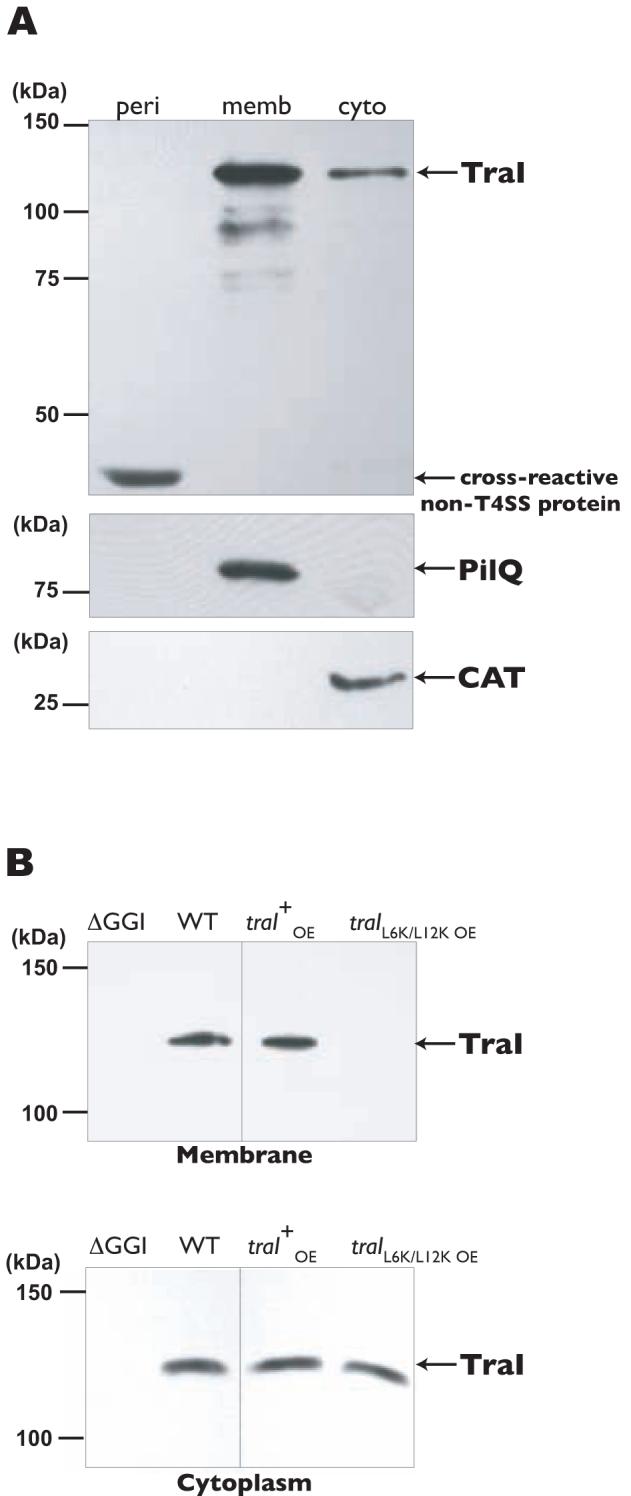
A secondary structure prediction of the unique N-terminal region of Ng TraI indicated that the hydrophobic region lies on an α helix and is interrupted by polar residues, resulting in the prediction of an amphipathic α helix (Fig. 4B). Amphipathic α helixes are characterized by the presence of a polar side and a hydrophobic side that makes proteins capable of interacting with membranes (Elazar et al., 2004). Therefore, an indication that TraI contains an amphipathic α helix would be membrane localization in N. gonorrhoeae. TraI was overexpressed in a wild-type gonococcal strain from an inducible construct on the chromosome. Overexpression was followed by cellular fractionation and detection of TraI by Western blot with TraI-specific polyclonal antisera. TraI was detected in the cytoplasmic and membrane fractions, but not in the periplasmic fraction (Fig. 6A). A faster-migrating band (∼35 kDa) that consistently cross-reacted with the TraI antisera was detected in the periplasmic fraction and barely detected in the cytoplasmic fraction (Fig. 6A, top panel), demonstrating proper extraction of periplasmic proteins. PilQ, a gonococcal outer membrane protein, was detected only in the membrane fraction (Fig. 6A, middle panel), and chloramphenicol acetyltransferase (CAT) was detected only in the cytoplasmic fraction (Fig. 6A, bottom panel). Lack of TraI detection in the periplasm further confirms that TraI is not transported by the Sec system. Fractionation of a ΔGGI strain overexpressing TraI also resulted in its detection only in the cytoplasmic and membrane fractions (data not shown), indicating that TraI’s association with the membrane is not due to interactions with other proteins of the T4SS. Furthermore, the membrane association of TraI is not a result of aggregation due to overexpression, since TraI was also found in the membrane fraction of wild-type cells (Fig 6B, left panels). These results suggest that TraI exists in two forms, soluble in the cytoplasm and associated with the membrane. Another observation that results from these experiments is that TraI migrates anomalously. TraI is predicted to have a molecular weight of ∼94 kDa, but it migrated at an apparent molecular weight of ∼125 kDa. DNaseI treatment of fractions prior to SDS-PAGE had no effect on TraI migration (data not shown), indicating that the aberrant migration is not due to bound DNA. TraI’s aberrant SDS-PAGE migration is more likely due to post-translational modifications, dimerization, or the protein adopting an unusual conformation.
Fig. 6.
Subcellular localization of TraI.
A. Full-length TraI overexpressed in a wild-type strain from an inducible construct on the chromosome. TraI was detected by Western blot with TraI-specific polyclonal antiserum. Periplasmic (peri), membrane (memb), and cytoplasmic (cyto) fractions are indicated. A cross-reactive protein (∼35kDa) was found in the periplasmic fractions from strains with or without the T4SS. Antiserum to the gonococcal outer membrane protein PilQ was used to detect membrane material. Chloramphenicol acetyltransferase (CAT) was detected to identify cytoplasmic material.
B. Top Panels. Membrane fractions from wild-type (WT) and ΔGGI strains (left panel) and strains overexpressing (oe) traI or traIL6K/L12K from the native site (right panel). Bottom Panels. Cytoplasmic fractions from wild-type (WT) and ΔGGI strains (left panel) and strains overexpressing (oe) traI or traIL6K/L12K from the native site (right panel). TraI was detected by Western blot with TraI-specific polyclonal antiserum.
To investigate if the predicted amphipathic nature of the N-terminal region is involved in mediating TraI’s association with the membrane, we introduced charged residues within the hydrophobic side of the predicted amphipathic α helix (L6K/L12K) (Fig. 4B). For ease of detection, we introduced an ermC cassette with a strong promoter (Hamilton et al., 2001) upstream of the traIL6K/L12K promoter region to drive traI transcription from its native site. Wild-type TraI overexpressed from its native site was detected in the membrane and cytoplasmic fractions (Fig. 6B, right panels). However, TraIL6K/L12K was detected only in the cytoplasmic fraction, suggesting that the N-terminal region mediates TraI’s association with the membrane, possibly through the formation of an amphipathic α helix (Fig. 6B, right panels). We tested DNA secretion in the strain carrying the L6K/L12K mutation and found significantly reduced levels of DNA in the culture medium (Fig. 1A). These results suggest that TraI’s association with the membrane is required for wild-type levels of DNA secretion and may represent a unique step that aids in the DNA transport process for type IV secretion.
Nuclease treatment results in degradation of secreted DNA only by nucleases that have single-strand 3′→5′ exonuclease activity or endonuclease activity
An additional approach to characterize the mechanism of DNA processing in N. gonorrhoeae is to test the susceptibility of the secreted DNA to specific nucleases. Two methods were used to assess the effects of nucleases on the secreted DNA: fluorometric detection of DNA in cell-free supernatants after incubation with nucleases, and transfer of an antibiotic resistance marker (cat) during co-culture transformation in the presence of nucleases. The nuclease treatment of supernatants was carried out under optimum conditions for each nuclease, while the medium for the co-culture assay was optimized to provide the best possible conditions that would allow enzyme activity and gonococcal growth. ExoIII is a double-strand specific exonuclease that converts double-stranded DNA to single-stranded DNA, by degrading one strand (Richardson et al., 1964). The addition of ExoIII did not significantly affect the fluorescence of culture supernatants (Fig. 7A) or transfer of the cat marker (Fig. 8), but resulted in the degradation of added double-stranded λ-HindIII DNA in culture medium (Fig. 7C). The marker transferred in the co-culture transformation assay, cat, carries an internal EcoRI site. EcoRI is a restriction enzyme that cuts double-stranded DNA (Greene et al., 1975). EcoRI cut plasmid DNA added to medium (data not shown), but did not affect transfer of the cat marker in co-culture (Fig. 8). By contrast, the 3′→5′ single-strand specific nuclease ExoI (Lehman and Nussbaum, 1964) significantly reduced the fluorescence of culture supernatants (Fig. 7) and reduced transfer of the cat marker in co-culture (Fig. 8), suggesting that the secreted DNA is single-stranded with a free 3′ end. However, the 5′→3′ single-strand specific exonuclease RecJf (Lovett and Kolodner, 1989) did not significantly affect the fluorescence of culture supernatants (Fig. 7A) or transfer of the cat marker (Fig. 8), but degraded single-stranded λ-HindIII DNA added to culture medium (Fig. 7B). Furthermore, the only other nucleases found to degrade secreted DNA were DNaseI and BAL-31 (Figs. 7 and 8). DNaseI is an endonuclease that acts on double- and single-stranded DNA (Vanecko and Laskowski, 1961), and BAL-31 is a double-strand specific exonuclease and single-strand specific endonuclease (Gray et al., 1975). Since ExoIII had no effect on the secreted DNA, any activity exhibited by BAL-31 was predicted to be due to its single-strand endonuclease activity. Both nucleases significantly reduced the fluorescence of culture supernatants (Fig. 7) and reduced transfer of the cat marker (Fig. 8). Altogether, these results are consistent with the hypothesis that the secreted DNA is single-stranded and bound to TraI at the 5′ end.
Fig. 7.
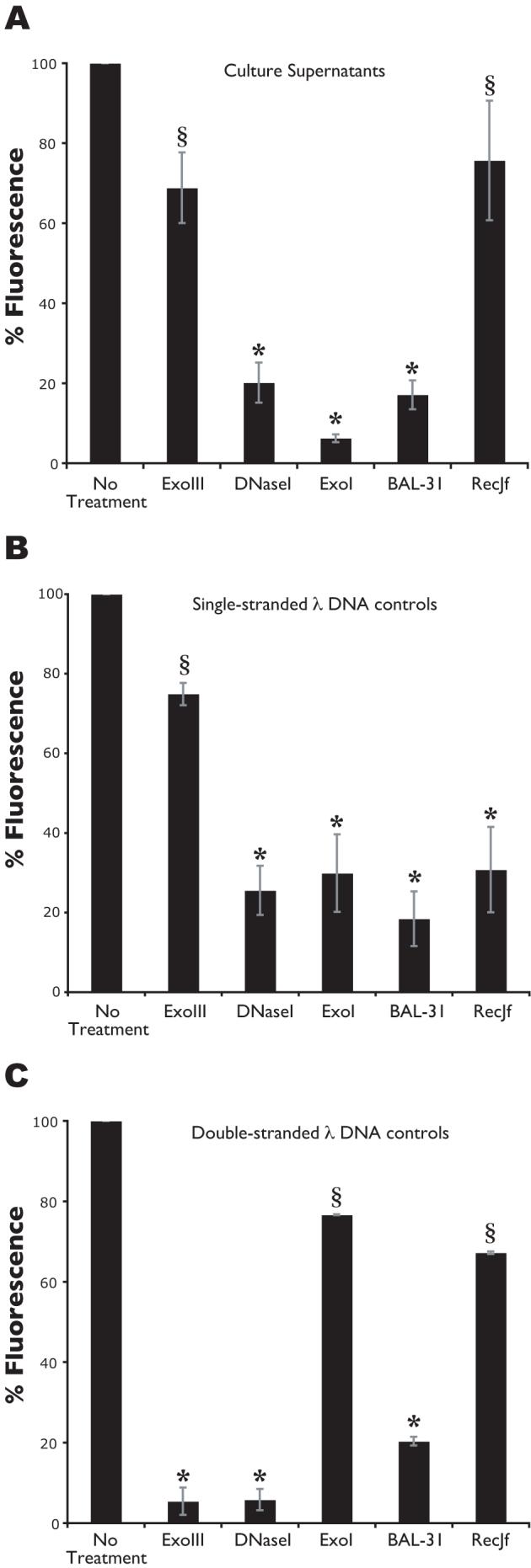
Nuclease treatment of culture supernatants. Fluorometric detection of DNA in cell-free supernatants after overnight incubation with nucleases. ExoIII is a double-strand specific exonuclease that converts double-stranded DNA to single-stranded DNA, by degrading one strand. ExoI is a 3′→5′ single-strand specific exonuclease. DNaseI and BAL-31 have endonuclease activity. BAL-31 also has double-strand specific exonuclease activity. RecJf is a 5′→3′ single-strand specific exonuclease. Culture supernatants collected from a wild-type strain (A), culture supernatants supplemented with HindIII-digested, single-stranded λ DNA (B) or with double-stranded λ DNA (C). *p-value<0.05. §Not significantly different from no treatment.
Fig. 8.
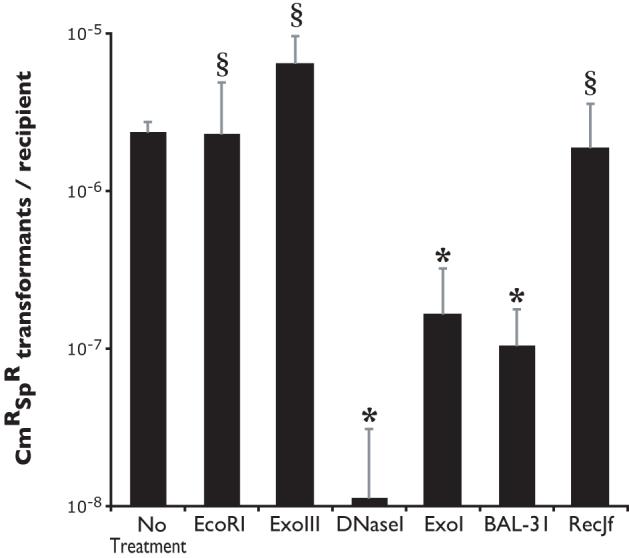
Co-culture transformation in the presence of nucleases. Transfer of an antibiotic resistance marker during co-culture transformation in the presence of nucleases. The transferred marker, cat, carries an internal EcoRI site. EcoRI is a restriction enzyme that cuts double-stranded DNA. ExoIII is a double-strand specific exonuclease. ExoI is a 3′→5′ single-strand specific exonuclease. RecJf is a 5′→3′ single-strand specific exonuclease. DNaseI has double- and single-strand endonuclease activities; BAL-31 has single-strand specific endonuclease and double-strand specific exonuclease activities. *p-value<0.05. §Not significantly different from no treatment.
Identification of a possible origin of transfer
Conjugative and mobilizable plasmids are recognized for transfer by the presence of a specific sequence known as the origin of transfer (oriT). Characterized oriT sequences contain an inverted repeat and are often found near the relaxase gene, between divergent transcriptional promoters, and in A-T rich regions (Lanka and Wilkins, 1995). We hypothesized that if a single oriT exists in the gonococcal chromosome, that it is likely to be located in the GGI. Therefore we searched the GGI sequence for inverted repeats that might mark the oriT. The only sequence meeting all the criteria characteristic of oriTs is the one that contains the inverted repeat found between the T4SS genes yaf and ltgX (Fig. 9A). Furthermore, the oriT in F-plasmid is adjacent to geneX (orf169) (Frost et al., 1994), the homologue of the gonococcal ltgX, as is the predicted oriT for the GGI. An insertion disrupting this inverted repeat, and well separated from the predicted transcriptional promoters for the two transcripts, was found to abolish DNA secretion (Fig. 9B). Complementation with the yaf-ltgX non-coding sequence containing an intact inverted repeat at a distant location on the chromosome restored DNA secretion (Fig. 9B). These results suggest that this region contains the oriT and that there is only one oriT in the chromosome.
Fig. 9.
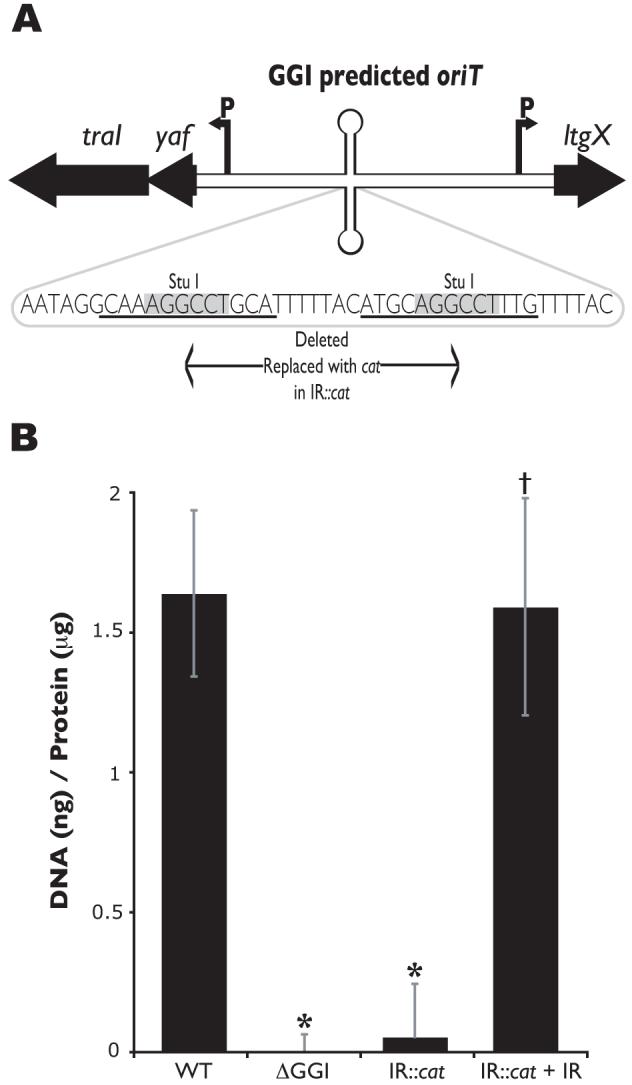
Predicted oriT sequence located in the GGI.
A. Map of the GGI region predicted to contain the oriT. traI encodes the putative relaxase, yaf encodes an unknown protein, and ltgX encodes a putative lytic transglycosylase. Arrows labeled “P” mark the promoter region for the transcripts. The sequence of the yaf-ltgX inverted repeat is shown. An insertion containing a cat marker was introduced at the StuI site of the inverted repeat, deleting the sequence in between the StuI sites.
B. Fluorometric detection of secreted DNA. Piliated gonococcal strains were grown for 2.5h in liquid culture. Cell-free culture supernatants were collected and DNA was detected with the fluorescent DNA-binding dye PicoGreen and normalized to total protein in the cell pellet. MS11 was used as the wild-type (WT) strain and ND500 (ΔGGI in MS11) as the negative control. The results are an average of at least four independent experiments. *p-value<0.003 when compared to wild-type. †p-value<0.003 when compared to its respective mutant.
Discussion
DNA donation in N. gonorrhoeae contributes to the high frequency of genetic exchange that results in the rapid spread of antibiotic resistance genes and in greater genetic diversity of alleles of surface molecules for evasion of the human immune response. To characterize DNA donation by N. gonorrhoeae, we examined the mechanism of DNA processing focusing on the putative relaxase TraI and the sensitivities of the secreted DNA to specific nucleases. Mutational analysis of TraI is consistent with its function as a relaxase, and the nuclease data indicate that the DNA is single-stranded with a free 3′ end. Therefore, we propose that N. gonorrhoeae secretes a nucleoprotein complex of single-stranded DNA protected at the 5′ end, presumably by TraI. Other proteins may also bind to the secreted DNA to aid in transformation. Conjugation systems have been found to transfer the relaxase as well as RecA along with plasmid DNA (Heinemann, 1999; Luo and Isberg, 2004), and A. tumefaciens transfers the single-stranded DNA binding protein VirE2 into host cells to aid in the establishment of the Ti DNA (Vergunst et al., 2000). Similarly, N. gonorrhoeae may secrete proteins that bind the secreted DNA facilitating transformation and/or enhancing DNA uptake or recombination in recipient cells.
In relaxases, the active-site amino acid residue is a tyrosine, where the hydroxyl group attacks a phosphate on the DNA backbone (Pansegrau et al., 1994). We searched for conserved tyrosines in the Ng TraI family and found that forty-two of the proteins conserve a tyrosine at position 93 (in Ng TraI), with the rest of the proteins conserving a tyrosine at positions -8, -5, or +3 from Tyr93 (Fig. S1). Substitution of Tyr93 with phenylalanine abolished DNA secretion, while substitution of Tyr201, conserved in all the proteins, resulted in intermediate levels of DNA secretion. The requirement of Tyr93 for the function of TraI in DNA secretion supports the hypothesis that TraI is a relaxase, and suggests a model where Tyr93 is involved in the initial cleavage of supercoiled DNA. The intermediate DNA secretion phenotype of the strain carrying the Y201F mutation suggests that Tyr201 may be required for a second cleavage for termination of DNA processing, analogous to the TrwC mechanism of DNA processing (Garcillan-Barcia et al., 2007; Gonzalez-Perez et al., 2007; Grandoso et al., 2000). Without termination, DNA might be secreted but remain attached to the cells. In our assay for DNA secretion, the cells are removed before the supernatants are assayed for fluorescence. Therefore, DNA in the supernatants might be reduced to the amount that is sheared from the cells. Tyrosines are not only involved in DNA cleavage reactions but are also known to contribute to non-specific binding interactions with the phosphodiester backbone for many enzymes involved in nucleic acid metabolism, including DNaseI and DNA polymerase beta (Mundle et al., 2004). Hence, it is possible that Tyr201 is required instead for the binding affinity of TraI to the DNA backbone.
The presence of the HD domain suggests an important difference in the mechanism of TraI function when compared to classical relaxases. The HD domain has been shown to be required for the metal-dependent phosphohydrolase activity of the E. coli dGTPase (Huber et al., 1988), the ppGpp(p)-hydrolase SpoT (Gentry and Cashel, 1996), and nucleotidyltransferases (Yakunin et al., 2004), providing evidence that the HD domain functions in nucleic acid metabolism (Aravind and Koonin, 1998). Furthermore, conservation of multiple histidine and aspartate residues in the HD domain (Fig. 2) indicates that one of the primary functions of this domain may be the coordination of divalent cations (Aravind and Koonin, 1998). If the HD domain of TraI performs this function, then the 3H motif of classical relaxases would be unnecessary, thus explaining the wild-type phenotypes of the traI H106S, H108S, and H106A/H108A mutants in DNA secretion. The H-rich motif sequence PASExHHH is a hallmark of the proteins in the Ng TraI family (Fig. 2 and Fig. S1). Therefore, the dispensability of the histidine residues in this motif was an unexpected result. The HD domain mutations H161S and D162N (HD motif II) also did not significantly reduce DNA secretion. However, Asp120 (HD motif I) was found to be required for wild-type levels of DNA secretion, providing evidence that the HD domain contributes to TraI function. Asp81 of F-plasmid TraI and Asp85 of TrwC have been shown to interact directly with tyrosines, presumably for activation of the catalytic tyrosine or for structural maintenance of the active site (Guasch et al., 2003; Larkin et al., 2005). Asp120 of the HD domain could have similar roles. Alternatively, Asp120 could function in metal coordination by a mechanism similar to Glu104 of the MbeA HEN motif (Varsaki et al., 2003). MbeA is the relaxase encoded by plasmid ColE1 that contains a variant of the canonical 3H motif, where His97, Glu104 and Asn106 of the HEN motif are required for the activity of the protein (Varsaki et al., 2003).
In addition to the presence of the HD domain, another unusual feature of TraI is a hydrophobic region at the N-terminus. This region was initially identified as a possible signal sequence, suggesting a two-step secretion mechanism for TraI, as described for pertussis toxin (Weiss et al., 1993) and some proteins of A. tumefaciens (Pantoja et al., 2002). However, the data does not support this hypothesis. The predicted signal peptide was not functional in transporting TraI into inverted E. coli inner membrane vesicles (Fig. 5), and TraI was not detected in the periplasm of N. gonorrhoeae (Fig. 6A). Further examination of the N-terminal region revealed that, while highly hydrophobic, it contains several polar residues resulting in the secondary structure prediction of an amphipathic α helix. Disruption of the amphipathic nature of the region (by substitution of two hydrophobic residues with charged residues, L6K/L12K) indicated that the N-terminal region is necessary not only for membrane association but also for wild-type levels of DNA secretion. Secondary structure prediction analysis of the N-terminal region of proteins in the Ng TraI family revealed that almost all of these proteins contain predicted amiphathic α helices (Fig. 3, Fig. S1). Therefore, association with the membrane may be a general property of relaxases in the Ng TraI family. The advantage of this association may be to facilitate interaction with the T4SS apparatus. Interaction with the coupling protein or other parts of the T4SS apparatus might still occur without membrane association, but might occur less often or be less stable. This idea is consistent with the reduced DNA secretion seen in the traI L6K/L12K mutant (Fig 1A). Alternatively, membrane association of TraI could be more important for the transformation steps after DNA secretion, possibly binding secreted DNA to the outer membrane or outer membrane vesicles of the donor for presentation to recipient cells or aiding in binding DNA to the outer membrane of recipients for DNA uptake.
DNA processing for type IV secretion is extremely specific and occurs only at the oriT (Lanka and Wilkins, 1995). All of the known genes for type IV secretion in N. gonorrhoeae are found in a 57-kb region of the chromosome called the GGI (Dillard and Seifert, 2001; Hamilton et al., 2005). We hypothesized that if a single oriT is present in the gonococcal chromosome that it would be present in this region. We found a sequence in the yaf-ltgX non-coding region of the GGI that has many of the characteristics of oriTs (Lanka and Wilkins, 1995). This sequence contains an inverted repeat, is located between divergently transcribed genes in an A-T rich region, and is close to traI. An insertion in the inverted repeat abolished DNA secretion, suggesting that this sequence is required for DNA processing for type IV secretion. Furthermore, complementation with the yaf-ltgX non-coding region at a distant location on the chromosome restored DNA secretion. Although it is possible that this region might encode a very small protein or a small RNA that we have not detected, we favor the hypothesis that this region is the oriT and may be cut by TraI to initiate DNA transfer.
To date, relaxases have been identified that contain an N-terminal relaxase domain or an N-terminal relaxase domain with a C-terminal helicase or primase domain (Francia et al., 2004). The TraI domain architecture, an N-terminal putative relaxase domain fused to the HD domain of phosphohydrolases (all within the first three-hundred amino acids), has not been previously described. However, the TraI N-terminal domain architecture is not unique to TraI; it is present in at least fifty-three uncharacterized proteins. These proteins conserve (i) Ng TraI Tyr93 and Tyr201, (ii) an H-rich motif of signature h(Q/H)xhPASExHHHx3GG(L/M)h (where x is any residue and h is a hydrophobic residue), (iii) the five motifs characteristic of the HD domain of phosphohydrolases, (iv) a predicted amphipathic α helix at the N-terminus proximal region, and (v) a conserved domain of unknown function (DUF1528) near the C-terminus. Therefore, we propose that these proteins are part of a previously undescribed family of relaxases or relaxase-like proteins and that their mechanism of DNA processing may be similar to TraI. The large majority of the proteins in the Ng TraI family are in transfer regions that are chromosomally encoded and most of these species also encode type IV pilus components, suggesting that they may be naturally transformable. Thus, it is likely that these relaxase homologues act in processing chromosomal DNA, and it is possible that some of them may export DNA by the same mechanism as N. gonorrhoeae. Regardless of whether DNA donation occurs by conjugation or direct secretion, the mechanism of TraI function appears to be a widespread process for chromosomal DNA donation.
Experimental procedures
Bacterial strains and growth conditions
The N. gonorrhoeae strain MS11 was used as the wild-type strain and for the construction of the gonococcal strains described in Table 2. Growth of gonococci on GCB (Difco) solid medium was performed as previously described (Dillard, 2006). E. coli strains were grown on Luria-Bertani (LB) agar plates or in LB broth (Sambrook et al., 1989). Graver-Wade (GW) medium (pH 6.8) was used for the growth of gonococci in liquid culture (Wade and Graver, 2007). When required, chloramphenicol (Cm) was used at a concentration of 10μg ml-1 (Cm10) or 25μg ml-1 (Cm25) for gonococci and E. coli, respectively. Erythromycin was used at a concentration of 500μg ml-1 (Em500) for E. coli and 10μg ml-1 (Em10) for gonococci.
Table 2.
Bacterial plasmids and strains
| Plasmid/Strain | Description | Reference |
|---|---|---|
| Plasmids | ||
| pIDN1 | Cloning vector (EmR), 2 kb | Hamilton et al. (2001) |
| pIDN2 | Cloning vector (EmR), 2 kb | Hamilton et al. (2001) |
| pIDN3 | Cloning vector (EmR), 2 kb | Hamilton et al. (2001) |
| pKH37 | Complementation vector (CmR), 6.5 kb | Kohler et al. (2007) |
| pKS94 | pIDN2 containing ∼7 kb of the GGI from traI’ to traB’, 9 kb | Hamilton et al. (2001) |
| pWSP3 | ‘yaf-traI’; ∼0.8 kb BamHI-MfeI fragment of pKS94 in pIDN2, 2.8 kb | This study |
| pWSP4 | traIY200F/Y201F (TAT to TTT); mutagenesis PCR of pWSP3, 2.8 kb | This study |
| pWSP5 | traIY93F (TAT to TTT); mutagenesis PCR of pWSP3, 2.8 kb | This study |
| pWSP14 | Full-length traI; PCR amplified from MS11, ClaI and SacI digested in pIDN1, 4.7 kb | This study |
| pWSP15 | traI 3′ end Flag tag; PCR of pWSP14 with primers containing the Flag tag; GAC TAC AAG GAC GAC GAC AAG, 4.7 kb | This study |
| pWSP30 | ‘traI-ltgX’; ∼5.2 kb PvuII-SpeI fragment of pKS94 | This study |
| pWSP31 | IR::cat; PmeI-FspI cat-containing fragment from pKH37 in StuI-digested pWSP30, ∼8.1 kb | This study |
| pWSP33 | traI 3′ end Flag & His10 tags; PCR of pWSP15 with primers containing the His10 tag, 4.7 kb | This study |
| pWSP34 | traI 3′ end Flag and His10 tags; ∼2.7 kb ClaI-SacI fragment of pWSP33 in pKH37, 9 kb | This study |
| pWSP36 | Full-length traI; PCR amplified from MS11 ClaI- and SacI-digested in pKH37, 9 kb | This study |
| pWSP37 | yaf-ltgX non-coding region; PCR amplified, ∼750 bp in pIDN1, 2.4 kb | This study |
| pWSP38 | 0.9 kb EcoRV-NheI ermC-containing fragment from pIDN3 in PsiI-digested pWSP37, 4.5 kb | This study |
| pWSP42 | traIY200F (TAT to TTC); mutagenesis PCR of pWSP3, 2.8 kb | This study |
| pWSP43 | traIY201F (TAT to TTC); mutagenesis PCR of pWSP3, 2.8 kb | This study |
| pWSP44 | traIR99S (AGG to AGC); mutagenesis PCR of pWSP3, 2.8 kb | This study |
| pWSP45 | traIH106S (CAT to TCG); mutagenesis PCR of pWSP3, 2.8 kb | This study |
| pWSP46 | traIH108S (CAT to AGC); mutagenesis PCR of pWSP3, 2.8 kb | This study |
| pWSP47 | traI’; ∼2.9 kb BseRI-XhoI fragment of pWSP14 | This study |
| pWSP48 | traIH106A/H108A (CAT to AGC); mutagenesis PCR of pWSP3, 2.8 kb | This study |
| pWSP49 | traIH161S (CAC to AGT); mutagenesis PCR of pWSP47, 2.9 kb | This study |
| pWSP50 | traID162N (GAC to AAC); mutagenesis PCR of pWSP47, 2.9 kb | This study |
| pWSP51 | traIY212F (TAT to TTT); mutagenesis PCR of pWSP47, 2.9 kb | This study |
| pWSP52 | traID120N (GAC to AGT); mutagenesis PCR of pWSP47, 2.9 kb | This study |
| pWSP54 | traIL6K/L12K (CTC and CTA to AAG); mutagenesis PCR of pWSP3, 2.8 kb | This study |
| pWSP58 | yaf-ltgX non-coding region in pKH37, 8.2 kb | This study |
| pSJ001 | traIΔ261-850; PCR amplified from MS11, BspHI and XhoI digested in pET15b | This study |
| pBSKftsQ | Plasmid encoding FtsQ | van der Laan et al. (2004) |
| pET147 | Plasmid encoding proOmpA | van der Wolk et al. (1997) |
| Strains | ||
| MS11 | Wild-type N. gonorrhoeae | Swanson et al. (1971) |
| ND500 | Gonococcal genetic island deletion in MS11 | Hamilton et al. (2005) |
| WSP4 | MS11 transformed with pWSP4; traIY200F/Y201F | This study |
| WSP5 | MS11 transformed with pWSP5; traIY93F | This study |
| WSP31 | MS11 transformed with pWSP31; IR::cat | This study |
| WSP34 | E. coli overexpressing TraIFLAG/His10 (C-terminus tags) | This study |
| WSP36 | MS11 transformed with pWSP36; traI+OE from an inducible construct on the chromosome | This study |
| WSP38 | MS11 transformed with pWSP38; traI+OE from native site | This study |
| WSP42 | MS11 transformed with pWSP42; traIY200F | This study |
| WSP43 | MS11 transformed with pWSP43; traIY201F | This study |
| WSP44 | MS11 transformed with pWSP44; traIR99S | This study |
| WSP45 | MS11 transformed with pWSP45; traIH106S | This study |
| WSP46 | MS11 transformed with pWSP46; traIH108S | This study |
| WSP48 | MS11 transformed with pWSP48; traIH106A/H108A | This study |
| WSP49 | MS11 transformed with pWSP49; traIH161S | This study |
| WSP50 | MS11 transformed with pWSP50; traID162N | This study |
| WSP51 | MS11 transformed with pWSP51; traIY212F | This study |
| WSP52 | MS11 transformed with pWSP52; traID120N | This study |
| WSP54 | MS11 transformed with pWSP54; traIL6K/L12K | This study |
| HH532 | TraI C-terminus truncation in MS11; traIΔ349-850 | Hamilton et al. (2005) |
| WSP532 | HH532 transformed with pWSP36; traI C-terminus truncation complement | This study |
| WSP3604 | WSP4 transformed with pWSP36; traIY200F/Y201F complement | This study |
| WSP3605 | WSP5 transformed with pWSP36; traIY93F complement | This study |
| WSP3643 | WSP43 transformed with pWSP36; traIY201F complement | This study |
| WSP3854 | WSP54 transformed with pWSP38; traIL6K/L12KOE from native site | This study |
| WSP5831 | WSP31 transformed with pWSP58; IR::cat complement | This study |
| MS11Sp | Spectinomycin resistant, recipient strain (MS11 background) | Dillard and Seifert (2001) |
| JD1545 | MS11 recA6 cnp::cat, donor strain (MS11 background) | Dillard and Seifert (2001) |
Plasmid construction and generation of gonococcal mutants
Plasmids used for mutagenesis and complementation are described in Table 2. The Bordo and Argos guidelines were used to determine which amino acids would be used for substitutions (Bordo and Argos, 1991). The traI mutations were introduced into gonococci by allelic exchange without selection (Dillard, 2006). Complementation was achieved by introduction of the wild-type copy of traI or the yaf-ltgX non-coding region into the gonococcal chromosome between aspC and lctP along with the cat marker via the pKH37 vector (Kohler et al., 2007).
DNA secretion assay
This assay is a modification of the DNA secretion assay reported by Hamilton et al. (Hamilton et al., 2005). Piliated (P+) gonococcal colonies from an overnight culture grown on GCB agar plates were harvested with a sterile dacron swab, resuspended in GW medium, diluted to an OD540 0.18 in a 3-ml culture, and grown for 2h at 37°C with aeration. After the first round of growth, the cultures were vortexed for 1min, a 600-μl volume was transferred to 2.4-ml fresh GW medium, and the cultures were grown for an additional 2.5h. Supernatants were collected at the beginning (t=0h) and end (t=2.5h) of the second round of growth in triplicate. DNA in culture supernatants was detected using the fluorescent DNA-binding dye PicoGreen (Invitrogen) and normalized to total cell protein (Bradford assay), as previously described (Hamilton et al., 2005). For consistency across strains, only the results for which the final protein concentration (at t=2.5h) was within the range of 20-40μg ml-1 were plotted. The results are an average of at least four independent experiments. An average background fluorescence, determined by performing the secretion assay with the ΔGGI N. gonorrhoeae strain (ND500), was subtracted from the average result of all the strains.
In Vitro Transcription/Translation and Transport Assay
The full-length TraI is relatively long to synthesize in an in vitro transcription/translation assay, and since sec-dependent protein translocation is only determined by the N-terminal signal peptide and normally not affected by C-terminal truncations, a truncated version of TraI (TraIΔ261-850) was constructed. To detect even small amounts of transported protein, inverted membrane vesicles derived from a strain overexpressing the SecYEG translocase were also used. SecA, SecB, and wild-type or SecYEG overexpressing inverted inner membrane vesicles (IMVs) were isolated as described (Manting et al., 2000). The in vitro transcription and translation reaction was performed as described by van der Laan et al. (van der Laan et al., 2004). Shortly, the RiboMax in vitro transcription kit (Promega) was used with plasmids pBSKftsQ (van der Laan et al., 2004), pET147 (van der Wolk et al., 1997) or pSJ001 to generate 35S-labeled proteins. To study co-translational transport, the in vitro translation reactions were carried out for 30 min at 37°C in the presence or absence of wild-type or SecYEG IMVs (0.16mg ml-1; (De Vrije et al., 1987). Reactions were started by the addition of the 35S-labeled methionine. After 30min at 37°C, a 10% synthesis control was removed and the remainder was treated with 0.4mg ml-1 proteinase K for 30 min on ice in the presence or absence of 1% Triton X-100. Samples were trichloroacetic acid-precipitated and analyzed by 12% SDS-PAGE and phosphorimaging and quantified using the LumiAnalyst software from Roche Applied Science. FtsQ was used as the positive control. FtsQ is a membrane protein whose translocation into the inverted vesicles is mediated by Sec components (van der Laan et al., 2004). A small domain is not transported and is accessible to degradation by proteinase K. To study post-translational transport, 35S-labeled TraIΔ261-850 and proOmpA (positive control, contains a signal peptide that transports the protein into the periplasm and is processed by leader peptidase) were synthesized in vitro, dissolved in 6M urea, and used in post-translational translocation reactions as described (Cunningham and Wickner, 1989). Shortly, reaction mixtures contained 1.6μg of SecB, 0.5μg of SecA, and wild-type or SecYEG E.coli IMVs in buffer [50mM HEPES-KOH, pH 7.5, 30mM KCl, 0.5mg ml-1 BSA, 2mM DTT, 2mM Mg(OAc)2] in an 80-μl volume. 35S-proOmpA and 35S-TraIΔ261-850 were diluted 50-fold from a solution containing 6 M urea. Translocation reactions were performed in the presence or absence of 2mM ATP and an ATP regenerating system (10mM creatine phosphate, 0.5μg of creatine kinase) at 37°C for 30min, chilled on ice, and treated with proteinase K (0.1mg ml-1) for 15 min. Transport of proteins into the E. coli IMVs was assayed by their accessibility to added proteinase K. Samples were analyzed by SDS-12% PAGE and autoradiography.
Generation of TraI-specific antibodies
Strain WSP34, an E. coli strain harboring a plasmid encoding traIFLAG/His10, was grown in a 30-ml LB liquid culture (Cm25) supplemented with 1M sorbitol at 37°C, from OD600 0.2 to 0.5, induced with 1mM IPTG, switched to 18°C, and grown to an OD600 1. Soluble protein was obtained by sonication and centrifugation (15,000 x g for 1min) of total cell extracts at 4°C. TraIFLAG/His10 was purified over a nickel column, according to the manufacturer’s instructions (Sigma), and verified by Western blot with an anti-FLAG M2 antibody (Sigma). TraI-specific polyclonal antiserum was generated in rabbits (Chemicon International).
Gonococcal growth for subcellular localization studies and detection of mutant proteins
Growth of strains that overexpress (OE) TraI from an inducible construct at a distant site on the chromosome: WSP36 (traI+OE ectopic) and WSP536 (ΔGGI + traIOE ectopic) were grown in 3-ml liquid cultures (Cm10) with 1mM IPTG from an OD600 0.1 for 4h at 37°C with aeration. Growth of strains that overexpress TraI from its native site on the chromosome: WSP38 (traI+OE native) and WSP3854 (traIL6K/L12KOE native) were grown in 15-ml liquid cultures (Em10) from an OD600 0.1 for 4h at 37°C with aeration. Growth of strains for detection of TraI without overexpression: ND500 (ΔGGI), MS11 (wild type), WSP5 (traIY93F), WSP4 (traIY200F/Y201F), and WSP52 (traID120N) were grown in 30-ml liquid cultures from an OD600 0.1 for 4h at 37°C with aeration. Approximately 0.5mg of total cell extracts were electrophoresed by SDS-PAGE in a 10% polyacrylamide gel.
Gonococcal fractionation
The Judd and Porcella protocol, specific for N. gonorrhoeae, was followed for the extraction of gonococcal periplasm (Judd and Porcella, 1993), and the membrane and soluble fractionations were performed as described (Gauthier et al., 2003). Briefly, for extraction of periplasmic proteins, 20-μl volume of chloroform was added to bacterial pellets collected from 3-ml log-phase cultures, vortexed, and incubated for 15min at room temperature. Periplasmic proteins were recovered by the addition of 100-μl volume of 0.01M Tris-HCl (pH 7.0), centrifugation (15,000 x g for 1min), and collection of the aqueous fraction. The remaining cell pellet was washed with cold PBS buffer, resuspended in 500-μl volume of cold 0.01M Tris-HCl, sonicated on ice, centrifuged at 4°C to remove unbroken cells, and cleared lysates ultracentrifuged for 1h at 50,000 x g. The cytoplasmic proteins were concentrated to 100-μl volume in a Nanosep centrifugal device (Pall, Ann Arbor, Michigan). The membrane pellet was washed with PBS and resuspended in 100-μl volume of 0.01M Tris-HCl (pH 7.0) with 0.5% N-lauroylsacosine (Sigma). Cellular fractions were electrophoresed by SDS-PAGE in 10% polyacrylamide gels. In addition to recognizing TraI, the TraI antiserum consistently identified a band of ∼35 KDa on Western blots. The band was found in the periplasmic fraction from gonococcal strains that produce TraI (WT) or strains that lack the traI gene (ND500), indicating that this cross-reactive band does not represent TraI. Detection of this band was used to identify periplasmic material.
Western blotting
Electrophoresed proteins were transferred onto a polyvinylidene difluoride (PVDF) membrane, blocked for 1h with 3% nonfat dry milk in Tris-buffered saline (TBS), incubated for 1h with protein-specific rabbit antisera, washed for 10min, incubated for 1h with anti-rabbit peroxidase-conjugated antibody (Santa Cruz Biotechnology, 1:10,000 dilution), and washed three times for 10 min before addition of the peroxidase substrate (Immune Star, BioRad). The working dilutions of the rabbit antisera used are as follows: anti-TraI (1:10), anti-PilQ (1:10,000), and anti-CAT-1 (1:1000, Sigma). Anti-PilQ serum was provided by Dr. H. S. Seifert.
Nuclease treatment
Wild-type N. gonorrhoeae was grown in liquid culture, essentially as described for the DNA secretion assay. However, a 600-μl volume of the initial culture was centrifuged (15,000 x g for 1min), the pellet resuspended in fresh medium, and transferred to a 3-ml culture for a second round of growth. Cultures were grown for 2h and bacterial cells removed by centrifugation (3,000 x g for 5 min). Culture supernatants were collected, filter sterilized (0.22m filter), and stored at -20°C. DNaseI (4U ml-1), Exonuclease III (400U ml-1), Exonuclease I (80U ml-1), RecJf (60U ml-1), or BAL-31 (4U ml-1) purchased from New England Biolabs were used to treat culture supernatants in a 250-μl reaction, with their respective buffers, at 37°C for 12h. GW medium alone or culture supernatants collected from the wild-type strain MS11 or the DNA-secretion deficient strain WSP5 supplemented with double- or single-stranded λ-HindIII DNA (0.625μg ml-1, NE Biolabs) were used to test for activity of the enzymes. This concentration of λ-HindIII DNA is ten-times the concentration of secreted gonococcal DNA in culture supernatants. Single-stranded λ-HindIII DNA was generated by boiling the DNA for 15min and quick cooling in an ice-water bath for 10 min. All the enzymes showed activity against their specific DNA substrate in GW medium alone (data not shown), WSP5 (traIY93F) conditioned medium (data not shown), or MS11 conditioned medium (Fig. 7B, C).
Co-culture transformation assay
We used a modification of the method of Dillard and Seifert to measure transformation in co-culture (Dillard and Seifert, 2001). Briefly, a RecA-deficient, chloramphenicol (Cm) resistant donor strain (JD1545) was grown together with a spectinomycin (Sp) resistant recipient strain (MS11 Sp) in a 2-ml culture (GW medium) in the presence or absence of nucleases, and the frequency of CmRSpR transformants after 2h of co-culture was determined. Co-culture growth conditions: DNaseI (1U ml-1), Exonuclease III (100U ml-1), and EcoRI (40U ml-1), pH 6.8 and 7mM MgSO4; Exonuclease I (20U ml-1), pH 7.4 and 6.7mM MgSO4; RecJf (15U ml-1), pH 7.4 and 10mM MgSO4; BAL-31 (4U ml-1), pH 7.4, 12mM MgSO4, and 12mM CaCl2. ExoI was reported to exhibit 20% activity at pH 7.5 (Lehman, 1960), which is approximately the expected activity of this nuclease in co-culture where the pH is 7.4. All the enzymes showed activity against their specific DNA substrate.
The LOOPP (Learning, Observing and Outputting Protein Patterns) server
Using the full-length TraI amino acid sequence, secondary-structure-based threading detected secondary structure similarities between TraI (amino acids 86 to 606, 61.53% of the sequence) and the endo/exocellulose E4 domain from Thermomonospora fusca (DOI 10.2210/pdb1tf4/pdb). Threading of TraI to the T. fusca endo/exocellulose E4 domain positioned Asp120 on an α-helix adjacent to the Tyr93 α-helix, where Asp120 and Tyr93 are facing each other.
Web-based programs used
NCBI PSI BLAST (http://www.ncbi.nlm.nih.gov/BLAST), score value of P=0.0001 as a threshold
CLUSTAL W (http://www.ebi.ac.uk/clustalw/index.html) (Thompson et al., 1994)
SignalP 3.0 (http://www.cbs.dtu.dk/services/) (Bendtsen et al., 2004)
Helical Wheel Custom Images (http://kael.net/helical.htm)
The LOOPP server (http://cbsuapps.tc.cornell.edu/loopp.aspx)
PROF - Secondary Structure Prediction System (http://www.aber.ac.uk/~phiwww/prof/)
Simple Interactive Statistical Analysis (SISA) t-test (http://home.clara.net/sisa/)
Supplementary Material
Acknowledgements
Part of this work was carried out by using the resources of the Computational Biology Service Unit from Cornell University, which is partially funded by Microsoft Corporation (The LOOPP). We thank H. Steven Seifert for his generous gift of the PilQ antisera, Marcin Filutowicz for critical reading of the manuscript, and Petra L. Kohler for technical assistance. This work was supported by NIH grant R01AI47958 to J.P.D. and traineeship on NIH grant T32 AI055397 to W.S.P.
References
- Aravind L, Koonin EV. The HD domain defines a new superfamily of metal-dependent phosphohydrolases. Trends Biochem Sci. 1998;23:469–472. doi: 10.1016/s0968-0004(98)01293-6. [DOI] [PubMed] [Google Scholar]
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- Biswas GD, Sparling PF. Entry of double-stranded deoxyribonucleic acid during transformation of Neisseria gonorrhoeae. J Bacteriol. 1981;145:638–640. doi: 10.1128/jb.145.1.638-640.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordo D, Argos P. Suggestions for “safe” residue substitutions in site-directed mutagenesis. J Mol Biol. 1991;217:721–729. doi: 10.1016/0022-2836(91)90528-e. [DOI] [PubMed] [Google Scholar]
- Christie PJ. Type IV secretion: intercellular transfer of macromolecules by systems ancestrally related to conjugation machines. Mol Microbiol. 2001;40:294–305. doi: 10.1046/j.1365-2958.2001.02302.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie PJ, Atmakuri K, Krishnamoorthy V, Jakubowski S, Cascales E. Biogenesis, architecture, and function of bacterial type IV secretion systems. Annu Rev Microbiol. 2005;59:451–485. doi: 10.1146/annurev.micro.58.030603.123630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham K, Wickner W. Specific recognition of the leader region of precursor proteins is required for the activation of translocation ATPase of Escherichia coli. Proc Natl Acad Sci U S A. 1989;86:8630–8634. doi: 10.1073/pnas.86.22.8630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vrije T, Tommassen J, De Kruijff B. Optimal posttranslational translocation of the precursor of PhoE protein across Escherichia coli membrane vesicles requires both ATP and the protonmotive force. Biochim Biophys Acta. 1987;900:63–72. doi: 10.1016/0005-2736(87)90278-1. [DOI] [PubMed] [Google Scholar]
- Dillard JP, Seifert HS. A variable genetic island specific for Neisseria gonorrhoeae is involved in providing DNA for natural transformation and is found more often in disseminated infection isolates. Mol Microbiol. 2001;41:263–277. doi: 10.1046/j.1365-2958.2001.02520.x. [DOI] [PubMed] [Google Scholar]
- Dillard JP. Genetic manipulation of Neisseria gonorrhoeae. In: Coico R, Kowalik T, Quarles JM, Stevenson B, Taylor RK, Simon AE, editors. Current Protocols in Microbiology. John Wiley and Sons; 2006. pp. 4A.2.1–4A.2.19. [DOI] [PubMed] [Google Scholar]
- Elazar M, Liu P, Rice CM, Glenn JS. An N-terminal amphipathic helix in hepatitis C virus (HCV) NS4B mediates membrane association, correct localization of replication complex proteins, and HCV RNA replication. J Virol. 2004;78:11393–11400. doi: 10.1128/JVI.78.20.11393-11400.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francia MV, Varsaki A, Garcillan-Barcia MP, Latorre A, Drainas C, de la Cruz F. A classification scheme for mobilization regions of bacterial plasmids. FEMS Microbiol Rev. 2004;28:79–100. doi: 10.1016/j.femsre.2003.09.001. [DOI] [PubMed] [Google Scholar]
- Frost LS, Ippen-Ihler K, Skurray RA. Analysis of the sequence and gene products of the transfer region of the F sex factor. Microbiol Rev. 1994;58:162–210. doi: 10.1128/mr.58.2.162-210.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcillan-Barcia MP, Jurado P, Gonzalez-Perez B, Moncalian G, Fernandez LA, de la Cruz F. Conjugative transfer can be inhibited by blocking relaxase activity within recipient cells with intrabodies. Mol Microbiol. 2007;63:404–416. doi: 10.1111/j.1365-2958.2006.05523.x. [DOI] [PubMed] [Google Scholar]
- Gauthier A, Puente JL, Finlay BB. Secretin of the enteropathogenic Escherichia coli type III secretion system requires components of the type III apparatus for assembly and localization. Infect Immun. 2003;71:3310–3319. doi: 10.1128/IAI.71.6.3310-3319.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentry DR, Cashel M. Mutational analysis of the Escherichia coli spoT gene identifies distinct but overlapping regions involved in ppGpp synthesis and degradation. Mol Microbiol. 1996;19:1373–1384. doi: 10.1111/j.1365-2958.1996.tb02480.x. [DOI] [PubMed] [Google Scholar]
- Gomis-Ruth FX, Sola M, de la Cruz F, Coll M. Coupling factors in macromolecular type-IV secretion machineries. Curr Pharm Des. 2004;10:1551–1565. doi: 10.2174/1381612043384817. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Perez B, Lucas M, Cooke LA, Vyle JS, de la Cruz F, Moncalian G. Analysis of DNA processing reactions in bacterial conjugation by using suicide oligonucleotides. Embo J. 2007;26:3847–3857. doi: 10.1038/sj.emboj.7601806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandoso G, Avila P, Cayon A, Hernando MA, Llosa M, de la Cruz F. Two active-site tyrosyl residues of protein TrwC act sequentially at the origin of transfer during plasmid R388 conjugation. J Mol Biol. 2000;295:1163–1172. doi: 10.1006/jmbi.1999.3425. [DOI] [PubMed] [Google Scholar]
- Gray HB, Jr., Ostrander DA, Hodnett JL, Legerski RJ, Robberson DL. Extracellular nucleases of Pseudomonas BAL 31. I. Characterization of single strand-specific deoxyriboendonuclease and double-strand deoxyriboexonuclease activities. Nucleic Acids Res. 1975;2:1459–1492. doi: 10.1093/nar/2.9.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene PH, Poonian MS, Nussbaum AL, Tobias L, Garfin DE, Boyer HW, Goodman HM. Restriction and modification of a self-complementary octanucleotide containing the EcoRI substrate. J Mol Biol. 1975;99:237–261. doi: 10.1016/s0022-2836(75)80143-4. [DOI] [PubMed] [Google Scholar]
- Grinter NJ. Analysis of chromosome mobilization using hybrids between plasmid RP4 and a fragment of bacteriophage lambda carrying IS1. Plasmid. 1981;5:267–276. doi: 10.1016/0147-619x(81)90004-4. [DOI] [PubMed] [Google Scholar]
- Guasch A, Lucas M, Moncalian G, Cabezas M, Perez-Luque R, Gomis-Ruth FX, de la Cruz F, Coll M. Recognition and processing of the origin of transfer DNA by conjugative relaxase TrwC. Nat Struct Biol. 2003;10:1002–1010. doi: 10.1038/nsb1017. [DOI] [PubMed] [Google Scholar]
- Hamilton HL, Schwartz KJ, Dillard JP. Insertion-duplication mutagenesis of Neisseria: use in characterization of DNA transfer genes in the gonococcal genetic island. J Bacteriol. 2001;183:4718–4726. doi: 10.1128/JB.183.16.4718-4726.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton HL, Dominguez NM, Schwartz KJ, Hackett KT, Dillard JP. Neisseria gonorrhoeae secretes chromosomal DNA via a novel type IV secretion system. Mol Microbiol. 2005;55:1704–1721. doi: 10.1111/j.1365-2958.2005.04521.x. [DOI] [PubMed] [Google Scholar]
- Hamilton HL, Dillard JP. Natural transformation of Neisseria gonorrhoeae: from DNA donation to homologous recombination. Mol Microbiol. 2006;59:376–385. doi: 10.1111/j.1365-2958.2005.04964.x. [DOI] [PubMed] [Google Scholar]
- Harley MJ, Toptygin D, Troxler T, Schildbach JF. R150A mutant of F TraI relaxase domain: reduced affinity and specificity for single-stranded DNA and altered fluorescence anisotropy of a bound labeled oligonucleotide. Biochemistry. 2002;41:6460–6468. doi: 10.1021/bi011969i. [DOI] [PubMed] [Google Scholar]
- Heinemann JA. Genetic evidence of protein transfer during bacterial conjugation. Plasmid. 1999;41:240–247. doi: 10.1006/plas.1999.1392. [DOI] [PubMed] [Google Scholar]
- Huber HE, Beauchamp BB, Richardson CC. Escherichia coli dGTP triphosphohydrolase is inhibited by gene 1.2 protein of bacteriophage T7. J Biol Chem. 1988;263:13549–13556. [PubMed] [Google Scholar]
- Judd RC, Porcella SF. Isolation of the periplasm of Neisseria gonorrhoeae. Mol Microbiol. 1993;10:567–574. doi: 10.1111/j.1365-2958.1993.tb00928.x. [DOI] [PubMed] [Google Scholar]
- Kohler PL, Hamilton HL, Cloud-Hansen K, Dillard JP. AtlA functions as a peptidoglycan lytic transglycosylase in the Neisseria gonorrhoeae type IV secretion system. J Bacteriol. 2007;189:5421–5428. doi: 10.1128/JB.00531-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanka E, Wilkins BM. DNA processing reactions in bacterial conjugation. Annu Rev Biochem. 1995;64:141–169. doi: 10.1146/annurev.bi.64.070195.001041. [DOI] [PubMed] [Google Scholar]
- Larkin C, Datta S, Harley MJ, Anderson BJ, Ebie A, Hargreaves V, Schildbach JF. Inter- and intramolecular determinants of the specificity of single-stranded DNA binding and cleavage by the F factor relaxase. Structure. 2005;13:1533–1544. doi: 10.1016/j.str.2005.06.013. [DOI] [PubMed] [Google Scholar]
- Lawley TD, Gilmour MW, Gunton JE, Standeven LJ, Taylor DE. Functional and mutational analysis of conjugative transfer region 1 (Tra1) from the IncHI1 plasmid R27. J Bacteriol. 2002;184:2173–2180. doi: 10.1128/JB.184.8.2173-2180.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawley TD, Klimke WA, Gubbins MJ, Frost LS. F factor conjugation is a true type IV secretion system. FEMS Microbiol Lett. 2003;224:1–15. doi: 10.1016/S0378-1097(03)00430-0. [DOI] [PubMed] [Google Scholar]
- Lehman IR. The deoxyribonucleases of Escherichia coli. I. Purification and properties of a phosphodiesterase. J Biol Chem. 1960;235:1479–1487. [PubMed] [Google Scholar]
- Lehman IR, Nussbaum AL. The Deoxyribonucleases of Escherichia coli V. On the Specificity of Exonuclease I (Phosphodiesterase) J Biol Chem. 1964;239:2628–2636. [PubMed] [Google Scholar]
- Llosa M, Gomis-Ruth FX, Coll M, de la Cruz Fd F. Bacterial conjugation: a two-step mechanism for DNA transport. Mol Microbiol. 2002;45:1–8. doi: 10.1046/j.1365-2958.2002.03014.x. [DOI] [PubMed] [Google Scholar]
- Llosa M, O’Callaghan D. Euroconference on the Biology of Type IV Secretion Processes: bacterial gates into the outer world. Mol Microbiol. 2004;53:1–8. doi: 10.1111/j.1365-2958.2004.04168.x. [DOI] [PubMed] [Google Scholar]
- Lovett ST, Kolodner RD. Identification and purification of a single-stranded-DNA-specific exonuclease encoded by the recJ gene of Escherichia coli. Proc Natl Acad Sci U S A. 1989;86:2627–2631. doi: 10.1073/pnas.86.8.2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo ZQ, Isberg RR. Multiple substrates of the Legionella pneumophila Dot/Icm system identified by interbacterial protein transfer. Proc Natl Acad Sci U S A. 2004;101:841–846. doi: 10.1073/pnas.0304916101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manting EH, van Der Does C, Remigy H, Engel A, Driessen AJ. SecYEG assembles into a tetramer to form the active protein translocation channel. Embo J. 2000;19:852–861. doi: 10.1093/emboj/19.5.852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard Smith JM, Smith NH, O’Rourke M, Spratt BG. How clonal are bacteria? Proc Natl Acad Sci U S A. 1993;90:4384–4388. doi: 10.1073/pnas.90.10.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundle ST, Fattal MH, Melo LF, Coriolan JD, O’Regan NE, Strauss PR. Novel role of tyrosine in catalysis by human AP endonuclease 1. DNA Repair (Amst) 2004;3:1447–1455. doi: 10.1016/j.dnarep.2004.06.009. [DOI] [PubMed] [Google Scholar]
- Pansegrau W, Ziegelin G, Lanka E. Covalent association of the traI gene product of plasmid RP4 with the 5′-terminal nucleotide at the relaxation nick site. J Biol Chem. 1990;265:10637–10644. [PubMed] [Google Scholar]
- Pansegrau W, Lanka E. Common sequence motifs in DNA relaxases and nick regions from a variety of DNA transfer systems. Nucleic Acids Res. 1991;19:3455. doi: 10.1093/nar/19.12.3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pansegrau W, Schroder W, Lanka E. Relaxase (TraI) of IncP alpha plasmid RP4 catalyzes a site-specific cleaving-joining reaction of single-stranded DNA. Proc Natl Acad Sci U S A. 1993;90:2925–2929. doi: 10.1073/pnas.90.7.2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pansegrau W, Schroder W, Lanka E. Concerted action of three distinct domains in the DNA cleaving-joining reaction catalyzed by relaxase (TraI) of conjugative plasmid RP4. J Biol Chem. 1994;269:2782–2789. [PubMed] [Google Scholar]
- Pantoja M, Chen L, Chen Y, Nester EW. Agrobacterium type IV secretion is a two-step process in which export substrates associate with the virulence protein VirJ in the periplasm. Mol Microbiol. 2002;45:1325–1335. doi: 10.1046/j.1365-2958.2002.03098.x. [DOI] [PubMed] [Google Scholar]
- Richardson CC, Lehman IR, Kornberg A. A Deoxyribonucleic Acid Phosphatase-Exonuclease from Escherichia coli Ii. Characterization of the Exonuclease Activity. J Biol Chem. 1964;239:251–258. [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1989. [Google Scholar]
- Snyder LA, Jarvis SA, Saunders NJ. Complete and variant forms of the ‘gonococcal genetic island’ in Neisseria meningitidis. Microbiology. 2005;151:4005–4013. doi: 10.1099/mic.0.27925-0. [DOI] [PubMed] [Google Scholar]
- Stein DC. Transformation of Neisseria gonorrhoeae: physical requirements of the transforming DNA. Can J Microbiol. 1991;37:345–349. doi: 10.1139/m91-056. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Laan M, Nouwen N, Driessen AJ. SecYEG proteoliposomes catalyze the Deltaphi-dependent membrane insertion of FtsQ. J Biol Chem. 2004;279:1659–1664. doi: 10.1074/jbc.M306527200. [DOI] [PubMed] [Google Scholar]
- van der Wolk JP, de Wit JG, Driessen AJ. The catalytic cycle of the Escherichia coli SecA ATPase comprises two distinct preprotein translocation events. Embo J. 1997;16:7297–7304. doi: 10.1093/emboj/16.24.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanecko S, Laskowski M., Sr. Studies of the specificity of deoxyribonuclease I. III. Hydrolysis of chains carrying a monoesterified phosphate on carbon 5′. J Biol Chem. 1961;236:3312–3316. [PubMed] [Google Scholar]
- Varsaki A, Lucas M, Afendra AS, Drainas C, de la Cruz F. Genetic and biochemical characterization of MbeA, the relaxase involved in plasmid ColE1 conjugative mobilization. Mol Microbiol. 2003;48:481–493. doi: 10.1046/j.1365-2958.2003.03441.x. [DOI] [PubMed] [Google Scholar]
- Vergunst AC, Schrammeijer B, den Dulk-Ras A, de Vlaam CM, Regensburg-Tuink TJ, Hooykaas PJ. VirB/D4-dependent protein translocation from Agrobacterium into plant cells. Science. 2000;290:979–982. doi: 10.1126/science.290.5493.979. [DOI] [PubMed] [Google Scholar]
- Wade JJ, Graver MA. A fully defined, clear and protein-free liquid medium permitting dense growth of Neisseria gonorrhoeae from very low inocula. FEMS Microbiol Lett. 2007;273:35–37. doi: 10.1111/j.1574-6968.2007.00776.x. [DOI] [PubMed] [Google Scholar]
- Weiss AA, Johnson FD, Burns DL. Molecular characterization of an operon required for pertussis toxin secretion. Proc Natl Acad Sci U S A. 1993;90:2970–2974. doi: 10.1073/pnas.90.7.2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakunin AF, Proudfoot M, Kuznetsova E, Savchenko A, Brown G, Arrowsmith CH, Edwards AM. The HD domain of the Escherichia coli tRNA nucleotidyltransferase has 2′,3′-cyclic phosphodiesterase, 2′-nucleotidase, and phosphatase activities. J Biol Chem. 2004;279:36819–36827. doi: 10.1074/jbc.M405120200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.