Abstract
Calcium (Ca2+) is a highly versatile second messenger that regulates various cellular processes. Previous studies showed that elevation of intracellular Ca2+ regulates the activity of Na+/H+ exchanger 3 (NHE3). However, the effect of Ca2+-dependent signaling on NHE3 activity varies depending on cell types. In this study, we report the identification of IP3 receptor-binding protein released with IP3 (IRBIT) as a NHE3 interacting protein and its role in regulation of NHE3 activity. IRBIT bound to the carboxyl-terminal domain of NHE3, which is necessary for acute regulation of NHE3. Ectopic expression of IRBIT resulted in Ca2+-dependent activation of NHE3 activity, whereas silencing of endogenous IRBIT resulted in inhibition of NHE3 activity. Ca2+-dependent stimulation of NHE3 activity was dependent on the binding of IRBIT to NHE3. Previously Ca2+-dependent inhibition of NHE3 was demonstrated in the presence of NHERF2. Co-expression of IRBIT was able to reverse the NHERF2-dependent inhibition of NHE3. We also showed that IRBIT-dependent activation of NHE3 involves exocytic trafficking of NHE3 to the plasma membrane and this activation was blocked by inhibition of calmodulin (CaM) or CaM-dependent kinase II. These results suggest that the overall effect of Ca2+ on NHE3 activity is balanced by IRBIT-dependent activation and NHERF2-dependent inhibition.
The calcium ion (Ca2+) is a highly versatile second messenger
that can regulate many different cellular functions
(1,
2). Ca2+ regulates
various cellular processes by activating or inhibiting cellular signaling
pathways and Ca2+-regulated proteins
(1,
2). A major intracellular store
of Ca2+ is the endoplasmic reticulum. Extracellullar stimuli,
including hormones, growth factors, and neurotransmitter, induce production of
the second messenger, inositol 1,4,5-triphosphate
(IP3),2
through phosphoinositide turnover, which engages with IP3 receptor
(IP3R) to release Ca2+ from endoplasmic reticulum
(1,
3). Ca2+ is often
viewed as a second messenger acting as a trigger for Ca2+-dependent
events. However, for Ca2+ to regulate multiple cellular processes
in different cells with different outcomes, Ca2+ homeostasis must
be tightly regulated temporally and spatially
(1,
2). Recently, an IP3
receptor binding protein, IP3 receptor-binding protein released
with IP3 (IRBIT), has been identified
(4). IRBIT inhibits activation
of IP3R and Ca2+ release by competing with
IP3 (5).
Interestingly, IRBIT binds to the pancreas-type electrogenic
Na+/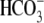 cotransporter 1
(pNBC1) and co-expression of IRBIT and pNBC1 in Xenopus oocytes
activated Na+/
cotransporter 1
(pNBC1) and co-expression of IRBIT and pNBC1 in Xenopus oocytes
activated Na+/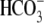 cotransporter activity (6).
cotransporter activity (6).
Na+/H+ exchange is a process present in all organisms
from prokaryotes to human (7).
Na+/H+ exchange in mammals is mediated by the
Na+/H+ exchange gene family (NHE) consisting of 9
members (7,
8). The NHE3 isoform is
expressed in the apical membrane of epithelial cells located in the renal
proximal tubule and intestine
(9,
10). In the kidney, NHE3 is
responsible for the majority of Na+ and
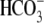 reabsorption in renal proximal
tubule as evidenced by the studies using NHE3-null mice
(11,
12). In the intestine, NHE3
mediates electroneutral NaCl absorption by coupling to a
Cl-/
reabsorption in renal proximal
tubule as evidenced by the studies using NHE3-null mice
(11,
12). In the intestine, NHE3
mediates electroneutral NaCl absorption by coupling to a
Cl-/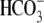 exchanger
(8). Through osmotic coupling
to passive water absorption, NHE3 is a major constituent in basal
Na+ absorption in intestine and a frequent target of inhibition in
many diarrheal diseases (8).
Among diverse stimuli and second messengers that regulate NHE3,
Ca2+ remains an enigmatic second messenger because the effect on
NHE3 by Ca2+ varies depending on cell types
(13-15).
exchanger
(8). Through osmotic coupling
to passive water absorption, NHE3 is a major constituent in basal
Na+ absorption in intestine and a frequent target of inhibition in
many diarrheal diseases (8).
Among diverse stimuli and second messengers that regulate NHE3,
Ca2+ remains an enigmatic second messenger because the effect on
NHE3 by Ca2+ varies depending on cell types
(13-15).
NHE3 and other mammalian Na+/H+ exchangers consist of two structurally and functionally distinct domains, the NH2 terminus and COOH terminus (16, 17). The COOH-terminal domain has been shown to be necessary for all identified acute regulation. Part of this necessity is the ability of the COOH-terminal domain to interact with an increasing number of cellular proteins (8, 16, 17). In this study, we report the identification of IRBIT as a NHE3 interacting protein. We provide evidence that IRBIT activates NHE3 activity in response to Ca2+.
EXPERIMENTAL PROCEDURES
Plasmid Constructs—The bait plasmid, pGBKT7-C832, was constructed by cloning the coding region of the cytoplasmic COOH-terminal domain of rabbit NHE3 (aa 475-832) into the yeast DNA-binding vector pGBKT7 vector (Clontech). NHE3 COOH-terminal domains, aa 475-591 and 475-696, were cloned in pGBKT7 to generate pGBKT7-C591 and pGBKT7-C696, respectively. pACT-IRBIT is the GAL4 activation domain plasmid containing the entire coding sequence of IRBIT. The cDNAs corresponding to aa 1-105, 1-224, and 105-530 of IRBIT were cloned into the GAL4-activation domain plasmid, pACT2, to generate pACT-105R, pACT-SB, and pACT-105T, respectively.
Yeast Two-hybrid—The bait plasmid, pGBKT7-C832, was transformed into the yeast strain AH109. The resulting AH109 (pGBKT7-C832) yeast strain was mated with >3 × 106 clones of the yeast strain Y187 pretransformed with a human kidney Matchmaker cDNA library (Clontech). The resulting yeast strain was plated on synthetic dropout plates lacking adenosine, histidine, leucine, and tryptophan (SD/-Ade/-His/-Leu/-Trp). Yeast colonies formed after 5 days were replated on SD/-Ade/-His/-Leu/-Trp plates containing 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-gal). Positive library plasmids were rescued from the yeast colonies and transformed together with pGBKT7-C832 into AH109 and double transformants were selected by growth on SD/-Leu/-Trp plates. Resulting yeast colonies were plated on SD/-Ade/-His/Leu/-Trp plates to reconfirm the interaction. Positive clones were sequenced to identify their cDNA inserts.
Cell Cultures—PS120 cells stably expressing rabbit NHE3 fused at the COOH terminus with an antibody epitope derived from vesicular stomatitis virus glycoprotein (VSVG), PS120/NHE3V, was previously described (18, 19). PS120/NHE3V truncation mutants with portions of the COOH-terminal cytoplasmic domain deleted, PS120/NHE3V-585 and PS120/NHE3V-689, were previously described and were kindly provided by Dr. Ming Tse, Johns Hopkins University (16). All PS120 fibroblasts were cultured in Dulbecco's modified Eagle's medium supplemented with 1 mm sodium pyruvate, 50 units/ml penicillin, 50 μg/ml streptomycin, and 10% fetal bovine serum in a 5% CO2 humidified incubator at 37 °C. Caco2BBE cells transfected with NHE3V, Caco2BBE/NHE3V, were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 25 mm NaHCO3, 50 μg/ml streptomycin, 50 units/ml penicillin, and 1% nonessential amino acids.
Transfection and Gene Silencing—cDNA encoding IRBIT was tagged with a hemagglutinin (HA) epitope at the NH2 terminus by PCR and was subcloned into pcDNA3.1Hygro (Invitrogen). PS120 and Caco2BBE cells and their variants were transfected with pcDNA-HA-IRBIT by using Lipofectamine 2000 (Invitrogen) according to the manufacturer's recommendation and selected with 600 μg/ml hygromycin. pLKO.1 harboring small hairpin RNA (shRNA) constructs for IRBIT were obtained from Sigma. PS120/NHE3 cells were transfected with shRNA plasmids and selected with 4 μg/ml puromycin.
Immunoprecipitation—PS120 cells were washed twice in cold phosphate-buffered saline (PBS), scraped, and lysed in lysis buffer (Cell Signaling, Danvers, MA), containing 20 mm Tris-HCl (pH 7.5), 150 mm NaCl, 1 mm β-glycerophosphate, 2.5 mm sodium pyrophosphate, 1 mm Na2EDTA, 1 mm EGTA, 1 mm Na3VO4, 1 μg/ml leupeptin, 1% Triton X-100, and protease inhibitor mixture tablets (Roche). The crude lysate was sonicated for 2 × 15 s and spun at 14,000 × g for 15 min. Protein concentration was determined by the bicinchoninic acid assay (Sigma). Lysate (500 μg) was pre-cleared by incubation with 30 μl of protein A-Sepharose beads for 1 h and the supernatant was then incubated overnight with either monoclonal anti-VSVG antibody or polyclonal anti-IRBIT antibody. Immunocomplex was purified by incubating with 50 μl of protein A-Sepharose beads for 1 h, followed by 3 washes in lysis buffer and 2 washes in PBS. All the above steps were performed at 4 °C or on ice. The bound immunocomplex was eluted by incubating the protein A beads in Laemmli sample buffer for 10 min at 95 °C and then separated by SDS-PAGE. The proteins were then transferred to nitrocellulose membrane for Western immunoblot.
Surface Biotinylation—Surface biotinylation of NHE3 was performed as previously described (20). Briefly, cells grown in 10-cm Petri dishes were treated with 100 nm thapsigargin, 500 nm ionomycin, or ethanol for 10 min, followed by rinsing twice in PBS and 10 min incubation in borate buffer composed of 154 mm NaCl, 7.2 mm KCl, 1.8 mm CaCl2, and 10 mm H3BO3 (pH 9.0). Cells were then incubated for 40 min with 0.5 mg/ml NHS-SS-biotin (Pierce) in borate buffer. Unbound NHS-SS-biotin was quenched with Tris buffer (20 mm Tris, 120 mm NaCl, pH 7.4). Cells were then rinsed with PBS, scraped, lysed in the lysis buffer described above, and sonicated for 2 × 15 s. The lysate was agitated for 30 min and spun at 14,000 × g for 15 min to remove the insoluble cell debris. An aliquot was retained as the total fraction representing the total cellular NHE3 and IRBIT. Protein concentration was determined and 1 mg of lysate was then incubated with streptavidin-agarose beads (Pierce) for 2 h. The strepavidin-agarose beads were washed 3 times in lysis buffer and twice in PBS. All the above procedures were performed at 4 °C or on ice. Biotinylated surface proteins were then eluted by boiling the beads at 95 °C for 10 min. Dilutions of the total and surface NHE3 were resolved by SDS-PAGE, and immunoblotted with anti-VSVG antibody or anti-HA monoclonal antibody. Densitometric analysis was performed using Scion Image software (National Institutes of Health).
Na+-dependent Intracellular pH
Recovery—The Na+-dependent changes in intracellular pH
(pHi) by NHE3 was determined with the use of the ratio
fluorometric, pH-sensitive dye
2′,7′-bis-(2-carboxyethyl)-5-carboxyfluorescein acetoxymethyl
ester (BCECF-AM) as previously described
(20,
21). Briefly, cells were
seeded on coverslips, grown to 70-80% confluence for PS120 or 5 days
post-confluence for Caco2 cells, and then serum starved overnight. Cells were
washed in Na+ buffer (30 mm NaCl, 20 mm
HEPES, 5 mm KCl, 1 mm tetramethylammonium-PO4
(TMA-PO4), 2 mm CaCl2, 1 mm
MgSO4, and 18 mm glucose) and then were dye-loaded by
incubating for 20 min with 6.5 μm BCECF-AM in the same solution.
The coverslips were mounted on a perfusion chamber mounted on an inverted
microscope, and were superfused with
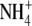 buffer (40 mm
NH4Cl, 90 mm NaCl, 20 mm HEPES, 5
mm KCl, 1 mm TMA-PO4, 2 mm
CaCl2, 1 mm MgSO4, and 18 mm
glucose) and subsequently with TMA+ buffer (130 mm
TMA-Cl, 20 mm HEPES, 5 mm KCl, 1 mm
TMA-PO4, 2 mm CaCl2, 1 mm
MgSO4, and 18 mm glucose), with each containing 100
nm thapsigargin, 500 nm ionomycin, or ethanol as a
control. Na+ buffer supplemented with (for Caco2BBE) or without
(for PS120) 50 μm HOE-694 was then reintroduced to drive
Na+-dependent pH recovery. Calibration of the fluorescence signal
was performed using the K+/H+ ionophore nigericin as
described previously (21). The
microfluorometry was performed on a Nikon TE200 inverted microscope with a
Nikon CFI Super Fluor ×40 objective, coupled to a Lambda 10-2 filter
wheel controller equipped with a multiwavelength filter set designed for
BCECF. Photometric data were acquired using the Metafluor software (Molecular
Devices). Na+/H+ exchange rate was described by the rate
of pHi recovery, which was calculated by determining
slopes along the pHi recovery by linear least-squares
analysis over a minimum of 9 s
(20).
buffer (40 mm
NH4Cl, 90 mm NaCl, 20 mm HEPES, 5
mm KCl, 1 mm TMA-PO4, 2 mm
CaCl2, 1 mm MgSO4, and 18 mm
glucose) and subsequently with TMA+ buffer (130 mm
TMA-Cl, 20 mm HEPES, 5 mm KCl, 1 mm
TMA-PO4, 2 mm CaCl2, 1 mm
MgSO4, and 18 mm glucose), with each containing 100
nm thapsigargin, 500 nm ionomycin, or ethanol as a
control. Na+ buffer supplemented with (for Caco2BBE) or without
(for PS120) 50 μm HOE-694 was then reintroduced to drive
Na+-dependent pH recovery. Calibration of the fluorescence signal
was performed using the K+/H+ ionophore nigericin as
described previously (21). The
microfluorometry was performed on a Nikon TE200 inverted microscope with a
Nikon CFI Super Fluor ×40 objective, coupled to a Lambda 10-2 filter
wheel controller equipped with a multiwavelength filter set designed for
BCECF. Photometric data were acquired using the Metafluor software (Molecular
Devices). Na+/H+ exchange rate was described by the rate
of pHi recovery, which was calculated by determining
slopes along the pHi recovery by linear least-squares
analysis over a minimum of 9 s
(20).
Production of Anti-IRBIT Antiserum—The cDNA encoding the NH2-terminal region (aa 1-104) of IRBIT was subcloned into the bacterial hexahistidine (His6) fusion vector pET-16b (Novagen, Gibbstown, NJ) to generate the IRBIT-(1-104)-His6 recombinant protein. The expressed IRBIT-(1-104)-His6 protein was then purified using nickel-nitrilotriacetic acid-agarose resin (Qiagen, Valencia, CA) and used for immunization of New Zealand White rabbits by Convance Research Products (Denver, PA) to produce anti-IRBIT serum.
Confocal Immunofluorescence—PS120/NHE3V/HA-IRBIT and PS120/NHE3V/pcDNA cells grown on coverslips were washed twice with cold PBS, fixed in 4% paraformaldehyde in PBS for 10 min at room temperature, permeabilized in 0.2% Triton X-100 in PBS for 5 min, and blocked in PBS containing 5% normal goat serum for 30 min at room temperature. Cells were then stained with polyclonal anti-HA (Sigma) or monoclonal anti-VSVG antibodies for 1 h at room temperature. Following three washes, 10 min each, with PBS, the cells were incubated with Alexa 488-conjugated donkey anti-mouse IgG or Alexa 555-conjugated goat anti-rabbit IgG (Invitrogen) for 1 h at room temperature. After three 10-min washes with PBS, the coverslips were mounted with ProLong Gold Antifade Reagent (Invitrogen) and observed under a Zeiss LSM510 laser confocal microscope (Zeiss Microimaging, Thornwood, NY) coupled to a Zeiss Axioplan2e with ×63 Pan-Apochromat oil lenses.
Statistical Analysis—Results were presented as mean ± S.E. Statistical significance was assessed by analysis of variance, with p < 0.05 considered significant.
RESULTS
IRBIT Interacts with NHE3—To identify proteins interacting with NHE3, we screened a human kidney cDNA library under high stringency with the GAL4 DNA-binding domain fused to the cytoplasmic domain of NHE3 as bait. Screening of >3 × 106 independent clones yielded 4 positive clones containing cDNA encoding calcineurin homologous protein, which was previously shown to interact with NHE3 (22). Six other clones contained cDNA inserts encoding S-adenosylhomocysteine hydrolase-like 1 (AdoHcyL1). The relative strength of interaction of calcineurin homologous protein and AdoHcyL1 with NHE3 was robust and comparable based on the size of yeast colonies and the β-galactosidase activity (data not shown). AdoHcyL1 was recently shown to interact with IP3R and was termed IRBIT (4). AdoHcyL1 will be referred to as IRBIT hereafter.
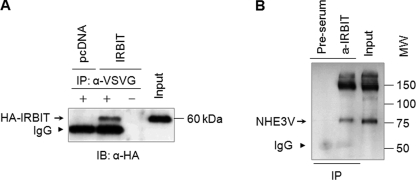
To confirm the interaction between NHE3 and IRBIT in a two-hybrid system, we tested co-immunoprecipitation of NHE3 and IRBIT in PS120/NHE3V fibroblasts transfected with HA-IRBIT. As shown in Fig. 1A, IRBIT co-immunoprecipitated with NHE3V in PS120/NHE3V/HA-IRBIT cells confirming the results from the two-hybrid screen. This interaction was specific as the presence of IRBIT was not detected in the absence of an anti-VSVG antibody or in sham-transfected cells. Fig. 1B shows an opposite experiment where IRBIT was pulled down with an anti-IRBIT antibody and co-immunoprecipitation of NHE3V was detected by Western blot.
FIGURE 1.
NHE3 interacts with IRBIT. A, PS120/NHE3V cells were transfected with HA-IRBIT or pcDNA. Cell lysates were incubated with or without monoclonal anti-VSVG antibodies and bound proteins were subjected to Western blotting using an anti-HA antibody. B, cell lysates from PS120/NHE3V/IRBIT were incubated with anti-IRBIT antibody or pre-serum and co-immunoprecipitated NHE3 was detected by Western blotting with an anti-VSVG antibody. Representative immunoblots from three independent experiments are shown.
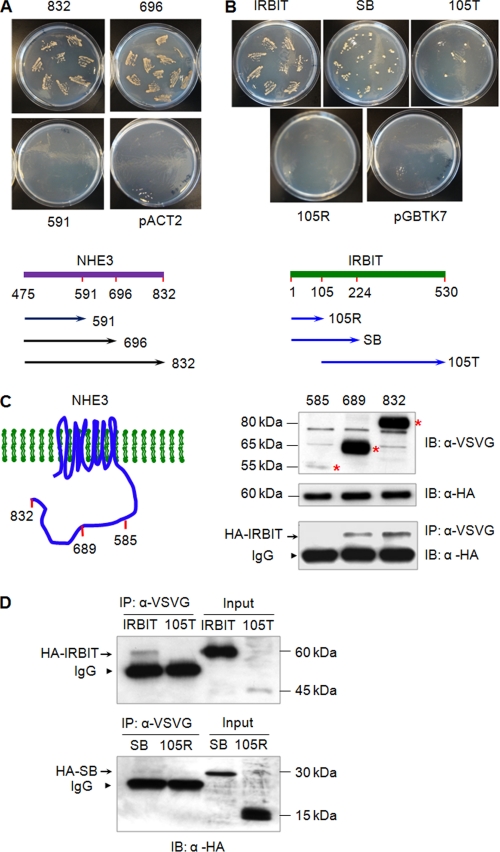
Having confirmed the interaction between NHE3 and IRBIT, we next determined domains of NHE3 and IRBIT mediating their interaction using yeast two-hybrid analysis. Results shown in Fig. 2A indicate that the truncation of a region distal to aa 696 of NHE3 (C696: aa 475-696) did not affect its interaction with IRBIT as compared with C832 (aa 475-832). A larger deletion of aa 592-832 of NHE3 (C591: aa 475-591) completely abrogated its ability to bind IRBIT, indicating that the region necessary for the interaction with IRBIT lies between aa 591 and 696. Fig. 2B shows that the binding of IRBIT to C832 was significantly attenuated by deletions of parts of IRBIT. The NH2-terminal 105 amino acids of IRBIT (105R) showed no interaction with C832. The 105T construct (aa 105-530) resulted in several isolated yeast colonies, suggesting a marginal capacity, if any, to bind NHE3. The SB construct of IRBIT (aa 1-224) weakly interacted with NHE3 compared with full-length IRBIT based on the number of yeast colonies. These results were reproducible in three independent matings.
FIGURE 2.
The region between aa 585 and 696 is necessary for the interaction with IRBIT. A, yeast two-hybrid assay was used to determine domains within the NHE3 COOH terminus that are necessary for the interaction with IRBIT. pACT2-IRBIT was co-transformed into host strain AH109 with C832, C696, or C591 in pGBKT7 and selected on SD/-Leu/-Trp for double transformants. 8-9 colonies were re-streaked on SD/-Ade/-His/-Leu/-Trp to verify the interaction between IRBIT and NHE3. A schematic diagram of IRBIT constructs is shown at the bottom. Similar results were obtained from three independent transformation and subsequent screens by auxotrophic growth. B, pGBKT7-C832 was co-transformed with pACT2 harboring truncated constructs of IRBIT into AH109 cells and protein interaction was determined by growth of double transformants on SD/-Ade/-His/-Leu/-Trp. A schematic diagram of IRBIT constructs is shown at the bottom. Similar results were obtained from three independent transformations and subsequent screen by auxotrophic growth. C, HA-IRBIT was expressed in PS120/NHE3V, PS120/NHE3V-689, or PS120/NHE3V-585 cells. Left panel illustrates NHE3V and its truncated constructs. NHE3V and truncated constructs were immunoprecipitated with an anti-VSVG antibody and co-immunoprecipitated IRBIT was detected by Western blotting using an anti-HA antibody. Top and middle panels show input NHE3V and the variants (*) and HA-IRBIT proteins, respectively, in the cell lysate. Bottom panel shows co-immunoprecipitated HA-IRBIT. Arrow, co-immunoprecipitated IRBIT. Arrowheads, IgG. D, HA-IRBIT and its variants were expressed in PS120/NHE3V cells. NHE3V was immunoprecipitated with an anti-VSVG antibody and co-immunoprecipitated HA-IRBIT and its truncated proteins were detected by an anti-HA antibody. Right two lanes show input and left two lanes are pulled down immunocomplex. Arrow, co-immunoprecipitated IRBIT. Arrowheads, IgG. Representative immunoblots from three separate experiments are shown.
The domains involved in NHE3-IRBIT association were further explored by co-immunoprecipitation in PS120 cells. HA-IRBIT was expressed in PS120/NHE3V, PS120/NHE3V-689, and PS120/NHEV-585 cells and NHE3 and its variants were immunoprecipitated with anti-VSVG, followed by Western blot using an anti-HA antibody to detect co-immunoprecipitated HA-IRBIT. The upper and middle panels of Fig. 2C show the expression of NHE3 variants and IRBIT, respectively, in each cell line. The bottom panel shows Western blot of the immunocomplex pulled down by an anti-VSVG antibody. The results shown in Fig. 2C confirmed that IRBIT bound to NHE3V-689 as well as full-length NHE3V. Although the expression level of NHE3V-585 was significantly lower, no apparent interaction was observed between IRBIT and NHE3V-585. We confirmed the absence of interaction between NHE3V-585 and IRBIT by using as much as 4 mg of lysate with no sign of co-immunoprecipitated IRBIT (data not shown).
Fig. 2D shows co-immunoprecipitation of IRBIT and its variants with NHE3V. The expression level of 105T protein in PS120 cells was extremely low relative to IRBIT and 105R, suggesting that the 105T protein may not be stable in these cells. Neither 105T nor 105R co-immunoprecipitated with NHE3V. On the other hand, a small amount of SB co-immunoprecipitated with NHE3V, but the relative intensity of the SB band compared with IRBIT indicated that its interaction was significantly compromised, which was consistent with the results from two-hybrid analysis.
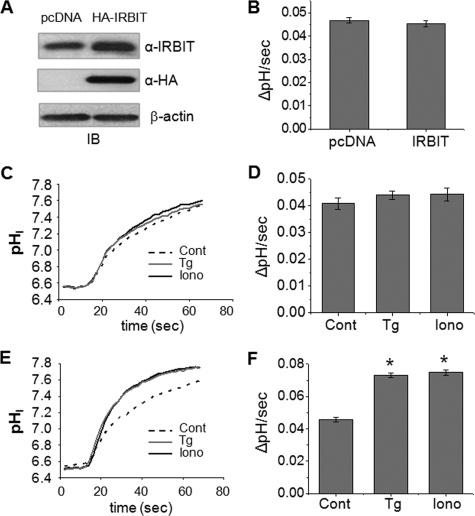
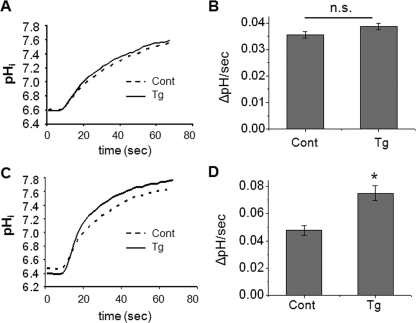
IRBIT-mediated Activation of NHE3 Dependent on Ca2+ Elevation—Recent studies have shown that IRBIT regulates intracellular Ca2+ via its ability to bind IP3R (4, 5). Ca2+ as a second messenger for regulation of NHE3 has been widely studied (14, 15, 23-25). Hence, we explored the possibility that IRBIT may regulate NHE3 in a Ca2+-dependent manner by overexpressing HA-IRBIT in PS120/NHE3V cells (Fig. 3A). Ectopic expression of IRBIT did not affect the basal transport activity of NHE3 of PS120/NHE3V cells (Fig. 3B). We next examined whether the endoplasmic/sarcoplasmic reticulum Ca2+-ATPase pump inhibitor thapsigargin (100 nm) regulates NHE3 activity in PS120/NHE3V fibroblasts. Fig. 3, C and D, show that thapsigargin had no effect on NHE3 activity in PS120/NHE3V cells. Similarly, the Ca2+ ionophore, ionomycin (500 nm), did not affect NHE3 activity. On the other hand, both thapsigargin and ionomycin increased NHE3 transporter activity by ∼60% in PS120/NHE3V/IRBIT cells (Fig. 3, E and F). These results show that IRBIT mediates Ca2+-dependent stimulation of NHE3 activity.
FIGURE 3.
Expression of IRBIT results in Ca2+-dependent activation of NHE3 activity. A, Western blot (IB, immunoblot) shows ectopic expression of HA-IRBIT in PS120/NHE3V cells. Total IRBIT was detected by anti-IRBIT and ectopic expression of IRBIT was determined with an anti-HA antibody. B, serum-starved PS120/NHE3V/pcDNA or PS120/NHE3V/IRBIT cells loaded with BCECF and NHE3 activity was determined fluorometically as described under “Experimental Procedures.” NHE3 activity was represented as the rate of Na+-dependent pH recovery, ΔpH/s. n = 8. C, representative traces of Na+-dependent pH recovery in PS120/NHE3V/pcDNA cells treated with 100 nm thapsigargin, 500 nm ionomycin, or carrier are shown. D, NHE3 activities in PS120/NHE3V/pcDNA cells treated with thapsigargin, ionomycin, or carrier are shown. The rates of pHi recovery at pHi 6.7 are shown. n = 8. E, representative traces of Na+-dependent pH recovery in PS120/NHE3V/HA-IRBIT cells are shown. F, NHE3 activities in PS120/NHE3V/IRBIT cells are shown. The rates of pHi recovery at pHi 6.7 are shown. n = 12 or more. *, p < 0.01 compare with the control.
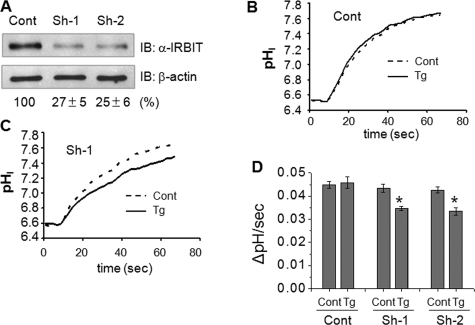
To affirm the importance of IRBIT in Ca2+-mediated activation of NHE3, we knocked down endogenous expression of IRBIT in PS120/NHE3V cells. Two shRNA constructs, sh-1 and sh-2, were able to knock down IRBIT expression by ∼75% in PS120/NHE3V cells (Fig. 4A). Importantly, in contrast to untransfected or control transfected cells in which thapsigargin exerted no effect on NHE3 activity, thapsigargin inhibited NHE3 activity in sh-1 or sh-2 transfected cells by ∼20% (Fig. 4B), confirming the role of IRBIT in up-regulation of NHE3 activity.
FIGURE 4.
Knockdown of IRBIT results in Ca2+-dependent inhibition of NHE3 activity. A, PS120/NHE3V cells were transfected with shRNA targeting IRBIT or control shRNA. Each effective shRNA (sh-1 or sh-2) knocked down IRBIT expression in PS120 cells by ∼75%. Representative traces of Na+-dependent pH recovery in PS120/NHE3V cells treated with control (B) or sh-1 (C) in the presence of absence of thapsigargin (Tg) are shown. D, NHE3 activities in PS120/NHE3V cells transfected with control, sh-1, or sh-2 determined at pHi 6.6 are shown. *, p < 0.01 compare with the control. IB, immunoblot.
Having shown the role of IRBIT in NHE3 regulation, we sought to determine whether binding of IRBIT to the COOH-terminal domain of NHE3 is necessary for the regulation of NHE3 activity. We examined the effect of thapsigargin on Na+/H+ exchange activity in PS120/NHE3V-585/IRBIT and PS120/NHE3V-689/IRBIT cells. Earlier we showed that IRBIT bound to NHE3V-689 but not to NHE3V-585 (Fig. 2). Consistent with the IRBIT binding, thapsigargin stimulated Na+/H+ exchange activity in PS120/NHE3V-689/IRBIT cells (Fig. 5B), but not in PS120/NHE3V-585/IRBIT (Fig. 5A), providing evidence that the interaction of IRBIT and NHE3 is necessary for IRBIT-dependent activation of NHE3 activity.
FIGURE 5.
The presence of IRBIT activates NHE3V-689 but not NHE3V-585. A and B, PS120/NHE3V-585; or C and D, PS120/NHE3V-689 cells were transfected with HA-IRBIT and the effect of thapsigargin (Tg) on NHE3 activity was determined. A and C show representative traces. B and D show the rate of pH recovery at pHi at 6.7. n = 8 or more. n.s., not significant. *, p < 0.01. IB, immunoblot.
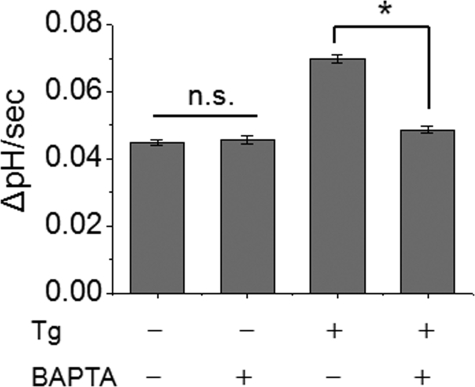
To verify that IRBIT-dependent regulation of NHE3 is dependent on the rise in intracellular Ca2+, we prevented the change in Ca2+ by pretreating cells with the Ca2+ chelator, BAPTA-AM. Pretreatment with BAPTA-AM (40 μm) for 20 min during dye loading showed no inhibitory effect on basal NHE3 activity, but completely blocked thapsigargin-induced stimulation of NHE3 activity (Fig. 6). These findings suggest that the IRBIT-dependent stimulatory effect on NHE3 activity requires a change in intracellular Ca2+.
FIGURE 6.
Ca2+-dependent activation of NHE3 is blocked by BAPTA. PS120/NHE3V/IRBIT cells were pretreated with the Ca2+ chelator, BAPTA (40 μm), for 20 min and NHE3 activity was determined with or without thapsigargin (Tg) treatment. The rates of pH recovery at pHi 6.6 are shown. n.s., not significant. n = 8 or more. *, p < 0.01.
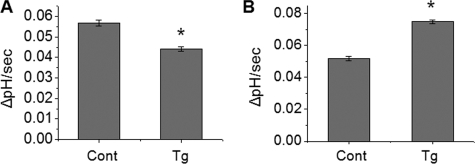
Expression of IRBIT Reverses NHERF2-dependent Ca2+ Inhibition of NHE3 Activity—Previous reports suggested that NHERF2 is required for Ca2+-mediated inhibition of NHE3 activity (15, 24). We were curious what would happen to NHE3 activity when IRBIT was co-expressed with NHERF2. Although thapsigargin did not affect NHE3 activity in PS120/NHE3V cells, which lack endogenous expression of NHERF2 (15, 26), ectopic expression of NHERF2 resulted in an inhibition of NHE3 activity when the cells were treated with thapsigargin (Fig. 7A). Thapsigargin treatment stimulated NHE3 transport activity by ∼40% in PS120/NHE3V/NHERF2/IRBIT (Fig. 7B) as opposed to ∼60% in PS120/NHE3V/IRBIT cells. These results indicate that NHERF2 and IRBIT impose opposite effects on NHE3 activity and the effects dependent on IRBIT or NHEFR2 are additive.
FIGURE 7.
Expression of IRBIT counteracts the effect of NHERF2 on NHE3 activity. A, PS120/NHE3V/NHERF2 cells were treated with thapsigargin (Tg) or carrier and NHE3 activity was determined. B, NHE3 activity in PS120/NHE3V/NHERF2/IRBIT cells with or without thapsigargin treatment was determined. The rates of pH recovery at pHi 6.7 are shown. n = 8. *, p < 0.01.
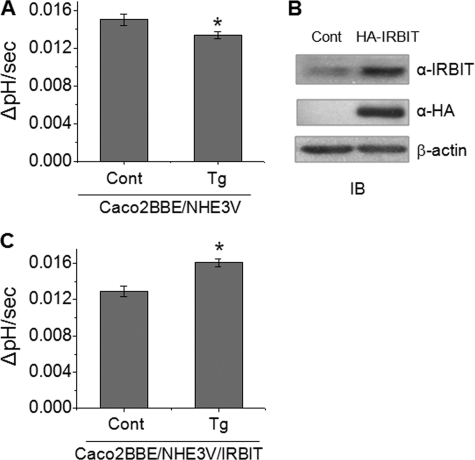
Ca2+ Elevation Enhances NHE3 Activity in Caco2BBE/NHE3V/IRBIT Cells—Having shown that IRBIT mediates stimulation of NHE3 transport activity in PS120 cells, we determined whether IRBIT exerts a similar effect on NHE3 in epithelial cells. To this end, we used Caco2BBE cells stably transfected with NHE3V to boost the overall NHE3 activity. NHE3 activity in Caco2BBE/NHE3V cells were measured in the presence of 50 μm HOE694, which blocked all Na+/H+ exchange activity of NHE1 and NHE2 (20). As shown in Fig. 8A, thapsigargin treatment attenuated NHE3 transport activity by ∼13%. It is noteworthy that the inhibition of NHE3 by thapsigargin in Caco2BBE/NHE3V cells is consistent with the presence of NHERF2 (19). The endogenous expression level of IRBIT in Caco2BBE cells was relatively low, which was significantly heightened by ectopic expression of HA-IRBIT (Fig. 8B). As in PS120 cells, ectopic expression of IRBIT resulted in stimulation of NHE3 activity in Caco2BBE/NHE3V cells (Fig. 8C), confirming its role in up-regulation of NHE3 activity.
FIGURE 8.
Expression of IRBIT induces Ca2+-dependent activation of NHE3 in Caco2BBE cells. A, NHE3 activity in Caco2BBE/NHE3V cells with or without thapsigargin (Tg) treatment was determined in the presence of 50 μm HOE694. The rate of pH recovery at pHi 6.9 are shown. B, HA-IRBIT was ectopically expressed in Caco2BBE/NHE3V cells. Total and exogenous IRBIT proteins were detected by anti-IRBIT and anti-HA antibodies, respectively. C, NHE3 activity, represented as the rate of pH recovery at pHi 6.9, was determined in Caco2BBE/NHE3V/IRBIT cells. n = 8. *, p < 0.05 compare with the control.
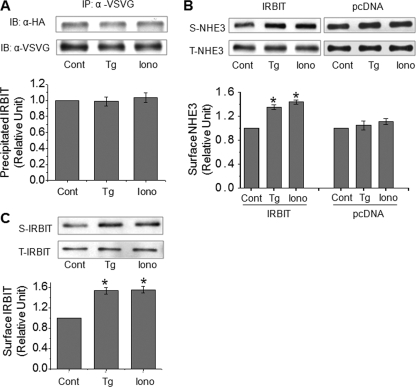
Ca2+ Elevation Increases Exocytosis of NHE3 in PS120/NHE3V/IRBIT Cells—Next, we explored the mechanism underlying stimulation of NHE3 activity by Ca2+ elevation in IRBIT-transfected PS120/NHE3V cells. We first examined whether the interaction between IRBIT and NHE3 was regulated by thapsigargin or ionomycin treatment, because a previous study demonstrated that binding of IRBIT to pNBC1 was increased by the presence of Ca2+ in lysate buffer (6). On the contrary, we found that the IRBIT-NHE3 interaction was not regulated by elevation of Ca2+ by either thapsigargin or ionomycin treatments (Fig. 9A). Thus, the enhanced NHE3 activity is unlikely due to an increase in IRBIT binding to NHE3.
FIGURE 9.
Elevated Ca2+ increases surface NHE3 protein abundance in the presence of IRBIT. A, NHE3V and IRBIT were co-immunoprecipitated with anti-VSVG in PS120/NHE3V/IRBIT cells treated with thapsigargin (Tg), ionomycin, or carrier. Relative amounts of co-immunoprecipitated HA-IRBIT proteins was quantified. B, PS120/NHE3V/pcDNA and PS120/NHE3V/IRBIT cells were treated with thapsigargin, ionomycin, or carrier for 10 min, and the amount of NHE3V protein on the plasma membrane was determined by surface biotinylation as detailed under “Experimental Procedures.” Aliquots of surface and total protein were resolved by SDS-PAGE and the amount of surface (S) NHE3 and total (T) NHE3 proteins were determined by Western blot using an anti-VSVG antibody. The amount of surface NHE3 was normalized to total NHE3 and relative changes are shown in the graph. C, the amount of IRBIT in surface and total fractions was determined. n = 4. *, p < 0.01 compare with the control. IB, immunoblot.
NHE3 protein recycles between the plasma membrane and intracellular compartments, which largely accounts for the acute regulation of NHE3 activity in a manner of endocytosis or exocytosis (15, 27-29). Therefore, we performed surface biotinylation studies aimed to determine whether the amount of NHE3 protein in the plasma membrane was increased in PS120/NHE3V/IRBIT cells with elevated Ca2+. As shown in Fig. 9B (left), both thapsigargin and ionomycin treatment increased the surface NHE3 protein abundance by ∼40% compared with the vehicle-treated control after normalizing to the total NHE3 protein expression level. In contrast, no significant change in NHE3 protein was observed in PS120/NHE3V/pcDNA cells by Ca2+ elevation (Fig. 9B, right). Interestingly, we found that the amount of IRBIT protein associated with the plasma membrane was concurrently increased by ∼50% (Fig. 9C). IRBIT does not contain a putative transmembrane helix (4) and the presence of IRBIT in the surface fraction must be mediated via interaction with membrane proteins such as NHE3. It is noteworthy that we estimated the amount of membrane-associated IRBIT to be less than 1% of total IRBIT in the cell. Together, these results suggest that elevation of Ca2+ enhances exocytic insertion of the NHE3-IRBIT complex into the plasma membrane without altering the dynamics of NHE3-IRBIT interaction.
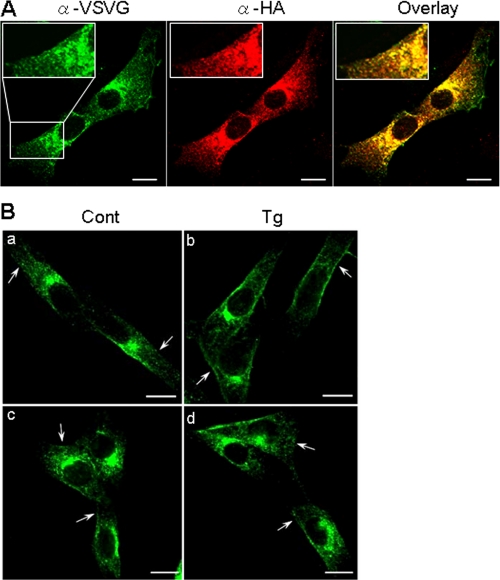
Co-localization of NHE3 and IRBIT in PS120 Cell—To further determine whether there is a spatial association between NHE3 and IRBIT in PS120/NHE3V/IRBIT cells, we performed confocal immunofluorescence analysis using monoclonal anti-VSVG and polyclonal anti-HA antibodies for detection of NHE3 and IRBIT, respectively. NHE3V (Fig. 10, green) was largely distributed at the juxtanuclear cytoplasmic compartment, representing the recycling endosomes as previously shown (27). In addition, distinct labeling of NHE3V at the plasma membrane was also evident. HA-IRBIT (red) was localized mainly to the cytoplasms with weak labeling of IRBIT at the plasma membrane. The weak membrane labeling of IRBIT is consistent with our estimation from surface biotinylation analysis that only a small fraction of IRBIT (<1%) is associated with the plasma membrane. However, significant overlap of NHE3V and IRBIT was observed throughout the cytoplasm, although the overlap was more prominent near the nuclei (Fig. 10A). Treatment of PS120/NHE3V/IRBIT cells with thapsigargin for 10 min increased the intensity of membrane staining by the anti-VSVG antibody, consistent with the increase in surface NHE3 protein as determined by surface biotinylation (Fig. 10B, a and b). Although the biochemical analysis showed an increase in membrane-associated IRBIT (Fig. 9), we could not detect a change in IRBIT surface expression by confocal microscopic analysis with any certainty (not shown) probably because of low membrane expression of IRBIT.
FIGURE 10.
NHE3 and IRBIT co-localize in PS120/NHE3V cells. A, cellular distribution of NHE3 and IRBIT in PS120/NHE3V/IRBIT cells was determined by indirect immunofluorescence using anti-VSVG and anti-HA antibodies, respectively. NHE3 (green) and IRBIT (red) were analyzed by a confocal microscope using a ×63 lens. The overlaid image of the double staining is shown on the right. Insets show magnified images. B, cellular localization of NHE3 in the presence or absence of IRBIT under basal conditions or after 10 min of thapsigargin (Tg) treatment was determined. Arrows point to NHE3 proteins on the plasma membrane. Bars, 10 μm.
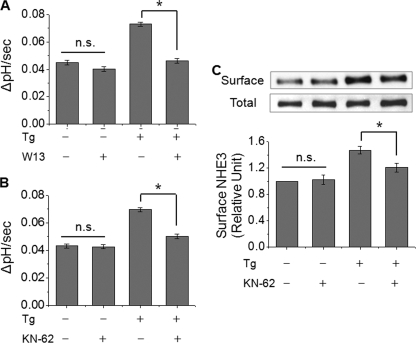
IRBIT-mediated Exocytosis of NHE3 and Increase in NHE3 Activity Are CaM-CaMKII Dependent—Calmodulin (CaM) is a representative protein that amplifies Ca2+ signaling (30). Considering that Ca2+ elevation is required for IRBIT-mediated activation of NHE3 transport activity, we next examined whether CaM was an intermediate protein in this cascade. Pre-incubation of PS120/NHE3V/IRBIT cells with the CaM inhibitor W13 (50 μm) for 20 min did not show a significant effect on the Na+/H+ exchange activity of NHE3 under the basal conditions (Fig. 11A). However, the presence of W13 completely abrogated the Ca2+-induced activation of NHE3 activity.
FIGURE 11.
Ca2+-mediated activation of NHE3 is dependent on CaM and CaMKII. The effect of thapsigargin (Tg) on NHE3 activity was determined with or without pretreatment of cells with (A) CaM inhibitor W13 (50 μm) or (B) CaMKII inhibitor KN-62 (20 μm). C, the effect of KN-62 on the amount of NHE3 protein at the surface membrane was determined by surface biotinylation. n.s., not significant. *, p < 0.05 compare with the absence of KN-62.
To determine whether CaM-dependent kinase II (CaMKII) is involved in this regulation, PS120/NHE3V/IRBIT cells were pretreated with the CaMKII inhibitor KN-62 (20 μm) for 20 min before measuring NHE3 activity. Similarly to W13, KN-62 itself had no effect on the basal activity of NHE3, but it attenuated the stimulatory effect of thapsigargin treatment on NHE3 activity (Fig. 11B).
We showed earlier that Ca2+-stimulated NHE3 transport activity by inserting NHE3 proteins in the plasma membrane. Thus, we examined whether the inhibition of CaMKII by KN-62 perturbed the exocytic insertion of NHE3 into the plasma membrane. In parallel with the effects on NHE3 transport activity, KN-62 pretreatment attenuated the change in surface NHE3 expression incurred by thapsigargin treatment (Fig. 11C).
DISCUSSION
In this work, we identified IRBIT as a NHE3-binding protein. Expression of IRBIT resulted in stimulation of NHE3 transport activity, whereas silencing of IRBIT inhibited NHE3 activity in response to increased Ca2+. We showed that an increase in Ca2+ by thapsigargin or ionomycin increased the surface NHE3 abundance, but the interaction between NHE3 and IRBIT was not regulated by Ca2+. Moreover, activation of NHE3 in the presence of IRBIT was CaM and CaMKII dependent.
S-Adenosylhomocysteine hydrolase (AdoHcy) is involved in cellular methylation (31). AdoHcy catalyzes the hydrolysis of S-adenosyl-l-homocysteine to adenine and l-homocystein. IRBIT/AdoHcyL1 shares 51% identity (74% similarity) to AdoHcy and it is ubiquitously expressed (4). The importance of IRBIT/AdoHcyL1 remains incompletely understood, but its expression is elevated during dendritic cell differentiation (32). Ectopic expression of IRBIT/AdoHcyL1 in zebrafish embryos induced dorsalization, whereas gene silencing of IRBIT/AdoHcyL1 induced ventralized morphologies, none of which could be mimicked by the manipulation of AdoHcy gene expression, suggesting distinct functions by IRBIT from AdoHcy (33). Ando et al. (4) using an IP3R fusion protein affinity column cloned IRBIT. The binding of IRBIT to IP3R is mediated by the NH2-terminal hydrophilic domain (aa 1-277) of IRBIT interacting with the IP3 binding core of IP3R (4). In our study, neither the NH2-terminal aa 1-105 (105R) nor aa 105-500 (105T) was able to bind NHE3, indicating that neither domain contains a single essential region for the interaction with NHE3. The NH2-terminal aa 1-224 (SB) bound to NHE3, but its interaction was significantly weaker compared with full-length IRBIT. Although whether aa 1-277 is capable of binding NHE3 was not examined, the inability of 105R (aa 1-105) or 105T (aa 105-530) alone implies that entire IRBIT is necessary for presumptive three-dimensional folding of IRBIT.
Another protein that interacts with IRBIT is NBC1 (6). Interestingly, IRBIT binds pancreas-type pNBC1 but not kidney-type kNBC1. pNBC1 differs from kNBC1 in that it has a unique NH2-terminal domain, which is enriched with positively charged residues clustered together between aa 40 and 60. It was proposed that the NH2-terminal domain of pNBC1 interacts with the serine-rich PEST domain of IRBIT (see the next paragraph) (6). In the present study, we showed that the region between aa 591 and 696 of NHE3 was essential for its interaction with IRBIT as shown by yeast two-hybrid and co-immunoprecipitation in PS120 cells. Accordingly, thapsigargin or ionomycin activated in Na+/H+ exchange activity in PS120/NHE3V-689 cells but not PS120/NHE3V-585, suggesting that binding of IRBIT is required for Ca2+-dependent activation of NHE3. Similarly to the NH2-terminal domain of pNBC1, the region between aa 591 and 696 of NHE3 is highly hydrophilic and has a propensity to form α-helices. Thus, we speculate that IRBIT-NHE3 association is in part mediated by these alkaline amino acid residues interacting with phosphorylated serine residues in the NH2-terminal domain of IRBIT.
A comparative analysis between IRBIT (AdoHcyL1) and AdoHcy indicates that the region distal to aa 105 of IRBIT is homologous to AdoHcy, but the NH2-terminal aa 1-104 of IRBIT is unique to IRBIT (34). This NH2-terminal region contains a PEST domain enriched with serine residues and phosphorylation of several serine residues is essential for the interaction with IP3R (5, 34). A recent study showed that a small region of IRBIT preceding the PEST domain binds protein phosphatase-1, which dephosphorylates Ser-68 of IRBIT, suggesting that IRBIT is regulated by phosphorylation and dephosphorylation (35). Phosphorylation at Ser-68 is apparently required for subsequent phosphorylation of Ser-71 and Ser-74. Although the protein kinase phosphorylating Ser-68 in vivo remains unknown, an in silico analysis suggested several candidate protein kinases, including CaMKII (35). Our present study showed that IRBIT-dependent regulation of NHE3 activity was sensitive to the inhibition of CaM and CaMKII. Both IRBIT and NHE3 contain CaMKII consensus phosphorylation sites, and it is tempting to speculate that CaMKII phosphorylates both IRBIT and NHE3. However, the IRBIT and NHE3 interaction was not affected by Ca2+, whereas Ca2+ increased the surface abundance of NHE3 in CaM- and CaMKII-dependent mechanisms. Therefore, we conclude that the Ca2+/CaM/CaMKII signaling axis regulates IRBIT-dependent trafficking of NHE3. This mechanism of NHE3 regulation differs from that of pNBC1 in which co-expression of IRBIT in Xenopus oocytes did not affect the surface expression of pNBC1 despite the activation of the transport activity (6).
IRBIT binds the IP3 binding pocket of IP3R acting as a “pseudoligand” that inhibits the activity of IP3R (5). IRBIT displaced from IP3RbyIP3 has been proposed to function as a downstream signal transducer of IP3R (4). However, it is unlikely that IRBIT regulates NHE3 as a downstream transducer of IP3R. We conclude that IRBIT and NHE3 form a microcomplex in the cytoplasm based on the observations that the IRBIT-NHE3 interaction occurs constitutively and is insensitive to elevation of Ca2+ and that NHE3 and IRBIT appear to be concurrently exocytosed to the surface membrane. In addition, significant fractions of NHE3 and IRBIT are present in the cytoplasm such that IRBIT can potentially interact with NHE3 and other interacting partners without having to translocate from the endoplasmic reticulum to the cytoplasm.
Ca2+-mediated regulation of NHE3 is complex. In rabbit ileal brush-border vesicles, elevated Ca2+ has been shown to inhibit brush-border Na+/H+ exchange through activation of protein kinase C (36). Using a peptide inhibitor of CaMKII, the same investigators showed that Ca2+ inhibits brush-border Na+/H+ exchange through a Ca2+/CaM/CaMKII-dependent mechanism in rabbit brush-border membrane vesicles (37). However, Ca2+ does not regulate Na+/H+ exchange activity of NHE3 in PS120 unless NHERF2 and α-actinin are co-expressed (15). On the contrary, lysophosphatidic acid, which activates phospholipase C through its cognate receptors, activates NHE3 in OK cells in a Ca2+-dependent manner (13). Chu et al. (14) showed that activation of endothelin-B receptor stimulates NHE3 activity by a Ca2+-dependent pathway in OKP cells. Therefore, elevation of Ca2+ is not the sole determinant of NHE3 regulation and the outcome varies dependent on cell types.
The stimulation of NHE3 in IRBIT-transfected PS120 cell and the inhibition of NHE3 by knockdown of IRBIT are consistent with the notion that IRBIT promotes NHE3 activity. That knockdown of IRBIT resulted in a decrease in NHE3 activity in PS120 cells suggests the propensity to inhibit NHE3 activity in the absence of IRBIT. Hence, elevation of Ca2+ is expected to inhibit NHE3 in cells with low expression of IRBIT, whereas in other cells with endogenous IRBIT, as in PS120 cells, this inhibition is offset by IRBIT-dependent activation.
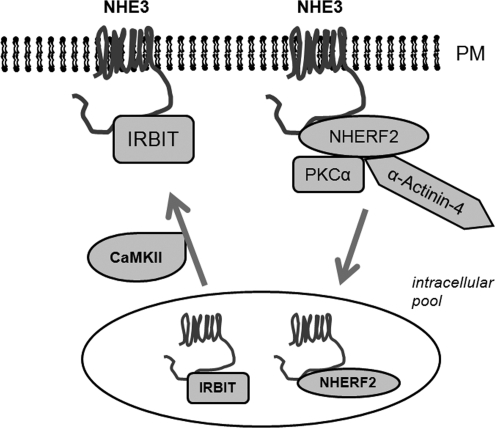
NHERF2 is a scaffold protein and its effect on signal transduction via its ability to assemble multiple proteins has been demonstrated (15, 21, 38-40). Previously, NHERF2 was shown to be necessary for the reconstitution of Ca2+-dependent inhibition of NHE3 activity in PS120 cells (15, 24) and we were able to reproduce the same results in the current study. Moreover, we showed that co-expression of IRBIT with NHERF2 reversed the direction of Ca2+-mediated NHE3 regulation from inhibition to stimulation, which suggests a competitive nature of NHERF2- and IRBIT-dependent effects. NHERF2 has been previously shown to bind aa 585-660 of NHE3 by an in vitro binding assay (18) and this NHERF2 binding domain overlaps with the IRBIT binding domain, aa 585-696. The magnitude of NHE3 stimulation in PS120/NHE3V/IRBIT/NHERF2 cells was ∼40% in comparison to ∼60% stimulation in PS120/NHE3V/IRBIT and ∼20% inhibition in PS120/NHE3V/NHERF2 cells. Based on the relative changes, it seems plausible that IRBIT may displace NHERF2, or vice versa. Alternatively, two separate pools of NHE3 may exist, one bound to NHERF2 and the other with IRBIT, and the overall effect on NHE3 activity is determined by the relative expression of NHERF2 and IRBIT. Although the exact relationship between NHERF2 and IRBIT cannot be deduced from the current study, we propose that the overall outcome of NHE3 regulation by Ca2+-elicited signaling is balanced by two opposing effects, positive regulation via IRBIT and negative regulation via NHERF2 (15, 24) (Fig. 12). Our preliminary study shows that the expression level of IRBIT protein is significantly higher in the kidney than in intestine.3 NHERF2 is expressed in the brush-border membrane of intestinal and renal proximal tubule epithelial cells (15, 41), although the functions of NHERF2 in regulation of NHE3 in vivo await a study using NHERF2 knock-out mice. The concurrent presence of IRBIT and NHERF2 suggests that IRBIT- or NHERF2-dependent regulation of NHE3 might occur in both intestine and kidney, but whether a similar mode of regulation occurs in kidney or intestine is yet to be investigated.
FIGURE 12.
A tentative model of regulation of NHE3 by Ca2+. Elevation of intracellular Ca2+ regulates NHE3 in two opposing pathways. NHE3 is retrieved into a cytoplasmic pool by a mechanism dependent on NHERF2-PKCα-Actinin-4. IRBIT stimulates NHE3 activity by enhancing exocytic trafficking of NHE3 to the surface membrane and this process is dependent of CaM/CaMKII. PM, plasma membrane.
A previous report showed that inhibition of CaM by W13 or CaMKII by KN-62 activated NHE3 activity in PS120 fibroblasts, suggesting that CaM and/or CaMKII plays a role in maintenance of basal NHE3 activity (16). Our studies using the same PS120 fibroblasts transfected with NHE3 did not show any effect on NHE3 transport activity by W13 or KN-62, although our cells were derived from different transfections, and the role of CaM and CaMKII on basal NHE3 activity remains contentious. Nevertheless, in agreement with our studies, these inhibitors did not affect basal NHE3 activity in OKP cells (14).
In summary, we showed that elevated Ca2+ stimulates NHE3 transport activity via IRBIT. The interaction of IRBIT is necessary for the stimulation of NHE3 activity, which resulted from enhanced translocation of NHE3 to the surface membrane. This exocytic trafficking of NHE3 is Ca2+ and CaMKII dependent.
Acknowledgments
We are grateful to Dr. Ming Tse for PS120 cells transfected with truncated NHE3. We also thank Dr. Didier Merlin for Caco2BBE cells.
This work was supported, in whole or in part, by National Institutes of Health Grant DK061418. This work was also supported by Emory Digestive Disease Research Development Center Grant DK064399. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: IP3, inositol 1,4,5-triphosphate;
IRBIT, IP3 receptor-binding protein released with IP3;
CaM, calmodulin; CaMKII, calmodulin-dependent kinase II; NHE,
Na+/H+ exchanger; NHERF2, Na+/H+
exchanger regulatory protein 2; NBC1,
Na+-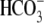 cotransporter 1;
VSVG, vesicular stomatitis virus glycoprotein; HA, hemagglutinin; AdoHcy,
S-adenosylhomocysteine hydrolase; AdoHcyL1,
S-adenosylhomocysteine hydrolase-like 1; PKC, protein kinase C;
shRNA, small hairpin RNA; aa, amino acid(s); PBS, phosphate-buffered saline;
TMA, tetramethylammonium; BCECF-AM,
2′,7′-bis-(2-carboxyethyl)-5-carboxyfluorescein acetoxymethyl
ester; BAPTA,
1,2-bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetic
acid; SD, synthetic dropout.
cotransporter 1;
VSVG, vesicular stomatitis virus glycoprotein; HA, hemagglutinin; AdoHcy,
S-adenosylhomocysteine hydrolase; AdoHcyL1,
S-adenosylhomocysteine hydrolase-like 1; PKC, protein kinase C;
shRNA, small hairpin RNA; aa, amino acid(s); PBS, phosphate-buffered saline;
TMA, tetramethylammonium; BCECF-AM,
2′,7′-bis-(2-carboxyethyl)-5-carboxyfluorescein acetoxymethyl
ester; BAPTA,
1,2-bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetic
acid; SD, synthetic dropout.
P. He and C. C. Yun, unpublished data.
References
- 1.Berridge, M. J., Bootman, M. D., and Roderick, H. L. (2003) Nat. Rev. Mol. Cell. Biol. 4 517-529 [DOI] [PubMed] [Google Scholar]
- 2.Berridge, M. J., Lipp, P., and Bootman, M. D. (2000) Nat. Rev. Mol. Cell. Biol. 1 11-21 [DOI] [PubMed] [Google Scholar]
- 3.Mikoshiba, K. (2007) J. Neurochem. 102 1426-1446 [DOI] [PubMed] [Google Scholar]
- 4.Ando, H., Mizutani, A., Matsu-ura, T., and Mikoshiba, K. (2003) J. Biol. Chem. 278 10602-10612 [DOI] [PubMed] [Google Scholar]
- 5.Ando, H., Mizutani, A., Kiefer, H., Tsuzurugi, D., Michikawa, T., and Mikoshiba, K. (2006) Mol. Cell 22 795-806 [DOI] [PubMed] [Google Scholar]
- 6.Shirakabe, K., Priori, G., Yamada, H., Ando, H., Horita, S., Fujita, T., Fujimoto, I., Mizutani, A., Seki, G., and Mikoshiba, K. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 9542-9547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brett, C. L., Donowitz, M., and Rao, R. (2005) Am. J. Physiol. 288 C223-C239 [DOI] [PubMed] [Google Scholar]
- 8.Zachos, N. C., Tse, M., and Donowitz, M. (2005) Annu. Rev. Physiol. 67 411-443 [DOI] [PubMed] [Google Scholar]
- 9.Biemesderfer, D., Rutherford, P. A., Nagy, T., Pizzonia, J. H., Abu-Alfa, A. K., and Aronson, P. S. (1997) Am. J. Physiol. 273 F289-F299 [DOI] [PubMed] [Google Scholar]
- 10.Hoogerwerf, W. A., Tsao, S. C., Devuyst, O., Levine, S., Yun, C. H. C., Yip, J. W., Cohen, M., Wilson, P. D., Lazenby, A. J., Tse, M., and Donowitz, M. (1996) Am. J. Physiol. 270 G29-G41 [DOI] [PubMed] [Google Scholar]
- 11.Schultheis, P. J., Clarke, L. L., Meneton, P., Miller, M. L., Soleimani, M., Gawenis, L. R., Riddle, T. M., Duffy, J. J., Doetschman, T., Wang, T., Giebisch, G., Aronson, P. S., Lorenz, J. N., and Shull, G. E. (1998) Nat. Genet. 19 282-285 [DOI] [PubMed] [Google Scholar]
- 12.Wang, T., Yang, C. L., Abbiati, T., Schultheis, P. J., Shull, G. E., Giebisch, G., and Aronson, P. S. (1999) Am. J. Physiol. 277 F298-F302 [DOI] [PubMed] [Google Scholar]
- 13.Choi, J. W., Lee-Kwon, W., Jeon, E. S., Kang, Y. J., Kawano, K., Kim, H. S., Suh, P.-G., Donowitz, M., and Kim, J. H. (2004) Biochim. Biophys. Acta 1683 59-68 [DOI] [PubMed] [Google Scholar]
- 14.Chu, T. S., Peng, Y., Cano, A., Yanagisawa, M., and Alpern, R. J. (1996) J. Clin. Investig. 97 1454-1462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim, J. H., Lee-Kwon, W., Park, J. B., Ryu, S. H., Yun, C. H., and Donowitz, M. (2002) J. Biol. Chem. 277 23714-23724 [DOI] [PubMed] [Google Scholar]
- 16.Levine, S. A., Nath, S., Yun, C. H. C., Yip, J. W., Montrose, M. H., Donowitz, M., and Tse, C. M. (1995) J. Biol. Chem. 270 13716-13725 [DOI] [PubMed] [Google Scholar]
- 17.Yun, C. H. C., Tse, C.-M., and Donowitz, M. (1995) Proc. Natl. Acad. Sci. U. S. A. 92 10723-10727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yun, C. C., Lamprecht, G., Forster, D. V., and Sidor, A. (1998) J. Biol. Chem. 273 25856-25863 [DOI] [PubMed] [Google Scholar]
- 19.Yun, C. C., Chen, Y., and Lang, F. (2002) J. Biol. Chem. 277 7676-7683 [DOI] [PubMed] [Google Scholar]
- 20.Wang, D., Sun, H., Lang, F., and Yun, C. C. (2005) Am. J. Physiol. 289 C802-C810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lamprecht, G., Weinman, E. J., and Yun, C. C. (1998) J. Biol. Chem. 273 29972-29978 [DOI] [PubMed] [Google Scholar]
- 22.Di Sole, F., Cerull, R., Babich, V., Quinones, H., Gisler, S. M., Biber, J., Murer, H., Burckhardt, G., Helmle-Kolb, C., and Moe, O. W. (2004) J. Biol. Chem. 279 2962-2974 [DOI] [PubMed] [Google Scholar]
- 23.Donowitz, M., Cohen, M. E., Gould, M., and Sharp, G. W. G. (1989) J. Clin. Investig. 83 1953-1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee-Kwon, W., Kim, J. H., Choi, J. W., Kawano, K., Cha, B., Dartt, D. A., Zoukhri, D., and Donowitz, M. (2003) Am. J. Physiol. 285 C1527-C1536 [DOI] [PubMed] [Google Scholar]
- 25.Cinar, A., Chen, M. M., Riederer, B., Bachmann, O., Manns, M., Kocher, O., and Seidler, U. (2007) J. Physiol. 581 1235-1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yun, C. C., Palmada, M., Embark, H. M., Fedorenko, O., Feng, Y., Henke, G., Setiawan, I., Boehmer, C., Weinman, E. J., Sandrasagra, S., Korbmacher, C., Cohen, P., Pearce, D., and Lang, F. (2002) J. Am. Soc. Nephr. 13 2823-2830 [DOI] [PubMed] [Google Scholar]
- 27.D'Souza, S., Garcia-Cabado, A., Yu, F., Teter, K., Lukacs, G., Skorecki, K., Moore, H.-P., Orlowski, J., and Grinstein, S. (1998) J. Biol. Chem. 273 2035-2043 [DOI] [PubMed] [Google Scholar]
- 28.Akhter, S., Cavet, M. E., Tse, C. M., and Donowitz, M. (2000) Biochemistry 39 1990-2000 [DOI] [PubMed] [Google Scholar]
- 29.Collazo, R., Fan, L., Hu, M. C., Zhao, H., Wiederkehr, M. R., and Moe, O. W. (2000) J. Biol. Chem. 275 31601-31608 [DOI] [PubMed] [Google Scholar]
- 30.Hoeflich, K. P., and Ikura, M. (2002) Cell 108 739-742 [DOI] [PubMed] [Google Scholar]
- 31.Kloor, D., and Osswald, H. (2004) Trends Pharmacol. Sci. 25 294-297 [DOI] [PubMed] [Google Scholar]
- 32.Dekker, J. W., Budhia, S., Angel, N. Z., Cooper, B. J., Clark, G. J., Hart, D. N., and Kato, M. (2002) Immunogenetics 53 993-1001 [DOI] [PubMed] [Google Scholar]
- 33.Cooper, B. J., Key, B., Carter, A., Angel, N. Z., Hart, D. N. J., and Kato, M. (2006) J. Biol. Chem. 281 22471-22484 [DOI] [PubMed] [Google Scholar]
- 34.Devogelaere, B., Nadif Kasri, N., Derua, R., Waelkens, E., Callewaert, G., Missiaen, L., Parys, J. B., and De Smedt, H. (2006) Biochem. Biophys. Res. Commun. 343 49-56 [DOI] [PubMed] [Google Scholar]
- 35.Devogelaere, B., Beullens, M., Sammels, E., Derua, R., Waelkens, E., van lint, J., Parys, J. B., Missiaen, L., Bollen, M., and De Smedt, H. (2007) Biochem. J. 407 303-311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cohen, M. E., Wesolek, J., McCullen, J., Rys-Sikora, K., Pandol, S., Rood, R. P., Sharp, G. W. G., and Donowitz, M. (1991) J. Clin. Investig. 88 855-863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cohen, M. E., Reinlib, L., Watson, A. J. M., Gorelick, F., Rys-Sikora, K., Tse, M., Rood, R. P., Czerik, A. J., Sharp, G. W. G., and Donowitz, M. (1990) Proc. Natl. Acad. Sci. U. S. A. 87 8990-8994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yun, C. C. (2003) Cell Physiol. Biochem. 13 29-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fam, S. R., Paquet, M., Castleberry, A. M., Oller, H., Lee, C. J., Traynelis, S. F., Smith, Y., Yun, C. C., and Hall, R. A. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 8042-8047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mahon, M. J., Donowitz, M., Yun, C. C., and Segre, G. V. (2002) Nature 417 858-861 [DOI] [PubMed] [Google Scholar]
- 41.Wade, J. B., Liu, J., Coleman, R. A., Cunningham, R., Steplock, D. A., Lee-Kwon, W., Pallone, T. L., Shenolikar, S., and Weinman, E. J. (2003) Am. J. Physiol. 285 C1494-C1503 [DOI] [PubMed] [Google Scholar]