Abstract
Proteasomes mediate the regulated degradation of Insig-1, a membrane protein of the endoplasmic reticulum (ER) that plays a crucial role in lipid metabolism. We showed previously that sterols inhibit this degradation by blocking ubiquitination of Insig-1. Here we show that unsaturated fatty acids stabilize Insig-1 without affecting its ubiquitination. Instead unsaturated fatty acids inhibit extraction of ubiquitinated Insig-1 from membranes, a process known to be mediated by valosin-containing protein and necessary for ER-associated degradation. Valosin-containing protein is recruited to Insig-1 through the action of another protein, Ubxd8. Unsaturated fatty acids block the binding between Ubxd8 and Insig-1, thereby abrogating the membrane extraction of Insig-1. Unsaturated fatty acid-mediated stabilization of Insig-1 enhances the ability of sterols to inhibit proteolytic activation of SREBP-1, which activates transcription of genes involved in fatty acid synthesis. The current study provides a molecular mechanism for regulation of proteasome-mediated ER protein degradation at a postubiquitination step.
The ubiquitin-proteasome system is the major pathway by which cells degrade intracellular proteins (1). Most proteins destined to be degraded by proteasomes are first modified by covalent attachment of polyubiquitin chains. Ubiquitination is mediated by a series of enzymes that include a ubiquitin activation enzyme (E1),3 ubiquitin-conjugating enzymes (E2s), and ubiquitin ligases (E3s), the last of which determine substrate specificity (1). To date, interactions between protein substrates and E3s have been considered to be the step primarily regulated by signals that control degradation of substrate proteins (2). The mechanism by which ubiquitinated proteins are delivered to the proteasome is less well characterized. In particular, it remains unclear whether proteasomal degradation can be regulated at a postubiquitination step.
The ubiquitin-proteasome system degrades membrane proteins of the endoplasmic reticulum (ER) through a process termed ER-associated degradation (ERAD) (3). In addition to the ubiquitin system and proteasomes, ERAD requires valosin-containing protein (VCP) (3, 4), which extracts ubiquitinated proteins from the ER membrane to make them accessible to proteasomes (5, 6). There is no published evidence to indicate that ERAD can be regulated through control of interaction between VCP and ubiquitinated protein substrates.
In mammalian cells, ERAD of several proteins can be regulated by small molecule metabolites. One such protein is Insig-1, an ER membrane protein that plays a crucial role in feedback regulation of cholesterol synthesis (7). When sterols build up in cells Insig-1 reduces cholesterol synthesis by binding to Scap, a polytopic membrane protein (7). Binding to Scap prevents the activation of SREBPs, membrane-bound transcription factors that enhance transcription of genes required for cholesterol synthesis and uptake (8, 9). Scap is an escort protein that transports SREBPs from ER to Golgi where the SREBPs are cleaved to release NH2-terminal fragments from the membrane, allowing them to enter the nucleus to activate their target genes. When Insig-1 binds to Scap, the Scap-SREBP complex is trapped in the ER, and transcription of SREBP target genes declines, leading to a reduction in cholesterol synthesis and uptake (10).
We have shown previously that Insig-1 is degraded by proteasomes in a process that is regulated by sterols. In sterol-depleted cells Insig-1 protein becomes ubiquitinated by gp78, an E3 ubiquitin ligase that binds to Insig-1 (11). Ubiquitinated Insig-1 is rapidly degraded by proteasomes (12). When cells are presented with sterols, Scap binds to Insig-1, which causes gp78 to dissociate from Insig-1. As a result, ubiquitination of Insig-1 is blocked and Insig-1 is stabilized (11, 12). Mammalian cells also contain a close relative of Insig-1, namely Insig-2 (13). In comparison with Insig-1, Insig-2 has a much longer half-life, and its degradation is not subject to sterol regulation (14, 15). In cultured fibroblasts, Insig-1 is by far the predominant Insig isoform (16).
In addition to sterols, unsaturated fatty acids have been shown to decrease the proteolytic processing of one isoform of SREBP, namely SREBP-1 (17). The mechanism of this effect is unknown. In the current study, we demonstrate that unsaturated fatty acids inhibit SREBP cleavage by increasing the amount of Insig-1 protein. This increase is attributable to an inhibition in the degradation of Insig-1. Surprisingly this inhibition occurs at a postubiquitination step. Indeed in the presence of unsaturated fatty acids Insig-1 is ubiquitinated but not extracted from the ER membrane, and therefore it is not degraded. We show that the block in the membrane extraction is attributable to the disruption of the complex between Insig-1 and VCP. The complex is disrupted because unsaturated fatty acids trigger the dissociation of Insig-1 from UBX domain-containing protein 8 (Ubxd8), a protein that mediates complex formation between Insig-1 and VCP. These data provide insight into a mechanism by which metabolites stabilize an ER membrane protein by blocking degradation at a postubiquitination step.
EXPERIMENTAL PROCEDURES
Materials—We obtained all fatty acids, cycloheximide, triacsin C, polyclonal anti-actin, monoclonal anti-FLAG IgG, and monoclonal anti-hemagglutinin (HA) IgG from Sigma; MG132 and Nonidet P-40 alternative (Nonidet P-40) from Calbiochem; 25-hydroxycholesterol from Steraloids, Inc.; polyclonal anti-Myc, anti-HA, and anti-T7 IgG from Bethyl Laboratories; monoclonal anti-T7 from Novagen; monoclonal anti-CFTR IgG from Upstate Biotechnology; monoclonal anti-transferrin receptor IgG from Zymed Laboratories Inc.; polyclonal anti-Ubxd8 from Novus Biologicals; monoclonal anti-VCP from BD Transduction Laboratories; horseradish peroxidase-conjugated donkey anti-mouse and anti-rabbit IgGs (affinity-purified) from Jackson ImmunoResearch Laboratories; and Mouse TrueBlot™ Ultra from eBioscience. Hybridoma cells producing IgG-9E10, a mouse monoclonal antibody against Myc tag, were obtained from the American Type Culture Collection (Manassas, VA). IgG-2179, a polyclonal antibody against SREBP-1 (18), a polyclonal anti-gp78 (19), and a polyclonal anti-Insig-1 IgG (12) were described in the indicated references. Delipidated fetal calf serum (FCS) was prepared from newborn calf serum by n-butyl alcohol and isopropyl ether extraction method (17). Solutions of sodium mevalonate (20) and sodium compactin (20) were prepared as described previously. All fatty acids added into culture media were conjugated to bovine serum albumin (17). The LXR agonist T0901317 was provided kindly by Bei Shan of Tularik, Inc.
Plasmid Constructs—The following plasmids were described in the indicated reference or obtained from the indicated sources: pTK-Insig1-Myc (12) and pTK-Insig2-Myc (15) encoding human Insig-1 and human Insig-2, respectively, followed by six tandem copies of a c-Myc epitope tag (EQKLISEEDL) under control of the thymidine kinase (TK) promoter; pCMV-Insig1-T7 encoding full-length versions of wild-type human Insig-1 followed by three tandem copies of a T7 epitope tag under control of the cytomegalovirus (CMV) promoter (12); pCMV-Flag-Insig1 encoding human Insig-1 with two tandem copies of a FLAG epitope tag (DYKDDDDK) inserted between amino acids 61 and 62 (21); pEF1a-HA-ubiquitin (provided by Dr. Zhijian Chen, University of Texas Southwestern Medical Center) encoding amino acids 1-76 of human ubiquitin preceded by an epitope tag derived from the influenza HA protein (YPYDVPDY) under control of the EF1a promoter; pCIneo-gp78 (obtained from Dr. Allan M. Weissman, National Cancer Center) encoding human gp78 under control of the CMV promoter; pCMV-gp78-Myc encoding human gp78 followed by five tandem copies of a Myc epitope tag under control of the CMV promoter (19); and pCMV-CFTR(ΔF508) (provided by Dr. Philip J. Thomas, University of Texas Southwestern Medical Center) encoding a mutant cystic fibrosis transmembrane conductance regulator that contains a deletion of phenylalanine at position 508 under control of the CMV promoter. pCMV-Insig1-T7-pA and pCMV-Insig2-T7-pA encode human Insig-1 and Insig-2, respectively, followed sequentially by three tandem copies of a T7 epitope tag, a tobacco etch virus protease cleavage site (ENLYFQG), and a protein A tag (NCBI accession number CAA65431) under control of the CMV promoter. Expressed sequence tag clones containing full-length human Ubxd8 (clone number 3942891), Faf-1 (clone number 6379855), and Ubxd2 (clone number 2813866) cDNA were obtained from Invitrogen. pCMV-Myc-Ubxd8, pCMV-Myc-Faf1, and pCMV-Myc-Ubxd2 encode full-length human Ubxd8, Faf-1, and Ubxd2, respectively, preceded by five tandem copies of a Myc epitope tag under control of the CMV promoter. They were generated by ligating AgeI-cleaved vector pcDNA3.1-(Myc)5 (22) with PCR fragments containing full-length Ubxd8, Fas-associated factor 1 (FAF1), or Ubxd2 amplified by primers containing AgeI overhangs using the expressed sequence tag clones as templates.
Cell Culture—CHO-7 cells are a clone of CHO-K1 cells selected for growth in lipoprotein-deficient serum (23). Monolayers of CHO-7 cells were grown in medium A (a 1:l mixture of Ham's F-12 medium and Dulbecco's modified Eagle's medium, 100 units/ml penicillin, and 100 μg/ml streptomycin) supplemented with 5% lipoprotein-deficient serum. SRD-13A cells are a clone of mutant CHO cells deficient in Scap (24). Monolayers of SRD-13A cells were grown in medium A supplemented with 5% (v/v) fetal calf serum, 5 μg/ml cholesterol, 1 mm sodium mevalonate, and 20 μm sodium oleate. Both CHO-7 and SRD-13A cells were maintained at 37 °C in 8% CO2. HEK-293S/pInsig1 and HEK-293S/pInsig2 cells are lines of HEK-293S cells (25) that stably express pCMV-Insig1-T7-pA and pCMV-Insig2-T7-pA, respectively. They were generated by transfecting HEK-293S cells with pCMV-Insig1-T7-pA or pCMV-Insig2-T7-pA followed by selection with 700 μg/ml G418. Monolayers of HEK-293S/pInsig1 and HEK-293S/pInsig2 cells were grown in 5% CO2 at 37 °C in medium B (Dulbecco's modified Eagle's medium (low glucose) containing 100 units/ml penicillin and 100 μg/ml streptomycin sulfate) supplemented with 10% (v/v) FCS and 400 μg/ml G418.
RNA Interference—Duplexes of small interfering RNA (siRNA) were synthesized by Dharmacon Research (Lafayette, CO). The two siRNA sequences targeting human Ubxd8 (NCBI accession number AB088120) are at nucleotide positions (relative to the codon for the initiating methionine) 831-849 and 915-933 for Ubxd8-1 and Ubxd8-2, respectively. The two siRNA sequences targeting human VCP (NCBI accession number NM_007126) are at nucleotides 464-482 and 1307-1335 for VCP-1 and VCP-2, respectively. The sequence for the siRNA targeting green fluorescent protein was reported previously (26). HEK-293S/pInsig1 cells cultured in a 60-mm dish were transfected with 400 pmol of siRNA duplexes using Lipofectamine™ RNAiMAX reagent (Invitrogen) as described by the manufacturer after which the cells were used for experiments as described in the figure legends.
Transient Transfection and Immunoblot Analysis—SRD-13A cells were transiently transfected with FuGENE 6 reagent (Roche Applied Science) according to the manufacturer's protocol. Conditions of incubation after transfection are described in the figure legends. After incubation, duplicate dishes of cells were pooled, harvested, and lysed in 0.1 ml of buffer A (25 mm Tris-HCl at pH 7.2, 0.15 m NaCl, 1% (v/v) Nonidet P-40, and a protease inhibitor mixture including 10 μg/ml leupeptin, 5 μg/ml pepstatin A, 10 μg/ml aprotinin, and 25 μg/ml N-acetylleucinal-leucinal-norleucinal). Aliquots of the lysate were subjected to SDS-PAGE and immunoblot analysis. Antibodies used in the current studies were IgG-9E10 (1 μg/ml), a polyclonal anti-Myc (0.2 μg/ml), a polyclonal anti-T7 (0.2 μg/ml), a monoclonal anti-T7 (1 μg/ml), a monoclonal anti-HA (1:1000 dilution), a polyclonal anti-gp78 (1:10,000 dilution), a monoclonal anti-CFTR (1:1000 dilution), a monoclonal anti-FLAG (2 μg/ml), and a monoclonal anti-transferrin receptor (1 μg/ml). Except for anti-T7 immunoblots shown in Figs. 3 and 5 in which we used Mouse TrueBlot Ultra (1:2000 dilution) as the secondary antibody, horseradish peroxidase-conjugated donkey anti-mouse and anti-rabbit IgGs (0.2 μg/ml) were used as the secondary antibody in all other immunoblot analysis. Bound antibodies were visualized by chemiluminescence using the SuperSignal substrate system (Pierce) according to the manufacturer's instructions. Filters were exposed to Kodak X-Omat Blue XB-1 films at room temperature. Densitometric quantification of the immunoblot was carried out as described previously (26).
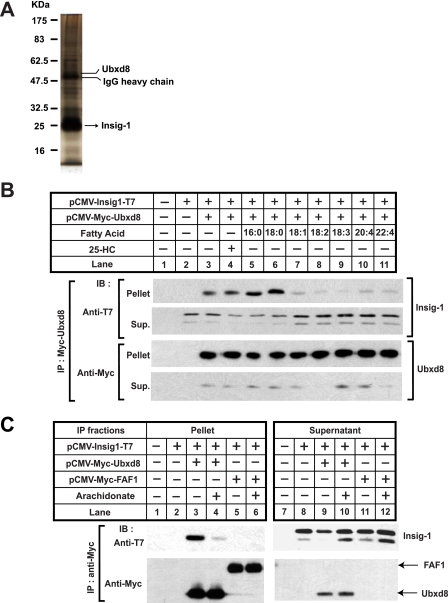
FIGURE 3.
Unsaturated fatty acids disrupt the binding between Insig-1 and Ubxd8. A, Insig-1 was immunoprecipitated from lysates of HEK-293S/pInsig1 cells and eluted from the beads as described under “Experimental Procedures.” The eluted sample was subjected to SDS-PAGE followed by silver staining. Bands corresponding to Insig-1 and Ubxd8 as determined by tandem mass spectroscopy are indicated. B and C, SRD-13A cells were seeded, transfected with 0.2 μg of the indicated plasmids, and incubated as described in Fig. 1B for the first 2 days. B, on day 3, cells were treated with a 50 μm concentration of the indicated fatty acids or 1 μg/ml 25-hydroxycholesterol (25-HC) in medium supplemented with 5% delipidated FCS and 10 μm MG132 for 6 h. Detergent lysates of these cells were subjected to immunoprecipitation with anti-Myc to precipitate transfected Ubxd8. Pellets (representing a 0.1 dish of cells) and supernatants (representing a 0.01 dish of cells) of the immunoprecipitation were subjected to SDS-PAGE followed by immunoblot analysis with polyclonal anti-Myc (against Ubxd8) and anti-T7 (against Insig-1). C, on day 3, cells were treated with 100 μm arachidonate in medium supplemented with 5% delipidated FCS and 10 μm MG132 for 6 h. Transfected Ubxd8 and FAF1 were immunoprecipitated, and pellets and supernatants (Sup.) of the immunoprecipitation (IP) were analyzed by immunoblot (IB) analysis as described in B. It appears that only the top band of Insig-1 was co-immunoprecipitated with Ubxd8 in this experiment. However, this observation was not reproduced in other similar experiments.
FIGURE 5.
Ubxd8 mediates the complex formation between VCP and Insig-1. A, HEK-293S/pInsig1 cells were seeded, transfected with indicated siRNA, and treated as described in Fig. 4B. Cell lysate was subjected to SDS-PAGE followed by immunoblot analysis with anti-VCP and anti-T7 (against Insig-1). B, SRD-13A cells were seeded, transfected, and treated with arachidonate as described in Fig. 3C. Detergent lysates of these cells were subjected to immunoprecipitation with anti-Myc IgG-coupled agarose beads to immunoprecipitate transfected Ubxd8. Pellets (representing a 0.1 dish of cells) and supernatants (representing a 0.01 dish of cells) of the immunoprecipitates were subjected to SDS-PAGE followed by immunoblot (IB) analysis with polyclonal anti-Myc (against Ubxd8), anti-T7 (against Insig-1), and anti-VCP. C, SRD-13A cells were seeded, transfected with 0.2 μg of the indicated plasmids, and treated with arachidonate as described in Fig. 3C. Detergent lysates of these cells were subjected to immunoprecipitation (IP) with anti-FLAG IgG-coupled agarose beads to immunoprecipitate transfected Insig-1. Pellets and supernatants of the immunoprecipitates were analyzed as described in B. D, HEK-293S/pInsig1 cells were seeded at 6.0 × 105/100-mm dish on day 0. On day 2, cells were switched to medium containing 10% delipidated FCS. On day 3, 10 μm MG132 was added into medium in the absence or presence of 100 μm arachidonate. After incubation for 6 h, cells were harvested, and the cellular lysate was subjected to immunoprecipitation with anti-T7 to precipitate Insig-1. Pellets (representing one dish of cells) and supernatants (representing a 0.067 dish of cells) of the immunoprecipitates were subjected to SDS-PAGE followed by immunoblot analysis with anti-VCP, anti-Ubxd8, and anti-T7 (against Insig-1). GFP, green fluorescent protein.
Immunoprecipitation and Detection of Ubiquitinated Insig-1—The pooled cell pellets from duplicate 60-mm dishes of cells were lysed and immunoprecipitated as described previously (27). Immunoprecipitation of Insig-1 followed by detection of Insig-1 ubiquitination was carried out as described previously (11). Briefly cells were directly lysed in buffer supplemented with 8 m urea. The lysate was then diluted with buffer to reduce the urea concentration to 2 m and subjected sequentially to immunoprecipitation and immunoblot analysis to determine the ubiquitination of Insig-1.
Cell Fractionation—The pooled cell pellets were fractionated into membranes, nuclear extracts, and cytosol fractions exactly as described previously (28).
Purification of Insig-1-associated Protein—HEK-293S/pInsig1 cells were seeded at a density of 2.0 ×105/ml in a round flask for suspension culture on day 0. On day 7, cells collected from 1 liter of the culture were lysed with 30 ml of buffer B (25 mm Tris-HCl at pH 7.2, 0.15 m NaCl, 10 μg/ml leupeptin, 5 μg/ml pepstatin A, 10 μg/ml aprotinin, and 25 μg/ml N-acetylleucinal-leucinal-norleucinal) supplemented with 0.1% Nonidet P-40. The lysates were then incubated with IgG-coupled agarose beads (Sigma) for 16 h at 4 °C to immunoprecipitate Insig-1 fused with a protein A tag, which binds to IgG. After washing the immunoprecipitates five times (for 10 min each) with buffer B at 4 °C, 70 units of tobacco etch virus protease (Invitrogen) were added to the immunoprecipitates for 5 h at 25 °C to release Insig-1 and Insig-1-associated proteins from the beads. The eluted materials were subjected to SDS-PAGE followed by silver staining. The identity of each band on the gel was determined by tandem mass spectrometry (Protein Chemistry Core Facility, University of Texas Southwestern Medical Center).
Triglyceride Synthesis Analysis—The amount of radiolabeled oleate incorporated into triglyceride was determined exactly as described previously (29).
RESULTS
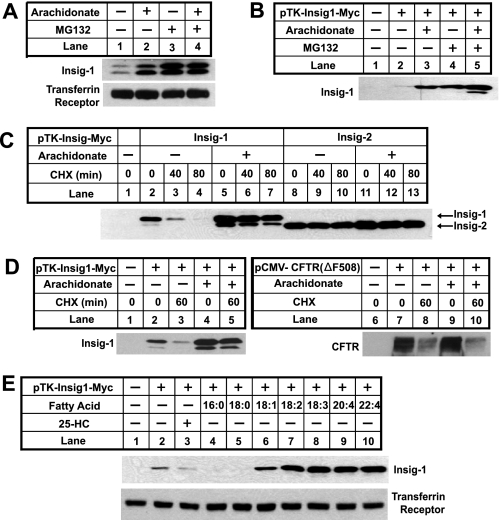
To examine the effect of unsaturated fatty acids on Insig-1, we incubated CHO-7 cells in medium supplemented with delipidated FCS, which lacks all fatty acids, in the absence or presence of exogenously added arachidonate, an unsaturated fatty acid, or MG132, a proteasome inhibitor. To avoid sterol-mediated stabilization of Insig-1, these cells were also treated with compactin, an inhibitor of hydroxymethylglutaryl-CoA reductase that blocks synthesis of cholesterol. The amount of endogenous Insig-1 was then determined by immunoblot analysis. As shown in Fig. 1A, arachidonate significantly increased the amount of Insig-1 protein (Fig. 1A, lanes 1 and 2). As observed previously, Insig-1 showed two bands that represent translational products initiated with different methionines (10). MG132 also raised the amount of Insig-1 (Fig. 1A, lane 3). Addition of arachidonate on top of MG132 did not further enhance the amount of Insig-1 protein (Fig. 1A, lane 4). The increase of Insig-1 protein upon treatment with arachidonate occurred in the absence of a change in Insig-1 mRNA as determined by quantitative real time PCR analysis (data not shown).
FIGURE 1.
Unsaturated fatty acids inhibit degradation of Insig-1. A, CHO-7 cells were seeded at 3.5 × 105/60-mm dish on day 0. On day 2, cells were switched to medium A supplemented with 5% delipidated FCS with 50 μm compactin and 50 μm mevalonate. On day 3, 100 μm arachidonate or 10 μm MG132 was added into the medium as indicated. After incubation for 6 h, cells were harvested. Cell lysate was subjected to SDS-PAGE followed by immunoblot analysis with anti-Insig and anti-transferrin receptor. B, SRD-13A cells were seeded as described in A. On day 2, cells were transfected with 0.4 μg of pTK-Insig1-myc. Total plasmid concentration was adjusted to 2 μg/dish by using the empty vector pcDNA3.1. Following incubation for 8 h, cells were switched to medium A supplemented with 5% delipidated FCS. On day 3, cells were treated as described in A, and transfected Insig-1 was detected by immunoblot analysis with IgG-9E10. C, SRD-13A cells were seeded, transfected with 0.4 μg of pTK-Insig1-myc or pTK-Insig2-myc, and incubated as described in B for the first 2 days. On day 3, cells were incubated in the absence or presence of 100 μm arachidonate in medium supplemented with 5% delipidated FCS for 6 h. These cells were then treated with 50 μm cycloheximide (CHX) for the indicated period of time. Cell lysate was subjected to SDS-PAGE followed by immunoblot analysis with IgG-9E10 to detect transfected Insig proteins. D, SRD-13A cells were seeded, transfected with 0.4 μg of pTK-Insig1-myc or 1.5 μg of pCMV-CFTR(ΔF508), and treated with arachidonate and cycloheximide (CHX) as described in C. Cell lysate was subjected to SDS-PAGE followed by immunoblot analysis with IgG-9E10 (against Insig-1) and anti-CFTR. E, SRD-13A cells were seeded, transfected with 0.4 μg of pTK-Insig1-myc, and incubated as described in B for the first 2 days. On day 3, cells were treated with a 50 μm concentration of the indicated fatty acids or 1 μg/ml 25-hydroxycholesterol (25-HC) in medium supplemented with 5% delipidated FCS for 6 h. Cells were then harvested, and cell lysate was subjected to SDS-PAGE followed by immunoblot analysis with IgG-9E10 (against Insig-1) and anti-transferrin receptor.
Scap is required for sterol-mediated stabilization of Insig-1 (12). To test whether Scap is required for the elevation of Insig-1 protein by arachidonate, we transfected a plasmid encoding human Insig-1 into SRD-13A cells, a mutant line of CHO cells that lack Scap (24). In these cells sterols fail to stabilize Insig-1 (12). As shown in Fig. 1B, arachidonate was as efficient as MG132 in raising the amount of Insig-1 in SRD-13A cells, and the effect was not additive. Thus, Scap is not required for Insig-1 stabilization by arachidonate.
To show more directly that arachidonate inhibits Insig-1 degradation, SRD-13A cells were incubated in the presence or absence of arachidonate and treated with cycloheximide to block the synthesis of Insig-1 (Fig. 1C). In cells that did not receive arachidonate, Insig-1 declined by more than 50% after 40 min and became barely detectable after 80 min of cycloheximide treatment (Fig. 1C, lanes 2-4). This disappearance was slowed in cells treated with arachidonate. Indeed the amount of Insig-1 barely changed after 80 min of cycloheximide treatment (Fig. 1C, lanes 5-7). In previous studies, we found that the time course of Insig-1 disappearance following cycloheximide treatment reflects its rate of degradation determined by pulse-chase analysis (12). Thus, these results indicate that arachidonate retards the degradation of Insig-1. As a control, we also examined the effect of arachidonate on Insig-2 degradation. As expected, the amount of Insig-2 protein did not change after 80 min of cycloheximide treatment regardless of incubation with arachidonate (Fig. 1C, lanes 8-13). In contrast to the results of Insig-1 (Fig. 1D, lanes 2-5), degradation of a mutant version of the cystic fibrosis transmembrane conductance regulator protein, CFTR(ΔF508), a well known substrate for ERAD (30), was not altered by treatment with arachidonate (Fig. 1D, lanes 7-10). This result indicates that ERAD in general is not affected by arachidonate.
Previous studies showed that unsaturated fatty acids including arachidonate strongly inhibit cleavage of SREBP-1, whereas saturated fatty acids have no effect (17). To examine whether these changes coincide with stabilization of Insig-1 protein, SRD-13A cells were transfected with a plasmid encoding Insig-1 and treated with sterols or fatty acids (Fig. 1E). Treatment of cells with several unsaturated fatty acids including oleate (18:1), linoleate (18:2), linolenate (18:3), arachidonate (20:4), and docosatetraenoate (22:4) increased the amount of Insig-1 protein (Fig. 1E, lanes 6-10). Palmitate (16:0) and stearate (18:0), two saturated fatty acids, failed to raise the amount of Insig-1 protein (Fig. 1E, lanes 4 and 5). We also added 25-hydroxycholesterol in the same experiment. As expected, 25-hydroxycholesterol did not raise the amount of Insig-1 protein in SRD-13A cells because sterol-mediated stabilization of Insig-1 requires Scap (Fig. 1E, lane 3) (12).
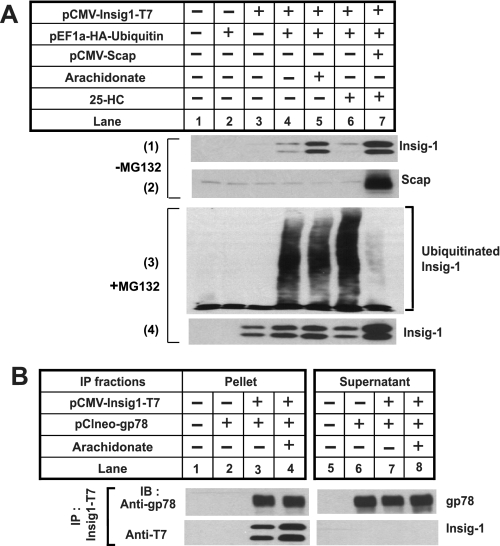
In previous studies we showed that in cells expressing Scap sterols stabilize Insig-1 by inhibiting its ubiquitination (12). To examine whether arachidonate also blocks ubiquitination of Insig-1, SRD-13A cells were transfected with a plasmid encoding Insig-1 and a plasmid encoding ubiquitin tagged with an epitope derived from the influenza HA protein with or without the cotransfection of a plasmid encoding Scap. The cells were incubated in the absence or presence of arachidonate or 25-hydroxycholesterol. Consistent with the results shown in Fig. 1, arachidonate stabilized Insig-1 in the absence of Scap (Fig. 2A, panel 1, lane 5). As reported previously (12), 25-hydroxycholesterol did not stabilize Insig-1 in the absence of Scap (Fig. 2A, panel 1, lane 6) but increased the amount of Insig-1 when Scap was cotransfected (Fig. 2A, panel 1, lane 7). Parallel dishes of cells were treated with MG132 to inhibit the degradation of ubiquitinated Insig-1. Following immunoprecipitation with transfected Insig-1, Insig-1 and ubiquitin in the immunoprecipitates were analyzed by immunoblot analysis (Fig. 2A, panels 3 and 4). Ubiquitinated Insig-1, which was visualized as high molecular weight smears in the anti-HA immunoblot, was observed in cells deprived of sterols and fatty acids (Fig. 2A, panel 3, lane 4). As reported previously, treatment with 25-hydroxycholesterol inhibited ubiquitination of Insig-1 but only when Scap was cotransfected (Fig. 2A, panel 3, lanes 6 and 7). Surprisingly arachidonate did not block the ubiquitination of Insig-1 (Fig. 2A, panel 3, lane 5). The total amount of Insig-1 in immunoprecipitates from these MG132-treated cells was not significantly affected by the addition of arachidonate or 25-hydroxycholesterol (Fig. 2A, panel 4). We did not observe the appearance of a smear of ubiquitinated Insig-1 in the immunoblot detecting Insig-1. This likely results from rapid deubiquitination of Insig-1 because of the strong activity of deubiquitinating enzymes in the lysates (11, 31).
FIGURE 2.
Arachidonate does not affect ubiquitination of Insig-1. A, SRD-13A cells were seeded; transfected with 0.1 μg of pEF1a-HA-ubiquitin, 0.1 μg of pCMV-Insig1-T7, and 0.4 μg of pCMV-Scap; and incubated as described in Fig. 1B for the first 2 days. On day 3, 100 μm arachidonate or 1 μg/ml 25-hydroxycholesterol (25-HC) was added to the cells as indicated. Some dishes of cells were harvested 6 h later, and cell lysate was subjected to SDS-PAGE followed by immunoblot analysis with anti-T7 (against Insig-1) and anti-Scap (-MG132). Parallel dishes of cells were treated with 10 μm MG132 5 h after the incubation (+MG132). Following continued incubation for 2 h, cells were harvested, and the cell lysate was subjected immunoprecipitation with anti-T7 to precipitate transfected Insig-1. Aliquots of immunoprecipitates were subjected to SDS-PAGE followed by immunoblot analysis with anti-HA (against ubiquitin) and anti-T7 (against Insig-1). The slight increase in Insig-1 in lane 4 compared with lane 3 in panel 1 was not observed in other experiments. B, SRD-13A cells were seeded, transfected with 0.1 μg of pCIneo-gp78 and 0.2 μg of pCMV-Insig1-T7, and incubated as described in Fig. 1B for the first 2 days. On day 3, cells were treated with or without 100 μm arachidonate in medium supplemented with 5% delipidated FCS and 10 μm MG132 for 6 h. Cells were then harvested and subjected to immunoprecipitation (IP) with anti-T7 to precipitate transfected Insig-1. Pellets (representing a 0.25 dish of cells) and supernatants (representing a 0.05 dish of cells) of the immunoprecipitation were subjected to SDS-PAGE and immunoblot (IB) analysis with anti-gp78 and anti-T7 (against Insig-1).
We then examined whether arachidonate affects the association between Insig-1 and gp78, an E3 ubiquitin ligase for Insig-1 (11). For this purpose, we transfected SRD-13A cells with plasmids encoding Insig-1 and gp78. These cells were incubated in the absence or presence of arachidonate and treated with MG132 to equalize the amount of Insig-1, and the interaction between Insig-1 and gp78 was determined by co-immunoprecipitation analysis (Fig. 2B). The amount of gp78 co-immunoprecipitated with Insig-1 was not reduced in cells treated with arachidonate (Fig. 2B, lanes 3 and 4). This result is consistent with the observation that Insig-1 ubiquitination is not affected by arachidonate (Fig. 2A).
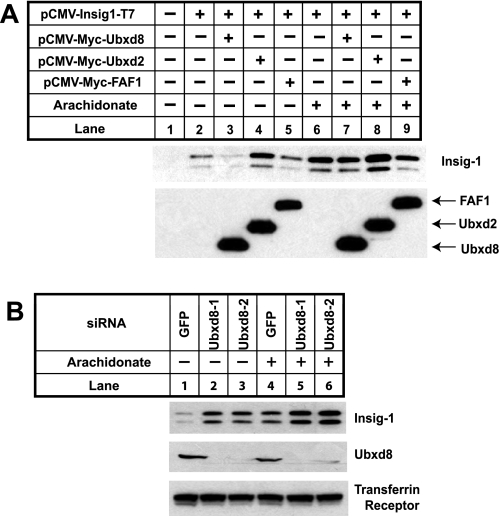
In Saccharomyces cerevisiae, proteasomal degradation of ubiquitinated ER proteins requires Ubx2, which is required to assemble the complex that brings together ubiquitinated proteins and VCP (32, 33). Ubx2 contains a UBX domain that binds to VCP (34). In mammalian cells, immunoprecipitation of Insig-1 brought down a protein called Ubxd8 (Fig. 3A), which shares a UBX domain with yeast Ubx2 (35). No published data address the function of Ubxd8. To determine whether Ubxd8 plays a role in unsaturated fatty acid-regulated proteasomal degradation of Insig-1, we first examined whether the binding between Insig-1 and Ubxd8 is regulated by unsaturated fatty acids. To this end, we transfected SRD-13A cells with plasmids encoding Insig-1 and Ubxd8. These cells were then cultured in the absence or presence of sterols or fatty acids and treated with MG132 to stabilize ubiquitinated Insig-1. The binding between Insig-1 and Ubxd8 was analyzed by co-immunoprecipitation analysis. As shown in Fig. 3B, Insig-1 was co-immunoprecipitated with Ubxd8 in control cells that did not receive any sterol or fatty acid (Fig. 3B, top panel, lane 3). The amount of Insig-1 co-immunoprecipitated with Ubxd8 was not reduced by addition of 25-hydroxycholesterol, palmitate, or stearate (Fig. 3B, top panel, lanes 4-6). However, unsaturated fatty acids including arachidonate markedly reduced the amount of Insig-1 co-immunoprecipitated with Ubxd8 (Fig. 3B, top panel, lanes 7-11).
In mammalian cells the closest relative of Ubxd8 is FAF1 (36), which also contains a UBX domain and is 27% identical to Ubxd8 (35). To test whether FAF1 also binds to Insig-1, we transfected SRD-13A cells with plasmids encoding Insig-1 together with those encoding Ubxd8 or FAF1 and immunoprecipitated Ubxd8 or FAF1. Immunoblot analysis revealed that Insig-1 was co-immunoprecipitated with Ubxd8 in the absence of arachidonate (Fig. 3C, top panel, lane 3). The amount of Insig-1 co-immunoprecipitated with Ubxd8 was diminished when cells were treated with arachidonate (Fig. 3C, top panel, lane 4). In contrast, Insig-1 was not co-immunoprecipitated with FAF1 (Fig. 3C, top panel, lanes 5 and 6).
We next examined whether increased expression of Ubxd8 would accelerate Insig-1 degradation. For this purpose, we transfected SRD-13A cells with plasmids encoding Insig-1, Ubxd8, or FAF1. Cotransfection of Ubxd8 significantly reduced the amount of Insig-1 in cells that were not treated with arachidonate (Fig. 4A, top panel, lanes 2 and 3). Cotransfection of FAF1 did not reduce the amount of Insig-1 (Fig. 4A, top panel, lane 5). Cotransfection of Ubxd2, another mammalian UBX domain-containing protein that is known to be involved in ERAD (37), also did not reduce the amount of Insig-1 (Fig. 4A, top panel, lane 4). Arachidonate prevented Ubxd8 from reducing the amount of Insig-1 (Fig. 4A, top panel, lanes 6 and 7) presumably because arachidonate prevents the association of Ubxd8 with Insig-1 (Fig. 3).
FIGURE 4.
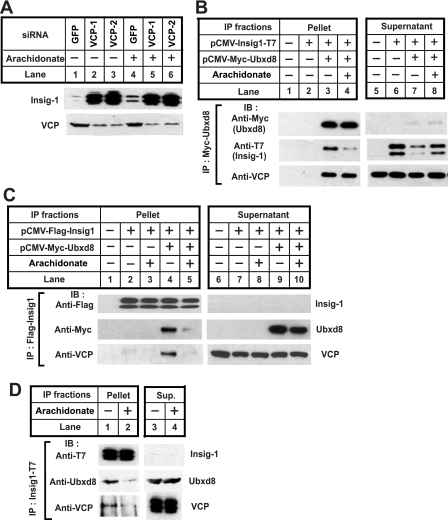
Ubxd8 is required for Insig-1 degradation in the absence of unsaturated fatty acids. A, SRD-13A cells were seeded, transfected with 0.2 μg of each indicated plasmid, and incubated as described in Fig. 1B for the first 2 days. On day 3, cells were incubated in the absence or presence of 100 μm arachidonate in medium supplemented with 5% delipidated FCS for 6 h. Cells were then harvested, and cell lysate was subjected to SDS-PAGE followed by immunoblot analysis with polyclonal anti-Myc (against Ubxd8, FAF1, and Ubxd2) and anti-T7 (against Insig-1). B, HEK-293S/pInsig1 cells were seeded at 2.0 × 105/60-mm dish on day 0. On day 2, cells were transfected with the indicated siRNA at 400 pmol/dish. On day 4, cells were switched to medium B supplemented with 10% delipidated FCS, 50 μm compactin, and 50 μm mevalonate. On day 5, 100 μm arachidonate was added into the medium as indicated. After incubation for 6 h, cells were harvested, and cell lysate was subjected to SDS-PAGE followed by immunoblot analysis with anti-Ubxd8, anti-T7 (against Insig-1), and anti-transferrin receptor. GFP, green fluorescent protein.
To determine whether Ubxd8 is required for Insig-1 degradation, we used RNA interference to reduce the amount of Ubxd8 expression. For this purpose, we studied HEK-293S/pInsig1 cells, a line of HEK-293-derived cells stably transfected with Insig-1. We transfected the cells with two different duplexes of siRNA targeting different regions of Ubxd8. As a control, we transfected HEK-293/pInsig1 cells with siRNA targeting green fluorescent protein, an mRNA not present in the cells. Inasmuch as these cells express Scap, they were treated with compactin to block cholesterol synthesis and prevent sterol-mediated stabilization of Insig-1. The two Ubxd8 siRNAs decreased expression of Ubxd8 as revealed by immunoblot analysis with anti-Ubxd8 (Fig. 4B, middle panel). Knockdown of Ubxd8 increased the amount of Insig-1 protein in cells that were not treated with arachidonate (Fig. 4B, upper panel, lanes 1-3). Such increase in arachidonate-treated cells was much less pronounced (Fig. 4B, upper panel, lanes 4-6).
Based on analogy with Ubx2 in yeast, Ubxd8 is likely to interact with VCP. Fig. 5A shows that Insig-1 accumulated in cells transfected with siRNAs targeting VCP, indicating that VCP is required for Insig-1 degradation. Knockdown of VCP was more potent than arachidonate in raising the amount of Insig-1 presumably because Insig-1 was stabilized for longer periods of time in cells transfected with the siRNA targeting VCP than those treated with arachidonate.
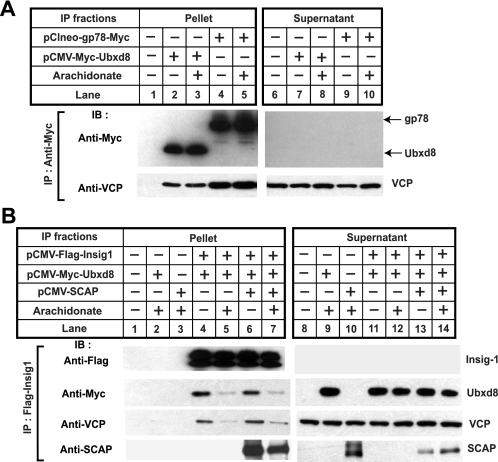
To examine the interaction between Ubxd8 and VCP, we transfected SRD-13A cells with plasmids encoding Insig-1 and Ubxd8. The cells were incubated in the absence or presence of arachidonate and treated with MG132 to prevent proteasomal degradation of Insig-1. Cells were harvested, and transfected Ubxd8 was immunoprecipitated. The amount of transfected Insig-1 and Ubxd8 and endogenous VCP in the immunoprecipitates was determined by immunoblot analysis (Fig. 5B). As expected, binding between Insig-1 and Ubxd8 was reduced by arachidonate (Fig. 5B, middle panel, lanes 3 and 4). VCP was also co-immunoprecipitated with Ubxd8, but this interaction was not inhibited by arachidonate (Fig. 5B, bottom panel, lanes 3 and 4).
To further investigate the interaction between Insig-1, Ubxd8, and VCP, we performed a reciprocal immunoprecipitation experiment similar to that described in Fig. 5B by precipitating Insig-1 instead of Ubxd8. As expected, Ubxd8 co-immunoprecipitated with Insig-1, and this interaction was disrupted by arachidonate (Fig. 5C, middle panel, lanes 4 and 5). In the absence of transfected Ubxd8, VCP was barely detectable in the immunoprecipitates (Fig. 5C, bottom panel, lanes 2 and 3). When Ubxd8 was cotransfected, an increased amount of VCP was found in the immunoprecipitates but only when cells were incubated in the absence of arachidonate (Fig. 5C, bottom panel, lanes 4 and 5). These results indicate that Ubxd8 mediates complex formation between Insig-1 and VCP, and arachidonate disrupts this complex by triggering the dissociation of Ubxd8 from Insig-1.
To avoid the massive overexpression inherent in transient transfection, we also examined the interaction between Insig-1, Ubxd8, and VCP in HEK-293S/pInsig1 cells by a similar co-immunoprecipitation approach. The amount of stably transfected Insig-1 mRNA in these cells is only 3 times as much as that of endogenous Insig-1 (data not shown). Inasmuch as the interaction between Insig-1 and VCP is indirect, the efficiency of the co-immunoprecipitation was expected to be low. Therefore, in this experiment we loaded 10 times more immunoprecipitated material as compared with the experiment shown in Fig. 5C. As shown in Fig. 5D, a small amount of Ubxd8 and VCP was co-immunoprecipitated with Insig-1 when cells were incubated in the absence of fatty acids (Fig. 5D, lane 1), and this interaction was inhibited by arachidonate (Fig. 5D, lane 2).
Previous studies have shown that gp78 also interacts with VCP (19, 38). This observation raises the question of why VCP is not recruited to Insig-1 via gp78, which remains associated with Insig-1 even in cells treated with arachidonate (Fig. 2B). One possibility is that the interaction between gp78 and VCP is also disrupted by arachidonate. To test this hypothesis we performed a co-immunoprecipitation experiment similar to that shown in Fig. 5B, and the result indicated that the binding of gp78 to VCP is not affected by arachidonate (Fig. 6A). This outcome suggests that Ubxd8 rather than gp78 plays a decisive role in recruiting VCP to Insig-1. This conclusion is further supported by an experiment similar to that shown in Fig. 5C, but samples from cells transfected with Scap and treated with sterols were also included (Fig. 6B). The result indicated that binding between Insig-1 and Ubxd8-VCP complex was not affected by the presence of Scap and sterols (Fig. 6B), a condition that prevents association of Insig-1 with gp78 (11). Thus, recruitment of VCP to Insig-1 appears to be gp78-independent.
FIGURE 6.
Recruitment of VCP to Insig-1 is gp78-independent. A, SRD-13A cells were seeded, transfected with 0.2 μg of pCMV-Myc-Ubxd8 and 0.05 μg of pCIneo-gp78-Myc, and incubated as described in Fig. 1B for the first 2 days. On day 3, cells were treated with or without 100 μm arachidonate in medium containing 5% delipidated FCS and 10 μm MG132 for 6 h. Detergent lysates of the cells were subjected to immunoprecipitation with anti-Myc to precipitate transfected Ubxd8 and gp78. Pellets (representing a 0.1 dish of cells) and supernatants (representing a 0.01 dish of cells) of the immunoprecipitation (IP) were subjected to SDS-PAGE followed by immunoblot (IB) analysis with anti-Myc (against Ubxd8 and gp78) and anti-VCP. B, SRD-13A cells were seeded; transfected with 0.2 μg of pCMV-Myc-Ubxd8, 0.2 μg of pCMV-Flag-Insig1, and 0.5 μg of pCMV-Scap; and incubated as described in Fig. 1B for the first 2 days. On day 3, cells were treated with or without 100 μm arachidonate in medium containing 5% delipidated FCS, 10 μm MG132, and 1 μg/ml 25-hydroxycholesterol for 6 h. Detergent lysates of the cells were subjected to immunoprecipitation with anti-FLAG IgG-coupled agarose beads. Pellets (representing a 0.1 dish of cells) and supernatants (representing a 0.01 dish of cells) of the immunoprecipitation were subjected to SDS-PAGE followed by immunoblot analysis with anti-Myc (against Ubxd8), anti-FLAG (against Insig-1), anti-Scap, and anti-VCP.
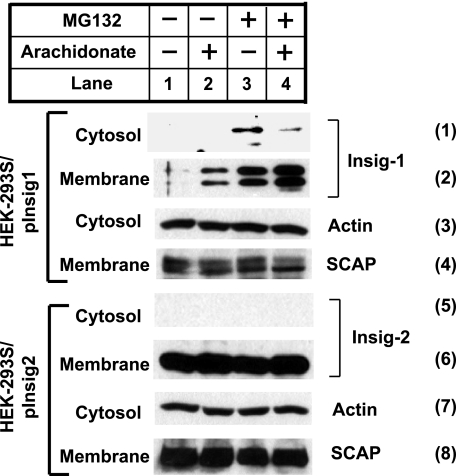
VCP is required to extract ubiquitinated proteins from the ER membranes so that these proteins can be delivered to proteasomes for degradation (5, 6). We thus examined the effect of arachidonate on the amount of Insig-1 that had been extracted into the cytosol. For this purpose, we incubated HEK-293S/pInsig1 cells in the absence or presence of arachidonate or MG132. The cells were harvested and separated into membrane and cytosol fractions. In cells that were not treated with MG132, Insig-1 was exclusively found in membrane fractions (Fig. 7, panels 1 and 2, lanes 1 and 2). In cells treated with MG132, Insig-1 was detected in the cytosol fraction. Arachidonate reduced the amount of cytosolic Insig-1 and increased the amount of Insig-1 in the membrane fraction (Fig. 7, panels 1 and 2, lanes 3 and 4). We also examined the subcellular localization of Insig-2, a protein that is not subject to rapid ERAD (15). For this purpose, we prepared the cytosol and membrane fractions from HEK-293S/pInsig2 cells, a line of HEK-293-derived cells stably transfected with T7-tagged Insig-2. In contrast to Insig-1, Insig-2 was not found in the cytosol fraction regardless of the treatment (Fig. 7, panels 5 and 6).
FIGURE 7.
Membrane extraction of Insig-1 is regulated by arachidonate. HEK-293S/pInsig1 and HEK-293S/pInsig2 cells were seeded at 5.0 × 105/60-mm dish on day 0. On day 2, the cells were switched to medium containing 10% delipidated FCS. On day 3, 100 μm arachidonate was added into medium as indicated. 6 h later, 10 μm MG132 was added to medium as indicated. After continued incubation for 2 h, cells were harvested and fractionated. Aliquots of membrane (representing a 0.04 dish of cells) and cytosol fraction (representing a 0.08 dish of cells) were subjected to SDS-PAGE followed by immunoblot analysis with anti-actin, anti-Scap, and anti-T7 (against stably transfected Insig-1 and Insig-2).
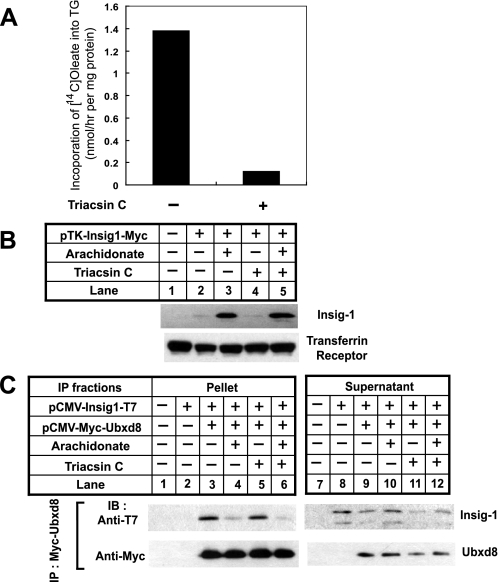
Results presented above provide a mechanism for unsaturated fatty acid-regulated ERAD of Insig-1. However, it remains unclear whether the effect is a direct response to free fatty acids or indirect through changing the lipid composition of the ER membrane. To address this question, we treated SRD-13A cells with triacsin C, an inhibitor of long chain acyl-coA synthetases (39) that catalyze the activation of fatty acids by conjugating them with coenzyme A, a reaction required for fatty acids to be incorporated into various lipids and proteins (40). The drug treatment was effective as it inhibited the incorporation of oleate into triglyceride (Fig. 8A). However, such treatment did not affect arachidonate-mediated stabilization of Insig-1 (Fig. 8B), nor did it have an effect on arachidonate-regulated interaction between Insig-1 and Ubxd8 (Fig. 8C).
FIGURE 8.
Triacsin C does not affect arachidonate-regulated degradation of Insig-1. A, SRD-13A cells were seeded at 3. 5 × 105/60-mm dish on day 0. On day 2, cells were switched to medium containing 5% delipidated FCS. On day 3, 10 μm triacsin C was added into the medium as indicated. Following incubation for 30 min, cells were labeled with 4 μCi of [1-14C]oleate (specific activity, 54.6 mCi/mmol) in the same medium (2 ml) for 6 h. Cells were harvested, and lipids were extracted from the cells. The amount of radioactivity found in triglyceride (TG) was determined as described under “Experimental Procedures.” B, SRD-13A cells were seeded, transfected, and treated as described in Fig. 1B for the first 2 days. On day 3, 100 μm arachidonic acid or 10 μm triacsin C was added into the medium as indicated. After incubation for 6 h, cells were harvested, and the amount of Insig-1 was determined as described in Fig. 1B. C, SRD-13A cells were seeded, transfected, and treated as described in Fig. 3B for the first 2 days. On day 3, the cells were treated with arachidonate and triacsin C as described in B together with 10 μm MG132. Myc-tagged Ubxd8 was immunoprecipitated, and the amount of Insig-1 and Ubxd8 in pellets and supernatants of the immunoprecipitation (IP) was analyzed as described in Fig. 3B. IB, immunoblot.
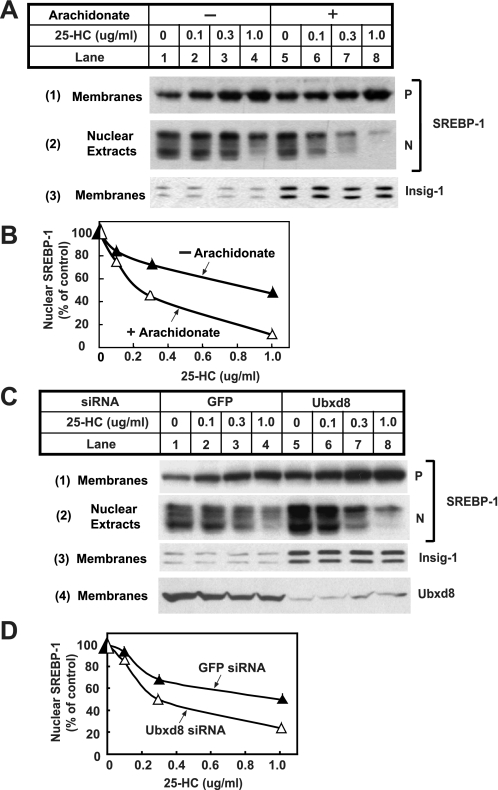
Fig. 9 shows an experiment in which we examined the functional significance of unsaturated fatty acid-mediated stabilization of Insig-1. To this end, we incubated HEK-293S/pInsig1 cells in medium supplemented with delipidated FCS and compactin, a condition that induces maximal cleavage of SREBPs. These cells were then incubated in the absence or presence of arachidonate and treated with the LXR agonist T0901317 to prevent arachidonate-mediated inhibition of SREBP-1 mRNA transcription (41). We treated the cells with various concentrations of 25-hydroxycholesterol, an oxysterol that inhibits cleavage of SREBPs (26). To avoid complications caused by sterol-induced stabilization of Insig-1, we treated the cells with 25-hydroxycholesterol for only a short period of time (1 h). As shown in Fig. 9, treatment of cells with arachidonate raised the amount of Insig-1 protein as expected (Fig. 9A, panel 3, compare lanes 5-8 with lanes 1-4). In cells that were not incubated with arachidonate, treatment with 25-hydroxycholesterol up to 1 μg/ml only slightly decreased the amount of cleaved nuclear SREBP-1 (Fig. 9A, panel 2, lanes 1-4). Treatment with arachidonate alone in the absence of sterols did not affect cleavage of SREBP-1 (Fig. 9A, panel 2, lanes 1 and 5). However, in cells receiving arachidonate, nuclear SREBP-1 decreased significantly upon 25-hydroxycholesterol treatment in a concentration-dependent manner (Fig. 9A, panel 2, lanes 5-8). Quantification of the results shown in Fig. 9A indicates that treatment of cells with arachidonate resulted in a roughly 5-fold increase in the sensitivity of SREBP-1 cleavage inhibited by 25-hydroxycholesterol (Fig. 9B). These results suggest that arachidonate increases the amount of Insig-1 protein, which in turn makes SREBP-1 more sensitive to the inhibitory effects of sterols. This conclusion is consistent with previous observations that elevation of Insig-1 protein increases the ability of sterols to inhibit proteolytic activation of SREBP-1 (10).
FIGURE 9.
Effects of arachidonate on proteolytic activation of SREBP-1. A, HEK-293S/pInsig1 cells were seeded at 4.0 × 105/60-mm dish on day 0. On day 2, cells were switched to medium B supplemented with 10% delipidated FCS, 50 μm compactin, and 50 μm mevalonate. On day 3, 2 μm LXR agonist T091317 was added into the medium in the absence or presence of 100 μm arachidonate. Following incubation for 6 h, the cells received additions of the indicated amount of 25-hydroxcholesterol (25-HC) and were further incubated for 1 h. Cells were then harvested and fractionated. Aliquots of membrane and nuclear extracts were subjected to SDS-PAGE followed by immunoblot analysis with anti-SREBP1 and anti-T7 (against Insig-1). P and N denote precursor and nuclear forms of SREBP-1, respectively. B, densitometric quantification of the result in A. The intensity of the cleaved nuclear form of SREBP-1 in the absence of 25-hydroxcholesterol was arbitrarily set at 100%. C, HEK-293S/pInsig1 cells were seeded and transfected with the indicated siRNA as described in Fig. 4B. On day 3, cells were switched to medium B containing 10% delipidated FCS with 50 μm compactin and 50 μm mevalonate. On day 4, 2 μm LXR agonist T091317 was added into the medium. Following incubation for 6 h, the cells received additions of the indicated amount of 25-hydroxycholesterol and were further incubated for 1 h. Cells were then harvested and analyzed as described in A. D, densitometric quantification of the result in C as described in B. GFP, green fluorescent protein.
If arachidonate affects SREBP-1 cleavage through stabilization of Insig-1, knockdown of Ubxd8 is expected to produce the same effect even in cells that are not treated with arachidonate. To test this hypothesis, we transfected HEK-293S/pInsig1 cells with duplexes of siRNA targeting Ubxd8 or green fluorescent protein as a control. Cells were then incubated in the absence of arachidonate, and proteolytic processing of SREBP-1 was analyzed as described in Fig. 9A. For reasons that we do not understand, knockdown of Ubxd8 raised the amount of cleaved nuclear SREBP-1 in cells that were not treated with 25-hydroxycholesterol (Fig. 9C, panel 2, lanes 1 and 5). The amount of Insig-1 protein was elevated in cells transfected with siRNA targeting Ubxd8 (Fig. 9C, panel 3, compare lanes 5-8 with lanes 1-4). Consequently cleavage of SREBP-1 was more sensitive to the inhibitory effects of 25-hydroxycholesterol in these cells (Fig. 9C, panel 2, compare lanes 5-8 with lanes 1-4). Quantification of the results shown in Fig. 9C indicates that knockdown of Ubxd8 led to a 3-fold increase in the sensitivity of SREBP-1 cleavage inhibited by 25-hydroxycholesterol (Fig. 9D).
DISCUSSION
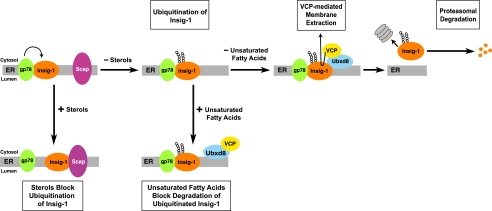
The current data establish a model for regulated degradation of Insig-1 by various lipids. The essential features of this model are illustrated in Fig. 10. In cells that are depleted of sterols and unsaturated fatty acids, Insig-1 is ubiquitinated by gp78 (11) and binds to Ubxd8, which is attached to VCP. The recruitment of VCP allows Insig-1 to be extracted from the ER membrane. The cytosolic Insig-1 is then degraded by the proteasome. In the presence of sterols, Scap binds to Insig-1 in a reaction that displaces gp78 (11). Ubiquitination of Insig-1 is thus prevented, and Insig-1 is stabilized (11). Treatment of cells with unsaturated fatty acids does not inhibit binding between Insig-1 and gp78, and therefore it does not block ubiquitination of Insig-1. Instead unsaturated fatty acids cause dissociation of the Ubxd8-VCP complex from Insig-1. As a result, Insig-1 is not extracted from membranes and thus is not delivered to proteasomes. When membrane dislocation of Insig-1 is blocked, the ubiquitinated Insig-1 appears to be rapidly deubiquitinated as suggested by the lack of accumulation of ubiquitinated Insig-1 in cells treated with arachidonate but not MG132 (data not shown). Indeed the deubiquitinating activity for Insig-1 is so strong that even in cells treated with MG132 we had to prepare denatured cell lysates as quickly as possible to observe ubiquitinated Insig-1 (11). Even under this circumstance, only a small fraction of Insig-1 is ubiquitinated as revealed by the observation that ubiquitinated Insig-1 was visible only in immunoblots detecting ubiquitin but not in those detecting Insig-1 (Fig. 2A).
FIGURE 10.
Model for sterol- and unsaturated fatty acid-mediated control of Insig-1 degradation. In cells that are depleted of sterols and unsaturated fatty acids, Insig-1 is ubiquitinated by gp78 and binds to Ubxd8 that mediates the interaction between Insig-1 and VCP. Recruitment of VCP allows Insig-1 to be extracted from the ER membrane and subsequently degraded by proteasomes. In the presence of sterols, Scap binds to Insig-1 in a reaction that displaces gp78. Ubiquitination of Insig-1 is thus prevented, and Insig-1 is stabilized. Treatment of cells with unsaturated fatty acids does not inhibit ubiquitination of Insig-1. Such treatment leads to dissociation of Ubxd8-VCP complex from Insig-1, a reaction that prevents membrane dislocation of Insig-1. Thus, unsaturated fatty acids stabilize Insig-1 at a postubiquitination step.
An important finding in the current study is that Ubxd8 is required for recruitment of VCP to Insig-1. If this is the case, then why cannot cytosolic VCP-Ufd1-Npl4 complex, which has been shown to interact with polyubiquitin chains (42), access ubiquitinated Insig-1? The answer to this question may lie in the complexity of polyubiquitin chains. Although the VCP complex is able to bind polyubiquitin chains, the exact polyubiquitin linkage bound to the VCP complex was not examined. We have found that Insig-1 is not ubiquitinated by the standard Lys-48-linked polyubiquitin chains (data not shown). It is possible that the novel polyubiquitin chains attached to Insig-1 are not recognized directly by the VCP complex, thereby requiring Ubxd8 for the complex formation between Insig-1 and VCP.
Ubxd8 belongs to a family of proteins that contain a UBX domain, which comprises about 80 amino acids with a structural similarity to ubiquitin (43). While this manuscript was under review, two reports were published to address the importance of UBX domain-containing protein in proteasomal degradation. In one report, Ubxd8 was shown to be required for human cytomegalovirus US11 protein-mediated membrane extraction and proteasomal degradation of Class I major histocompatibility complex heavy chains (44). In another report, Ubxd7 was shown to accelerate proteasomal degradation of hypoxia-inducible factor 1α by recruiting VCP to ubiquitinated hypoxia-inducible factor 1α (45). In the current studies, we presented evidence showing that interaction between Ubxd8 and Insig-1 is regulated by fatty acids, and this regulation accounts for fatty acid-mediated stabilization of Insig-1. Thus, UBX domain-containing proteins may not only be required but may also regulate proteasomal degradation.
Besides Ubxd8, Ubxd2 is the only other mammalian UBX domain-containing protein that is known to be involved in ERAD (37). Ubxd2 accelerates degradation of CD3δ (37), which is also ubiquitinated by gp78 (38). Our results show that Ubxd8 but not Ubxd2 accelerates degradation of Insig-1 (Fig. 4A). Although both gp78 and Ubxd2 play an important role in recruiting VCP to CD3δ (37, 38), recruitment of VCP to Insig-1 depends solely on Ubxd8 (Fig. 6). The difference between Ubxd2 and Ubxd8 suggests that these two proteins are not functionally redundant. Thus, proteasomal degradation of a certain membrane protein may require a specific UBX domain-containing protein.
The current study provides a mechanism for unsaturated fatty acid-regulated ERAD of Insig-1. However, it remains unclear how cells sense the change in the amount of unsaturated fatty acids. The observation that treatment with the acyl-coA inhibitor triacsin C did not affect arachidonate-mediated stabilization of Insig-1 suggests that free fatty acids may be directly sensed (Fig. 8). However, our studies do not rule out the possibility that arachidonate might alter the membrane lipid composition or physical property even in the presence of triacsin C. Future studies will be required to address this important question.
The results from the current study might also explain a previous observation that cleavage of SREBP-1 is inhibited by unsaturated fatty acids (17). Unsaturated fatty acids inhibit synthesis of SREBP-1 mRNA (41) as well as proteolytic activation of SREBP-1 protein (17). Here we show that when arachidonate-mediated inhibition of SREBP-1 transcription was blocked by the treatment with the LXR agonist T0901317 arachidonate alone did not inhibit SREBP-1 cleavage (Fig. 9A). Instead treatment with arachidonate enhanced the ability of sterols to inhibit cleavage of SREBP-1 (Fig. 9A). Previously, increased expression of Insig-1 was shown to make sterols more active in inhibiting cleavage of SREBPs (10). We believe that this sterol-sensitizing effect of arachidonate is attributable to an increase in Insig-1. This notion is further supported by the correlation between fatty acids that inhibit SREBP-1 cleavage and those that stabilize Insig-1 (Fig. 1E).
Stearoyl-CoA desaturase-1 is the main enzyme that synthesizes unsaturated fatty acids in mammalian cells (46). It is known that transcription of stearoyl-CoA desaturase-1 in mouse livers is inhibited by dietary unsaturated fatty acids (47). This feedback regulation in synthesis of unsaturated fatty acids most likely arises from inhibition of the proteolytic processing of SREBP-1, which is crucial for full activation of transcription of stearoyl-CoA desaturase-1 (48). Thus, unsaturated fatty acid-triggered dissociation of Insig-1 from the Ubxd8-VCP complex and subsequent stabilization of Insig-1 may play an important role in this feedback regulation. Interestingly, the feedback regulation in synthesis of unsaturated fatty acids in S. cerevisiae also requires the ubiquitin-proteasome system and VCP (49). Unlike mammalian cells, S. cerevisiae do not contain orthologues of Insig-1, SREBPs, and proteases that cleave SREBPs. Instead they produce a membrane-bound transcription factor, Spt23 (50). In the absence of unsaturated fatty acids, ubiquitinated Spt23 binds to VCP, a reaction that leads to limited degradation of Spt23 by proteasomes (50). After the proteasomal digestion, the remaining NH2-terminal fragment of Spt23 is liberated from the ER membrane and enters the nucleus to drive transcription of OLE1 (50), the orthologue of mammalian stearoyl-CoA desaturase-1 (51). In the presence of exogenously added unsaturated fatty acids, proteolytic activation of Spt23 by proteasomes does not occur (50). However, it is currently unknown whether unsaturated fatty acids inhibit ubiquitination of Spt23 or block cleavage of Spt23 at a postubiquitination step. Inasmuch as both yeast and mammalian cells use the ubiquitin-proteasome system to regulate synthesis of unsaturated fatty acids, a common factor in the ubiquitinproteasome pathway may be a sensor for unsaturated fatty acids. Whether this protein is Ubxd8 or some other protein(s) will require further study.
Acknowledgments
We thank Drs. Michael S. Brown and Joseph L. Goldstein for continued support and advice; Zhijian Chen and George DeMartino for critical evaluation of the manuscript; Lisa Beatty, Angela Carroll, and Shomanike Head for invaluable help with tissue culture; and Saada Abdalla for excellent technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grant HL-20948. This work was also supported by the Perot Family Foundation. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: E1, ubiquitin activation enzyme; E2, ubiquitin-conjugating enzyme; E3, ubiquitin ligase; ER, endoplasmic reticulum; ERAD, ER-associated degradation; VCP, valosin-containing protein; FCS, fetal calf serum; Ubxd8, UBX domain-containing protein 8; FAF1, Fas-associated factor 1; siRNA, small interfering RNA; SREBP, sterol-regulatory element-binding protein; HA, hemagglutinin; CFTR, cystic fibrosis transmembrane conductance regulator; LXR, liver X receptor; CMV, cytomegalovirus; CHO, Chinese hamster ovary; UBX, ubiquitin regulatory X.
References
- 1.Hershko, A., and Ciechanover, A. (1998) Annu. Rev. Biochem. 67 425-479 [DOI] [PubMed] [Google Scholar]
- 2.Fang, S., and Weissman, A. M. (2004) CMLS Cell. Mol. Life Sci. 61 1546-1561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meusser, B., Hirsch, C., Jarosch, E., and Sommer, T. (2005) Nat. Cell Biol. 7 766-772 [DOI] [PubMed] [Google Scholar]
- 4.Halawani, D., and Latterich, M. (2006) Mol. Cell 22 713-717 [DOI] [PubMed] [Google Scholar]
- 5.Carlson, E., Pitonzo, D., and Skach, W. (2006) EMBO J. 25 4557-4566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakatsukasa, K., Huyer, G., Michaelis, S., and Brodsky, J. L. (2008) Cell 132 101-112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goldstein, J. L., DeBose-Boyd, R. A., and Brown, M. S. (2006) Cell 124 35-46 [DOI] [PubMed] [Google Scholar]
- 8.Brown, M. S., and Goldstein, J. L. (1999) Proc. Natl. Acad. Sci. U. S. A 96 11041-11048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horton, J. D., Shah, N. A., Warrington, J. A., Anderson, N. N., Park, S. W., Brown, M. S., and Goldstein, J. L. (2003) Proc. Natl. Acad. Sci. U. S. A 100 12027-12032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang, T., Espenshade, P. J., Wright, M. E., Yabe, D., Gong, Y., Aebersold, R., Goldstein, J. L., and Brown, M. S. (2002) Cell 110 489-500 [DOI] [PubMed] [Google Scholar]
- 11.Lee, J. N., Song, B., DeBose-Boyd, R. A., and Ye, J. (2006) J. Biol. Chem. 281 39308-39315 [DOI] [PubMed] [Google Scholar]
- 12.Gong, Y., Lee, J. N., Lee, P. C. W., Goldstein, J. L., Brown, M. S., and Ye, J. (2006) Cell Metab. 3 15-24 [DOI] [PubMed] [Google Scholar]
- 13.Yabe, D., Brown, M. S., and Goldstein, J. L. (2002) Proc. Natl. Acad. Sci. U. S. A 99 12753-12758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee, J. N., and Ye, J. (2004) J. Biol. Chem. 279 45257-45265 [DOI] [PubMed] [Google Scholar]
- 15.Lee, J. N., Gong, Y., Zhang, X., and Ye, J. (2006) Proc. Natl. Acad. Sci. U. S. A 103 4958-4963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sever, N., Lee, P. C. W., Song, B. L., Rawson, R. B., and DeBose-Boyd, R. A. (2004) J. Biol. Chem. 279 43136-43147 [DOI] [PubMed] [Google Scholar]
- 17.Hannah, V. C., Ou, J., Luong, A., Goldstein, J. L., and Brown, M. S. (2001) J. Biol. Chem. 276 4365-4372 [DOI] [PubMed] [Google Scholar]
- 18.Shimano, H., Horton, J. D., Hammer, R. E., Shimomura, I., Brown, M. S., and Goldstein, J. L. (1996) J. Clin. Investig. 98 1575-1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Song, B. L., Sever, N., and DeBose-Boyd, R. A. (2005) Mol. Cell 19 829-840 [DOI] [PubMed] [Google Scholar]
- 20.Kita, T., Brown, M. S., and Goldstein, J. L. (1980) J. Clin. Investig. 66 1094-1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feramisco, J. D., Goldstein, J. L., and Brown, M. S. (2004) J. Biol. Chem. 279 8487-8496 [DOI] [PubMed] [Google Scholar]
- 22.Wang, C., Gale, J., Keller, B. C., Huang, H., Brown, M. S., Goldstein, J. L., and Ye, J. (2005) Mol. Cell 18 425-434 [DOI] [PubMed] [Google Scholar]
- 23.Metherall, J. E., Goldstein, J. L., Luskey, K. L., and Brown, M. S. (1989) J. Biol. Chem. 264 15634-15641 [PubMed] [Google Scholar]
- 24.Rawson, R. B., DeBose-Boyd, R., Goldstein, J. L., and Brown, M. S. (1999) J. Biol. Chem. 274 28549-28556 [DOI] [PubMed] [Google Scholar]
- 25.Reeves, P. J., Thurmond, R. L., and Khorana, H. G. (1996) Proc. Natl. Acad. Sci. U. S. A 93 11487-11492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adams, C. M., Reitz, J., De Brabander, J. K., Feramisco, J. D., Li, L., Brown, M. S., and Goldstein, J. L. (2004) J. Biol. Chem. 279 52772-52780 [DOI] [PubMed] [Google Scholar]
- 27.Sakai, J., Nohturfft, A., Cheng, D., Ho, Y. K., Brown, M. S., and Goldstein, J. L. (1997) J. Biol. Chem. 272 20213-20221 [DOI] [PubMed] [Google Scholar]
- 28.Sakai, J., Duncan, E. A., Rawson, R. B., Hua, X., Brown, M. S., and Goldstein, J. L. (1996) Cell 85 1037-1046 [DOI] [PubMed] [Google Scholar]
- 29.Yao, H., and Ye, J. (2008) J. Biol. Chem. 283 849-854 [DOI] [PubMed] [Google Scholar]
- 30.Ward, C. L., Omura, S., and Kopito, R. R. (1995) Cell 83 121-127 [DOI] [PubMed] [Google Scholar]
- 31.Sever, N., Song, B. L., Yabe, D., Goldstein, J. L., Brown, M. S., and DeBose-Boyd, R. A. (2003) J. Biol. Chem. 278 52479-52490 [DOI] [PubMed] [Google Scholar]
- 32.Neuber, O., Jarosch, E., Volkwein, C., Walter, J., and Sommer, T. (2005) Nat. Cell Biol. 7 993-998 [DOI] [PubMed] [Google Scholar]
- 33.Schuberth, C., and Buchberger, A. (2005) Nat. Cell Biol. 7 999-1006 [DOI] [PubMed] [Google Scholar]
- 34.Schuberth, C., Richly, H., Rumpf, S., and Buchberger, A. (2004) EMBO Rep. 5 818-824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Imai, Y., Nakada, A., Hashida, R., Sugita, Y., Tanaka, T., Tsujimoto, G., Matsumoto, K., Akasawa, A., Saito, H., and Oshida, T. (2002) Biochem. Biophys. Res. Commun. 297 1282-1290 [DOI] [PubMed] [Google Scholar]
- 36.Song, E. J., Yim, S. H., Kim, E., Kim, N. S., and Lee, K. J. (2005) Mol. Cell. Biol. 25 2511-2524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liang, J., Yin, C., Doong, H., Fang, S., Peterhoff, C., Nixon, R. A., and Monteiro, M. J. (2006) J. Cell Sci. 119 4011-4024 [DOI] [PubMed] [Google Scholar]
- 38.Zhong, X., Shen, Y., Ballar, P., Apostolou, A., Agami, R., and Fang, S. (2004) J. Biol. Chem. 279 45676-45684 [DOI] [PubMed] [Google Scholar]
- 39.Tomoda, H., Igarashi, K., Cyong, J. C., and Omura, S. (1991) J. Biol. Chem. 266 4214-4219 [PubMed] [Google Scholar]
- 40.Coleman, R. A., Lewin, T. M., and Muoio, D. M. (2000) Annu. Rev. Nutr. 20 77-103 [DOI] [PubMed] [Google Scholar]
- 41.Ou, J., Tu, H., Shan, B., Luk, A., DeBose-Boyd, R. A., Bashmakov, Y., Goldstein, J. L., and Brown, M. S. (2001) Proc. Natl. Acad. Sci. U. S. A 98 6027-6032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meyer, H., Wang, Y., and Warren, G. (2002) EMBO J. 21 5645-5652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Buchberger, A. (2002) Trends Cell Biol. 12 216-221 [DOI] [PubMed] [Google Scholar]
- 44.Mueller, B., Klemm, E. J., Spooner, E., Claessen, J. H., and Ploegh, H. L. (2008) Proc. Natl. Acad. Sci. U. S. A 105 12325-12330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alexandru, G., Graumann, J., Smith, G. T., Kolawa, N. J., Fang, R., and Deshaies, R. J. (2008) Cell 134 804-816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dobrzyn, A., and Ntambi, J. M. (2005) Prostaglandins Leukot. Essent. Fatty Acids 73 35-41 [DOI] [PubMed] [Google Scholar]
- 47.Ntambi, J. M. (1992) J. Biol. Chem. 267 10925-10930 [PubMed] [Google Scholar]
- 48.Liang, G., Yang, J., Horton, J. D., Hammer, R. E., Goldstein, J. L., and Brown, M. S. (2002) J. Biol. Chem. 277 9520-9528 [DOI] [PubMed] [Google Scholar]
- 49.Hitchcock, A. L., Krebber, H., Frietze, S., Lin, A., Latterich, M., and Silver, P. A. (2001) Mol. Biol. Cell 12 3226-3241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hoppe, T., Matuschewski, K., Rape, M., Schlenker, S., Ulrich, H. D., and Jentsch, S. (2000) Cell 102 577-586 [DOI] [PubMed] [Google Scholar]
- 51.Martin, C. E., Oh, C. S., and Jiang, Y. (2007) Biochim. Biophys. Acta 1771 271-285 [DOI] [PubMed] [Google Scholar]