Abstract
Pigment epithelium-derived factor (PEDF) is a multifunctional serpin with antitumorigenic, antimetastatic, and differentiating activities. PEDF is found within tissues rich in the glycosaminoglycan hyaluronan (HA), and its amino acid sequence contains putative HA-binding motifs. We show that PEDF coprecipitation with glycosaminoglycans in media conditioned by human retinoblastoma Y-79 cells decreased after pretreatments with hyaluronidase, implying an association between HA and PEDF. Direct binding of human recombinant PEDF to highly purified HA was demonstrated by coprecipitation in the presence of cetylpyridinium chloride. Binding of PEDF to HA was concentration-dependent and saturable. The PEDF-HA interactions were sensitive to increasing NaCl concentrations, indicating an ionic nature of these interactions and having affinity higher than PEDF-heparin. Competition assays showed that PEDF can bind heparin and HA simultaneously. PEDF chemically modified with fluorescein retained the capacity for interacting with HA but lacked heparin affinity, suggesting one or more distinct HA-binding regions on PEDF. The HA-binding region was examined by site-directed mutagenesis. Single-point and cumulative alterations at basic residues within the putative HA-binding motif K189A/K191A/R194A/K197A drastically reduced the HA-binding activity without affecting heparin- or collagen I binding of PEDF. Cumulative alterations at sites critical for heparin binding (K146A/K147A/R149A) decreased HA affinity but not collagen I binding. Thus these clusters of basic residues (BXBXXBXXB and BX3AB2XB motifs) in PEDF are functional regions for binding HA. In the spatial PEDF structure they are located in distinct areas away from the collagen-binding site. The HA-binding activity of PEDF may contribute to deposition in the extracellular matrix and to its reported antitumor/antimetastatic effects.
Hyaluronan (hyaluronic acid, HA)2 is a non-sulfated glycosaminoglycan consisting of 2,000–25,000 repeating disaccharide subunits of glucuronic acid and N-acetylglucosamine. It was discovered in the vitreous of the bovine eye, but is now known to be ubiquitously distributed and is recognized as the major glycosaminoglycan type present in the extracellular matrix (1). HA serves as a scaffold for other molecules and participates in diverse physiological and cellular functions by interacting with specific binding domains in proteins. Moreover, alterations in HA metabolism, distribution, and function have been documented in diseases such as arthritis, immune and inflammatory disorders, pulmonary and vascular disease, and cancer. HA is well recognized as an important determinant of cancer cell behavior, and increased production of HA is associated with tumors (2). In addition, soluble HA-binding proteins have been shown to have antitumorigenic properties (3).
Pigment epithelium-derived factor (PEDF) belongs to the serine protease inhibitor (serpin) superfamily of proteins, which are related through a highly conserved folded protein conformation (4). Unlike most serpins, PEDF does not inhibit serine proteases, but like most of them it is secreted and found extracellularly in a variety of matrixes, body fluids, and tissues (e.g. the vitreous, interphotoreceptor matrix, aqueous humor, serum, cerebral spinal fluid, bone, and cartilage) (5–9). Although PEDF was discovered as a retinal pigment epithelium-derived differentiation factor for human retinoblastoma cells (10), it exhibits several other activities in and outside of the eye. Numerous reports have demonstrated the involvement of PEDF in neuronal differentiation and survival, as well as inhibition of angiogenesis (11, 12). Its efficacy in preventing tumor growth and metastasis has been shown on neuroblastomas (13, 14), prostate carcinoma cells (15), and in vivo models for hepatocellular carcinoma and lung carcinoma (16). PEDF also induces differentiation of tumor cells (e.g. neuronal phenotype on human retinoblastoma Y-79 and Weri cells in vitro (17, 18), and neuroendocrine differentiation in prostate cancer cells (19)). The antitumor effects of PEDF are believed to involve the induction of differentiation or apoptosis of tumor cells (20). The molecular mechanisms of PEDF as a neurotrophic and antiangiogenic factor are known to be mediated by binding to receptors on the surface of cells (4), but little is known about the mechanisms that govern the cellular events for the PEDF antitumor activity.
Previous studies have described the association of PEDF with extracellular matrix components, such as collagens and sulfated glycosaminoglycans (heparin, heparan sulfate, and chondroitin sulfates) (21–24). Mapping of collagen- and heparin-binding sites on mouse PEDF revealed that the acidic amino acid residues Asp255, Asp257, and Asp299 are critical to collagen I binding, and that three clustered basic amino acid residues Arg145, Lys146, and Arg148 are necessary for heparin binding (25). It is also apparent that the biological activities of PEDF require the participation of its extracellular matrix-binding properties. Mutational analysis supports the involvement of the collagen I binding site in the anti-angiogenic activity of PEDF (26). Experiments with heparin- and heparan sulfate-depleted cells support the importance of heparin/heparan sulfate in efficient cell surface receptor binding of PEDF (27). Recent studies provide evidence that heparin induces a conformational change in PEDF that could facilitate its interactions with a receptor on the cell surface (28).
Given that PEDF colocalizes with HA in extracellular matrixes, and both are highly concentrated in the interphotoreceptor matrix (5, 29) we compared and aligned the amino acid sequences of PEDF to known HA-binding proteins for structure-function studies. We found that PEDF from several species contains regions with strong homology to the HA-binding motifs of known HA-binding proteins CD44, RHAMM, hyaluronidase, and BH-P (see Table 1). Linear basic residue clusters span between residues Lys134–Lys151 and Lys189–Lys197 of the human PEDF sequence. They contain motifs that conform to the sequence pattern BX7B, in particular BX3AB2XB and BXBX2BX2B, where B represents lysine or arginine residues separated by amino acids, excluding aspartic or glutamic acid in the latter, but having only one glutamic acid residue (A) in the former homologous motif. However, it is not known yet whether PEDF binds to HA. In this study, we used retinoblastoma-derived or highly purified HA and recombinant human or bovine PEDF, as well as chemically modified and genetically engineered PEDF, to investigate the possible interactions between PEDF and HA. We found that PEDF binds to HA, provide evidence for an HA-binding site, and discuss implications in PEDF biology.
TABLE 1.
Comparison and alignment of PEDF amino acid sequences and HA-binding proteins at HA-binding regions
A linear HA-binding motif conforms to the sequence pattern BX7B, where B represents basic amino acids separated by residues (X) excluding acidic amino acids (51). For PEDF the sequence patterns are BX4B2XB and BXBX2BX2B
A 42-amino acid peptide (designated as BH-P) that contains several HA-binding motifs BX7B from human brain HA-binding protein (3)
EXPERIMENTAL PROCEDURES
Materials—Hyaluronate lyase (EC 4.2.2.1) purified from Streptomyces hyalurolyticus was purchased from ICN Pharmaceuticals, Inc. Toluidine Blue-O was purchased from Sigma. 2-Aminoacridone HCl (AMAC) was from Molecular Probes. d-Glucose, d-mannose, mercuric acetate, glacial acetic acid (99.99+%), DMSO (99.9%), sodium cyanoborohydride (95%), and glycerol (99.5%) were from Sigma-Aldrich. Proteinase K and phenol red (0.5% w/v) were from Invitrogen. MONO™ composition gels (#60100) and MONO™ gel running buffer (#70100) were from Glyko Inc. Dowex AG50W-X8 (200–400 mesh) was from Bio-Rad Laboratories. Highly purified hyaluronan (Healon®, the trade name for highly purified HA; patient grade without detectable GAG contaminants) was from Pharmacia, Maersk Medicall Ltd., UK. Rabbit polyclonal antibody to PEDF, Ab-rPEDF, was as described before (8). Collagen type I purified from rat tail was purchased from BD Biosciences. Gradient 10–20% polyacrylamide gels with Tricine were from Novex (Invitrogen). Recombinant human PEDF was purified from baby hamster kidney (BHK) cells containing an expression vector with human PEDF cDNA (24). Fluorescein-5EX succinimidyl ester was from Molecular Probes. Fluoresceinated PEDF (Fl-PEDF) was prepared as described before (30). Determination of the concentration and the degree of labeling was performed following the manufacturer's instructions.
Preparation of Conditioned Media—Human retinoblastoma Y-79 cells (0.45–5 × 106 cells/ml) were cultured in defined media (minimal essential medium containing 10 mm HEPES, 1 mm sodium pyruvate, 0.1 mm non-essential amino acids, 1 mm l-glutamine, 1% penicillin/streptomycin (Invitrogen)) at 37 °C for 16–24 h. The conditioned media was separated from the cells by centrifugation at 1,000 × g for 5 min at 4 °C and concentrated by ultrafiltration using membrane filters with a molecular weight cut off of 10,000 (Amicon YM10 filters).
Analysis of HA and Chondroitin Glycosaminoglycans—The glycosaminoglycans in the media conditioned by Y-79 cells were analyzed by fluorophore-assisted carbohydrate electrophoresis, a procedure for the qualitative and quantitative analysis of glycosaminoglycans. The details of methods used for sample preparation, carbohydrate analysis, and quantitation have been previously described (31–33). Culture medium was treated with proteinase K to free glycosaminoglycans from their core protein and boiled to inactivate proteinase K. The digests were concentrated and ethanol-precipitated to pellet any macromolecular material, including the HA and chondroitin chains. The supernatant solution containing the low molecular weight molecules, such as glucose and salts, was aspirated. The precipitate was re-suspended in 100 μl of 0.0005% phenol red, 100 nm ammonium acetate, pH 7. The ammonium acetate extracts were digested at 37 °C with 100 milliunits/ml of Streptococcus dysgalactiae hyaluronidase for 1 h, followed by 3 h with the addition of 100 milliunits/ml of Proteus vulgaris chondroitinase ABC. The enzyme digests were dried on a vacuum concentrator and derivatized by addition of 20 μl of 12.5 mm 2-aminoacrodone HCl (AMAC) in 85% DMSO/15% acetic acid followed by incubation for 15 min at room temperature. Then, 20 μl of 1.25 m sodium cyanoborohydride in ultrapure water were added followed by incubation for 16 h at 37 °C. After derivatization, 10 μl of glycerol (20% final concentration) was added to each sample prior to electrophoresis. All derivatized samples were stored in the dark at -70 °C. The derivatized unknown samples to be run in separate lanes were spiked with one AMAC-derivatized chondroitin ΔDi2S as an internal standard. Media did not contain ΔDi2S standard, thus suitable as spiked standard. Samples were applied on MONO™ composition gels with MONO™ gel running buffer. Electrophoresis was at 4 °C for 80 min with a constant 500 V, with a starting current of 25 mA/gel and a final current of 10 mA/gel. The gels in their glass supports were illuminated with UV light (365 nm) from an Ultra Lum Transilluminator, and imaged with a Quantix cooled charge-coupled device camera (Roper Scientific/Photometrics). The images were analyzed using the Gel-Pro Analyzer™ program version 3.0 (Media Cybernetics). The digital images shown under “Results” depict oversaturated pixel intensity for the major derivatized structures to allow visualization of less abundant derivatized carbohydrates. Quantitation was performed on gel images having all pixels within a linear 12-bit intensity depth range.
Cetylpyridinium Chloride Precipitation—To demonstrate HA binding, we used a protocol previously described (34, 35). Solutions of PEDF and HA were mixed as indicated and incubated at 4 °C for 60 min. The glycosaminoglycans were precipitated in the presence of 1.25% cetylpyridinium chloride (CPC) by incubation at 37 °C for 1 h. The precipitate was separated by centrifugation in an Eppendorf centrifuge at 14,000 rpm for 15 min. at 4 °C, washed with 200 μl of 1% CPC in phosphate-buffered saline, and finally mixed with 30 μl of sample buffer for SDS-PAGE. Protein detection in gels was performed with Coomassie Blue staining or immunostaining after Western blotting.
Conditioned media from several batches of retinoblastoma cell cultures (total ∼50 ml; 1.25 × 105 cells/ml) were tested for PEDF binding to glycosaminoglycans by coprecipitation of PEDF by CPC. Media were concentrated to a final protein concentration in the range of 1–20 μg/ml. Half of each sample was treated with hyaluronidase. BSA was added to each so that the final protein concentration in each medium batch was 100 μg/ml. Purified recombinant human PEDF (10–12 μg) was added to each concentrated medium (500 μl) and the mixture incubated at 4 °C for 60 min. CPC precipitation was followed as described above.
Site-directed Mutagenesis—Single point mutations to convert each Lys189, Lys191, Lys197, Arg194, Asp256, Asp258, and Asp300, and three cumulative alterations Lys146/Lys147/Arg149, Lys189/Lys191/Lys197/Arg194, and Asp256/Asp258/Asp300 into alanine residues were performed by PCR-based site-directed mutagenesis by Lofstrand Labs. The human PEDF cDNA in plasmid πFS17 (18) was transferred to pBK-CMV (Stratagene) to be used as template for mutagenesis. Four primers were designed for the preparation of each construct: Nhe/EcoR primer (S) (5′-ACG GCT AGC GAA TTC AAG CGG ACG CTG GAT TAG-3′), HinDIII primer (A) (5′-GGG AAG CTT TTT TTT TTT TTA GGG ATA AAG CTC-3′), and the sense (S) and antisense (A) primers for each desired mutation. The Nhe/EcoR primer is in the upstream of the PEDF starting codon and the HinDIII primer is in the downstream of PEDF stop codon. The sequences of the primer sets for each desired mutation are annotated below with the base changes underlined: K146/147/149A (S), TTT GAG GCG GCG CTG GCC ATA AAA TCC AGC TTT GTG GC; K146/147/149A (A), TTT TAT GGC CAG CGC CGC CTC AAA GAC GAT CCG GGA GGC; K189A (S), GCG CAG ATG GCA GGG AAG CTC GCC AGG TCC; K189A (A), GAG CTT CCC TGC CAT CTG CGC CTG CAC CC; K191A (S), ATG AAA GGG GCG CTC GCC AGG TCC ACA AAG; K191A (A), CCT GGC GAG CGC CCC TTT CAT CTG CGC CTG; R194A (S), AAG CTC GCC GCG TCC ACA AAG GAA ATT CCC; R194A (A), CTT TGT GGA CGC GGC GAG CTT CCC TTT CAT C; K197A (S), AGG TCC ACA GCG GAA ATT CCC GAT GAG ATC; K197A (A), GGG AAT TTC CGC TGT GGA CCT GGC GAG CTT C; K189/191/194/197A (S), GCA GGG GCG CTC GCC GCG TCC ACA GCG GAA ATT CCC GAT GAG ATC AGC ATT C; K189/191/194/197A (A), CGC TGT GGA CGC GGC GAG CGC CCC TGC CAT CTG CGC CTG CAC CCA GTT GTT G; D256/258A (S), GGC TTG GCT TCA GCT CTC AGC TGC AAG ATT G; D256/258A (A), GCT GAG AGC TGA AGC CAA GCC ATA GCG TAA AAC; D300A (S), TTC ATT CAT GCC ATA GAC CGA GAA CTG AAG; and D300A (A), TCG GTC TAT GGC ATG AAT GAA CTC GGA GG. Two primary PCRs were performed using pBKCMV/PEDF as the DNA template with Nhe/EcoR primer and antisense primer (A) for desire mutation or HinDIII primer and sense primer (S) for desire mutation. The products from two primary PCR reactions were annealed and used as the template for secondary PCR with the Nhe/EcoR primer/HinDIII primer set. The full length of PEDF with the appropriate mutation from secondary PCR and pBKCMV/PEDF were digested with NheI/HinDIII and ligated together. The mutation was confirmed by DNA sequencing. All constructs were prepared following this strategy using the respective primer sets.
Site-directed mutagenesis to convert the Lys134, Lys137, Lys189, and Lys191 into Ala was performed using the QuikChange site-directed mutagenesis kit (Stratagene) following instructions by the manufacturer. The template was plasmid phbPEDF (36), which contains the first 43 codons of the human PEDF cDNA (codons 1–20 correspond to a secretion signal peptide) followed by the 42–416 codons of the bovine PEDF cDNA. Amino acid numbers for this set of mutants refer to alignment with the human sequence. Cumulative mutations were obtained sequentially starting from the template to prepare pPEDF(K134A), which served as template to prepare pPEDF(K134/137A), which served as template for pPEDF(K134A/K137A/K189A), which served as template to prepare pPEDF(K134A/K137A/K189A, K191A). Sequences of primers used to generate desired mutations are annotated with the base changes underlined: 5′-CGTCACCGCCCCCCAGGCGAACCTTAAGAGTGC-3′ and 5′-GCACTCTTAAGGTTCGCCTGGGGGGCGGTGACG-3′ for pPEDF(K134A); 5′-GCCCCCCAGGCGAACCTTGCGAGTGCTTCCCGG-3′ and 5′-CCGGGAAGCACTCGCAAGGTTCGCCTGGGGGGC-3′ for pPEDF(K134/137A); 5′-GTGCAGGCCCAGATGGCAGGGAAAGTCGCTAGG-3′ and 5′-CCTAGCGACTTTCCCTGCCATCTGGGCCTGCAC-3′ for pPEDF(K134A/K137A/K189A); and 5′-GCCCAGATGGCAGGGGCAGTGCCTAGGTCCACG-3′ and 5′-CGTGGACCTAGCGACTGCCCCTGCCATCTGGGC-3′ for pPEDF(K134A/K137A/K189A and K191A). Original codons for lysines (AAG or AAA), in the sense strand, were substituted by the GCG or GCA alanine codons. The mutations were confirmed by DNA sequencing. The PEDF coding sequences with the desired mutations in all expression vectors were under the control of the CMV transcriptional promoter within the pBK-CMV (Stratagene) containing a neomycin-resistant gene sequence.
Recombinant Expression of PEDF in BHK Cells—DNA expression vectors were introduced into BHK cells. PEDF protein is undetectable in media of BHK cells and in their corresponding cell extracts (36). Selection of transfected cells was performed by culturing them in the presence of 400 μg/ml neomycin as described before (36, 37). Transfected BHK cells were incubated at 37 °C in Dulbecco's modified Eagle's medium with 10% serum for 24 h and then without serum for 24 h in repetitive cycles as described before (24). Serum-free conditioned media from each cycle were harvested, filtered, and concentrated before using for HA binding assays.
Cation-exchange Column Chromatography—Cation-exchange column chromatography of modified proteins was performed as described before (24). Media of transfected cells, as above, were dialyzed against buffer S (20 mm sodium phosphate, 1 mm dithiothreitol, 10% glycerol, pH 6.5) containing 50 mm NaCl, and the clear dialysates were injected into a strong cation-exchange resin (POROS HS™, 1.7-ml bed volume) in a column attached to an automated perfusion chromatography work station (BioCad 700E, Applied BioSystems) that had been equilibrated with buffer S plus 50 mm NaCl with a flow rate of 3 ml/min. The column was washed with equilibration buffer to remove unbound material (25 column volumes). Elution of bound PEDF protein was accomplished with 15 column volumes of a linear NaCl gradient (50–500 mm) in buffer S, collected in 1-ml fractions. The A280 nm, column pressure, pH, and conductivity gradients were recorded automatically in a real-time chromatogram. Aliquots from peak fractions were subjected to SDS-PAGE.
Other Methods—Proteins in samples were resolved by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) in Tricine/SDS buffer as described by the manufacturer (Novex). Protein detection was by Coomassie Blue, Ponceau Red, or immunostaining after Western blotting immunoreactions against Ab-rPEDF performed as described previously (9). The protein concentration was determined using a Bio-Rad Protein Assay. Collagen I-binding assays for PEDF were performed as described before (23). Heparin affinity column chromatography for PEDF was performed as described previously (21). HA-affinity Sepharose was prepared as described in the supplemental materials, and the chromatography was performed as for heparin affinity column chromatography (21). The determination of uronic acid content in HA solutions was performed as described in the supplemental materials.
RESULTS
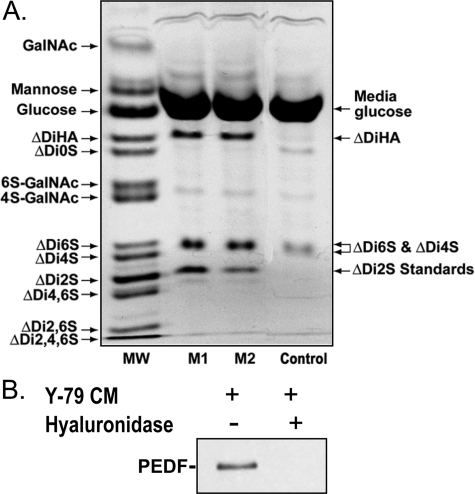
PEDF Affinity for HA Secreted by Retinoblastoma Y-79 Cells— We examined the presence of HA in media conditioned by retinoblastoma Y-79 cells by fluorophore-assisted carbohydrate electrophoresis, a procedure for the qualitative and quantitative analysis of glycosaminoglycans. Electrophoretic patterns of AMAC-labeled carbohydrates from media from two different batches of Y-79 cells (Fig. 1A) showed the presence of ΔDiHA, specific products of HA, which were absent in media not conditioned by Y-79 cells (negative control media). The single band midway between the ΔDi4S and ΔDi6S standards consists of these two chondroitin disaccharides that do not resolve in these preparations. The lack of separation between the two chondroitin disaccharides in retinal pigment epithelial cell cultures and matrices has been previously reported (33). The unsulfated chondroitin disaccharide was at or below detection levels. The disaccharide concentrations estimated from fluorophore-assisted carbohydrate electrophoresis for HA, and chondroitin 6S and 4S were 70 ± 3 pmol/ml media and 30.3 ± 4.4 pmol/ml media, respectively, indicating release of HA, and chondroitin 6S and 4S into the medium of Y-79 cells.
FIGURE 1.
PEDF association to hyaluronan secreted by human retinoblastoma Y-79 cells. A, human retinoblastoma Y-79 cells (1.25 × 105 cells/ml) were cultured in serum-free medium for 16 h, and then the culture conditioned media were collected and concentrated 10-fold before being subjected to fluorophore-assisted carbohydrate electrophoresis. The fluorogram shows the separation of saccharides recovered from Y-79-conditioned media from two different batches of Y-79 cell cultures (M1 and M2). The image shown depicts oversaturated pixel intensity for the major derivatized components to allow visualization of less abundant components. The lane marked MW shows the resolution of a standard mixture of 13 AMAC-derivatized saccharides. The Control lane shows a derivatized medium sample not conditioned by human retinoblastoma Y-79 cells for background corrections. ΔDi2S standards at 62.5 and 31.5 pmol (as determined by hexuronic acid analysis) were added to lanes marked M1 and M2, respectively, for the quantification of resolved disaccharides. B, PEDF coprecipitates with glycosaminoglycans secreted into the Y-79-conditioned media. Serum-free medium from human retinoblastoma cell cultures (1.25 × 106 cells/ml) maintained at 37 °C for 16 h was concentrated (25-fold), and 500-μl aliquots were either left untreated or treated with Streptomyces hyluronidase (10 turbidity reducing units/ml) for 10 min at 37 °C. Recombinant human PEDF (20 μg/ml) and BSA (100 μg/ml) were added, and the mixtures were incubated at 4 °C for 120 min. Glycosaminoglycans were precipitated with CPC. A Western blot of the precipitates was immunostained with Ab-rPEDF, and is shown. Treatments are indicated at the top of each lane.
We then examined whether PEDF associates with glycosaminoglycans in the Y-79-conditioned media by using CPC to specifically precipitate glycosaminoglycans. CPC is a cationic detergent that binds to, and precipitates anionic glycosaminoglycans leading to coprecipitation of binding partners (38). Proteins bound to glycosaminoglycans can be detected in the precipitate. Recombinant human PEDF was mixed with Y-79-conditioned media obtained from different batches of cells and incubated for 1 h at 4 °C before CPC precipitation. We detected PEDF in the CPC precipitates of each and every tested conditioned media, except when media were pre-treated with Streptomyces hyaluronidase, as illustrated in Fig. 1B. These results show that PEDF binding to glycosaminoglycans from Y-79-conditioned media was lost when the media was depleted of HA, indicating that HA was the main PEDF-binding glycosaminoglycan secreted by Y-79 cells.
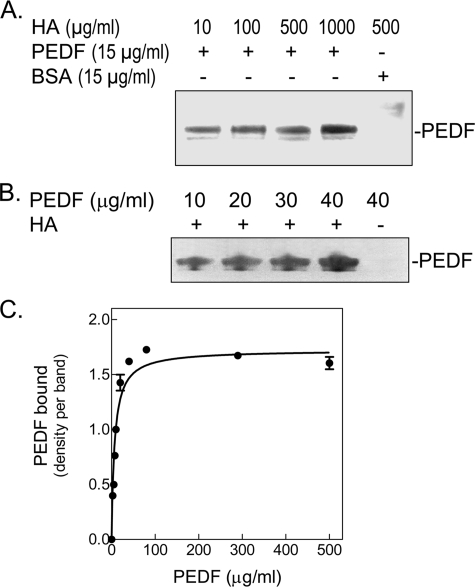
Direct Binding of PEDF to HA—Purified human recombinant PEDF protein and commercial highly pure HA (Healon®) were mixed in different buffers and incubated for 1 h at 4 °C to allow for binding. As shown in Fig. 2, PEDF was readily detected in the CPC precipitate of binding reactions in phosphate buffers at pH 6.2–6.4, or culture media at pH 7.5, in contrast to bovine serum albumin (negative control protein). Note that PEDF did not precipitate when HA was not present (Figs. 2B). A concentration-response curve performed with a given concentration of HA and increasing concentrations of PEDF showed that the binding of PEDF to HA was concentration-dependent and saturable (Fig. 2C) with a KD = 141 nm PEDF. Real-time binding of HA to PEDF was also performed using surface plasmon resonance (SPR) with immobilized PEDF on sensor chips. The observed KD values for steady-state affinity analyses of the SPR HA-PEDF interactions were ∼32 nm considering a molecular weight of 4 × 106 for HA Healon® (see Fig. S4).3 Similar analyses performed with bovine vitreous HA (with an estimated molecular weight of 3 × 106) revealed higher KD values (∼130 nm).
FIGURE 2.
Direct binding of PEDF to HA. Purified recombinant human PEDF and highly purified hyaluronan (Healon®) were incubated at 4 °C for 1 h. HA was precipitated with CPC, and the pellets were resuspended in SDS-PAGE sample buffer and applied to 10–20% polyacrylamide gels for detection of PEDF. A, binding reactions were performed with recombinant human PEDF and increasing concentrations of HA (indicated at the top of each lane) in 200 μl of buffer H (20 mm sodium phosphate, pH 6.5, 20 mm NaCl, and 10% glycerol). BSA was the negative control. Proteins in the precipitates were resolved by SDS-PAGE and visualized with Silver Stain. A photograph of a stained gel is shown. B, binding reactions were performed with 100 μg/ml HA and increasing concentrations of PEDF (indicated at the top of each lane) in 500 μl of Dulbecco's modified Eagle's medium plus 100 μg/ml BSA. PEDF in the precipitates was detected by Western blotting with antiserum Ab-rPEDF. C, concentration curve of PEDF binding to HA. Binding reactions were performed with 200 μg/ml HA and increasing PEDF concentrations in 200 μl of 150 mm NaCl in Buffer S (20 mm sodium phosphate, pH 6.4, 10% glycerol, 1 mm dithiothreitol) plus 100 μg/ml BSA (carrier protein). One-third of each reaction mixture was resolved by SDS-PAGE followed by silver staining, and two remaining thirds were analyzed by Western blotting with Ab-rPEDF. The density of each PEDF band was determined from scanned images using Stratagene Eagle Eye II. Data were analyzed by non-linear regression and one-site binding using a GraphPad Prism version 3.0 software program (plot shown). The best-fit values obtained from data combined from silver-stained gels and Western transfers were Bmax = 1.723 bound PEDF, and KD = 7.027 μg/ml PEDF (= 140.54 nm), with an R2 = 0.9715.
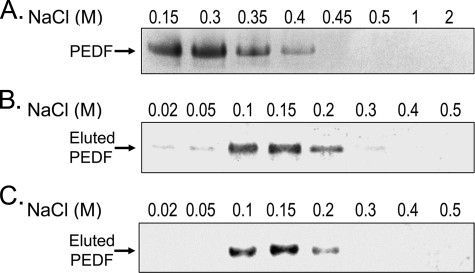
Effect of Ionic Strength on PEDF-HA Interactions—To elucidate the type of interactions between PEDF and HA, we examined the effect of increasing NaCl concentrations on the reactions. Fig. 3A shows that the binding of PEDF to HA decreased with increasing NaCl concentrations, and was completely lost with ≥450 mm NaCl. In contrast, binding to heparin-affinity resin was lost with 100–200 mm NaCl (Fig. 3B), similar to results reported before with bovine PEDF protein (21), and implying that PEDF has lower affinity for heparin-resin than for HA. We note that CPC coprecipitation of PEDF with heparin from a variety of sources and sizes was not sensitive to detect PEDF in the pellets,4 probably due to the low affinity of PEDF for heparin. HA-affinity column chromatography performed in identical conditions as the heparin-affinity chromatography showed that PEDF lost binding with 100–200 mm NaCl (Fig. 3C) similar to that of heparin-resin. These results showed that the HA binding occurred at physiological NaCl concentrations and was sensitive to increasing ionic strength, implying an ionic nature for the PEDF-HA interactions.
FIGURE 3.
Effect of increasing NaCl concentrations on the binding of PEDF to HA (A) and heparin (B). A, binding reactions were performed with recombinant human PEDF (14.5 μg/ml) and HA (200 μg/ml) in buffer S containing increasing concentrations of NaCl. After precipitation with CPC, proteins were resolved by SDS-PAGE and visualized with Coomassie Blue. A photograph of a gel is shown. B, heparin-affinity column chromatography was performed with heparin affinity beads in buffer H. Recombinant human PEDF (1.75 μg) was added to 0.5 ml of buffer H containing 17.5 μg of BSA and loaded onto a column with heparin-affinity beads (0.5 ml). The flow-through was collected and reloaded, repeating these steps three times. The column was washed with 20 column-volumes of buffer H. The bound proteins eluted with a NaCl step-gradient at 4 column-volume/fraction (numbers at top of each lane correspond to NaCl concentration). Fractions were concentrated to 40 μl, and an aliquot of 15 μl from each was resolved by SDS-PAGE followed by Western blotting. PEDF was detected by immunostaining with anti-PEDF. C, HA-affinity column chromatography was performed with HA affinity Sepharose in buffer H as in panel B, except that then the load was 10 μg of PEDF, and elutions were with one column-volume. Aliquots of the fractions (12 μl) were resolved by SDS-PAGE and Western blotting followed by immunostaining with Anti-PEDF.
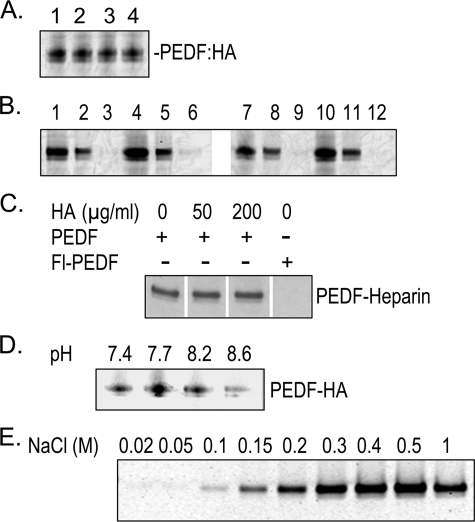
Binding of Fluorescein Conjugated PEDF to HA—Recombinant human PEDF protein was chemically modified by fluorescein conjugation using an activated N-hydroxysuccinimide ester form of fluorescein that reacts with primary amines such as those in lysines and amino terminus of polypeptides. The estimated number of modified lysine amino acids was ∼2 mol of fluorescein/mol of PEDF. We have reported that this type of modification decreases the affinity of PEDF for heparin (30), but increases that for collagen I (23). We performed HA binding assays with fluorescein-conjugated PEDF proteins, which had been prepared using buffers with different pH. In all the cases, the modified PEDF proteins (Fl-PEDF) retained the binding activity to HA (Fig. 4A, lanes 1 and 3, and supplemental Fig. S1), while they lost the capacity for binding heparin (see Fig. 4B, lanes 1–3 and 7–9, and Fig. 4C, right lane). HA-affinity column chromatography also showed that Fl-PEDF bound HA (Fig. 4E). The modified protein eluted with higher concentrations of NaCl than the unmodified PEDF (compare Figs. 3C and 4E). These results indicate that the modified lysines in the Fl-PEDF that were crucial for heparin binding were not essential for HA binding and imply a distinct HA binding region on PEDF.
FIGURE 4.
Binding of chemically modified PEDF to HA and heparin. Exposed lysines of recombinant human PEDF were modified with fluorescein (Fl) using Sulfo-NHS-LC-fluorescein. PEDF (0.4 mg/ml) was preincubated without or with HA (1.6 mg/ml) in buffer H at pH 7.7 or 8.6 at 25 °C for 15 min followed by addition of Sulfo-NHS-LC-fluorescein (87.5 μg/ml) and incubation at 25 °C for 1 h. Ethanolamine was added to the reactions at (150 mm) and incubated for 2 h at 25 °C. Reaction mixtures without the glycosaminoglycan were supplemented with HA to match those with HA. Proteins were separated from unbound fluorescein by ultrafiltration using centricon-30 devices and concentrated in phosphate-buffered saline. A, CPC coprecipitation of HA and chemically modified PEDF. Proteins (2 μg) were mixed with HA (6 μg) in 20 μl of buffer H, pH 7.7, containing 5 μg of BSA, and incubated at 37 °C for 1 h. PEDF was coprecipitated with HA using CPC and resolved by SDS-PAGE. Fluoresceinated PEDF was detected by Typhoon scanning. Reactions were applied to lanes as follows: lane 1, Fl-PEDF preincubated without HA, pH 7.7; lane 2, Fl-PEDF preincubated with HA, pH 7.7; lane 3, Fl-PEDF preincubated without HA, pH 8.6; lane 4, Fl-PEDF preincubated with HA, pH 8.6. B, heparin-affinity column chromatography of chemically modified PEDF. Proteins (2 μg) were mixed with BSA (15 μg) in 500 μl of buffer H, pH 6.4, and applied to a heparin-affinity column (0.5-ml bead-volume). The flow-through (FT) was collected and reloaded three times, before washing with 12 column volumes of buffer H, pH 6.4. Bound material was eluted with 500 mm NaCl in buffer H, pH 6.4. Load, FT, and eluates were concentrated and applied to a gel at equivalent volumes and resolved by SDS-PAGE. Fluoresceinated PEDF was detected by Typhoon scanning. Lanes 1–3, Fl-PEDF preincubated without HA, pH 7.7; lanes 4–6, Fl-PEDF preincubated with HA, pH 7.7; lanes 7–9, Fl-PEDF preincubated without HA, pH 8.6; lanes 10–12, Fl-PEDF preincubated with HA, pH 8.6. Load was applied in lanes 1, 4, 7, and 10;FTin lanes 2, 5, 8, and 11; and eluate in lanes 3, 6, 9, and 12. C, heparin-affinity column chromatography of PEDF and HA mixtures. Unmodified PEDF (30 μg) was added to 2 ml of buffer H containing 0, 50, or 200 μg/ml HA and preincubated at 4 °C before mixing with heparin-affinity beads (1 ml) with gentle rotation in a column at 4 °C for 1 h. PEDF conjugated to fluorescein (Fl-PEDF) in bicarbonate buffer, pH 9, was also subjected to heparin-affinity chromatography. Unbound material was washed with 10 column-volumes of buffer H. Bound proteins were eluted with 500 mm NaCl, concentrated, and equivalent volumes were resolved by SDS-PAGE followed by Coomassie Blue staining. Photographs of the lanes with eluates are shown with components of each reaction mixture indicated to the top. D, PEDF-HA binding and pH. PEDF (17.5 μg/ml) was mixed with HA (60 μg/ml) in 100 μl of buffer H at pH ranging from 7.4 to 8.6, containing BSA (50 μg/ml), and incubated at 25 °C for 1 h. PEDF was coprecipitated with HA using CPC and resolved by SDS-PAGE followed by Western blot. PEDF was immunodetected with anti-PEDF antibodies. The pH of each reaction is indicated at the top of each lane. E, HA-affinity column chromatography was performed with HA affinity Sepharose in buffer H as in Fig. 3C, except that then the load was 5 μg of Fl-PEDF. Aliquots of the fractions (18 μl) were resolved by SDS-PAGE, and the fluoresceinated PEDF was detected by Typhoon scanning. The numbers at top of each lane correspond to NaCl concentration for each fraction.
Competition Assays for HA and Heparin Binding to PEDF— Unmodified PEDF was preincubated with HA to allow binding and then assayed for heparin binding. Heparin-affinity column chromatography demonstrated that, even in the presence of high concentrations of HA, PEDF bound to heparin and with similar affinity as that without HA (Fig. 4C). These results show that HA did not compete with heparin for binding to PEDF and imply that both glycosaminoglycans can simultaneously interact with distinct regions of the PEDF molecule.
Furthermore, PEDF was preincubated with HA to allow binding and then conjugated with fluorescein in the presence of HA. The resulting proteins were used to investigate if interactions with HA would affect the chemical modification(s) of the heparin binding site. As shown in Fig. 4B, the fluoresceinated proteins lost heparin-binding activity, even with PEDF preincubated with HA prior to Fl conjugation. All of them bound to HA (Fig. 4A). These results demonstrate that HA could not protect the heparin-binding region of PEDF from undergoing the one or more modifications that abolish interactions with heparin. However, a very small amount of Fl-PEDF signal was detected in eluates of protein preincubated at pH 7.7 (compare lanes 3 and 6 of Fig. 4B), but not at pH 8.6 (compare lanes 9 and 12). Given that HA-PEDF binding assays in buffers at pH ranging from pH 7.4 to 8.6 revealed higher HA-binding activity at pH 7.7 than at pH 8.6 (Fig. 4D), the above observations suggested that at pH 7.7 HA might have protected one or more critical lysine residues for heparin binding in a very small population of PEDF molecules. Overall, these results point again to a separate HA-binding site in PEDF and insinuate a second site of low binding affinity shared with heparin.
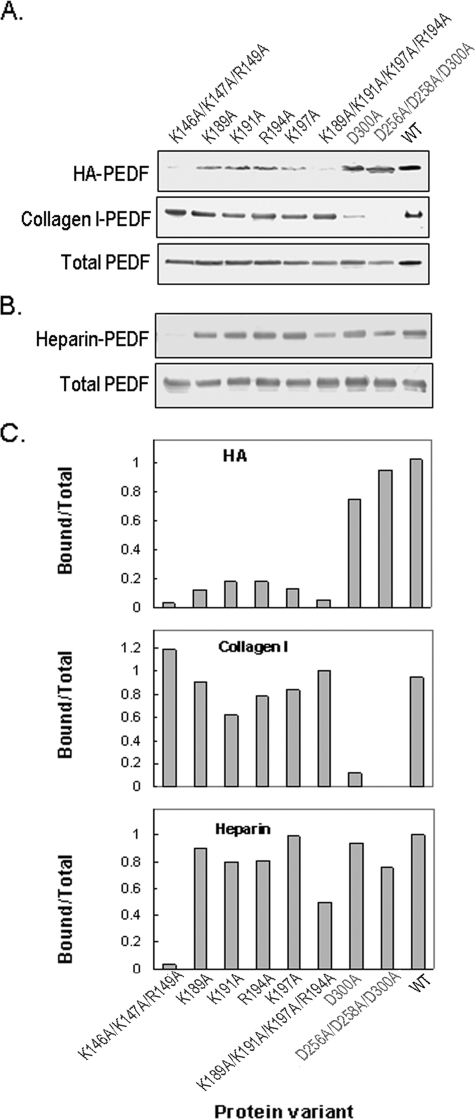
Determination of the HA-binding Sites on PEDF by Site-directed Mutagenesis—The sequences of PEDF from several species reveal BX7B signature motifs for HA-binding proteins that contain basic residues (Table 1). One homologous HA-binding motif is within positions 134–151 (Lys146, Lys147, and Arg149, human sequence), and conforms to BX3AB2XB. The other is located at positions 189–197 and conforms to BXBX2BX2B (Lys189–Lys197). To find out if these regions contain determinants for HA binding of PEDF, we altered the lysine and arginine residues within these regions. Alterations were from the charged amino acids to uncharged alanine residues. As controls, we also altered aspartic acid residues of the collagen I binding site located at Asp256, Asp258, and Asp300 in human PEDF. All of the altered proteins were readily obtained in soluble forms and were subjected to the HA-binding assay. In comparison to unmodified PEDF, proteins with single amino acid alterations showed a decrease in HA-binding activity. Cumulative alterations of amino acids of both HA-binding motifs (K189A/K191A/R194A/K197A and K146A/K147A/R149A) showed a marked reduction in HA-binding activity (Fig. 5A). The modified proteins were also subjected to collagen I binding assays as controls. No loss of collagen I-binding activity was observed for any of the proteins with basic amino acid alterations. As expected, however, collagen I binding was lost when acidic residues at the collagen-binding site were altered to alanine (Fig. 5A), implying that the modifications were specific and did not affect activities determined by other domains of the PEDF protein.
FIGURE 5.
Binding of genetically modified PEDF proteins to HA. BHK cells were transfected with mutated PEDF cDNA expression plasmids. A, culture media (100 μl) of stably transfected cells was concentrated and used in PEDF binding assays with HA (30 μg) and collagen I (2 μg) in 500 μl and 200 μl of phosphate-buffered saline, pH 7.5, respectively. HA-PEDF was isolated by CPC precipitation, and collagen I-PEDF by size exclusion ultrafiltration using centricon-100. PEDF was detected by immunoblotting versus anti-PEDF. HA-PEDF corresponds to PEDF bound to HA; Collagen I-PEDF to PEDF bound to collagen I; and Total PEDF to PEDF in media (20 μl). B, proteins in culture media of stably transfected cells were concentrated to ∼350 μg/ml protein, exchanged to buffer H (0.5 ml) and loaded onto heparin-affinity beads column (0.5 ml bed-volume). The flow-through was reloaded, and the unbound material was washed with 20 column-volumes of buffer H. Bound proteins were eluted with 2 column volumes of 1 m NaCl in buffer H, concentrated by ultrafiltration using centricon-30 devices, and resolved by SDS-PAGE. PEDF was detected by immunoblotting versus anti-PEDF. Heparin-PEDF corresponds to PEDF bound to heparin, and Total PEDF to PEDF in load (6 μg of protein per lane). C, quantification of PEDF bound to HA, collagen I, and heparin. PEDF-immunoreactive bands, as from above, were scanned using UNSCAN-IT software. Values from bound were divided by values from Total protein, plotted as a function of the genetically modified PEDF variant using EXCEL, Microsoft, and are shown.
The proteins were also subjected to heparin binding assays (Fig. 5B). Three critical residues for heparin binding in mouse PEDF (25) are conserved in human within the BX3AB2XB motif (Lys146, Lys147, and Arg149, human sequence). As expected, the human PEDF with cumulative alterations in these residues (K146A/K147A/R149A) abolished the heparin-binding activity. In contrast, proteins with single amino acid alterations at the HA-binding motif BXBX2BX2B (Lys189–Lys197) retained heparin-binding activity similar to that of the wild type, and cumulative alterations retained only partial activity probably due to the likely large change in surface electrostatic potential generated by altering four positions in the protein.
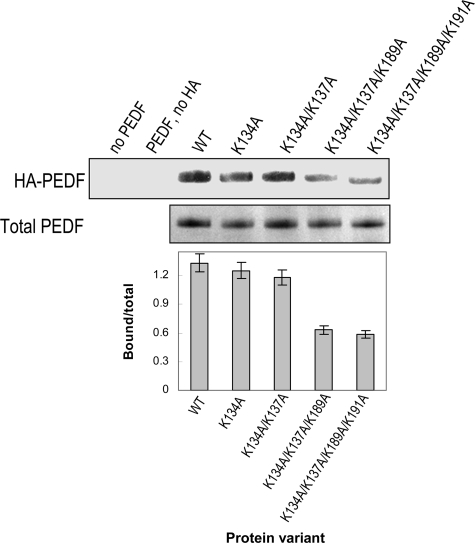
The spatial structure of PEDF shows that the Lys189–Lys197 motif is within a region at the center of β-sheet A-strands 2 and 3 and α-helix F that has a basic electrostatic surface potential and is densely populated with lysines exposed to the surface (Lys134, Lys137, Lys189, Lys191, His212, and Lys214) that are available to interact with polyanions (21, 39). The lysines at positions 134 and 137 are located in the vicinity of Lys189 and Lys191 and were mutated to determine their contributions to HA binding. The change in surface potential was followed by cation-exchange column chromatography (supplemental Fig. S2). Alteration of Lys134 to Ala did not change the elution profile relative to unaltered PEDF, whereas PEDF with double alterations K134A/K137A eluted from the column at lower NaCl concentration. This result indicated that alteration of Lys137 to neutral alanine changed the surface charge of PEDF. However, none of these alterations had an effect on HA binding (Fig. 6). In contrast, PEDF altered in three (K134A/K137A/K189A) and four (K134A/K137A/K189A/K191A) positions, which include Lys189 and Lys191 of the HA-binding motif, had significantly decreased HA binding and eluted much earlier from the cation-exchange column than unaltered PEDF. These results indicate that, although Lys134/Lys137 are in the vicinity of the HA-binding region in the folded PEDF protein and contribute to the basic surface potential of PEDF, they do not participate in HA binding.
FIGURE 6.
Binding to HA of PEDF modified on putative exposed lysines. Altered PEDF proteins were purified and concentrated. Purified altered PEDF proteins and HA were mixed in buffer H and incubated at 4 °C for 1 h. Bound protein was coprecipitated with CPC. Western blot of CPC precipitates immunostained with anti-PEDF is shown at the top (HA-PEDF). SDS-PAGE of purified PEDF altered proteins stained with Coomassie Brilliant Blue is shown at the bottom (Total PEDF). A plot of bound PEDF quantification was performed as in Fig. 5C.
Altogether these results demonstrate that two clustered groups of basic amino acids (Lys189/Lys191/Lys197/Arg194 and Lys146/Lys147/Arg149) in the human PEDF polypeptide are essential residues for HA binding. They conform to HA-binding motifs BXBX2BX2B and BX3AB2XB. The former one is distinct, and the latter one partially overlaps with the PEDF heparin-binding site (Lys146/Lys147/Arg149). They represent a novel domain for HA binding in a serpin, which in PEDF is distinct and non-overlapping from its neurotrophic and antiangiogenic active regions (19, 40) and the collagen I binding site (25).
DISCUSSION
In this study we show that PEDF has binding affinity for hyaluronan, that the interactions have an ionic component, and that the basic amino acids in the BX7B (BXBX2BX2B and BX3AB2XB) motifs of PEDF play a crucial role in HA binding. The association between PEDF and HA occurs under physiological conditions, which suggests that it may occur in vivo, (e.g. vitreous, interphotoreceptor matrix) where PEDF and HA deposit, and may play an important role in regulating the local availability of PEDF. In particular, we provide evidence for binding of PEDF to HA produced by tumor cells (e.g. retinoblastoma Y-79 cells). This is of relevance because 1) tumors are known to increase production of HA (2), 2) peptides enriched in HA-binding motif BX7B possess antitumor activity (3), and 3) PEDF has demonstrable antitumorigenic effects (17, 18, 20). These results demonstrate for the first time a specific binding between HA and PEDF and suggest that the HA-binding activity of PEDF may contribute to the antitumor effects exerted by this protein.
The facts that Y-79 cells release HA and chondroitin sulfates to the media and that PEDF binding is significantly decreased upon HA depletion (Fig. 1) indicate that HA is the glycosaminoglycan in the Y-79 media with the strongest affinity for PEDF and that Y-79 chondroitin sulfates have little if any affinity for PEDF. These observations are in agreement with loss of PEDF binding to chondroitin sulfates with >100 mm NaCl (21), as the media contains ∼150 mm NaCl. The fact that the PEDF interactions with HA (Healon®) have an apparent KD, which is ≥28-fold higher than those with heparin and heparan sulfate (apparent KD =∼4 μm) suggests a higher PEDF affinity for HA than for heparin/heparin sulfate. Moreover, PEDF did not precipitate with CPC treatment of PEDF-heparin complexes and no heparin binding to SPR PEDF chips could be observed, in agreement with PEDF having higher affinity for HA than for heparin.
It must be pointed out that PEDF binding to HA (Healon®) measured by CPC precipitation was less sensitive to NaCl (still observed with 300 mm NaCl) than binding to bovine vitreous HA-affinity Sepharose (lost with ≥150 mm NaCl) (Fig. 3C). Observations of SPR binding of HA Healon® and bovine vitreous to PEDF sensor chips lead to similar conclusions about NaCl sensitivity (supplemental Figs. S5B and S5C), which are in agreement with higher PEDF affinity for the highly purified HA (Healon®) than for bovine vitreous sample (supplemental Fig. S4). We suspect that the differences in KD and NaCl sensitivity between these two sources of HA may be due to the degree of purity of the HA sample preparations. Some preparations may contain salts and/or protein contaminants, or smaller fragments of HA that add multivalency effects. SDS-PAGE of the HA samples revealed one or more extra silver stain bands for the bovine vitreous HA, which were undetected in Healon® and in HA Hylumed®, highly purified HA from bacteria.5 Differences in the KD values for PEDF:HA (Healon®) interactions obtained by CPC treatment (∼141 nm) and SPR (∼32 nm) (Figs. 2C and S4C) must be pointed out also. In addition, some Fl-PEDF molecules were less NaCl-sensitive to binding bovine vitreous HA-affinity Sepharose than the unmodified protein (Figs. 3C and 4E). The chemistry used to immobilize PEDF on SPR sensor chips is similar to that used to conjugate Fl-PEDF. These observations imply an increase in HA affinity upon these types of chemical modifications of PEDF.
Chemical modification and competition assays point to the presence of a distinct HA-binding region in PEDF (Fig. 4). Two HA-binding motifs are found in PEDF, one at Arg141–Arg149 with the BX3AB2XB signature except for one negatively charged amino acid (A) in the X string that is conserved in three species, and the other at Lys189–Arg185 (BXBX2BX2B) with stronger homology to characteristic HA-binding regions (see Table 1). In addition, mutagenesis demonstrates that the basic residues within these two motifs of PEDF are crucial for its association with HA. Single point and cumulative alterations of basic residues within Lys189–Arg194 eliminate the HA-binding activity (Fig. 5) but retain the heparin-binding activity of PEDF (25), indicating that this HA-binding site is distinct from the heparin-binding site. In contrast, cumulative alterations at Lys146, Lys147, and Arg149 abolish both HA and heparin binding (Fig. 5). Single point alterations in mouse PEDF at these conserved sites also abolished heparin binding. These observations strongly imply that these positions participate in interacting with both glycosaminoglycans with a likely lower affinity than the above site. In the spatial PEDF structure the Lys146, Lys147, and Arg149 residues are located in a turn between β-strand s2A and α-helix E, and the motif Lys189–Arg194 is located in another turn between α-helix F and β-strand s3A (Fig. 7). The location in turns may be required to present the motif in the appropriate conformation for HA binding. These two HA-binding regions of PEDF can provide a positively charged surface and argue for a larger area on PEDF available to interact with HA than with heparin that may also contribute to higher affinity for HA than heparin. The presence of the A in the BX3AB2XB motif might also argue for a lower affinity with a decrease in basic potential. The fact that modified PEDF at K134A/K137A had a change in surface ionic potential but no loss of HA binding (Figs. 6 and supplemental Fig. S2) indicates that basic ionic potential on the PEDF surface does not suffice for binding HA. Note that Lys137 is in the vicinity of the HA-binding motif Lys189–Lys197 (see supplemental Fig. S3).
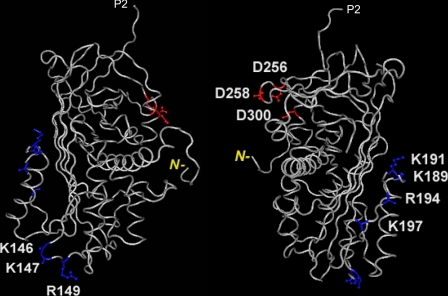
FIGURE 7.
Three-dimensional structure of human PEDF (from Protein Data Bank ID 1IMV) to illustrate the location of the HA-binding site. The two structures are rotated about 180° from each other with highlighted positions of single alterations made in this study. In blue are basic amino acids Lys146, Lys147, and Arg149 residues located in a turn between β-strand s2A and α-helix E, and Lys189, Lys191, Lys194, and Arg197 located in another turn between α-helix F and β-strand s3A, both within BX7B HA-binding sites; in red are acidic amino acids Asp256, Asp258, and Asp300 corresponding to the collagen-binding site. P2 corresponds to the residue next to the homologous serpin reactive site, P1; and N– in yellow indicates the position of the amino-end terminus of the polypeptide in the three-dimensional structure corresponding to position 26. Structures were visualized and reproduced using Cn3D (NCBI).
We also compared the binding affinity of interactions between PEDF and collagen, another extracellular matrix component. The affinity of PEDF is similar for collagen I (KD = 123 nm) and for HA, but the location of their binding sites on the three-dimensional structure of PEDF is distinct, and opposite in charge, i.e. the collagen I binding site is formed by negatively charged amino acids Asp256, Asp258, and Asp300. Thus, the HA-binding sites of PEDF are clearly distinct from the site for collagen I binding, and are topologically separated from it and the neurotrophic (78–121) and antiangiogenic (44–77) active regions of PEDF.
From results of heparin and HA binding assays, the one or more sites in PEDF that were chemically modified by fluorescein in our preparations can be deduced as one of the known three critical residues for heparin binding: Lys146, Lys147, and Arg149 (see Fig. 5B and Ref. 25). These conclusions are derived from clear demonstrations that this chemical modification abolished the binding to heparin but not to HA (see Figs. 4 and supplemental Fig. S1). Given that our Fl-PEDF had an estimated ∼2 sites modified per PEDF molecule, one of them would be Lys146 or Lys147; the second site would be the N terminus of the polypeptide, which is considered to have higher affinity for Fl conjugation than the primary amines in lysines.
Because PEDF is a member of the serpin superfamily of proteins, which are related through their highly conserved folded conformation, we compared an aligned list of 20 serpin sequences (41) around the turn between β-strand s2A and α-helix E, and between α-helix F and β-strand s3A of PEDF. No other serpin satisfied the BX7B motifs, and only a region of thyroxine-binding globulin had homology with Lys146–Arg149 of PEDF. It is not known if this serpin binds glycosaminoglycans. Thus our work identifies PEDF as the only member of the serpin superfamily of proteins for which HA binding has been described and characterized to date.
To our knowledge, this is the first report on the production and secretion of HA by retinoblastoma cells. It has been shown that most malignant solid tumors contain elevated levels of HA, thought to be produced by either the tumor cells themselves or by surrounding stromal cells (42, 43). Our findings are consistent with the former source. Our data are also in agreement with previous evidence for the production of high levels of HA by retinal cells in culture (44) and the presence of HA in the retina in vivo (6, 45). HA is a prominent constituent of the interphotoreceptor matrix, where it may serve to organize the matrix by functioning as a basic scaffold to which other macromolecules in the insoluble interphotoreceptor matrix are attached. The fact that HA and PEDF are both synthesized by the retinal pigment epithelium and are secreted preferentially from the apical cell surface (5, 33) suggests that these two molecules are available to associate intracellularly prior concerted secretion from this tissue into the interphotoreceptor matrix. The HA-binding activity of PEDF may represent the molecular basis for the directional cellular secretion and deposition in the interphotoreceptor matrix and vitreous, as well as other extracellular matrixes.
Our results also have important biological implications, as the HA-binding motifs in HA-binding proteins exhibit antitumor effects. Binding to cell-surface receptors is considered the first step for neurotrophic activities of PEDF, and heparin and heparan sulfate can act as positive modulators of the ligand-receptor interactions (27, 40). Because enzymatic depletion of HA has no effect on the interactions between PEDF and cell surfaces (27), the formation of an extracellular PEDF·HA complex may have a functional role in blocking the HA biological effects. HA contributes to cancer progression by providing a loose matrix for migrating tumor cells and by mediating adhesion of cancer cells (46). The HA receptor, CD44, is known to promote tumor growth and metastasis (47). Furthermore, the efficacy of soluble members of the HA-binding protein family (e.g. CD44 and RHAMM) to inhibit tumor formation has been demonstrated. Truncated HA receptors like soluble recombinant CD44, which disrupts the interaction between membrane-bound CD44 and HA, can inhibit tumor formation and metastasis (48, 49). Similarly, a soluble form of RHAMM can block the ability of tumor cells to form lung metastases (50). More interestingly, a 42-amino acid peptide (designated as BH-P; see Table 1) that contains several HA-binding motifs BX7B from human brain HA-binding protein inhibits tumor growth and induces apoptosis (3). These results suggest that the HA-binding motif is responsible for the antitumor effect. Simultaneous up-regulation of HA, its receptor CD44, and the heparan sulfate degrading enzyme heparanase, which generate a microenvironment that promotes tumor progression and metastasis, has been observed in many cancers. Recent evidence shows that HA, CD44, and heparanase regulate cancer cell proliferation, migration, and invasion, as well as tumor-associated angiogenesis, and they also seem to predict the clinical outcome of breast cancer patients (47). We propose that PEDF may disrupt the interaction between HA and CD44, thus blocking the promotion of tumor formation, growth, and metastasis. A concomitant increase in enzyme heparanase often observed in tumors (47) would contribute to diminish the positive effect of heparin/heparan sulfate on the conformation of PEDF for stronger ligand-cell surface receptor interactions (27, 28), permitting the availability of more free PEDF molecules to interact with the HA, which may lead to antitumorigenic effects. Thus, in this way, the biological effects of PEDF as a tumor suppressor may depend on its interactions with HA.
Supplementary Material
Acknowledgments
We thank Inigo Izal-Azcarate for assistance with constructions of plasmids and DNA sequencing and Christina Meyer and Natalia Balko for assistance with HA binding and collagen binding assays with Fl-PEDF and mutants.
This work was supported, in whole or in part, by National Institutes of Health NEI Intramural Research Program. This work was also supported by the Educational and Cultural Department of the “Gobierno de Navarra,” Spain (to A. P.-M.); by Grants EY14240-1 and EY 15638, Foundation for Fighting Blindness, and a Challenge Grant from Research to Prevent Blindness to the Department of Ophthalmology, Cleveland Clinic Lerner College of Medicine (to J. G. H.); and by Grant RO1 CA64472 (to V. N.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental text and Figs. S1–S5.
Footnotes
The abbreviations used are: HA, hyaluronan; PEDF, pigment epithelium-derived factor; CPC, cetylpyridinium chloride; BSA, bovine serum albumin; AMAC, 2-aminoacridone HCl; Tricine, N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine; BHK, baby hamster kidney; Fl-PEDF, fluoresceinated PEDF; CMV, cytomegalovirus; SPR, surface plasmon resonance.
S. P. Becerra, L. A. Perez-Mediavilla, J. E. Weldon, S. Locatelli-Hoops, P. Senanayake, L. Notari, V. Notario, and J. G. Hollyfield, unpublished observations.
S. Locatelli-Hoops, personal observations.
References
- 1.McDonald, J., and Hascall, V. C. (2002) J. Biol. Chem. 277 4575-4579 [DOI] [PubMed] [Google Scholar]
- 2.Toole, B. P., Wight, T. N., and Tammi, M. I. (2002) J. Biol. Chem. 277 4593-4596 [DOI] [PubMed] [Google Scholar]
- 3.Xu, X. M., Chen, Y., Chen, J., Yang, S., Gao, F., Underhill, C. B., Creswell, K., and Zhang, L. (2003) Cancer Res. 63 5685-5690 [PubMed] [Google Scholar]
- 4.Becerra, S. P. (2006) Exp. Eye Res. 82 739-740 [DOI] [PubMed] [Google Scholar]
- 5.Becerra, S. P., Fariss, R. N., Wu, Y. Q., Montuenga, L. M., Wong, P., and Pfeffer, B. A. (2004) Exp. Eye Res. 78 223-234 [DOI] [PubMed] [Google Scholar]
- 6.Hollyfield, J. G., Rayborn, M. E., Tammi, M., and Tammi, R. (1998) Exp. Eye Res. 66 241-248 [DOI] [PubMed] [Google Scholar]
- 7.Ortego, J., Escribano, J., Becerra, S. P., and Coca-Prados, M. (1996) Invest Ophthalmol. Vis. Sci. 37 2759-2767 [PubMed] [Google Scholar]
- 8.Wu, Y. Q., and Becerra, S. P. (1996) Invest Ophthalmol. Vis. Sci. 37 1984-1993 [PubMed] [Google Scholar]
- 9.Wu, Y. Q., Notario, V., Chader, G. J., and Becerra, S. P. (1995) Protein Expr. Purif 6 447-456 [DOI] [PubMed] [Google Scholar]
- 10.Tombran-Tink, J., Chader, G. G., and Johnson, L. V. (1991) Exp. Eye Res. 53 411-414 [DOI] [PubMed] [Google Scholar]
- 11.Bouck, N. (2002) Trends Mol. Med. 8 330-334 [DOI] [PubMed] [Google Scholar]
- 12.Tombran-Tink, J., and Barnstable, C. J. (2003) Nat. Rev. Neurosci. 4 628-636 [DOI] [PubMed] [Google Scholar]
- 13.Crawford, S. E., Stellmach, V., Ranalli, M., Huang, X., Huang, L., Volpert, O., De Vries, G. H., Abramson, L. P., and Bouck, N. (2001) J. Cell Sci. 114 4421-4428 [DOI] [PubMed] [Google Scholar]
- 14.Streck, C. J., Zhang, Y., Zhou, J., Ng, C., Nathwani, A. C., and Davidoff, A. M. (2005) J. Pediatr. Surg. 40 236-243 [DOI] [PubMed] [Google Scholar]
- 15.Doll, J. A., Stellmach, V. M., Bouck, N. P., Bergh, A. R., Lee, C., Abramson, L. P., Cornwell, M. L., Pins, M. R., Borensztajn, J., and Crawford, S. E. (2003) Nat. Med. 9 774-780 [DOI] [PubMed] [Google Scholar]
- 16.Wang, L., Schmitz, V., Perez-Mediavilla, A., Izal, I., Prieto, J., and Qian, C. (2003) Mol. Ther. 8 72-79 [DOI] [PubMed] [Google Scholar]
- 17.Seigel, G. M., Tombran-Tink, J., Becerra, S. P., Chader, G. J., Diloreto, D. A., Jr., del Cerro, C., Lazar, E. S., and del Cerro, M. (1994) Growth Factors 10 289-297 [DOI] [PubMed] [Google Scholar]
- 18.Steele, F. R., Chader, G. J., Johnson, L. V., and Tombran-Tink, J. (1993) Proc. Natl. Acad. Sci. U. S. A 90 1526-1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Filleur, S., Volz, K., Nelius, T., Mirochnik, Y., Huang, H., Zaichuk, T. A., Aymerich, M. S., Becerra, S. P., Yap, R., Veliceasa, D., Shroff, E. H., and Volpert, O. V. (2005) Cancer Res. 65 5144-5152 [DOI] [PubMed] [Google Scholar]
- 20.Fernandez-Garcia, N. I., Volpert, O. V., and Jimenez, B. (2007) J. Mol. Med. 85 15-22 [DOI] [PubMed] [Google Scholar]
- 21.Alberdi, E., Hyde, C. C., and Becerra, S. P. (1998) Biochemistry 37 10643-10652 [DOI] [PubMed] [Google Scholar]
- 22.Kozaki, K., Miyaishi, O., Koiwai, O., Yasui, Y., Kashiwai, A., Nishikawa, Y., Shimizu, S., and Saga, S. (1998) J. Biol. Chem. 273 15125-15130 [DOI] [PubMed] [Google Scholar]
- 23.Meyer, C., Notari, L., and Becerra, S. P. (2002) J. Biol. Chem. 277 45400-45407 [DOI] [PubMed] [Google Scholar]
- 24.Stratikos, E., Alberdi, E., Gettins, P. G., and Becerra, S. P. (1996) Protein Sci. 5 2575-2582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yasui, N., Mori, T., Morito, D., Matsushita, O., Kourai, H., Nagata, K., and Koide, T. (2003) Biochemistry 42 3160-3167 [DOI] [PubMed] [Google Scholar]
- 26.Hosomichi, J., Yasui, N., Koide, T., Soma, K., and Morita, I. (2005) Biochem. Biophys. Res. Commun. 335 756-761 [DOI] [PubMed] [Google Scholar]
- 27.Alberdi, E. M., Weldon, J. E., and Becerra, S. P. (2003) BMC Biochem. 4 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Valnickova, Z., Petersen, S. V., Nielsen, S. B., Otzen, D. E., and Enghild, J. J. (2007) J. Biol. Chem. 282 6661-6667 [DOI] [PubMed] [Google Scholar]
- 29.Hollyfield, J. G. (1999) Invest. Ophthalmol. Vis. Sci. 40 2767-2769 [PubMed] [Google Scholar]
- 30.Aymerich, M. S., Alberdi, E. M., Martinez, A., and Becerra, S. P. (2001) Invest. Ophthalmol. Vis. Sci. 42 3287-3293 [PubMed] [Google Scholar]
- 31.Calabro, A., Benavides, M., Tammi, M., Hascall, V. C., and Midura, R. J. (2000) Glycobiology 10 273-281 [DOI] [PubMed] [Google Scholar]
- 32.Calabro, A., Hascall, V. C., and Midura, R. J. (2000) Glycobiology 10 283-293 [DOI] [PubMed] [Google Scholar]
- 33.deS Senanayake, P., Calabro, A., Nishiyama, K., Hu, J. G., Bok, D., and Hollyfield, J. G. (2001) J. Cell Sci. 114 199-205 [DOI] [PubMed] [Google Scholar]
- 34.Goodstone, N. J., Hascall, V. C., and Calabro, A. (1998) Arch. Biochem. Biophys. 350 26-35 [DOI] [PubMed] [Google Scholar]
- 35.Lee, T. H., Wisniewski, H. G., and Vilcek, J. (1992) J. Cell Biol. 116 545-557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perez-Mediavilla, L. A., Chew, C., Campochiaro, P. A., Nickells, R. W., Notario, V., Zack, D. J., and Becerra, S. P. (1998) Biochim. Biophys. Acta 1398 203-214 [DOI] [PubMed] [Google Scholar]
- 37.Wigler, M., Silverstein, S., Lee, L. S., Pellicer, A., Cheng, Y., and Axel, R. (1977) Cell 11 223-232 [DOI] [PubMed] [Google Scholar]
- 38.Acharya, S., Rodriguez, I. R., Moreira, E. F., Midura, R. J., Misono, K., Todres, E., and Hollyfield, J. G. (1998) J. Biol. Chem. 273 31599-31606 [DOI] [PubMed] [Google Scholar]
- 39.Simonovic, M., Gettins, P. G., and Volz, K. (2001) Proc. Natl. Acad. Sci. U. S. A 98 11131-11135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alberdi, E., Aymerich, M. S., and Becerra, S. P. (1999) J. Biol. Chem. 274 31605-31612 [DOI] [PubMed] [Google Scholar]
- 41.Huber, R., and Carrell, R. W. (1989) Biochemistry 28 8951-8966 [DOI] [PubMed] [Google Scholar]
- 42.Knudson, W. (1996) Am. J. Pathol. 148 1721-1726 [PMC free article] [PubMed] [Google Scholar]
- 43.Knudson, W., Biswas, C., Li, X. Q., Nemec, R. E., and Toole, B. P. (1989) Ciba Found. Symp. 143 150-159; discussion 159-169, 281-285 [DOI] [PubMed] [Google Scholar]
- 44.Normand, G., Hicks, D., and Dreyfus, H. (1998) Glycobiology 8 1227-1235 [DOI] [PubMed] [Google Scholar]
- 45.Hollyfield, J. G., Rayborn, M. E., and Tammi, R. (1997) Exp. Eye Res. 65 603-608 [DOI] [PubMed] [Google Scholar]
- 46.Fjeldstad, K., and Kolset, S. O. (2005) Curr. Drug Targets 6 665-682 [DOI] [PubMed] [Google Scholar]
- 47.Gotte, M., and Yip, G. W. (2006) Cancer Res. 66 10233-10237 [DOI] [PubMed] [Google Scholar]
- 48.Guo, Y. J., Liu, G., Wang, X., Jin, D., Wu, M., Ma, J., and Sy, M. S. (1994) Cancer Res. 54 422-426 [PubMed] [Google Scholar]
- 49.Sy, M. S., Guo, Y. J., and Stamenkovic, I. (1992) J. Exp. Med. 176 623-627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mohapatra, S., Yang, X., Wright, J. A., Turley, E. A., and Greenberg, A. H. (1996) J. Exp. Med. 183 1663-1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen, Q., Cai, S., Shadrach, K. G., Prestwich, G. D., and Hollyfield, J. G. (2004) J. Biol. Chem. 279 23142-23150 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.