Abstract
Human APOBEC3G (hA3G) is a cytidine deaminase active on HIV single-stranded DNA. Small angle x-ray scattering and molecular envelope restorations predicted a C-terminal dimeric model for RNA-depleted hA3G in solution. Each subunit was elongated, suggesting that individual domains of hA3G are solvent-exposed and therefore may interact with other macromolecules even as isolated substructures. In this study, co-immunoprecipitation and in-cell quenched fluorescence resonance energy transfer assays reveal that hA3G forms RNA-independent oligomers through interactions within its C terminus. Residues 209–336 were necessary and sufficient for homoligomerization. N-terminal domains of hA3G were unable to multimerize but remained functional for Gag and viral infectivity factor (Vif) interactions when expressed apart from the C terminus. These findings corroborate the small angle x-ray scattering structural model and are instructive for development of high throughput screens that target specific domains and their functions to identify HIV/AIDS therapeutics.
Human APOBEC3G (hA3G)4 is a cytidine deaminase that catalyzes the deamination of cytidine to uridine in the context of single-stranded (ss) DNA (1–3). This activity is critical to the function of hA3G as a host defense factor against HIV infection (4). When HIV replicates in cells, it expresses its own protein defense against hA3G known as viral infectivity factor or Vif. The N terminus of Vif interacts with the N terminus of hA3G (5–10), thereby promoting hA3G degradation via the ubiquitin-dependent proteasomal pathway and mitigating hA3G host defense (11–14).
In the absence of functional Vif, hA3G hypermutates proviral DNA during HIV replication (15–21). In resting CD4+ T cells, hA3G attacks incoming virions as a low molecular mass complex (21). However, once activated, CD4+ T cells suppress hA3G antiviral activity through the formation of high molecular mass complexes bridged by RNA (21, 22). Newly translated hA3G can exert its antiviral activity but only by assembling with viral particles (21, 22). This encapsidation enables hA3G immediate access to viral replication complexes and leads to inhibition of reverse transcription and hypermutation of minus-strand HIV DNA in the target cell (15–20). Viral particle assembly involves hA3G interaction with RNA and the nucleocapsid portion of HIV Gag during the viral assembly process (23–33). An N-terminal region of 50 amino acids following the first zinc-dependent deaminase (ZDD) motif (2, 34) within hA3G is necessary for its incorporation into virus-like particles (29, 30). There is no consensus on whether the hA3G and Gag interaction is a direct protein-protein interaction or whether it is mediated by a short RNA bridge (23–33).
Small angle x-ray scattering (SAXS) and advanced envelope restoration methods revealed the shape of full-length and catalytically active hA3G in solution (35). The resulting molecular envelope suggested that the native conformation of hA3G is minimally a dimer with subunit contacts through the C termini. The protein has an elongated morphology and substantially exposed surface area along the contour length of the dimer. This structural model predicted a limited contact between hA3G subunits but did not have resolution sufficient to identify the residues involved in dimerization. In support of these predictions, the crystal structure of human APOBEC2 also had an elongated shape comprised of dimers with subunit contacts through the N termini and C-terminal contacts between dimers, reminiscent of a dimer of dimers (36).
In contrast to these structural models, an NMR study demonstrated that a fragment of hA3G containing the C-terminal catalytic domain was a monomer and retained deaminase activity in an Escherichia coli reporter system (37). These findings bring into question the biological relevance of multimeric versus monomeric structural models for hA3G. In the current study, we demonstrate through in vitro and in-cell studies that hA3G forms multimers through protein-protein contacts within the ZDD domain of the C terminus. Consistent with our previous SAXS model, domains within the N terminus of hA3G did not form protein-protein multimers (35). The proposed elongated conformation of hA3G was further supported by the finding that domains retained their unique attributes of known functions and interactions when expressed as individual monomeric domains.
EXPERIMENTAL PROCEDURES
Plasmid Construction—Full-length hA3G and domains ΔCD1, C1/2, N1/2, CD1, CD2, NCD1, and NCD2 were cloned into the pIRES-P (38) mammalian expression vector with EGFP-HA, EGFP-V5, REACh2-HA, HA, or V5 tags in the N or C terminus. HIV-1 expression construct pDHIV3-GFP is a pNL4-3-derived vector that contains a deletion of the env gene and in which the nef gene is replaced with EGFP. The Δvif HIV-1 expression construct pDHIV3-GFP/Δvif contains a 12-bp insert containing two termination codons that reside near residue 89 of Vif, thereby leading to the production of a truncated and non-functional vif gene product (both HIV constructs were gifts from Prof. Baek Kim, Dept. of Immunology and Microbiology, University of Rochester). The vif gene was also PCR-amplified from pDHIV3-GFP and inserted into the pIRES-P vector containing an N-terminal EGFP-HA tag.
Co-immunoprecipitation Analysis—Various combinations of alternatively tagged hA3G domains, HIV-1 expression constructs, and EGFP-HA-Vif were transfected into 293T cells with FuGENE® 6 (Roche Applied Science). Twenty-four hours after transfection, cell extracts were harvested in Nonidet P-40 lysis buffer (50 mm Tris, pH 7.4, 150 mm NaCl, 0.1% Nonidet P-40) with Complete® Mini EDTA free protease inhibitors (Roche Applied Science). Cell extracts were treated with 0, 40, or 400 μg/ml RNase A for 30 min at 37 °C and subsequently precleared with protein A-agarose beads (Roche Applied Science) tumbling for 1 h at 4 °C. The precleared extracts were split and added to a protein A bead slurry ± V5 (Invitrogen) or GFP (Clontech) antibodies for immunoprecipitation by tumbling overnight at 4 °C. The beads were washed three times with Nonidet P-40 lysis buffer and eluted three times with 1× Treat (50 mm Tris, pH 7.4, 10 mm dithiothreitol, 0.1% SDS). Elutions were acetone-precipitated and resuspended in SDS-PAGE gel loading buffer and separated by 10.5% SDS-PAGE. The protein was transferred to nitrocellulose (Bio-Rad) and Western blotted with V5 (Invitrogen), HA (Covance), GFP (Clontech), or p24 (antibody number 3537, National Institutes of Health AIDS Research and Reference Reagent Program) (39) antibodies. Trueblot™ ULTRA anti-mouse secondary antibody (eBioscience), which does not bind to unfolded antibody, was used to prevent immunoglobulin heavy chain from being detected on Western blots.
RNA UV Cross-linking—To evaluate hA3G non-selective RNA binding activity, a 448-nucleotide transcript of apoB RNA was in vitro transcribed with 32P-radiolabeled ATP and CTP (PerkinElmer Life Sciences) using T7 polymerase (Promega). The radiolabeled RNA was gel-isolated and added to 500 μl of cell extracts made from 293T cells expressing V5-tagged hA3G or EGFP-V5-CD2 for 30 min at 30 °C. The RNA was UV cross-linked to the proteins in the cell extract using short wave UV light in quartz cuvettes at 4 °C for 7 min as described previously (40). Immediately after UV cross-linking, cell extracts were treated with 5 μg/ml each of RNases T1 and A for 1 h at 37 °C followed by preclearing and immunoprecipitation as described above. The nitrocellulose transferred protein was exposed to Biomax XAR film (Eastman Kodak Co.) to identify radiolabeled bands from nucleotides covalently cross-linked to hA3G or CD2. The nitrocellulose was subsequently Western blotted with V5 antibody (Invitrogen) to overlay with the radiolabeled band.
In Vivo FqRET—EGFP served as donor whose fluorescence signal was quenched by the resonance energy-accepting chromoprotein 2 (REACh2). In this fluorescence-quenched resonance energy transfer (FqRET) assay, REACh2 (a variant of yellow fluorescent protein) absorbs light at the emission wavelength of EGFP but has no fluorescence emission itself (41).
EGFP-V5-tagged N1/2, C1/2, and CD2 were co-transfected with REACh2-HA-tagged N1/2, C1/2, and CD2 or empty vector at a 1:8 μg ratio of cDNA for EGFP to REACh2 into 293T cells. In co-transfected cells, the EGFP chimera fluorescence signal will be quenched only if the REACh2 chimera is within close proximity (i.e. due to the interaction between hA3G domains).
Twenty-four hours after transfection, a 10 μm final concentration of Hoechst 33342 (Anaspec Inc., San Jose, CA) was added to the cell culture media, and cells were imaged at 500-ms exposures by a QICIM-IR fast 12 bit monochrome camera viewed by Q capture software (Q-Imaging) through a ×20 Olympus objective with an Olympus IX 70 inverted fluorescence microscope and label-specific chrome filters. The gray value for each cell in three separate fields was determined with the ImageJ software (National Institutes of Health). A linear regression model was used to specify the relationship between the measure of fluorescence intensity (gray value) for the EGFP-tagged domains in the presence and absence of REACh2-tagged domains. χ2 tests were performed to evaluate each domain comparison of interest at α = 0.05. All analyses were performed using SAS 9.1 (SAS Institute Inc., Cary, NC) on a Windows XP platform.
RESULTS
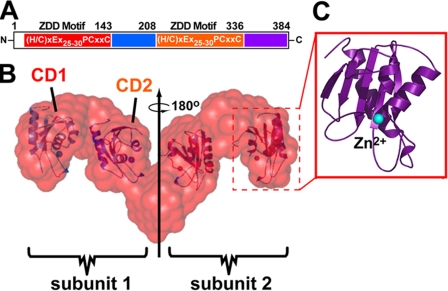
Rationale Guiding the Selection of hA3G Domains—SAXS analysis revealed the nm-resolution structure of catalytically active hA3G in solution. The size and mass suggested a dimer comprising four discrete, relatively large volumes arranged in an elongated and linear manner (Fig. 1B) (35). Each large volume of the molecular envelope accommodates the approximate volume of a representative ZDD domain (Fig. 1, B and C). Based on this distribution of ZDD domains within the SAXS envelope as well as the location of each ZDD motif in a single polypeptide (Fig. 1A), it was posited that hA3G subunits dimerize via the C-terminal domain (1, 35). Gel filtration of RNase-digested, recombinant full-length hA3G purified from baculovirus-infected Sf9 cells also supported a dimeric mass with an elution time corresponding to 95 kDa (35). Such a mass was consistent with a dimeric peak in other chromatography studies (21, 42, 43). However, the SAXS model does not have the resolution to determine exactly where within the C-terminal half-multimerization occurs, thus prompting a more comprehensive analysis in this study.
FIGURE 1.
Schematic diagram of hA3G domain boundaries. A, hA3G primary structure showing residues of CD1 (red), NCD1 (blue), CD2 (orange), and NCD2 (violet) with the consensus ZDD motif shown within CD1 and CD2. N, N terminus; C, C terminus. B, the molecular envelope of the tail-to-tail dimer of hA3G based on SAXS analysis with docked CD domains indicated within the model. C, the relation between the hA3G volumes and the minimal catalytic domain from a related crystal structure. Dashed red boxes (B) show how volumes in the SAXS envelope accommodate a representative CD domain located by computational analysis in our previous publication (35).
Alternatively tagged (HA and V5) forms of hA3G and domain combinations were expressed in 293T cells to probe for the existence of specific protein-protein interactions. With an initial emphasis on regions composed of a minimal ZDD fold found in all APOBEC related proteins (1), catalytic domain 1 (CD1, residues 1–143) and catalytic domain 2 (CD2, residues 209–336) were expressed. Subdomains adjacent to CD1 and CD2 were also expressed; these regions were called non-catalytic domain 1 (NCD1, 144–208) and non-catalytic domain 2 (NCD2, 337–384) (Fig. 1, A and B). Constructs containing two or more predicted domains were expressed that incorporated CD1-NCD1 (N1/2, 1–208), CD2-NCD2 (C1/2, 209–384), and a C-terminal domain lacking only CD1 (ΔCD1, 144–384).
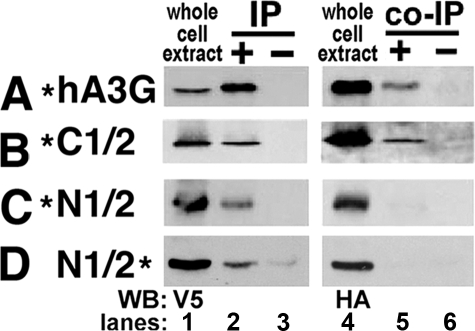
Multimerization Occurs via the C-terminal Half of hA3G—Cell extracts were prepared and analyzed by co-immunoprecipitation (co-IP). Extracts were treated with 40 μg/ml RNase A prior to co-IP to assuage interactions due to RNA bridging. All constructs were well expressed (Fig. 2, lanes 1 and 4), and each was recovered by IP utilizing the V5 tag (Fig. 2, lane 2). IPs performed with protein A beads alone indicated that none of the proteins associated non-specifically with protein A (Fig. 2, lanes 3 and 6). Co-IP analysis of N-terminal tagged hA3G and its half-domains revealed that full-length hA3G and the C1/2 domain were co-immunoprecipitated (Fig. 2, A and B), but there was no interaction in the N1/2 domain (Fig. 2C), consistent with predictions from the SAXS molecular envelope (Fig. 1). The lack of N-terminal interactions was confirmed using C-terminal HA- and V5-tagged constructs and co-IP analysis (Fig. 2D).
FIGURE 2.
Co-IPs of alternatively tagged hA3G and hA3G halves. A–D, the hA3G constructs co-transfected into 293T cells are represented in the left column with a * indicating the location of the tags (N- or C-terminal HA or V5) for each construct. Lanes 1 and 4 are blots showing the expression of each transfected protein in whole cell extracts. Lanes 2 and 5 are blots showing immunoprecipitated and co-immunoprecipitated proteins; all immunoprecipitations were performed with V5 antibody. Lanes 3 and 6 are controls for nonspecific binding to protein A beads. The antibody used for Western blotting (WB) is indicated on the bottom left for Western blots.
Although the N1/2 domain was incapable of homoligomerization, it was fully competent for its established interactions with Gag and Vif. N1/2 domain, including CD1-NCD1, was required for viral encapsidation and Gag binding (supplemental Fig. S1, A and B), whereas the CD1 domain alone was capable of interaction with Vif (supplemental Fig. S1C). These data were consistent with the literature (5–10, 23–33) and established that these autonomously expressed domains maintained fold and functionality, although they did not multimerize.
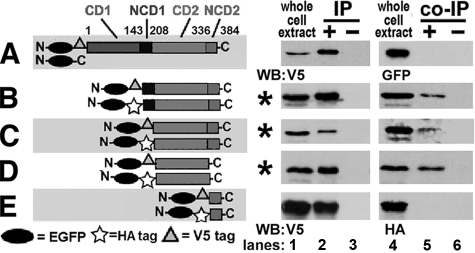
CD2 Is Necessary and Sufficient for hA3G Multimerization— The interaction of domains was further evaluated using smaller domains tagged with EGFP-V5 and EGFP-HA to increase their mass and thereby facilitate their analysis by SDS-PAGE. EGFP-V5-hA3G did not co-IP EGFP and therefore did not influence the analyses (Fig. 3A).
FIGURE 3.
Self-association of C-terminal domains of hA3G. In A–E, the hA3G domains co-transfected into 293T cells are represented in the left column as horizontal gray bars for CD1, NCD1, CD2, and NCD2 with symbols for EGFP, HA, and V5 tags. On the right, lanes are depicted as in Fig. 2. A, negative control for EGFP and EGFP-V5-hA3G. N, N terminus; C, C terminus. B–E, co-IPs of alternatively tagged hA3G domains with conditions in which co-immunoprecipitation was successful indicated by *. WB, Western blotting.
Co-IP analysis of hA3G domains revealed that ΔCD1, C1/2, and CD2 co-immunoprecipitated their alternatively tagged versions (Fig. 3, B–D), but NCD2 did not interact with itself (Fig. 3E). Combinations of N1/2 with C1/2, CD1 with CD2, and NCD1 with NCD2 were also unable to co-IP (supplemental Fig. S2). The data suggested that CD2 was necessary and sufficient for hA3G multimerization.
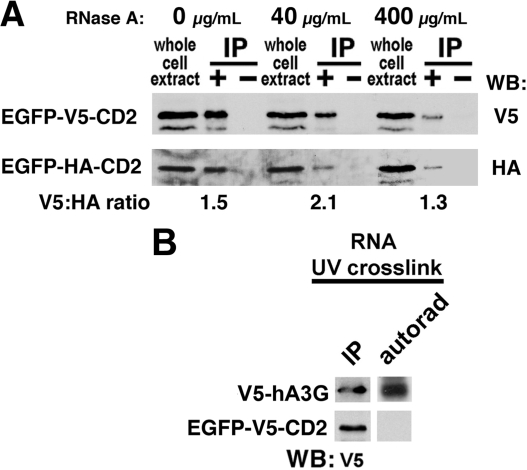
To strengthen our assertion that CD2 interacted via a protein-protein interaction and not an RNA bridge protected from RNase digestion, we analyzed the HA epitope to V5 epitope signal ratio by blotting EGFP-HA-CD2 co-immunoprecipitated by EGFP-V5-CD2 from cell extracts treated with 0, 40, and 400 μg/ml of RNase A. The V5:HA signal ratio was 1.5, 2.1, and 1.3 for the treatments, respectively (Fig. 4A), consistent with hA3G dimerization independent of an RNA bridge. Although the signal varied among treatments, the recovery of the HA epitope with the V5 pull-down remained strong, indicating that most of the interaction between CD2 domains depended on protein-protein contacts. In agreement with this proposal, EGFP-V5-CD2 did not UV cross-link to radiolabeled RNA (Fig. 4B). In contrast, full-length V5-hA3G did UV cross-link (Fig. 4B). This was consistent with the ability of full-length hA3G to bind RNA in the literature (25, 44, 45) and in our recent report showing that only full-length hA3G UV cross-linked RNA, whereas N1/2 and C1/2 could not (46).
FIGURE 4.
Evidence for protein-protein self-association within CD2. A, co-immunoprecipitations of CD2 domain with increasing amounts of RNase A (0, 40, and 400 μg/ml). The ratio of V5 and HA Western signals from the same exposure for each condition is indicated below each immunoprecipitation lane with densitometry of each band determined using ImageJ software (National Institutes of Health). WB, Western blotting. B, 32P-radiolabeled RNA was analyzed for UV cross-linking to V5-hA3G or EGFP-V5-CD2 in cell extracts followed by RNase digestion and immunoprecipitation, as described under “Experimental Procedures.” On the left are V5 blots detecting the full-length hA3G and the CD2 domain after immunoprecipitation. On the right, the autorad lane reveals the presence or absence of radioactivity associated with the matched Western band. The results are indicative of low or no RNA binding for CD2 relative to intact hA3G.
RNase-digested extracts of transfected 293T cells expressing EGFP-C1/2, EGFP-CD2, and EGFP were also analyzed by gel filtration for evidence that hA3G is multimeric and not monomeric. Fractions were collected and blotted with anti-GFP antibodies. EGFP eluted from the column as a monomer with a peak fraction corresponding to 27 kDa (supplemental Fig. S3). No significant protein was observed in fractions corresponding to monomers of EGFP-C1/2 or EGFP-CD2 (46 and 41 kDa, respectively), but significant protein appeared in fractions corresponding to dimers as well as higher order complexes (supplemental Fig. S3). Although RNase-digested, recombinant hA3G purified from Sf9 cells was clearly a dimer (35), higher molecular mass hA3G complexes in gel filtration of cell extracts have been described by several laboratories (42, 44, 45) and could be due to interactions with other proteins in the extract, multimerization of a higher order than dimers, or aggregation formed in cell extracts due to RNase digestion. As both the co-IP and the gel filtration analyses involved cell extracts, we utilized an additional approach of in vivo FqRET to confirm an interaction in the hA3G C terminus within living cells.
Interaction within C-terminal Domains Is Observed in Vivo by FqRET—EGFP is a FqRET donor, and REACh2 is a non-fluorescent FqRET acceptor (41). The non-fluorescent REACh2 is able to quench EGFP signal in a distance-dependent manner when they are linked to interacting domains. However, if there is no interaction, and EGFP and REACh2 are not proximal, quenching will not occur.
293T cells were co-transfected with EGFP-V5- and REACh2-HA-tagged versions of N1/2, C1/2, or CD2. EGFP-V5-tagged domains and an empty vector were also imaged to compare EGFP signals in the presence or absence of REACh2 to test protein-protein interactions in live cells.
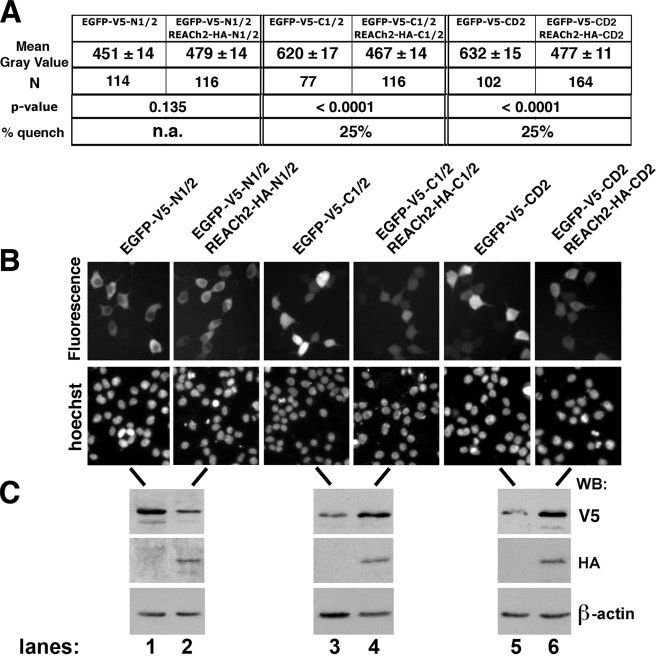
As expected, cells expressing the N1/2 domain showed no significant difference in fluorescence intensity with and without REACh2 yielding a p value equal to 0.135 (Fig. 5, A and B, lanes 1 and 2), suggesting no interaction in vivo. Western blotting for V5 and HA confirmed good expression of full-length EGFP and REACh2 chimeras (Fig. 5C, lanes 1 and 2). On the other hand, EGFP signal was quenched by 25% when linked to C1/2 or CD2 co-expressed with the REACh2-tagged C1/2 or CD2 counterparts relative to signal in the absence of the REACh2 linked domains (Fig. 5, A and B, lanes 3–6). p values of <0.0001 for both C1/2 and CD2 suggested that the difference in mean gray values were highly statistically significant (Fig. 5A). Comparisons between domains (adjusting for REACh2 expression) revealed highly significant differences for C1/2 and CD2 when compared with N1/2 (p < 0.0001) but not for the C1/2 versus CD2 comparison (p = 0.9457), suggesting that both domains quenched equivalently and that CD2 alone was sufficient for dimerization.
FIGURE 5.
In vivo FqRET of hA3G domains. A, as a measure of fluorescence intensity, the gray values from individual cells (number (N) indicated) in three separate fields were determined using ImageJ software (National Institutes of Health). The mean gray value is shown ± the S.E. The p values were determined by a linear regression model used to specify the relationship between the measure of fluorescence intensity in the presence and absence of REACh2-tagged domains as described under “Experimental Procedures.” The percentage of quench was measured as the difference in mean gray values of EGFP-tagged domains without and with REACh2, divided by the mean gray value for EGFP-tagged domains when expressed alone for C1/2 and CD2 domains. A quench value was not applicable (n.a.) for N1/2 because its p value revealed no significant difference (p > 0.05) with and without REACh2, suggesting that no quenching occurred. B, representative fields from images used to calculate fluorescence in A. EGFP fluorescence is shown above with Hoechst staining of the same field shown below representing live cell nuclei. C, Westerns with V5 and HA antibodies show relative tagged protein abundance for the transfected constructs, and the Western blot (WB) with β-actin antibody shows an equivalent load of cell extract for each condition. The transfected constructs indicated above B are the same for the lanes in B and C.
In the original study that identified the REACh2 protein, REACh1 (a variant of REACh2 with similar properties) quenched GFP signal by about 50% when covalently attached to GFP (41). Similarly, in our hands, transfection into 293T cells of a covalently tethered construct of EGFP-HA-REACh2 had a fluorescence signal that was about 50% quenched when compared with EGFP-HA-EGFP (data not shown). However, when the two FqRET proteins are expressed separately in the cell, there are three possible interactions if multimerization is occurring, which are: EGFP to EGFP, REACh2 to REACh2, and EGFP to REACh2 (quenching). Given that 50% quenching is the maximal anticipated result for end-to-end tethered donor and acceptor, 25% quenching of hA3G C-terminal domains is consistent with homoligomerization.
The intracellular distribution of EGFP-tagged domains suggested that the proteins were appropriately expressed in cells and not aggregated. In our previous study, we demonstrated a cytoplasmic retention signal in the N1/2 of hA3G (46). Thus the cytoplasmic fluorescence of EGFP-V5-N1/2 is consistent with a functionally folded chimera (Fig. 5B, lanes 1 and 2). On the other hand, there is no cytoplasmic retention signal in the C1/2 domain (46); therefore fluorescence throughout the cells for the C1/2 and CD2 domain chimeras is anticipated for non-aggregated proteins (Fig. 5B, lanes 3–6).
It is also important to note that the relative expression of each chimeric protein (as revealed by Western blotting, Fig. 5C) demonstrated that the reduction in fluorescence was due to quenching and not due to lower abundance of EGFP-tagged proteins. In fact EGFP-tagged C1/2 and CD2 were higher in abundance when their REACh2 counterparts were co-expressed when compared to when each was expressed alone (Fig. 5C, compare lane 3 with lane 4 and lane 5 with lane 6). Taken together, the data strongly support that hA3G is a multimer in vivo and that the intersubunit interaction predicted by the SAXS model occurs via RNA-independent interactions involving amino acid residues within CD2.
DISCUSSION
The SAXS molecular envelope represents the only experimentally derived structural model for full-length and catalytically active hA3G (35). The model predicted an elongated tail-to-tail dimer with a sparse amount of buried surface area. As such, the functional domains of hA3G were exposed along the length of each subunit with the exception of a C-terminal domain that formed intersubunit contacts. Although the nm-resolution of SAXS revealed that the interaction was likely to involve the C terminus, the domain or domains responsible for this interaction could not be determined given resolution limitations of the SAXS model.
In this study, we tested the predictions from SAXS by evaluating which domain in the C terminus of hA3G formed multimers and whether individual domains of hA3G expressed as monomers retained their function. We tested the domain organization using multiple methods to predict domain boundaries. We then evaluated interactions and functions of expressed domains using known functional interactions established for residues within these domains.
An important conclusion from this investigation is that domains of hA3G could be expressed individually as soluble proteins that retained many of their functional characteristics outside of the context of the full-length protein. This finding confirmed the prediction of an elongated structure of hA3G suggested by SAXS. The data also revealed that CD2 was sufficient for hA3G subunit multimerization.
Extensive RNase digestion did not disrupt CD2 subunit interactions. Most notably, under similar conditions of RNase digestion and immunoprecipitation, all other domains or combinations of domains lacking CD2 showed no capability to multimerize. Although we cannot rule out that a protected RNA fragment facilitated bridging between CD2 subunits, the immunoprecipitation data were consistent with dimerization of hA3G through what may be predominantly protein-protein interactions.
These findings compelled us to ask whether hA3G multimerization could be demonstrated in living cells. To this end, a FqRET assay was established using EGFP chimeras as fluorescence donors and REACh2 chimeras as fluorescence energy acceptors and quenchers. Quenching of fluorescence requires close proximity of EGFP and REACh2, making FqRET and fluorescence microscopy ideally suited for evaluating the ability of domains of hA3G to multimerize.
Consistent with the immunoprecipitation data, co-expression of the EGFP-hA3G N-terminal half with the REACh2-hA3G N-terminal half did not result in fluorescence quenching. In contrast, co-expression of the EGFP-hA3G C-terminal half with the REACh2-hA3G C-terminal half resulted in statistically significant fluorescence quenching. Co-expression of EGFP and REACh2 CD2 chimeras was also equally sufficient to induce fluorescence quenching. This report along with prior studies has shown that only full-length hA3G could bind to RNA (46). Therefore the quenched fluorescence of the C-terminal half of hA3G and CD2 was most likely due to protein-protein homoligomerization within CD2 rather than an indirect association through RNA bridging and ribonucleoprotein particle formation. Further fine structure mapping and high resolution structural analysis will be necessary to identify the residues involved in multimerization.
Catalytically active enzymes that target cytidine in nucleic acid substrates demonstrated modest site selectivity as evidenced by differences in their nearest neighbor preferences (47–49). RNA is the preferred substrate of APOBEC1, whereas ssDNA is the only known substrate for activation induced deaminase (AID), hA3G, and APOBEC3F (1–3). Moreover, the ZDD motif of APOBEC2 and the N termini of hA3G and APOBEC3F were determined to be catalytically inactive (49–51). The absence of deaminase activity may be a result of their inability to form catalytically productive protein-protein contacts. This is a likely hypothesis because trans-complementation of subunits is a characteristic of the cytidine deaminase active site architecture (1).
Biochemical and genetic studies suggested that APOBEC1 was a dimer (52–54) and that dimerization was required for RNA editing activity in vivo (55). Crystal structures for adenosine deaminases active on tRNA (ADAT) revealed dimeric interfaces as well (56, 57). More controversial is whether AID dimerization is required for class switch recombination and somatic hypermutation of immunoglobulin genes (58, 59).
The ZDD motif (H/CXEX(25–30)PCXXC) has been characterized in crystal structures of E. coli cytidine deaminase (60), the yeast enzyme Cdd1 (34), and APOBEC2 (34, 36). Several proposals have been made for the hA3G fold (2, 10, 61, 62). Higher order multimers of hA3G have been proposed as being functionally required for deaminase activity on ssDNA based on biochemical assays and atomic force microscopy (63). In contrast, mutagenesis of a C-terminal fragment of hA3G (amino acids 198–384) enabled this domain to be expressed as a soluble, monomeric protein that retained its ability to bind ssDNA and exhibited deaminase activity in E. coli when fused to GST (37). This finding indicated that there were conditions that may affect the ability of the C terminus to multimerize but begged the question of whether monomers of full-length hA3G were sufficient for catalytic and antiviral activities.
Biochemical studies have suggested that hA3G deaminase activity involved a protein dimer in RNA-depleted cell extracts (21). Chelico et al. (42) showed that the peak of hA3G deaminase activity corresponded to an RNA-depleted dimer by gel filtration and that the processive nature of the deaminase could be explained if two active sites are joined as a dimer. The Goodman laboratory (63) has predicted that hA3G monomers may lack the processivity of dC to dU deamination on HIV ssDNA that was a characteristic of hA3G activity in vivo. A recent report revealed that a dimer or monomer of hA3G was capable of deaminase activity; however, the most abundant form of the protein analyzed by fast protein liquid chromatography was clearly dimeric or in a deaminase inactive multimeric form with monomers nearly undetectable by Western blot (43). The authors concluded, however, that the full-length monomeric hA3G had a higher specific activity than dimeric hA3G. It was suggested that hA3G is multimeric in cells but is reduced to monomers in viral particles in which hA3G abundance is low (43). A focus for future studies is to gain clarity as to the role hA3G multimerization plays in antiviral activity in cells and viral particles.
Host defense activity of hA3G has been linked to formation of hA3G complexes consistent with dimerization, whereas inactivation occurred through RNA-mediated tetramer formation (35) and high order multimerization (21, 35, 44, 65, 66). Although these data strongly supported an RNA-independent, protein-protein-mediated C-terminal multimerization of hA3G (minimally a dimer) that can be compounded to higher order multimers through RNA bridging, they did not exclude the possibility that a small, protected segment of nucleic acid contributed to the formation of a dimeric interface of hA3G subunits, as noted previously (35).
The SAXS structural model also implied that hA3G dimers had an extensive extended surface readily accessible for macromolecular interactions. Support for this hypothesis was obtained in this study by expressing discrete hA3G domains outside the context of full-length protein and demonstrating that specific regions retained their biologically relevant interactions. For example, N1/2 retained the ability to bind to Gag and became encapsidated. The CD1 region was responsible for Vif binding in the context of full-length hA3G (5–10) and retained comparable binding activity when expressed alone. Given that the N terminus could not multimerize, these functional endpoints implied that domains within the N terminus could function autonomously as monomers.
Validation of the hA3G SAXS model is relevant because many investigators have considered modeling hA3G onto high resolution crystal structures of other deaminases, many of which are globular proteins (2, 10, 61, 62). However, it is anticipated that subdomains expressed from a protein with a globular fold would be poorly soluble and inactive due to the exposure of large numbers of hydrophobic residues that are normally buried in the protein core. A comparison between the experimentally measured x-ray scattering profile of dimeric hA3G in solution and the comparable profiles derived from the crystallographic coordinates of APOBEC family members demonstrated that the structure of other deaminases were illsuited for comparative modeling to hA3G (supplemental Fig. S4). This was because there is a topological restraint on the orientation of the ZDD domains within intact hA3G, which places the C terminus of N1/2 in close proximity to the N terminus of C1/2. APOBEC2 does not have this covalent tether because its polypeptide is half that of hA3G. Moreover, each subunit of hA3G contains two non-identical ZDD motifs, one in each half of the polypeptide chain (again unlike an APOBEC2 dimer). In addition, the amino acid sequence of hA3G is only ∼30% identical to APOBEC2 and ∼16% identical to yeast Cdd1 (1). From a functional perspective, the expected structure of a subunit of hA3G should be asymmetric because each of its halves are known to confer distinct capacities for host defense and viral protein interactions (24, 50, 51, 64).
The NMR structure of the C-terminal half of hA3G revisited the structure of the deaminase domain and demonstrated chemical shifts for those residues that bound ssDNA substrate (37). The parallel organization of the last β-strand in the NMR structure of the C-terminal domain was comparable with that of APOBEC2 (36) but differed from E. coli cytidine deaminase as well as yeast Cdd1 (reviewed in Ref. 1). The bulge between β2 and β2′ was an unexpected aspect of the structure that was noted as a possible N1/2 to C1/2 protein interface (37). Unfortunately, the NMR data only half-satisfied the requirements for understanding the link between structure and function for the full-length hA3G.
Through immunobiochemical techniques and live cell analyses, we have evaluated the veracity of the molecular envelope of hA3G determined by SAXS. We revealed that hA3G multimerizes through CD2 in the C terminus, thus extending our structural understanding of the homoligomerization of hA3G predicted by SAXS. We also corroborated the elongated nature of the SAXS structure by showing that the N1/2 did not multimerize, but it remained biologically competent in its known interactions. An important goal for future research is to obtain an atomic resolution crystal structure of full-length hA3G and to use structural information to design experiments to test whether dimerization of hA3G is crucial for deaminase and antiviral functions. Answers to these questions will provide us with a more complete understanding to create viable therapeutics that allow hA3G to be fully functional as an antiviral factor.
Supplementary Material
Acknowledgments
We thank Dr. Jay Reeder for use of a fluorescence microscope and for helpful discussions on quantification of data and software manipulations and Dr. Baek Kim for providing the HIV viral DNA constructs. The following reagents were obtained from the National Institutes of Health AIDS Research and Reference Reagent Program, Division of AIDS, NIAID: HIV-1 p24 monoclonal antibody (183-H12-5C) from Dr. B. Chesebro and K. Wehrly and HIV-1 Vif monoclonal antibody (antibody number 319) from Dr. M. H. Malim.
This work was supported, in whole or in part, by National Institutes of Health from the NIAID Grants AI058789 (to H. C. S.) and AI076085 (to J. E. W.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental “Experimental Procedures” and four supplemental figures.
Footnotes
The abbreviations used are: hA3G, human APOBEC3G; APOBEC, apolipoprotein B mRNA-editing enzyme, catalytic polypeptide; HIV, human immunodeficiency virus; Vif, viral infectivity factor; SAXS, small angle x-ray scattering; ssDNA, single-stranded DNA; ZDD, zinc-dependent deaminase; GFP, green fluorescent protein; EGFP, enhanced GFP; HA, hemagglutinin; REACh2, resonance energy-accepting chromoprotein 2; co-IP, co-immunoprecipitation; CD, catalytic domain; NCD, non-catalytic domain; FqRET, fluorescence-quenched resonance energy transfer.
References
- 1.MacElrevey, C., and Wedekind, J. E. (2008) in RNA and DNA Editing: Molecular Mechanisms and Their Integration into Biological Systems (Smith, H. C., ed) pp. 369-419, Wiley, New York
- 2.Wedekind, J. E., Dance, G. S., Sowden, M. P., and Smith, H. C. (2003) Trends Genet. 19 207-216 [DOI] [PubMed] [Google Scholar]
- 3.Jarmuz, A., Chester, A., Bayliss, J., Gisbourne, J., Dunham, I., Scott, J., and Navaratnam, N. (2002) Genomics 79 285-296 [DOI] [PubMed] [Google Scholar]
- 4.Sheehy, A., Gaddis, N., Choi, J., and Malim, M. (2002) Nature 418 646-650 [DOI] [PubMed] [Google Scholar]
- 5.Mehle, A., Wilson, H., Zhang, C., Brazier, A. J., McPike, M., Pery, E., and Gabuzda, D. (2007) J. Virol. 81 13235-13241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Russell, R. A., and Pathak, V. K. (2007) J. Virol. 81 8201-8210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schrofelbauer, B., Senger, T., Manning, G., and Landau, N. R. (2006) J. Virol. 80 5984-5991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mangeat, B., Turelli, P., Liao, S., and Trono, D. (2004) J. Biol. Chem. 279 14481-14483 [DOI] [PubMed] [Google Scholar]
- 9.Schrofelbauer, B., Chen, D., and Landau, N. R. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 3927-3932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huthoff, H., and Malim, M. H. (2007) J. Virol. 81 3807-3815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mariani, R., Chen, D., Schrofelbauer, B., Navarro, F., Konig, R., Bollman, B., Munk, C., Nymark-McMahon, H., and Landau, N. R. (2003) Cell 114 21-31 [DOI] [PubMed] [Google Scholar]
- 12.Mehle, A., Goncalves, J., Santa-Marta, M., McPike, M., and Gabuzda, D. (2004) Genes Dev. 18 2861-2866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stopak, K., De Noronha, C., Yonemoto, W., and Greene, W. C. (2003) Mol. Cell 12 591-601 [DOI] [PubMed] [Google Scholar]
- 14.Yu, X., Yu, Y., Liu, B., Luo, K., Kong, W., Mao, P., and Yu, X. F. (2003) Science 302 1056-1060 [DOI] [PubMed] [Google Scholar]
- 15.Bishop, K. N., Holmes, R. K., and Malim, M. H. (2006) J. Virol. 80 8450-8458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo, F., Cen, S., Niu, M., Saadatmand, J., and Kleiman, L. (2006) J. Virol. 80 11710-11722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mangeat, B., Turelli, P., Caron, G., Friedli, M., Perrin, L., and Trono, D. (2003) Nature 424 99-103 [DOI] [PubMed] [Google Scholar]
- 18.Harris, R. S., Bishop, K. N., Sheehy, A. M., Craig, H. M., Petersen-Mahrt, S. K., Watt, I. N., Neuberger, M. S., and Malim, M. H. (2003) Cell 113 803-809 [DOI] [PubMed] [Google Scholar]
- 19.Yu, Q., Konig, R., Pillai, S., Chiles, K., Kearney, M., Palmer, S., Richman, D., Coffin, J. M., and Landau, N. R. (2004) Nat. Struct. Mol. Biol. 11 435-442 [DOI] [PubMed] [Google Scholar]
- 20.Zhang, H., Yang, B., Pomerantz, R. J., Zhang, C., Arunachalam, S. C., and Gao, L. (2003) Nature 424 94-98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chiu, Y. L., Soros, V. B., Kreisberg, J. F., Stopak, K., Yonemoto, W., and Greene, W. C. (2005) Nature 435 108-114 [DOI] [PubMed] [Google Scholar]
- 22.Soros, V. B., Yonemoto, W., and Greene, W. C. (2007) PLoS Pathog. 3 e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burnett, A., and Spearman, P. (2007) J. Virol. 81 5000-5013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Navarro, F., Bollman, B., Chen, H., Konig, R., Yu, Q., Chiles, K., and Landau, N. R. (2005) Virology 333 374-386 [DOI] [PubMed] [Google Scholar]
- 25.Svarovskaia, E. S., Xu, H., Mbisa, J. L., Barr, R., Gorelick, R. J., Ono, A., Freed, E. O., Hu, W. S., and Pathak, V. K. (2004) J. Biol. Chem. 279 35822-35828 [DOI] [PubMed] [Google Scholar]
- 26.Khan, M. A., Kao, S., Miyagi, E., Takeuchi, H., Goila-Gaur, R., Opi, S., Gipson, C. L., Parslow, T. G., Ly, H., and Strebel, K. (2005) J. Virol. 79 5870-5874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schafer, A., Bogerd, H. P., and Cullen, B. R. (2004) Virology 328 163-168 [DOI] [PubMed] [Google Scholar]
- 28.Zennou, V., Perez-Caballero, D., Gottlinger, H., and Bieniasz, P. D. (2004) J. Virol. 78 12058-12061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luo, K., Liu, B., Xiao, Z., Yu, Y., Yu, X., Gorelick, R., and Yu, X. F. (2004) J. Virol. 78 11841-11852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cen, S., Guo, F., Niu, M., Saadatmand, J., Deflassieux, J., and Kleiman, L. (2004) J. Biol. Chem. 279 33177-33184 [DOI] [PubMed] [Google Scholar]
- 31.Alce, T. M., and Popik, W. (2004) J. Biol. Chem. 279 34083-34086 [DOI] [PubMed] [Google Scholar]
- 32.Khan, M. A., Goila-Gaur, R., Opi, S., Miyagi, E., Takeuchi, H., Kao, S., and Strebel, K. (2007) Retrovirology 4 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bogerd, H. P., and Cullen, B. R. (2008) RNA (Cold Spring Harbor) 14 1228-1236 [Google Scholar]
- 34.Xie, K., Sowden, M. P., Dance, G. S., Torelli, A. T., Smith, H. C., and Wedekind, J. E. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 8114-8119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wedekind, J. E., Gillilan, R., Janda, A., Krucinska, J., Salter, J. D., Bennett, R. P., Raina, J., and Smith, H. C. (2006) J. Biol. Chem. 281 38122-38126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prochnow, C., Bransteitter, R., Klein, M. G., Goodman, M. F., and Chen, X. S. (2007) Nature 445 447-451 [DOI] [PubMed] [Google Scholar]
- 37.Chen, K. M., Harjes, E., Gross, P. J., Fahmy, A., Lu, Y., Shindo, K., Harris, R. S., and Matsuo, H. (2008) Nature 452 116-119 [DOI] [PubMed] [Google Scholar]
- 38.Hobbs, S., Jitrapakdee, S., and Wallace, J. C. (1998) Biochem. Biophys. Res. Commun. 252 368-372 [DOI] [PubMed] [Google Scholar]
- 39.Chesebro, B., Wehrly, K., Nishio, J., and Perryman, S. (1992) J. Virol. 66 6547-6554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith, H. C. (1998) Methods (San Diego) 15 27-39 [DOI] [PubMed] [Google Scholar]
- 41.Ganesan, S., Ameer-Beg, S. M., Ng, T. T., Vojnovic, B., and Wouters, F. S. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 4089-4094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chelico, L., Pham, P., Calabrese, P., and Goodman, M. F. (2006) Nat. Struct. Mol. Biol. 13 392-399 [DOI] [PubMed] [Google Scholar]
- 43.Nowarski, R., Britan-Rosich, E., Shiloach, T., and Kotler, M. (2008) Nat. Struct. Mol. Biol. 15 1059-1066 [DOI] [PubMed] [Google Scholar]
- 44.Chiu, Y. L., Witkowska, H. E., Hall, S. C., Santiago, M., Soros, V. B., Esnault, C., Heidmann, T., and Greene, W. C. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 15588-15593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kozak, S. L., Marin, M., Rose, K. M., Bystrom, C., and Kabat, D. (2006) J. Biol. Chem. 281 29105-29119 [DOI] [PubMed] [Google Scholar]
- 46.Bennett, R. P., Presnyak, V., Wedekind, J. E., and Smith, H. C. (2008) J. Biol. Chem. 283 7320-7327 [DOI] [PubMed] [Google Scholar]
- 47.Beale, R. C., Petersen-Mahrt, S. K., Watt, I. N., Harris, R. S., Rada, C., and Neuberger, M. S. (2004) J. Mol. Biol. 337 585-596 [DOI] [PubMed] [Google Scholar]
- 48.Dang, Y., Wang, X., Esselman, W. J., and Zheng, Y. H. (2006) J. Virol. 80 10522-10533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harris, R. S., Petersen-Mahrt, S. K., and Neuberger, M. S. (2002) Mol. Cell 10 1247-1253 [DOI] [PubMed] [Google Scholar]
- 50.Hache, G., Liddament, M. T., and Harris, R. S. (2005) J. Biol. Chem. 280 10920-10924 [DOI] [PubMed] [Google Scholar]
- 51.Newman, E. N., Holmes, R. K., Craig, H. M., Klein, K. C., Lingappa, J. R., Malim, M. H., and Sheehy, A. M. (2005) Curr. Biol. 15 166-170 [DOI] [PubMed] [Google Scholar]
- 52.Lau, P. P., Zhu, H. J., Baldini, A., Charnsangavej, C., and Chan, L. (1994) Proc. Natl. Acad. Sci. U. S. A. 91 8522-8526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Navaratnam, N., Fujino, T., Bayliss, J., Jarmuz, A., How, A., Richardson, N., Somasekaram, A., Bhattacharya, S., Carter, C., and Scott, J. (1998) J. Mol. Biol. 275 695-714 [DOI] [PubMed] [Google Scholar]
- 54.Smith, H. C. (2007) Methods Enzymol. 424 389-416 [DOI] [PubMed] [Google Scholar]
- 55.Oka, K., Kobayashi, K., Sullivan, M., Martinez, J., Teng, B. B., Ishimura-Oka, K., and Chan, L. (1997) J. Biol. Chem. 272 1456-1460 [DOI] [PubMed] [Google Scholar]
- 56.Kuratani, M., Ishii, R., Bessho, Y., Fukunaga, R., Sengoku, T., Shirouzu, M., Sekine, S., and Yokoyama, S. (2005) J. Biol. Chem. 280 16002-16008 [DOI] [PubMed] [Google Scholar]
- 57.Macbeth, M. R., Schubert, H. L., Vandemark, A. P., Lingam, A. T., Hill, C. P., and Bass, B. L. (2005) Science 309 1534-1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang, J., Shinkura, R., Muramatsu, M., Nagaoka, H., Kinoshita, K., and Honjo, T. (2006) J. Biol. Chem. 281 19115-19123 [DOI] [PubMed] [Google Scholar]
- 59.Brar, S. S., Sacho, E. J., Tessmer, I., Croteau, D. L., Erie, D. A., and Diaz, M. (2008) DNA Repair (Amst.) 7 77-87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Betts, L., Xiang, S., Short, S. A., Wolfenden, R., and Carter, C. W., Jr. (1994) J. Mol. Biol. 235 635-656 [DOI] [PubMed] [Google Scholar]
- 61.Zhang, K. L., Mangeat, B., Ortiz, M., Zoete, V., Trono, D., Telenti, A., and Michielin, O. (2007) PLoS ONE 2 e378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huthoff, H., and Malim, M. H. (2005) Virology 334 147-153 [DOI] [PubMed] [Google Scholar]
- 63.Chelico, L., Sacho, E. J., Erie, D. A., and Goodman, M. F. (2008) J. Biol. Chem. 283 13780-13791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen, K. M., Martemyanova, N., Lu, Y., Shindo, K., Matsuo, H., and Harris, R. S. (2007) FEBS Lett. 581 4761-4766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stopak, K. S., Chiu, Y. L., Kropp, J., Grant, R. M., and Greene, W. C. (2007) J. Biol. Chem. 282 3539-3546 [DOI] [PubMed] [Google Scholar]
- 66.Wang, X., Dolan, P. T., Dang, Y., and Zheng, Y. H. (2007) J. Biol. Chem. 282 1585-1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.